A plant SUMO ligase regulates the protein stability of a chromatin remodeling factor in root development.
Abstract
Chromatin remodeling is essential for gene expression regulation in plant development and response to stresses. Brahma (BRM) is a conserved ATPase in the SWI/SNF chromatin remodeling complex and is involved in various biological processes in plant cells, but the regulation mechanism on BRM protein remains unclear. Here, we report that BRM interacts with AtMMS21, a SUMO ligase in Arabidopsis (Arabidopsis thaliana). The interaction was confirmed in different approaches in vivo and in vitro. The mutants of BRM and AtMMS21 displayed a similar defect in root development. In the mms21-1 mutant, the protein level of BRM-GFP was significantly lower than that in wild type, but the RNA level of BRM did not change. Biochemical evidence indicated that BRM was modified by SUMO3, and the reaction was enhanced by AtMMS21. Furthermore, overexpression of wild-type AtMMS21 but not the mutated AtMMS21 without SUMO ligase activity was able to recover the stability of BRM in mms21-1. Overexpression of BRM in mms21-1 partially rescued the developmental defect of roots. Taken together, these results supported that AtMMS21 regulates the protein stability of BRM in root development.
The structure of chromatin is critical for the interaction between DNA and proteins involved in replication, transcription, and other processes in nucleus (Müller and Leutz, 2001). ATP-dependent chromatin remodeling complexes regulate the accessibility of chromatin by altering the positions of histone octamers on DNA. The SWI/SNF complex, one of major classes in chromatin remodeling, has been shown to play a crucial function in various biological processes in different organisms. Brahma (BRM), a conserved DNA-dependent ATPase, has central catalytic activity in the SWI/SNF complex (Smith and Peterson, 2005). Recent studies suggested that BRM is primarily expressed in tissues with active cell division (Farrona et al., 2004) and plays important roles in embryonic, vegetative, and reproductive development of plants (Hurtado et al., 2006; Kwon et al., 2006; Tang et al., 2008; Archacki et al., 2009; Farrona et al., 2011; Li et al., 2015), possibly a consequence of altered gene expression. A recent study also indicated that BRM acts in the PLETHORA (PLT) pathway to maintain the root stem cell niche by directly altering the expression of PINs (Yang et al., 2015). In addition, BRM is involved in the signaling transduction in responses to several hormones (Han et al., 2012; Archacki et al., 2013; Efroni et al., 2013). Although some proteins such as LEAFY, SEPALLATA3, TCP4, ANGUSTIFOLIA3, BREVIPEDICELLUS, HD2C, SnRKs, and REF6 have been identified as BRM-associated proteins (Wu et al., 2012; Efroni et al., 2013; Vercruyssen et al., 2014; Zhao et al., 2015; Buszewicz et al., 2016; Li et al., 2016; Peirats-Llobet et al., 2016), most of them are cooperative factors with BRM in the control of chromatin dynamics and gene expression. Collectively, many processes in plants have been reported to be regulated by BRM, however, the regulation mechanism on the BRM protein itself remains unclear.
Ubiquitination and SUMOylation are two types of important posttranslational modifications that transfer a polypeptide ubiquitin or SUMO onto protein substrates, regulating protein trafficking, stability, and activity (Gill, 2004). The ubiquitin-proteasome system has been characterized as a major regulatory mechanism for protein degradation. Although the structures of ubiquitin and SUMO are similar, they may have antagonistic effects on the target proteins (Ulrich, 2005). Similar with ubiquitination, SUMOylation is mediated by a cascade containing an activating enzyme (E1), a conjugating enzyme (E2), and usually a ligase (E3), which may enhance the reaction in physiological conditions (Wilkinson and Henley, 2010). MMS21 is a conserved SUMO ligase and important for genome stability (Stephan et al., 2011). The Sugimoto group and our group discovered that the homolog of MMS21 in Arabidopsis (Arabidopsis thaliana; AtMMS21, also named HPY2) is critical for stem cell niche maintenance in root development (Huang et al., 2009; Ishida et al., 2009; Xu et al., 2013). AtMMS21 is also involved in DNA damage response, meiosis, and stress response (Zhang et al., 2013; Liu et al., 2014; Yuan et al., 2014). Recently, we discovered that AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation (Liu et al., 2016). Because AtMMS21 is a SUMO ligase in the SMC5/6 complex (Xu et al., 2013), which targets onto chromatin for DNA repair, an interesting question is whether it regulates the protein factors on DNA, such as components in the chromatin modification and remodeling complexes.
In the screening for AtMMS21-interacting proteins, we found that BRM interacts with AtMMS21 in a yeast two-hybrid system. The interaction was further confirmed in vitro and in vivo. Attachment of SUMO3 on BRM is enhanced by AtMMS21. The protein level of BRM is decreased in the roots of the AtMMS21 mutant, and the stability of BRM is controlled by SUMOylation and 26S proteasome-mediated degradation. Overexpression of BRM partially rescues the root defect phenotype in the AtMMS21 mutant, supporting that AtMMS21 regulates the stability of BRM in root development.
