SOC1 inhibits dark-induced leaf degreening and senescence by directly binding to the CArG box of PPH, NYE1, and SAG113 promoters and inhibiting their expression at the transcriptional level in Arabidopsis.
Abstract
Although the biochemical pathway of chlorophyll (Chl) degradation has been largely elucidated, how Chl is rapidly yet coordinately degraded during leaf senescence remains elusive. Pheophytinase (PPH) is the enzyme for catalyzing the removal of the phytol group from pheophytin a, and PPH expression is significantly induced during leaf senescence. To elucidate the transcriptional regulation of PPH, we used a yeast (Saccharomyces cerevisiae) one-hybrid system to screen for its trans-regulators. SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), a key flowering pathway integrator, was initially identified as one of the putative trans-regulators of PPH. After dark treatment, leaves of an SOC1 knockdown mutant (soc1-6) showed an accelerated yellowing phenotype, whereas those of SOC1-overexpressing lines exhibited a partial stay-green phenotype. SOC1 and PPH expression showed a negative correlation during leaf senescence. Substantially, SOC1 protein could bind specifically to the CArG box of the PPH promoter in vitro and in vivo, and overexpression of SOC1 significantly inhibited the transcriptional activity of the PPH promoter in Arabidopsis (Arabidopsis thaliana) protoplasts. Importantly, soc1-6 pph-1 (a PPH knockout mutant) double mutant displayed a stay-green phenotype similar to that of pph-1 during dark treatment. These results demonstrated that SOC1 inhibits Chl degradation via negatively regulating PPH expression. In addition, measurement of the Chl content and the maximum photochemical efficiency of photosystem II of soc1-6 and SOC1-OE leaves after dark treatment suggested that SOC1 also negatively regulates the general senescence process. Seven SENESCENCE-ASSOCIATED GENES (SAGs) were thereafter identified as its potential target genes, and NONYELLOWING1 and SAG113 were experimentally confirmed. Together, we reveal that SOC1 represses dark-induced leaf Chl degradation and senescence in general in Arabidopsis.
Leaf senescence is an integral part of plant development, an active process regulated by environmental factors and phytohormones, during which hundreds of SENESCENCE-ASSOCIATED GENES (SAGs) are differentially expressed (Lim et al., 2007; Fischer, 2012; Li et al., 2014). During leaf senescence, macromolecules (proteins, carbohydrates, and lipids) and chlorophylls (Chls) are degraded, and the resultant nutrients, as well as minerals, are recycled from senescing leaves to nascent tissues and organs, especially reproductive organs, to support their rapid development (Himelblau and Amasino, 2001; Guiboileau et al., 2012). The degreening phenotype caused by rapid Chl degradation in chloroplasts is often considered a visual marker of leaf senescence (Christ and Hörtensteiner, 2014).
During leaf senescence, the biochemical pathway of Chl degradation has been largely elucidated by the identification of Chl catabolic genes in Arabidopsis (Arabidopsis thaliana; Christ and Hörtensteiner, 2014). Before being degraded, Chl b is converted to Chl a via a two-step consecutive reduction catalyzed by Chl b reductase (NYC1 and NOL) and hydroxymethyl Chl a reductase (Kusaba et al., 2007; Horie et al., 2009; Meguro et al., 2011). The chelated magnesium ion in Chl a is then removed from the center of the porphyrin macrocycle by an enzymatic or nonenzymatic reaction to form pheophytin a. Pheophytinase (PPH) catalyzes pheophytin a to produce pheophorbide a (Morita et al., 2009; Schelbert et al., 2009; Ren et al., 2010). Then, the porphyrin macrocycle of pheophorbide a is oxygenolytically opened by pheophorbide a oxygenase (PAO) to generate a red Chl catabolite (RCC), and the RCC is further reduced by RCC reductase (RCCR) to a primary fluorescent Chl catabolite (pFCC; Pruzinská et al., 2003, 2007). Finally, the pFCC is transported from chloroplasts into the vacuole, where it is further degraded to diverse Chl catabolites (Christ et al., 2012, 2013). NONYELLOWING1 (NYE1), also known as STAY-GREEN1 (SGR1), which is responsible for the green/yellow cotyledon color trait of Mendel’s pea (Pisum sativum), is identified as a key regulator of Chl degradation (Armstead et al., 2007; Ren et al., 2007; Sakuraba et al., 2012). Because of its physical interaction with Chl catabolic enzymes at light-harvesting complex II, NYE1 was proposed as a recruiter of Chl catabolic enzymes in senescing chloroplasts to promote efficient Chl degradation (Sakuraba et al., 2012). In addition, NYE2/SGR2, a stand-by paralog of NYE1, also was identified as a positive regulator of Chl degradation (Wu et al., 2016). Remarkably, Shimoda et al. (2016) recently showed that NYE1 acts as a magnesium dechelatase during senescence in Arabidopsis.
Over the past few years, a number of Chl degradation regulators have been reported to be involved in the processes of seed maturation, fruit ripening (degreening), and leaf senescence. In rice (Oryza sativa), an NAC transcription factor (TF), OsNAP, could positively regulate Chl degradation by directly activating the expression of SGR1, NYC1, NYC3, and RCCR1 (Liang et al., 2014). In Citrus sinensis, CitERF13, an ethylene-responsive factor, binds directly to the CitPPH promoter and enhances its expression during fruit degreening (Yin et al., 2016). In Arabidopsis, ABSCISIC ACID INSENSITIVE3 (ABI3), a B3 domain TF, promotes seed degreening during maturation via directly up-regulating the expression of NYE1 and NYE2 (Delmas et al., 2013). Two bZIP TFs, ABI5 and ENHANCED EM LEVEL, the direct targets of PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5, bind to NYE1 and NYC1 promoters and activate their expression during leaf senescence (Sakuraba et al., 2014). More recently, we found that EIN3 and MYC2/3/4 bind directly to NYE1, NYC1, and PAO promoters to accelerate ethylene- or jasmonic acid-triggered Chl degradation (Qiu et al., 2015; Zhu et al., 2015). The NAC family TFs ORE1, ANAC019, ANAC046, and ANAC072 bind to NYC1, NOL, NYE1, NYE2, and PAO promoters and stimulate their expression (Qiu et al., 2015; Zhu et al., 2015; Li et al., 2016; Oda-Yamamizo et al., 2016).
During senescence, Chl degradation is regarded primarily as a detoxification process to facilitate the massive nutrient remobilization (Christ and Hörtensteiner, 2014). However, before the initiation of senescence, the content of Chl, as the light-harvesting pigment, has to be maintained relatively constant, being strictly protected from large-scale degradation (Breeze et al., 2011). PPH is a senescence-induced hydrolase, yet its transcriptional regulation remains largely unknown. In this study, we initially performed a yeast (Saccharomyces cerevisiae) one-hybrid (Y1H) screen and identified SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), a multifunctional protein regulating flowering time, floral meristem development, cold tolerance, greening, annual growth habit, and stomatal opening in Arabidopsis (Samach et al., 2000; Melzer et al., 2008; Liu et al., 2009; Seo et al., 2009; Richter et al., 2013; Kimura et al., 2015; Davin et al., 2016), as one of the putative trans-regulators of PPH. By physiological, molecular, and genetic analyses, we demonstrated that SOC1 negatively regulates Chl degradation by binding directly to the CArG box of the PPH promoter to inhibit its expression at the transcriptional level. SOC1 also trans-inhibits the expression of other SAGs (e.g. NYE1 and SAG113) by binding directly to their promoters, leading to a negative regulation of the general senescence process. Our work reveals a novel regulatory module of SOC1-suppressed leaf degreening as well as senescence in general.