RESULTS
BRM Interacts with AtMMS21 in Vivo and in Vitro
As protein modifications are important for chromatin structure regulation, we are interested in whether the SUMO ligase AtMMS21 targets the components associated with this process in plants. A yeast two-hybrid screen was used to identify AtMMS21-interacting proteins involved in chromatin remodeling and modification. Eight proteins including JMJ15, HDA6, HDA19, GCN5, ADA2B, BRM, SWI3B, and SWI3C were examined their interaction with AtMMS21. Most of these regulators did not interact with AtMMS21, except BRM, which is a critical DNA-dependent ATPase in the SWI/SNF complex for chromatin remodeling (Supplemental Fig. S1).
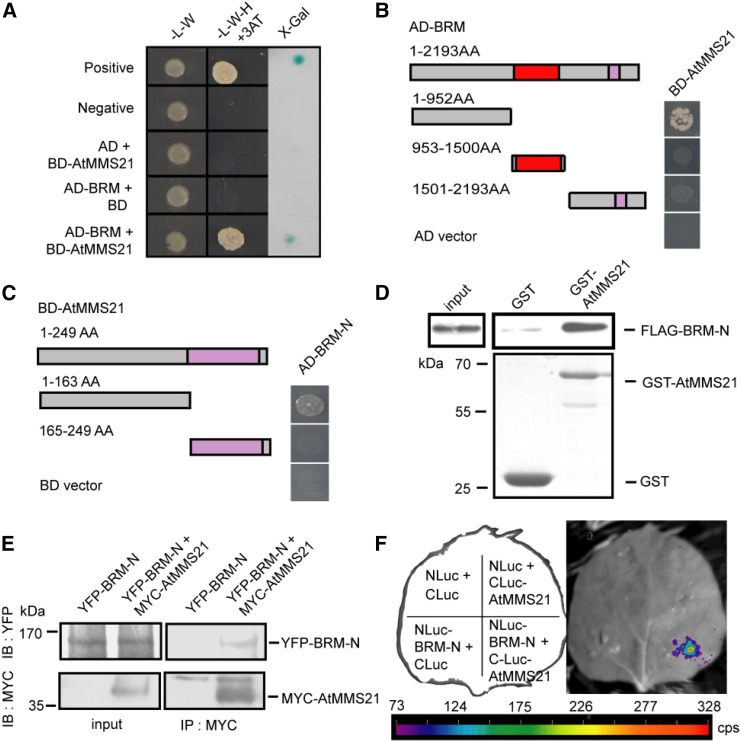
An independent yeast two-hybrid assay was used to confirm that BRM interacts with AtMMS21 (Fig. 1A) and identify the functional domains required for their interaction in yeast. The results indicated that the interaction is dependent on the N termini of both proteins (Fig. 1, B and C). The physical interaction between BRM and AtMMS21 was further confirmed by an in vitro pull-down assay. Compared with the control sample, the N terminus of BRM fused with a FLAG tag was pulled down by GST-AtMMS21, indicating that BRM directly interacts with AtMMS21 (Fig. 1D). To confirm their interaction in plant cells, the N terminus of BRM was fused with YFP and expressed in the wild-type or MYC-AtMMS21 overexpression plants and subjected for coimmunoprecipitation. The result showed that the N terminus of BRM specifically interacted with AtMMS21 (Fig. 1E), consistent with the in vitro results. The evidence from a luciferase complementary assay supported that BRM and AtMMS21 associates with each other in plant cells (Fig. 1F). Conclusively, all the data supported that BRM interacts with AtMMS21 in vivo and in vitro.
Figure 1.
BRM interacts with AtMMS21. A, The interaction between BRM (fused to the GAL4 activation domain (AD)) and AtMMS21 (fused to the GAL4 DNA binding domain (BD)) was detected in a yeast two-hybrid assay. B, Identification of the domain on BRM required for its interaction with AtMMS21 by yeast two-hybrid analysis. The SNF2 ATPase domain is indicated in red, and the Bromodomain is indicated in pink. C, Identification of the domain on AtMMS21 required for its interaction with BRM by yeast two-hybrid analysis. The SP-RING domain is shown in pink. D, The interaction between BRM and AtMMS21 was confirmed by in vitro pull-down assay. The N terminus of BRM (amino acids 1–952) fused with a FLAG tag was incubated with immobilized GST-AtMMS21 or GST (control). The precipitated N-BRM was detected by anti-FLAG antibody in immunoblotting. The protein levels of GST-AtMMS21 and GST were shown in the bottom. E, The interaction between BRM-N (amino acids 1–952) and AtMMS21 in an in vivo coimmunoprecipitation assay. 35S:YFP-BRM-N was transformed and expressed in protoplasts from wild-type or 35S:MYC-AtMMS21 plants. Total protein extracts were immunoprecipitated with the immobilized anti-MYC antibody. The proteins from lysates (left) and immunoprecipitated proteins (right) were detected using anti-YFP or anti-MYC antibody. F, Agrobacterium-mediated LCI assay for the interaction between BRM and AtMMS21. The Agrobacterium carrying the indicated construct pairs were injected into tobacco leaves, and the luciferase activities were measured 3 d after injection. The data are a representative from three independent experiments.
The Protein Level of BRM-GFP Is Decreased in mms21-1
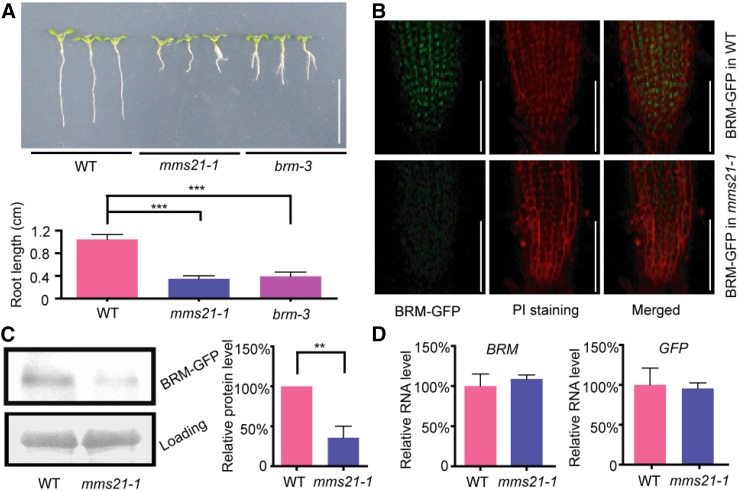
Previous reports from the Sugimoto group and our laboratory showed that the depletion of AtMMS21 results in a short root phenotype in Arabidopsis (Huang et al., 2009; Ishida et al., 2009). A recent result showed that the mutations on BRM also have effect on root development (Yang et al., 2015). The root length of mms21-1 and brm-3 were compared with that of wild type. Consistent with the previous data, both mutants have similar root growth retardation (Fig. 2A), supporting the possibility that AtMMS21 and BRM act in the same signaling pathway required for root development.
Figure 2.
The protein level of BRM-GFP is decreased in mms21-1. A, The phenotypes of root developmental defect in mms21-1 and brm-3. The photo was taken 5 d after germination. Bar = 1 cm. The measurement of root length of 5-d-old wild type (WT), mms21-1, and brm-3 were shown in the bottom. The data are means ± sd from at least 30 seedlings in three biological independent experiments. ***P < 0.001, Student’s t test. B, The BRM-GFP signals in the roots of 5-d-old wild-type and mms21-1 seedlings. The GFP (in green), PI staining (in red), and the merged images are shown. Bar = 100 µm. The data were collected on the same tissue layer under microscopy with same settings. The images are a representative from at least 30 seedlings in three independent experiments. C, The protein levels of BRM-GFP from 5-d-old wild type and mms21-1. The result from immunoblotting with anti-GFP antibody is shown in the top. The loading control from Coomassie Blue staining is shown in the bottom. The statistic results (right) are from three biological independent experiments quantified by the ImageJ software. **P < 0.01, Student’s t test. D, The RNA levels of BRM-GFP from 5-d-old wild type and mms21-1. The primer pairs for BRM or GFP were used in the qRT-PCR reactions. The data are means ± se from triplicated experiments.
Because AtMMS21 is a ligase in SUMOylation, an interesting question is whether the interaction between AtMMS21 and BRM has any effect on the localization, stability, or activity of BRM. PROBRM:BRM-GFP (Zhao et al., 2015) was introduced into mms21-1 by genetic cross, and the homologous plants for both genotypes were used for further analysis. The GFP signals were measured in the roots of wild type or mms21-1 expressing BRM-GFP. As a result, the nuclear localization of BRM-GFP did not change in the absence of AtMMS21. However, the fluorescence of BRM-GFP was much lower in mms21-1 than that in wild type (Fig. 2B), suggesting that the level of BRM-GFP was decreased in this mutant. The protein-level change of BRM-GFP in wild type and mms21-1 was confirmed by protein immunoblot using a GFP antibody (Fig. 2C). To exclude the possibility that the decrease of protein level is a result from variation on RNA level, a real-time quantitative PCR (qRT-PCR) reaction was performed using similar materials. The data indicated there was no difference on the RNA levels of BRM or GFP between wild type and mms21-1 (Fig. 2D), suggesting that AtMMS21 has effects on the protein level of BRM.
Given protein degradation via 26S proteasome is important for the regulation of protein stability, the level of BRM-GFP was measured under the treatment of MG132, an inhibitor specific for 26S proteasome. Compared with the control samples, BRM-GFP was increased in the roots of wild type, while the increase was more significant in mms21-1 (Supplemental Fig. S2), suggesting that the protein level of BRM may be regulated by 26S proteasome.