RESULTS
SOC1 Is a Putative Trans-Regulator of PPH
To study the transcriptional regulation of PPH, we initially performed a YIH assay to screen for its putative trans-regulators. A 1.11-kb fragment upstream of its start codon was used to drive its expression to complement the pph-1 mutant. We found that the stay-green phenotype of pph-1 was largely restored in p1110:PPH pph-1 compared with nontransgenic plants, and the expression of PPH was induced after dark treatment, a conventional way of inducing leaf degreening and senescence (Supplemental Fig. S1). This result suggested that the 1.11-kb fragment contains the core PPH promoter.
Then, we used this promoter fragment as the bait to screen for putative trans-regulators of PPH against an Arabidopsis leaf cDNA library. We identified several positive clones encoding SOC1, a MADS box (yeast, MCM1; plant, AGAMOUS and DEFICIENS; mammal, SERUM RESPONSE FACTOR) family TF involved in the multiple regulations of plant development (Shore and Sharrocks, 1995; West et al., 1998; Samach et al., 2000; Lee and Lee, 2010; Immink et al., 2012; Tao et al., 2012). MADS box family TFs were reported to be able to bind a special DNA motif, the CArG box (Riechmann et al., 1996; de Folter and Angenent, 2006). Expectedly, a CArG-box motif was indeed identified in the PPH promoter. To use the Y1H assay to initially examine the molecular relationship between SOC1 and the CArG-box motif, we cloned the full-length coding region of SOC1 into the vector pGADT7. It was shown that SOC1 interacted strongly with a 317-bp PPH promoter fragment containing the CArG box (between −847 and −531 bp upstream of the PPH start codon; Supplemental Fig. S2). This result preliminarily suggested that SOC1 is a putative trans-regulator of PPH.
SOC1 Represses Chl Degradation in Dark-Induced Senescence
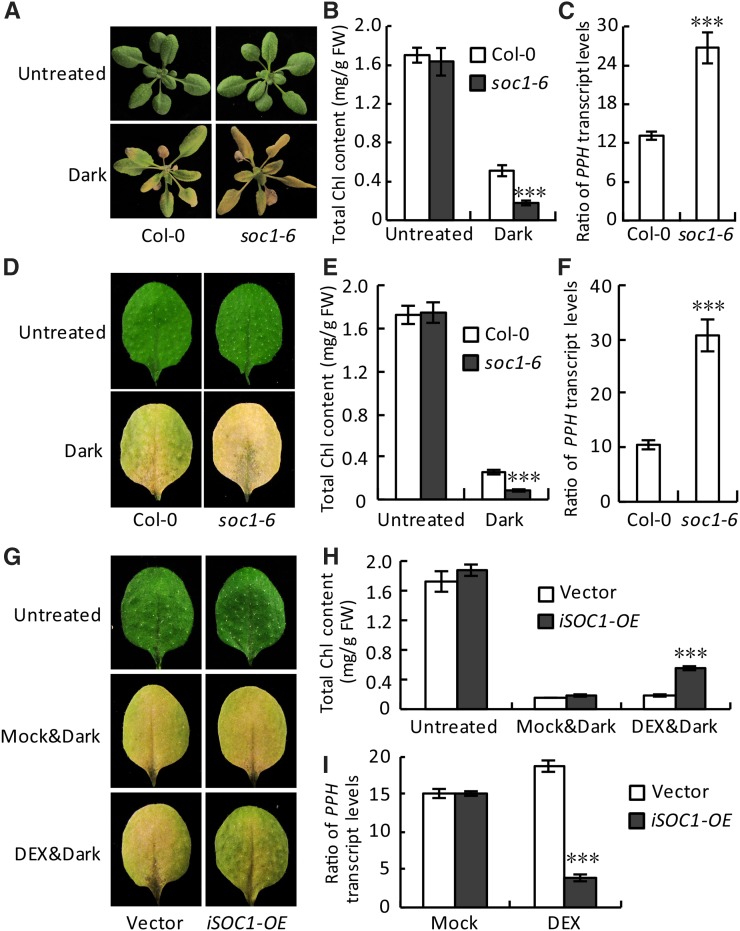
To determine whether SOC1 is actually involved in the regulation of Chl degradation, a T-DNA insertion mutant of SOC1 (soc1-6 [SALK_138131C], a late-flowering mutant) was initially analyzed (Supplemental Fig. S3; Immink et al., 2012). Remarkably, after dark treatment, both attached and detached leaves of soc1-6 showed accelerated yellowing phenotypes, which was verified by measuring the Chl content. It was further found that the relative expression level of PPH was enhanced significantly in the soc1-6 mutant (Fig. 1, A–F). In light of that a similar phenotype was observed in both the attached and the detached leaves, the detached leaves were then used for the following analyses for their convenience of preparation.
Figure 1.
SOC1 is a negative regulator of Chl degradation. A, Attached leaves of soc1-6 exhibited an accelerated yellowing phenotype during dark treatment. Three-week-old plants were treated in darkness for 7 d before being photographed. B, Chl contents in the fifth and sixth rosette leaves of the plants shown in A. C, Ratios (dark-untreated) of PPH transcript levels in the fifth and sixth rosette leaves of the plants shown in A. D, Detached leaves of soc1-6 exhibited an accelerated yellowing phenotype during dark treatment. Detached fifth and sixth rosette leaves of 3-week-old plants were treated in darkness for 5 d. E, Chl contents in the detached leaves shown in D. F, Ratios of PPH transcript levels after dark treatment over those in the untreated leaves shown in D. G, Induced overexpression of SOC1 (iSOC1-OE) resulted in a partial stay-green phenotype during dark treatment. Detached fifth and sixth rosette leaves of 3-week-old plants were treated in darkness for 6 d. Mock, Water; DEX, 30 µm DEX water solution; vector, empty vector control. H, Chl contents in the detached leaves shown in G. I, Ratios of PPH transcript levels after dark treatment over those in the untreated leaves shown in G. Data are means ± sd of three biological repeats. ***, P < 0.001 (Student’s t test). FW, Fresh weight.