Modification of BRM by SUMO3 Mediated by AtMMS21 Regulates the Stability of BRM
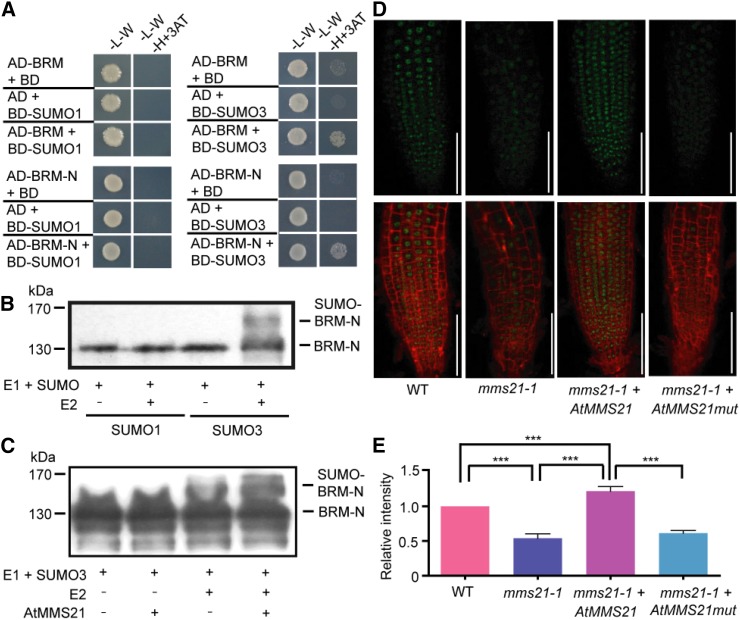
Because SUMO has been reported as an antagonist of ubiquitin (Ulrich, 2005), the next question is whether the regulation of BRM stability is dependent on the SUMOylation mediated by AtMMS21. Given there are several SUMO homologs in Arabidopsis, a yeast two-hybrid assay was used to identify whether BRM is associated any type of SUMO molecules. Surprisingly, the result showed that BRM interacted with SUMO3, but not SUMO1. Because the N terminus of BRM (amino acids 1–952) interacts with AtMMS21, this fragment was also tested in the assay. The result suggested that at least the N terminus of BRM is specifically associated with SUMO3 in yeast cells (Fig. 3A).
Figure 3.
Modification of BRM by SUMO3 mediated by AtMMS21 regulates the stability of BRM. A, The results from a yeast two-hybrid assay to detect the association between SUMO and BRM. SUMO1 or SUMO3 (fused to the GAL4 DNA binding domain (BD)) and BRM (full length, fused to the GAL4 activation domain (AD)) or BRM-N (amino acids 1–952, fused to AD) were used for detection. B, The SUMO conjugation of BRM-N was detected in a reconstituted SUMOylation system in E. coli. In the presence of E1 and SUMO1 or SUMO3, with or without E2, the unconjugated and SUMO-conjugated FLAG-BRM-N were detected by anti-FLAG antibody. C, AtMMS21 mediates the SUMOylation of BRM-N. GST-AtMMS21 was added in the in vitro system for reaction. The SUMOylation levels of BRM-N in different samples were detected in an immunoblot using anti-FLAG antibody. D, The BRM-GFP signals in the AtMMS21 complementation lines. 35S:AtMMS21 or 35S: AtMMS21mut (C178S and H180A mutations in the SP-RING motif, without ligase activity) were introduced into the mms21-1 plants carrying BRM-GFP. The BRM-GFP signals were measured in the roots of 5-d-old seedlings. The data are a representative from independent transgenic lines. Bar = 100 µm. E, The quantification of GFP signals in D is from 100 nuclei in three independent experiments. ***P < 0.001, Student’s t test. WT, Wild type.
To detect whether BRM is covalently modified by SUMO3, the reaction was performed in a reconstituted Escherichia coli system with Arabidopsis SUMOylation machinery proteins (Okada et al., 2009). Given BRM is an unstable protein with very high molecular mass (more than 240 kD) and is difficult for expression, the N terminal fragment of BRM (amino acids 1–952) was used for determination. In the presence of E1, E2, a FLAG-BRM-N signal with higher molecular mass, was detected in the sample with SUMO3 but not SUMO1 in the immunoblot, suggesting the N terminus of BRM is covalently modified by SUMO3 (Fig. 3B). To detect the effect of AtMMS21 on SUMO modification of BRM, purified GST-AtMMS21 was included in an in vitro reaction. The SUMOylation of BRM-N was enhanced with AtMMS21, suggesting that AtMMS21 facilitates the attachment of SUMO3 to BRM (Fig. 3C).
To determine whether the SUMOylation of BRM mediated by AtMMS21 has effect on the stability of BRM in plant cells, the wild-type or SP-RING mutation forms of AtMMS21 was expressed in mms21-1 with BRM-GFP (Supplemental Fig. S3). Because the SP-RING domain is required for the SUMO ligase activity of AtMMS21, the Cys-to-Ser (C178S) and His-to-Ala (H180A) mutations in this domain abolish its SUMO E3 activity (Ishida et al., 2009). When the wild-type form of AtMMS21 was overexpressed in mms21-1, the BRM-GFP level was even higher than that in wild-type plants. In contrast, the SP-RING mutant without SUMO ligase activity was unable to recover the BRM stability (Fig. 3, D and E). These results supported that AtMMS21 stabilizes BRM via SUMOylation.
Overexpression of BRM in mms21-1 Partially Rescues Its Root Developmental Defect
If decrease of BRM protein is a reason for the root developmental defect of mms21-1, artificial improvement of BRM expression in mms21-1 may recover the root development (Supplemental Fig. S4). Our experiment showed that overexpression of BRM in wild type had no significant effect on root growth. However, when BRM was overexpressed in mms21-1, the root length was significantly increased (Fig. 4, A and C). Although compared with wild type, overexpression of BRM did not completely recover the root growth of mms21-1, the partial rescue provided evidence for our hypothesis that the protein level of BRM is at least one of important factors regulated by AtMMS21 in root development.
Figure 4.
Overexpression of BRM in mms21-1 partially rescues its root developmental defect. A, The root length for 5-d-old seedlings of wild type (WT), mms21-1, 35S:BRM in mms21 (two independent lines), and 35S:BRM in wild type (two independent lines). The photo is a representative from three independent experiments. Bar = 1 cm. B, The meristem regions for 5-d-old roots of the indicated seedlings stained by PI. The meristem region is indicated by arrowheads; quiescent center is indicated by the lower arrowhead. Bar = 100 µm. C, The statistical data of root length and meristem cell number for 5-d-old seedlings. The data for root length are means ± sd from at least 30 seedlings in an experiment. Three biological independent experiments displayed a similar pattern. The meristem cell number (n = 15) are from at least three independent experiments. ***P < 0.001, Student’s t test.
Our previous study showed that the cell number in the root meristem of mms21-1 is lower than that in wild type (Huang et al., 2009; Xu et al., 2013), a potential reason for its defect in root development. Thus, the cell number in this region was compared in the plants with or without BRM overexpression. As a result, the meristem cell number did not change when BRM was overexpressed in wild type. However, when BRM under the control of 35S promoter was introduced into mms21-1, the size of meristem was significantly increased (Fig. 4, B and C), suggesting that the effect of the excess amount of BRM on root development is specific in the absence of AtMMS21. Conclusively, increasing expression of BRM partially rescues the root developmental defect of mms21-1.
AtMMS21 may regulate different pathways by SUMOylation, and the stability of BRM is one of the mechanisms for root development regulation. Given the protein level of BRM is low in mms21-1, the effect when BRM was completely removed in mms21-1 was detected in the mms21-1 brm-3 plants. The result indicated that when BRM was deleted in the mms21-1 background, the root was slightly shorter (Supplemental Fig. S5), compatible with our conclusion that the stability of BRM is one of regulation mechanism for root development via AtMMS21.
DISCUSSION
SUMOylation is important in regulation of the chromatin remodeling complex, for instance, ATP-dependent chromatin remodeler Mi-2 and pontin have been reported to play their functions in a SUMOylation-dependent manner in animal cells (Kim et al., 2007; Stielow et al., 2008). Here, we showed the stability of BRM, a critical ATPase in the SWI/SNF complex, is regulated by a SUMO ligase AtMMS21. The protein level of BRM is increased in the presence of MG132, suggesting that it is potentially controlled through the 26S proteasome pathway. The previous study in human cells provided evidence that BRM can be ubiquitinated and degraded by proteasome (Biggs et al., 2001), supporting our conclusion and implying a conserved regulation mechanism on BRM in different species. SUMO and ubiquitin are shown to have antagonistic effects on the target proteins (Ulrich, 2005), for example, SUMO and ubiquitin target to the same site on PCNA and play a competition in DNA damage response (Papouli et al., 2005). Our results showed that the protein level of BRM is decreased in the absence of AtMMS21, while the stability of BRM is dependent on the SUMO ligase activity of AtMMS21. Our biochemical results indicated that SUMOylation of BRM is enhanced by AtMMS21. Therefore, the degradation of BRM may be prevented by SUMOylation mediated via AtMMS21 in plant cells. However, more biochemical approaches need to be used to answer the question whether SUMOylation and ubiquitination have a direct competition on the same sites of BRM. BRM has been also reported to be modified by acetylation and phosphorylation (Bourachot et al., 2003; Peirats-Llobet et al., 2016), the interaction between different types of posttranslational modification in the regulation of BRM is interesting for further investigation.
Given chromatin remodeling regulates gene expression in different signaling pathways, the mutant of BRM has various effects on many processes, including the development of root, leaf, and inflorescence (Farrona et al., 2004; Hurtado et al., 2006; Kwon et al., 2006; Tang et al., 2008; Zhao et al., 2015). Similarly, AtMMS21 also regulates various processes in development and stress responses (Huang et al., 2009; Xu et al., 2013; Zhang et al., 2013; Liu et al., 2014; Yuan et al., 2014). Here, we focused on the regulation in root development and provided evidence to support that BRM and AtMMS21 acts in the same pathway in this process. Loss-of-function mutations in BRM or AtMMS21 result in defective maintenance of the root stem cell niche and stunted root, as well as decrease levels of the stem cell transcription factor genes PLT1and PLT2 (Xu et al., 2013; Yang et al., 2015), implying the possibility that BRM and AtMMS21 take action in the same pathway. The interaction between AtMMS21 and BRM provides a direct connection between these two components in root development. Given BRM is a chromatin remodeling factor involved in regulation of chromatin structure and gene expression, while AtMMS21 is a SUMO ligase for posttranslational modification of protein substrates, it is more possible that BRM controls the expression of genes in root development and AtMMS21 regulates BRM via its SUMO ligase activity. Our results that the protein level of BRM is decreased in mms21-1, and overexpression of BRM partially rescues the root defect of mms21-1 supported the hypothesis that AtMMS21 regulates BRM in root development.