SOC1-overexpressed transgenic lines were generated using a dexamethasone (DEX)-inducible system, and a representative line, iSOC1-OE #4, was chosen for further analysis (Supplemental Fig. S4). Dark treatment caused Chl degradation in both the vector control and iSOC1-OE #4 without DEX treatment (Fig. 1, G and H). As expected, DEX induced the expression of SOC1 in iSOC1-OE #4 (Supplemental Fig. S4), and, importantly, its induced expression suppressed the expression of PPH, leading to a reduced Chl degradation (Fig. 1, H and I; Supplemental Fig. S4B). As a result, iSOC1-OE #4 showed a partial stay-green phenotype compared with the vector control line (Fig. 1, G–I). These results suggested that SOC1 is a negative regulator of Chl degradation in dark-induced leaf senescence.
SOC1 Negatively Regulates the Transcription of PPH
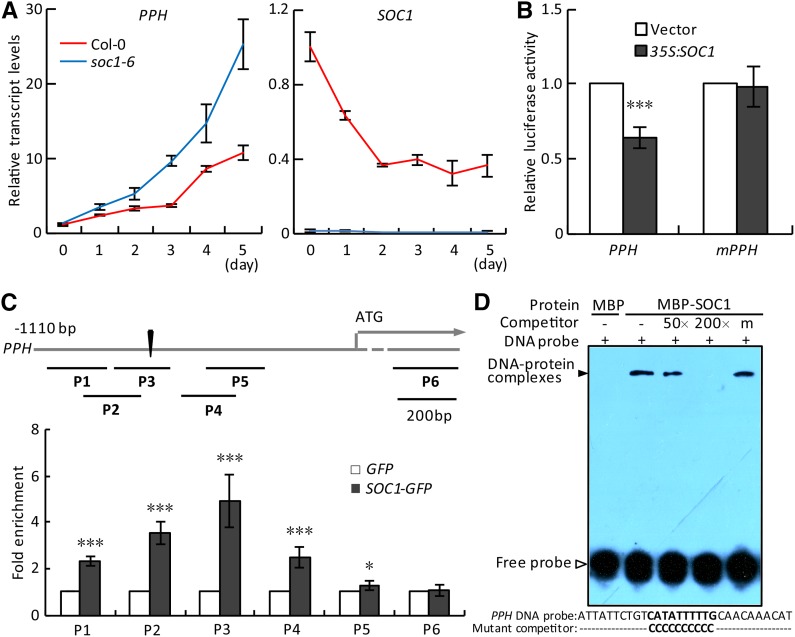
During dark treatment, a consecutive daily analysis revealed that the expression of PPH was gradually induced while the expression of SOC1 was sharply reduced in detached leaves of wild-type plants; an obviously enhanced expression of PPH was detected at each corresponding time point examined in the soc1-6 mutant (Fig. 2A). A consistent result was obtained in the detached leaves of iSOC1-OE #4, where an induced SOC1 expression negatively correlated with a reduced PPH expression in a short time window during dark treatment (Supplemental Fig. S5). These results indicated that SOC1 negatively regulates PPH expression in planta during dark-induced senescence. During the normal growth phase, we detected that the expression level of SOC1 in the fifth and sixth rosette leaves of wild-type plants increased steadily from 14 to 22 d after germination (DAG; Supplemental Fig. S6, A and B). This finding is consistent with a previous report that showed a similar SOC1 expression pattern from 23 to 31 d after sowing in the seventh rosette leaves of wild-type plants through a microarray analysis of global gene expression (Breeze et al., 2011). However, we found that the SOC1 transcript level became leveled off between 22 and 34 DAG and, afterward, declined dramatically from 34 to 42 DAG (Supplemental Fig. S6, A and B). This finding was seemingly different from the result reported by Breeze et al. (2011), who found that the expression of SOC1 became more or less leveled off after 31 to 39 d after sowing. This difference could be caused by multiple factors, including incompletely matched spectrum of the leaf age and systemic errors in the experimental design or data collection/analysis. Moreover, our data showed that SOC1 expression negatively correlated to PPH expression from 34 to 42 DAG, which was associated with a marked decrease in Chl content (Supplemental Fig. S6, B and D). This detection corroborated an in vivo negative regulatory relationship between SOC1 and PPH.
Figure 2.
SOC1 negatively regulates the expression of PPH by binding to its promoter. A, Relative transcript levels of PPH (left) and SOC1 (right) were analyzed by reverse transcription (RT)-quantitative PCR (qPCR) in the detached fifth and sixth rosette leaves of Columbia-0 (Col-0) and soc1-6 during dark treatment. The levels of PPH and SOC1 in Col-0 at day 0 were set to 1. B, PPH promoter activity was inhibited directly by overexpressing SOC1 in protoplasts in a dual-luciferase assay. Vector, Empty vector control; mPPH, the PPH promoter with mutations in the CArG box. C, ChIP-qPCR analysis of the association of SOC1 with the PPH promoter in vivo. Top, A schematic diagram of the PPH promoter showing the positions of the CArG box (black triangle) and six ChIP amplicons (P1–P6). Bottom, Fold enrichments of six amplified fragments were quantified by qPCR assay with chromatins isolated from 35S:SOC1-GFP as well as 35S:GFP lines. D, EMSA verification of the direct binding of SOC1 to the PPH promoter in vitro. MBP, Recombinant maltose-binding protein; MBP-SOC1, recombinant MBP-SOC1 protein; DNA probe, a 30-bp biotin-labeled PPH promoter fragment containing the wild-type CArG box was used as the probe; Competitor, nonlabeled wild-type fragment (50- or 200-fold excess) or the fragment with the CArG box mutated (200-fold excess). The black arrowhead points to DNA-protein complexes; the white arrowhead points to free probe. − and + represent absence and presence, respectively; m represents the mutated competitor. The 30-bp probe and mutated competitor sequence are shown below the EMSA image, with wild-type and mutated CArG boxes in boldface. The signal of biotin-labeled DNA was exposed to x-ray film. In A and C, data are means ± sd of two biological repeats; in B, data are means ± sd of three biological repeats. *, P < 0.05 and ***, P < 0.001 (Student’s t test).
To directly examine the regulatory relationship between SOC1 and PPH, we performed a dual-luciferase assay in Arabidopsis protoplasts. The reporter construct pPPH:LUC was cotransferred into protoplasts with the effector 35S:SOC1 or vector (control). It was found that the overexpression of SOC1 significantly inhibited the activity of the PPH promoter (Fig. 2B). However, once the CArG box in the PPH promoter was mutated, SOC1 was not able to affect the activity of the PPH promoter anymore (Fig. 2B). These results confirmed that SOC1 trans-inhibits the expression of PPH at the transcriptional level.
SOC1 Specifically Binds to the CArG Box of the PPH Promoter
The Y1H assay suggested that SOC1 may bind to the PPH promoter in vitro (Supplemental Fig. S2). To determine whether SOC1 interacts with the PPH promoter in vivo, we performed a chromatin immunoprecipitation (ChIP)-qPCR assay using a 35S:SOC1-GFP transgenic line, 35S:SOC1-GFP #15, with 15.8-fold increase in SOC1 expression (Supplemental Fig. S7). Immunoprecipitated DNA fragments from 10-d-old 35S:SOC1-GFP #15 rosette leaves as well as those from the 35S:GFP control line were used as templates to examine the fold enrichment of specific regions of the PPH promoter. Compared with the control line, about 5-fold enrichment of the PPH promoter region covering the CArG box, between −820 and −630 bp upstream of its start codon, was detected in the 35S:SOC1-GFP #15 line (Fig. 2C). In contrast, there was no enrichment in the coding region of the PPH gene far from the CArG box (Fig. 2C). This result suggested that SOC1 associates with the PPH promoter containing the CArG box in vivo.
We then performed an electrophoretic mobility shift assay (EMSA) to determine whether SOC1 could bind directly to the CArG box of the PPH promoter in vitro. Thirty-base-pair DNA fragments containing the wild-type CArG box were biotin labeled as the probe, whereas nonlabeled DNA fragments with or without mutations in the CArG box were used as competitors. SOC1 protein was found to be able to bind to the probe, and increasing amounts of the competitors without mutations in the CArG box competed with the binding (Fig. 2D; Supplemental Fig. S8A). However, the CArG-box mutated competitors could not compete with the probe for binding to SOC1 protein (Fig. 2D; Supplemental Fig. S8A). This result indicated that the SOC1 protein could bind specifically to the CArG box of the PPH promoter in vitro.
A Functional PPH Is Required for SOC1 Inhibition of Chl Degradation
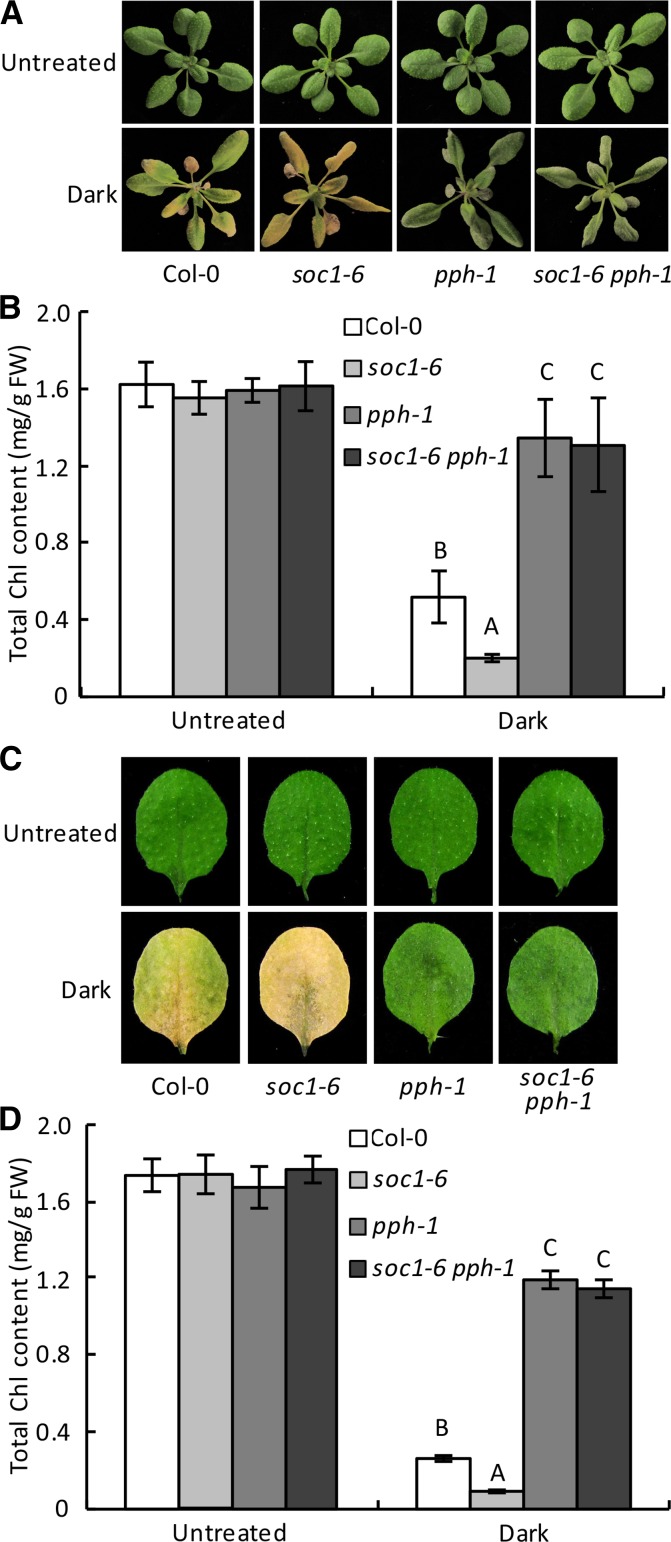
To determine whether or to what extent SOC1 inhibition of Chl degradation requires PPH, soc1-6 pph-1 double mutant was constructed. During dark treatment, both the attached and detached leaves of the soc1-6 pph-1 double mutant showed stay-green phenotypes similar to that of pph-1 but not anywhere close to the accelerated yellowing phenotype of soc1-6. The phenotypic observation was verified by the measurement of Chl content (Fig. 3). This result suggested that pph-1 is epistatic to soc1-6 and that a functional PPH is a prerequisite for SOC1 to effectively inhibit Chl degradation during leaf senescence.
Figure 3.
SOC1 inhibition of Chl degradation depends on a functional PPH. A, Dark-induced phenotypes of the attached leaves of different genotypes. soc1-6 pph-1 showed a stay-green phenotype similar to that of pph-1. Three-week-old plants were treated in darkness for 7 d before being photographed. B, Chl contents in the fifth and sixth rosette leaves of the plants shown in A. C, Dark-induced phenotypes of the detached leaves from different genotypes. soc1-6 pph-1 showed a stay-green phenotype similar to that of pph-1. The detached fifth and sixth leaves of 3-week-old plants were treated in darkness for 5 d. D, Chl contents in the detached leaves shown in C. In B and D, data are means ± sd of two biological repeats. Different letters indicate significant differences at P < 0.001 (one-way ANOVA). FW, Fresh weight.
SOC1 Negatively Regulates the Expression of Other SAGs
In Arabidopsis, the maximum photochemical efficiency of PSII (Fv/Fm) of wild-type rosette leaves is decreased at an accelerated rate during dark treatment (Ren et al., 2010). Interestingly, after dark treatment, the Fv/Fm ratio of the 3-week-old detached fifth and sixth rosette leaves was significant higher in SOC1-OE than in the vector control line, while the Fv/Fm ratio of soc1-6 was significantly lower compared with that in the wild type (Supplemental Fig. S9). These results prompted us to speculate that SOC1 may negatively regulate the expression of other SAGs in addition to PPH.