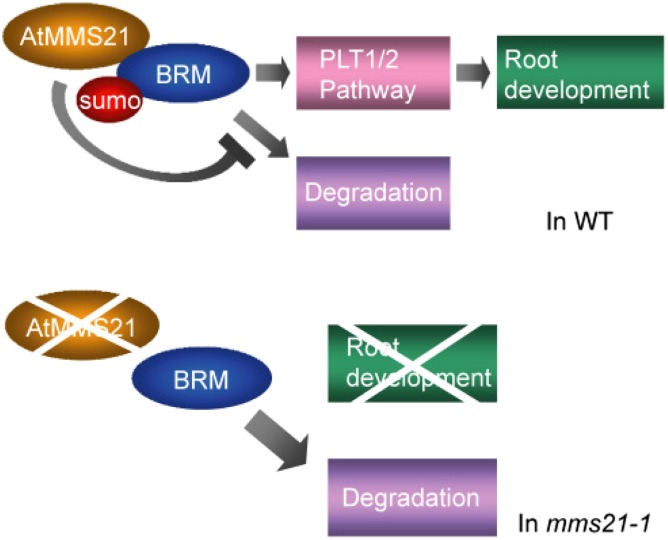
As a model, in the root of mms21-1, SUMOylation of BRM may be abolished, while the degradation of BRM is enhanced. As a result, the protein level of BRM is decreased and the downstream of pathway of BRM is attenuated similarly with that in the BRM mutants, resulting in the defect of root development (Fig. 5). Overexpression of BRM in mms21-1 may compensate the protein level of BRM in the absence of AtMMS21 and partially recover the root development of mms21-1. However, the root growth is unable to be completely rescued; possibly there are other factors that are also targets for AtMMS21 in this process. Among our screening, BRM is the only chromatin-associated component interacting with AtMMS21, suggesting a specific association between these proteins. Given both BRM and MMS21 are conserved proteins in different species from yeast, plant to human, it is interesting to investigate whether the interaction between BRM and MMS21 as well as the regulation mechanism on BRM are conserved among species. The characterization of association between BRM and MMS21 would improve our understanding on the mechanism of how protein modification regulates chromatin remodeling.
Figure 5.
A predicted model for the functional interaction between AtMMS21 and BRM in root development. The situations in wild type (WT) and mms21-1 are shown, respectively.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Columbia-0 (wild type), mms21-1 (CS848340), and brm-3 (SALK_088462) were described previously. The seeds of ProBRM:BRM-GFP are from the Wu laboratory (Zhao et al., 2015). The seeds with ProBRM:BRM-GFP/mms21-1 were generated by genetic cross, and the homologous plants for both genotypes were used for further analysis. Seeds were surface sterilized for 2 min in 75% ethanol followed by 5 min in 1% NaClO solution, rinsed five times with sterile water, plated on Murashige and Skoog medium with 1.5% Suc and 0.8% agar, and then stratified at 4°C in the dark for 2 d. Plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C.
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were performed according to the manufacturer’s instructions for the Matchmaker GAL4-based two-hybrid system 3 (Clontech). AtMMS21, SUMO1, and SUMO3 were cloned into the pGBKT7 vector. The coding sequences (CDSs) of JMJ15, HDA6, HDA19, GCN5, ADA2b, BRM, SWI3B, and SWI3C were cloned into the pGADT7 vector. For the interaction domain characterization, the truncated products were generated as described in the figure legends. Protein interactions were tested by stringent (SD/-Leu/-Trp/-His) selection supplied with 3-amino-1,2,4-triazole and β-galactosidase activity measurement (Clontech).
In Vitro Pull-Down Assay
The CDS of AtMMS21 was cloned into pGEX4T-1; the CDS of BRM-N (amino acids 1–952) fused with a FLAG tag was cloned into pCDFDuet-1; the proteins were expressed in BL21, respectively. GST or GST-AtMMS21 recombinant proteins were incubated with GST resins (GE Healthcare) in a binding buffer (50 mm Tris, pH 7.4, 120 mm NaCl, 5% glycerol, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and 1 mm β-mercaptoethanol), for 2 h at 4°C, then were collected and mixed with supernatant containing His6-FLAG-BRM-N and incubated at room temperature for 60 min. After being rinsed five times with the washing buffer (50 mm Tris, pH 7.4, 120 mm NaCl, 5% glycerol, and 0.5% Nonidet P-40), the bound proteins were boiled in sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting.