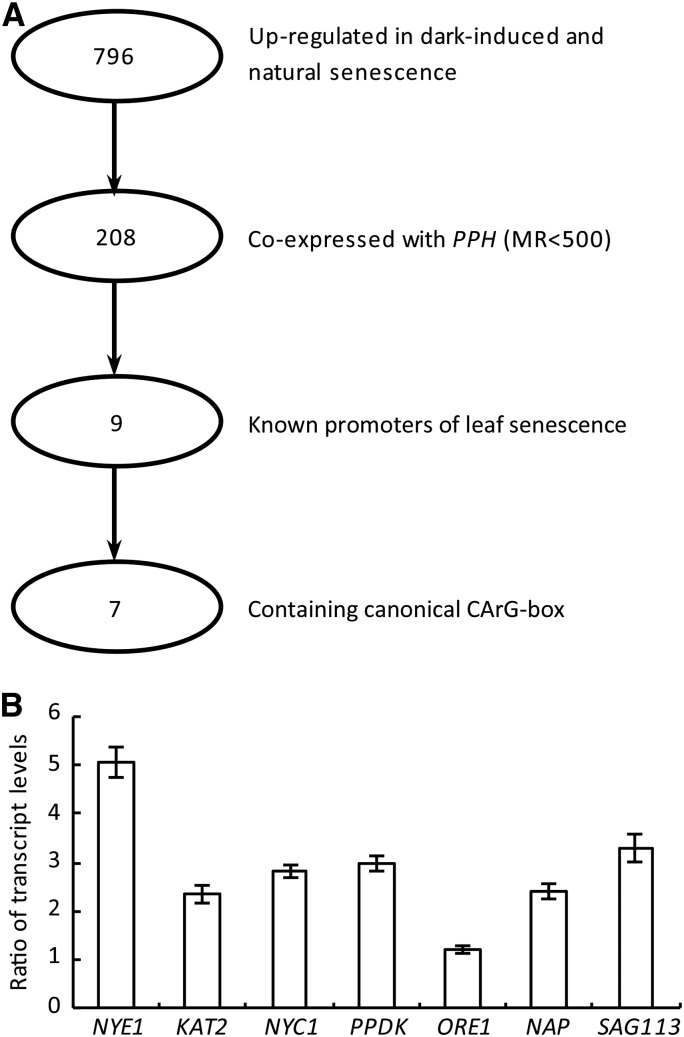
To test this hypothesis, we used a bioinformatics approach to identify other possible SOC1 targets during leaf senescence. Seven hundred ninety-six genes were demonstrated as being up-regulated in the processes of natural and dark-induced leaf senescence (van der Graaff et al., 2006). Among them, 208 genes were coexpressed with PPH (mutual rank value less than 500; Obayashi et al., 2007). So far, nine of them were reported to be able to promote leaf senescence (Pruzinská et al., 2003; Guo and Gan, 2006, 2011; Ren et al., 2007; Castillo and León, 2008; Kim et al., 2009; Horie et al., 2009; Taylor et al., 2010; Zhang and Gan, 2012), and seven of the nine genes contained the CArG box in the 1,000-bp regions upstream of their translation initiation sites (The Arabidopsis Information Resource [https://www.arabidopsis.org/]; Fig. 4A; Supplemental Table S1). To clarify whether the expression of the seven SAGs (NYE1, KAT2, NYC1, PPDK, ORE1, NAP, and SAG113) is indeed regulated by SOC1, we determined the ratios of their transcript levels in soc1-6 over that in Col-0 after dark treatment. It was found that the transcription of NYE1 and SAG113 was most induced by dark treatment in the soc1-6 mutant (Fig. 4B).
Figure 4.
Identification of putative SAG targets of SOC1 during leaf senescence. A, Diagram of the bioinformatics analysis procedure. MR, Mutual rank value. B, Ratios of the transcript levels of seven SAGs (NYE1, KAT2, NYC1, PPDK, ORE1, NAP, and SAG113) in soc1-6 over that in Col-0 after dark treatment (Fig. 1D), which were calculated as [soc1-6 (dark/untreated)/Col-0 (dark/untreated)]. Data are means ± sd of three biological repeats.
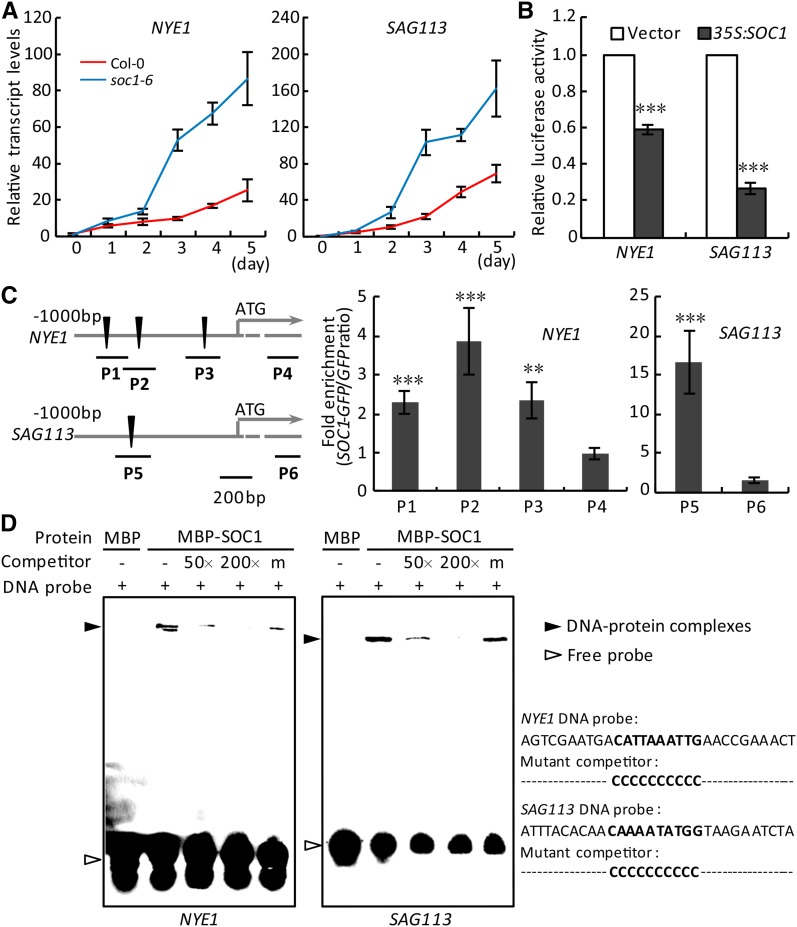
We then examined the transcript levels of NYE1 and SAG113 in the leaves of soc1-6 and Col-0 during dark treatment. It was found that the expression of NYE1 and SAG113 was induced by dark treatment and negatively correlated with that of SOC1. Moreover, their transcripts were more significantly accumulated in the soc1-6 mutant compared with the wild type (Figs. 2A and 5A). Conversely, after dark treatment, induced SOC1 inhibited NYE1 and SAG113 expression in the detached leaves of iSOC1-OE #4 (Supplemental Fig. S5). NYE1 and SAG113 expression showed negative correlations with that of SOC1 in the wild type from 34 to 42 DAG (Supplemental Fig. S6, E and F). We subsequently conducted a dual-luciferase assay and revealed that the overexpression of SOC1 significantly reduced the promoter activity of both NYE1 and SAG113 (Fig. 5B). Rosette leaves of 10-d-old 35S:SOC1-GFP #15 were then used for ChIP assays to examine the association of SOC1 with the promoters of NYE1 and SAG113 in vivo. It was demonstrated that the promoter regions containing the CArG box, but not their coding sequence (CDS) regions, were enriched significantly, suggesting that SOC1 indeed binds to the promoters of both NYE1 and SAG113 in vivo (Fig. 5C). The direct binding of SOC1 to the NYE1 and SAG113 promoters was further verified by EMSA (Fig. 5D; Supplemental Fig. S8, B and C). Collectively, SOC1 negatively regulates the expression of not only PPH but also other SAGs, including NYE1 and SAG113.
Figure 5.
SOC1 negatively regulates the expression of other SAGs. A, Relative transcript levels of NYE1 (left) and SAG113 (right) were analyzed by RT-qPCR in the detached fifth and sixth rosette leaves of Col-0 and soc1-6 during dark treatment. The levels of NYE1 and SAG113 in Col-0 at day 0 were set to 1. B, Overexpression of SOC1 inhibited the promoter activity of NYE1 and SAG113 in protoplasts. Vector, Empty vector control. C, ChIP-qPCR analysis of the association of SOC1 with the promoters of NYE1 and SAG113 in vivo. Left, Schematic diagrams of the promoters of NYE1 and SAG113. Black triangles, the CArG box; P1 to P6, ChIP-examined regions. Right, Fold enrichments of six amplified fragments quantified by qPCR assay with chromatins isolated from 35S:SOC1-GFP and 35S:GFP lines. D, EMSA verification of the direct binding of SOC1 to the promoters of NYE1 and SAG113 in vitro. MBP, Recombinant MBP protein; MBP-SOC1, recombinant MBP-SOC1 protein. Biotin-labeled NYE1 or SAG113 promoter fragment containing the wild-type CArG box was used as the probe, whereas nonlabeled wild-type fragment (50- or 200-fold excess) or the fragment with the CArG box mutated (200-fold excess) was used as the competitor. Black arrowheads point to DNA-protein complexes, and white arrowheads point to free probes. − and + represent absence and presence, respectively; m represents the mutated competitor. The 30-bp probe and competitor sequences are shown on the right side of the EMSA images, with wild-type and mutated CArG boxes in boldface. Biotin-labeled DNAs were exposed with the ChemiScope 3500 Mini Imaging System. In A and C, data are means ± sd of two biological repeats; in B, data are means ± sd of three biological repeats. **, P < 0.01 and ***, P < 0.001 (Student’s t test).
DISCUSSION
The molecular regulation of Chl degradation has been actively explored over the past few years, and numerous TFs have been reported to directly regulate Chl catabolic genes as well as other SAGs in Arabidopsis, rice, and citrus (Delmas et al., 2013; Liang et al., 2014; Sakuraba et al., 2014; Song et al., 2014; Qiu et al., 2015; Zhu et al., 2015; Gao et al., 2016; Li et al., 2016; Oda-Yamamizo et al., 2016; Yin et al., 2016). Intriguingly, all the reported TFs are positive regulators. In this study, we reveal that SOC1, a MADS-box protein, acts as a negative regulator of dark-induced leaf degreening and senescence in general in Arabidopsis. After dark treatment, the soc1-6 mutant exhibited an accelerated yellowing phenotype; conversely, SOC1-OE lines (iSOC1-OE) displayed a partial stay-green phenotype (Fig. 1). Importantly, a decline of SOC1 expression was accompanied by an increase of PPH expression during both dark-induced and developmental leaf degreening and senescence (Fig. 2A; Supplemental Fig. S6, B and D), and an induced SOC1 inhibited PPH expression in a short time window during dark treatment in the detached leaves (Supplemental Fig. S5). A dual-luciferase assay showed that SOC1 may trans-inhibit the expression of PPH at the transcriptional level in Arabidopsis protoplasts (Fig. 2B). Substantially, SOC1 protein could bind specifically to the CArG box of the PPH promoter in vitro and in vivo (Fig. 2, C and D; Supplemental Fig. S2). Importantly, mutation of PPH suppressed the accelerated yellowing phenotype of the soc1-6 mutant during dark treatment (Fig. 3), suggesting that the SOC1 inhibition of Chl degradation is PPH dependent. The characteristic changes in the Chl content and Fv/Fm ratio of soc1-6 and SOC1-OE leaves after dark treatment (Fig. 1; Supplemental Fig. S9) suggested that SOC1 might also negatively regulate the general senescence process. Seven SAGs were then identified as its potential target genes (Fig. 4). SOC1 binding to the CArG box in the promoters of its two putative target genes, NYE1 and SAG113, was confirmed experimentally, and its negative regulatory identity was validated by its trans-inhibition of NYE1 and SAG113 expression in protoplasts (Fig. 5). These results collectively demonstrate that SOC1 negatively regulates dark-induced leaf degreening (Chl degradation) and senescence in general in Arabidopsis.
PPH, an α/β-hydrolase located in chloroplasts, is a key enzyme for catalyzing the second step of Chl a degradation (Schelbert et al., 2009), and its expression is induced significantly during leaf senescence (Fig. 2A; Supplemental Fig. S6D; Ren et al., 2010; Breeze et al., 2011). Even though multiple components of major hormonal signaling pathways were reported recently to positively regulate Chl degradation in Arabidopsis, none is implicated in directly mediating the expression of PPH. Consistently, no abscisic acid response element motif, EIN3-binding site [A(C/T)G(A/T)A(C/T)CT], MYC-binding site (G box), or NAC-binding site (CACG) is present in the PPH promoter (Sakuraba et al., 2014; Qiu et al., 2015; Zhu et al., 2015; Oda-Yamamizo et al., 2016). In this report, we set out to study the transcriptional regulation of PPH and, using Y1H, identified several positive clones encoding SOC1 (Supplemental Fig. S2).
SOC1, a member of the MIKC-type MADS-box TF family, plays multiple roles in plant development (Lee and Lee, 2010). As the key flowering pathway integrator, SOC1 converges multiple flowering signals from diverse flowering regulatory pathways, induces LEAFY expression at the shoot apical meristem, and consequently determines the phase transition from vegetative to reproductive growth (Lee et al., 2008; Liu et al., 2008). It is also involved in regulating the development of the floral meristem by repressing the expression of SEPALLATA3 in emerging floral meristems to prevent precocious expression of the B and C genes (according to the ABC model) that maintain floral meristematic activity at the early developmental stage (Liu et al., 2009; Immink et al., 2012). SOC1 negatively regulates cold responses through trans-inhibiting the expression of C-REPEAT/DEHYDRATION RESPONSE ELEMENT-BINDING FACTORS and two GATA factors, GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM INVOLVED (GNC) and GNC-LIKE (GNL), by directly binding to the CArG box of their promoters (Seo et al., 2009; Richter et al., 2013). In addition, SOC1 also can affect the annual growth habit and stomatal opening in Arabidopsis (Melzer et al., 2008; Kimura et al., 2015; Davin et al., 2016). Intriguingly, SOC1 is also able to down-regulate Chl biosynthesis and consequently affect the greening process of 10-d-old Arabidopsis seedlings by directly repressing the expression of GNC and GNL (Richter et al., 2013). However, we detected neither an obvious change in the Chl content (Fig. 1, A, B, D, and E) nor a huge fold change in the expression of GNC and GNL in the leaves of the 3-week-old soc1-6 mutant before dark treatment (Supplemental Fig. S10). Therefore, it is likely that SOC1 exerts differential effects on the greening process at different developmental stages.