Coimmunoprecipitation
The 35S:YFP-BRM-N plasmid was constructed by fusing YFP with the CDS fragment encoding amino acids 1 to 952 of BRM in a pBluescript-based transient expression vector with a 35S promoter. Then, the plasmid was transformed into the protoplasts of wild-type or 35S:MYC-AtMMS21 transgenic plants (Liu et al., 2016). Forty-eight hours after transformation, the protoplasts were collected for coimmunoprecipitation. Proteins were extracted in extraction buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, and 1% Nonidet P-40) containing protease inhibitor cocktail (Roche). After centrifugation at 13,000g for 10 min, the supernatant was incubated with anti-MYC antibody (Clontech) at 4°C for 3 h. Then, 50 μL of protein A agarose beads (Sigma) was added. After 3 h of incubation at 4°C, the beads were centrifuged and washed four times with washing buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, and 0.1% Nonidet P-40). Proteins were eluted with SDS sample buffer and analyzed by immunoblotting by anti-GFP (TransGen Biotech) or anti-MYC (Clontech) antibodies.
LCI Assay
Luciferase complementation imaging (LCI) assays were performed as described previously (Chen et al., 2008). The CDS of BRM-N (amino acids 1–952) or AtMMS21 was cloned into pCAMBIA-NLuc and pCAMBIA-CLuc, respectively. All the constructs were transformed into Agrobacterium tumefaciens strain EHA105. An equal volume of A. tumefaciens harboring pCAMBIA-NLuc and pCAMBIA-CLuc (or their derivative constructs) was mixed to a final concentration of OD600 = 1.5. Four different combinations of A. tumefaciens were infiltrated into four different positions at the same leaves of Nicotiana benthamiana. Plants were placed in 23°C and allowed to recover for 60 h. A low-light cooled CCD imaging apparatus (NightOWL II LB983 with indiGO software) was used to capture the LUC image.
Root Length Analysis and Microscopy
The primary roots of plants incubated vertically on Murashige and Skoog medium were photographed using a camera, and the lengths of the primary roots at indicated day were determined using Digimizer 3.2 software. For confocal laser imaging of roots, roots were counterstained with 10 μg/mL propidium iodide (PI; Sigma) for 1 min and mounted in water for confocal microscopy. Confocal images were taken using a Zeiss LSM 710 laser scanning microscope with the following excitation/emission wavelengths: 561 nm/591 to 635 nm for PI, and 488 nm/505 to 530 nm for GFP. For comparison of GFP signals, same microscopy settings and scanning layers were used for analysis in different samples. The fluorescence intensity of nuclei was measured by ImageJ software.
RNA and Protein Analysis
Total RNA was extracted using the Plant RNAprep Pure Kit (TIANGEN) with DNase I treatment following the manufacturer’s instructions. The RNA was and reversely transcribed by a PrimeScript RT reagent kit (Takara) and was subjected for regular PCR. ACTIN2 was used as a reference gene. For qRT-PCR, the cDNA template was subjected to PCR using SYBR Premix Ex Taq (Takara) in a Bio-Rad CFX 96 system (C1000 Thermal Cycler) and detected by Bio-Rad CFX Manager software. For the analysis for protein level, the total proteins were extracted by adding 2× protein sample buffer into the grilled materials, boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis. The amounts of total proteins were quantified by Coomassie Blue staining (control), and the levels of BRM-GFP were detected in immunoblotting by an anti-GFP antibody. The relative intensity of proteins from three biological independent experiments was measured by ImageJ software.
SUMOylation Assay
The SUMOylation assay in Escherichia coli was performed as previously described (Okada et al., 2009; Liu et al., 2016). The CDS encoding amino acids 1 to 952 of BRM fused with a FLAG tag was cloned into pCDFDuet-1 (Novagen) and expressed in the bacteria carrying pET28-SAE1a-His-AtSAE2 (E1; Budhiraja et al., 2009) with pACYCDuet-1-SCE1-SUMO1GG or pACYCDuet-1-SCE1-SUMO3GG (E2 and SUMO). The transformed cells were cultured in LB medium to OD600 of 0.5 and induced by 0.5 mm isopropylthio-β-galactoside. After incubation for 12 h at 25°C, cells were harvested and used for immunoblotting by anti-FLAG antibody (Sigma). To detect the effect of AtMMS21 on the SUMOylation of BRM-N, the method was following the in vitro system described in the previous study with 1 μg purified GST-AtMMS21 (Liu et al., 2016).
Generation of Transgenic Plants
For the 35S:MYC-AtMMS21 construct in the complementation experiment, the CDS of AtMMS21 was obtained by PCR amplification and cloned into the PCanG-MYC vector. To generate the 35S:MYC-AtMMS21mut construct, site-directed mutagenesis was performed using the MutanBest kit (Takara). For the 35S:BRM construct, the CDS of BRM was cloned into pCanG-myc. The constructs were transformed into Agrobacterium EHA105, which was then used to transform Arabidopsis (Columbia) by the floral-dip method. Homozygous lines of transgenic plants were used in this study.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AtMMS21 (At3g15150), BRM (At2g46020), JMJ15 (At2g34880), HDA6 (AT5G63110), HDA19 (At4g38130), GCN5 (At3g54610), ADA2B (At4g16420), SWI3B (At2g33610), SWI3C (At1g21700), SUMO1 (At4G26840), and SUMO3 (At5G55170).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The yeast two-hybrid results for interaction between AtMMS21 and the indicated chromatin remodeling and modification components.