Here, we reveal a novel function of SOC1 in trans-inhibiting the expression of PPH, NYE1, and SAG113 to negatively regulate dark-induced leaf degreening (Chl degradation) and senescence (Figs. 2 and 5). NYE1 encodes a magnesium dechelatase during Chl degradation (Ren et al., 2007; Shimoda et al., 2016). SAG113, a phosphatase 2C protein, is responsible for accelerating the water loss during leaf senescence (Zhang and Gan, 2012). In addition to PPH, NYE1, and SAG113, a few other putative SOC1 target genes were also sensitive to dark treatment in soc1-6 (Fig. 4). Among them, NYC1 encodes a membrane-localized short-chain dehydrogenase/reductase that catalyzes the first step of Chl b degradation (Kusaba et al., 2007; Horie et al., 2009); PPDK codes for pyruvate,orthophosphate dikinase that is responsible for interconverting pyruvate and phosphoenolpyruvate and is involved in a nitrogen remobilization pathway during leaf senescence (Taylor et al., 2010); KAT2 is the gene encoding 3-KETOACYL-COENZYME A THIOLASE2, a key enzyme for jasmonic acid biosynthesis that is required for the timely onset of natural and dark-induced leaf senescence (Castillo and León, 2008); and NAP encodes a NAC TF that positively regulates senescence and abscisic acid level by enhancing the transcription of the abscisic acid biosynthesis gene ABSCISIC ALDEHYDE OXIDASE3 (Guo and Gan, 2006; Liang et al., 2014; Yang et al., 2014). These data suggest that SOC1 is involved in the negative regulation of Chl degradation and leaf senescence at multiple levels of the senescence regulatory network. Importantly, we also noticed that, with DEX-induced expression of SOC1, PPH was still up-regulated to some extent by dark treatment (Fig. 1I), implying that there likely exist other negative regulatory pathways of PPH expression. It could not be ruled out, however, that the induced expression of SOC1 was not high enough to fully inhibit PPH expression.
In Arabidopsis, whole-plant senescence and death are generally affected by reproduction, but the senescence of individual leaves cannot be associated specifically with reproduction, flowering initiation in particular (Noodén and Penney, 2001). In this study, after having revealed a role of SOC1 in regulating the degreening and senescence processes of fifth and sixth rosette leaves during dark treatment, we further analyzed the developmental degreening/senescence process and PPH transcription in the first 10 rosette leaves that share similar ages between their counterparts in the 6-week-old soc1-6 and iSOC1-OE plants. Unexpectedly, there were no obvious differences detected in the two parameters between the SOC1 mutant/overexpression lines and their corresponding controls, as indicated by Chl content measurements and RT-qPCR data (Supplemental Figs. S11 and S12). Thus, it can be tentatively concluded that SOC1 acts specifically as a negative regulator of the dark-induced Chl degradation and leaf senescence. Immediate efforts are required to explore the molecular mechanism underlying the differential effect of the SOC1-PPH/NYE1/SAG113 regulatory module on dark-induced degreening/senescence and developmentally regulated degreening/senescence.
MATERIALS AND METHODS
Plant Materials
All plant materials were derived from Arabidopsis (Arabidopsis thaliana) ecotype Col-0. The soc1-6 (SALK_138131C) mutant was obtained from the Arabidopsis Biological Resource Center Stock Center. The pph-1 mutant was described previously (Schelbert et al., 2009; Ren et al., 2010). The soc1-6 pph-1 double mutant was generated by crossing soc1-6 to pph-1. PCR-based genotyping was used to identify homozygous lines. All primer sequences are listed in Supplemental Table S2.
To obtain p1110:PPH pph-1 transgenic lines, the PPH CDS driven by the 1,110-bp promoter fragment (p1110) upstream of the PPH start codon was transferred into the pph-1 mutant. To generate iSOC1-OE transgenic lines, the full-length SOC1 CDS was cloned into pTA7002 vector and then transferred into Col-0. The full-length CDS (without stop codon) of SOC1 was cloned into pCAMBIA1302 (GFP-tagged) vector, which was then transferred into Col-0 to generate 35S:SOC1-GFP transgenic lines. The 35S:GFP line was used as a negative control (Zhu et al., 2015). Plants were grown at 22°C to 24°C, and light intensity was 100 μmol m−2 s−1 under a 16-h-light/8-h-dark photoperiod.
Induction Treatments
For dark treatment of plants, whole plants were placed in dark boxes in the same growth room where they had been grown. For dark treatment of leaves, detached fifth and sixth rosette leaves were incubated on wet filter papers in darkness. For DEX treatment, whole plants were sprayed with 30 µm DEX water solution, while individual leaves were soaked with 30 µm DEX water solution (Ren et al., 2010).
Plasmid Constructs
For the Y1H screening assay, the 1,110-bp fragment upstream of the PPH start codon was cloned into pAbAi vector (Clontech). For the Y1H retransformation assay, the SOC1 CDS was cloned into pGADT7 vector and the 317-bp PPH promoter fragment containing the CArG box was cloned into pAbAi vector. For the dual-luciferase assay, the PPH, mPPH (with mutations in the CArG box), NYE1, or SAG113 promoter was cloned into the pGreen II 0800-LUC vector individually. For EMSA, the SOC1 CDS was cloned into the pMAL-c5G vector (New England Biolabs).
Chl and Fv/Fm
Chl content was measured as described previously (Ren et al., 2007). The Fv/Fm ratio was determined using LI-COR6400 according to the manufacturer’s instructions (LI-COR).
RT-qPCR
Total RNAs were extracted using TRIzol reagent (TaKaRa) and digested with recombinant RNase-free DNase I (TaKaRa). First-strand cDNAs were synthesized by RT and used as the templates for RT-qPCR after making a 1:3 dilution. The RT-qPCR MyiQ2 Real Time PCR Detection System (Bio-Rad) and SYBR Premix Ex Taq II (TaKaRa) were used to perform RT-qPCR assays. β-ACTIN2 was used as an internal control. All primer sequences are listed in Supplemental Table S2.
Y1H Screening
The Matchmaker Gold Y1H Library Screening system (Clontech) was used in the Y1H screening. The construct driven by the 1,110-bp PPH promoter fragment was linearized, and the bait-reporter yeast (Saccharomyces cerevisiae) strain was obtained by integrating the linearized construct into the Y1HGold strain genome. The cDNA library was constructed with mRNAs isolated from the rosette leaves of 3-week-old Col-0 plants. The bait-reporter strain and cDNA library were used to screen for putative trans-regulators of PPH. Positive colonies were screened on the selective medium (synthetic dextrose/-Leu) with 100 ng mL−1 aureobasidin A (AbA), and the prey fragments were identified by DNA sequencing and BLAST.