Supplemental Figure S2. The treatment of MG132 increases the protein level of BRM-GFP.
Supplemental Figure S3. The expression levels of AtMMS21 in the complementary lines for BRM-GFP/mms21-1.
Supplemental Figure S4. The expression levels of BRM in the BRM overexpression lines.
Supplemental Figure S5. The effect of BRM deletion in the mms21-1 background.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the seeds and thank Professor Andreas Bachmair for the SUMO E1 plasmid used in this study.
Footnotes
This work was supported by the National Natural Science Foundation of China (31670286, 31400314, and U1201212), Major Project from the Education Department of Guangdong Province (2014), the University Innovation Program from the Department of Education of Guangdong Province (2014), the Natural Science Foundation of Guangdong (S2012020011032), Guangzhou Scientific and Technological Program (201607010377), and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2010).
References
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka-Nowicka R, et al. (2013) BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS One 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Sarnowski TJ, Halibart-Puzio J, Brzeska K, Buszewicz D, Prymakowska-Bosak M, Koncz C, Jerzmanowski A (2009) Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta 229: 1281–1292 [DOI] [PubMed] [Google Scholar]
- Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A (2009) Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 149: 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs JR, Yang J, Gullberg U, Muchardt C, Yaniv M, Kraft AS (2001) The human brm protein is cleaved during apoptosis: The role of cathepsin G. Proc Natl Acad Sci USA 98: 3814–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, Muchardt C (2003) Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J 22: 6505–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanicka-Nowicka R, Sosnowska K, Kuciński J, Pupel P, Olędzki J, et al. (2016) HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ 39: 2108–2122 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D (2013) Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC (2004) The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975 [DOI] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, March-Díaz R, Schmitz RJ, Florencio FJ, Turck F, Amasino RM, Reyes JC (2011) Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS One 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. (2004) SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev 18: 2046–2059 [DOI] [PubMed] [Google Scholar]
- Han SK, Sang Y, Rodrigues A, BIOL425 F2010, Wu M-F, Rodriguez PL, Wagner D, Wagner D (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, et al. (2009) The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J 60: 666–678 [DOI] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol 62: 291–304 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee JM, Nam HJ, Choi HJ, Yang JW, Lee JS, Kim MH, Kim SI, Chung CH, Kim KI, et al. (2007) SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proc Natl Acad Sci USA 104: 20793–20798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Li C, Chen C, Gao L, Yang S, Nguyen V, Shi X, Siminovitch K, Kohalmi SE, Huang S, Wu K, et al. (2015) The Arabidopsis SWI2/SNF2 chromatin Remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet 11: e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, et al. (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lai J, Yu M, Wang F, Zhang J, Jiang J, Hu H, Wu Q, Lu G, Xu P, et al. (2016) The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation. Plant Cell 28: 2225–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi S, Zhang S, Xu P, Lai J, Liu Y, Yuan D, Wang Y, Du J, Yang C (2014) SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis. BMC Plant Biol 14: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Leutz A (2001) Chromatin remodeling in development and differentiation. Curr Opin Genet Dev 11: 167–174 [DOI] [PubMed] [Google Scholar]
- Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K (2009) Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: Functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Peirats-Llobet M, Han SK, Gonzalez-Guzman M, Jeong CW, Rodriguez L, Belda-Palazon B, Wagner D, Rodriguez PL (2016) A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol Plant 9: 136–147 [DOI] [PubMed] [Google Scholar]
- Smith CL, Peterson CL (2005) ATP-dependent chromatin remodeling. Curr Top Dev Biol 65: 115–148 [DOI] [PubMed] [Google Scholar]
- Stephan AK, Kliszczak M, Morrison CG (2011) The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett 585: 2907–2913 [DOI] [PubMed] [Google Scholar]
- Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, Boutros M, Suske G (2008) Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell 29: 742–754 [DOI] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, Reyes JC, Wang A, Keller WA, Harada JJ, et al. (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. (2005) Mutual interactions between the SUMO and ubiquitin systems: A plea of no contest. Trends Cell Biol 15: 525–532 [DOI] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, Yao N, Yang C (2013) AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol 161: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M, Chen CY, Liu X, Luo M, Cui Y, et al. (2015) The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27: 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D, Lai J, Xu P, Zhang S, Zhang J, Li C, Wang Y, Du J, Liu Y, Yang C (2014) AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Repair (Amst) 21: 140–147 [DOI] [PubMed] [Google Scholar]
- Zhang S, Qi Y, Liu M, Yang C (2013) SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F). J Integr Plant Biol 55: 83–95 [DOI] [PubMed] [Google Scholar]
- Zhao M, Yang S, Chen CY, Li C, Shan W, Lu W, Cui Y, Liu X, Wu K (2015) Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet 11: e1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.