The bait-reporter strain was made using the 317-bp PPH promoter fragment containing the CArG box for the retransformation assay. The SOC1 CDS was cloned into pGADT7 vector and then transformed into the bait-reporter yeast strain. Transformed yeast cells were plated onto synthetic dextrose/-Leu/AbA solid medium (100 ng mL−1 AbA).
Dual-Luciferase Reporter Assay
Protoplasts prepared from the rosette leaves of wild-type plants were transformed using the polyethylene glycol-mediated method as described previously (Zhu et al., 2015). PPH, mPPH, NYE1, and SAG113 promoters were cloned individually into the pGreen II 0800-LUC vector to generate the reporters. The 35S:SOC1 construct was used as the effector.
After incubation for 16 h in darkness, protoplasts were spun down and lysed with cell lysis buffer. The Synergy 2 Multi-Mode Microplate Reader (Bio-Tek) and a dual-luciferase assay kit (Promega) were used to detect the firefly and Renilla reniformis luciferase activities according to the manufacturer’s instructions.
ChIP Assay
Rosette leaves of 10-d-old (after germination) 35S:SOC1-GFP transgenic plants were cross-linked with 1% (v/v) formaldehyde. The ChIP assay was performed as described previously (Zhu et al., 2015). Chromatins were sonicated to produce 0.2- to 0.5-kb DNA fragments. GFP-Trap (ChromoTek) was used to immunize specific protein-DNA complexes. After elution and reverse cross-linking, the ChIP DNA Clean & Concentrator kit (Zymo Research) was used to purify the products of ChIP. The immunoprecipitated DNA samples were quantified by qPCR with the primers listed in Supplemental Table S2. Fold enrichments of promoter regions in the 35S:SOC1-GFP transgenic line were calculated by comparison with the control 35S:GFP line.
EMSA
The full-length SOC1 CDS was cloned into pMAL-c5G vector to fuse with MBP and transferred into the Rosetta (DE3) Escherichia coli strain (Merck). The MBP-SOC1 protein and the MBP protein were induced by 0.5 mm isopropyl thio-β-d-galactoside at 20°C for 10 h. Amylose resin (New England Biolabs) was used to purify MBP-SOC1 and MBP according to the manufacturer’s instructions. The DNA probes used in EMSA were synthesized by SBS and are listed in Supplemental Table S2.
EMSA was performed as described previously (Qiu et al., 2015) with the following modifications. Each EMSA binding reaction (20 μL) contained 1 μg of purified recombinant protein, which was roughly confirmed by western immunoblotting with anti-MBP antibody (New England Biolabs), 1 μL of 200 fmol of biotin-labeled probe DNA, and 2 μL of 10× binding buffer. Transferred DNA and protein were cross-linked using a UV lamp at 312 nm. The biotin-labeled DNA was determined using the Thermo Scientific chemiluminescence kit and exposed to X-OMAT BT film (Carestream Health) or the ChemiScope 3500 Mini Imaging System (Clinx Science Instruments) according to the manufacturer’s instructions.
Accession Numbers
Sequence data in this report can be found in The Arabidopsis Information Resource or the GenBank/EMBL databases: SOC1 (AT2G45660), PPH (AT5G13800), NYE1/SGR1 (AT4G22920), SAG113 (AT5G59220), NYC1 (AT4G13250), NAP (AT1G69490), KAT2 (AT2G33150), PPDK (AT4G15530), ORE1 (AT5G39610), GNC (AT5G56860), GNL (AT4G26150), and β-ACTIN2 (AT3G18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The 1.11-kb fragment upstream of the PPH start codon contains the core PPH promoter.
Supplemental Figure S2. The SOC1 protein interacts with the PPH promoter in a Y1H assay.
Supplemental Figure S3. soc1-6 is a late-flowering mutant.
Supplemental Figure S4. DEX treatment could induce the transcription of SOC1 in iSOC1-OE transgenic lines (T3).
Supplemental Figure S5. An induced SOC1 inhibits PPH, NYE1, and SAG113 transcription in a short time window during dark treatment.
Supplemental Figure S6. Developmental senescence phenotype of the wild-type Arabidopsis leaves.
Supplemental Figure S7. Relative transcript levels of SOC1 in 35S:SOC1-GFP transgenic lines (T3).
Supplemental Figure S8. Western immunoblot analysis of the purified MBP and MBP-SOC1 proteins.
Supplemental Figure S9. Fv/Fm ratios of the detached fifth and sixth rosette leaves of 3-week-old soc1-6 and iSOC1-OE plants during dark treatment.
Supplemental Figure S10. Relative transcript levels of GNC and GNL in the fifth and sixth rosette leaves of 3-week-old soc1-6 plants.
Supplemental Figure S11. Developmental senescence phenotypes of soc1-6 leaves.
Supplemental Figure S12. Developmental senescence phenotypes of iSOC1-OE leaves.
Supplemental Table S1. Putative senescence-associated target genes of SOC1.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Juan Lin (Fudan University) for providing the soc1-6 (SALK_138131C) mutant, Dr. Jianxiang Liu (Fudan University) for the pGreen II 0800-LUC vector, and Dr. Aiwu Dong (Fudan University) for the pTA7002 vector.
Glossary
- Chl
chlorophyll
- TF
transcription factor
- Y1H
yeast one-hybrid
- DEX
dexamethasone
- DAG
days after germination
- ChIP
chromatin immunoprecipitation
- qPCR
quantitative PCR
- EMSA
electrophoretic mobility shift assay
- Col-0
Columbia-0
- CDS
coding sequence
- RT
reverse transcription
- AbA
aureobasidin A
Footnotes
This work was supported by the Science and Technology Commission of Shanghai Municipality (grant no. 13JC1400900 to B.K.).
References
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2007) Cross-species identification of Mendel’s I locus. Science 315: 73. [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo MC, León J (2008) Expression of the β-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot 59: 2171–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B, Hörtensteiner S (2014) Mechanism and significance of chlorophyll breakdown. J Plant Growth Regul 33: 4–20 [Google Scholar]
- Christ B, Schelbert S, Aubry S, Süssenbacher I, Müller T, Kräutler B, Hörtensteiner S (2012) MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol 158: 628–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B, Süssenbacher I, Moser S, Bichsel N, Egert A, Müller T, Kräutler B, Hörtensteiner S (2013) Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis. Plant Cell 25: 1868–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin N, Edger PP, Hefer CA, Mizrachi E, Schuetz M, Smets E, Myburg AA, Douglas CJ, Schranz ME, Lens F (2016) Functional network analysis of genes differentially expressed during xylogenesis in soc1ful woody Arabidopsis plants. Plant J 86: 376–390 [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC (2006) trans meets cis in MADS science. Trends Plant Sci 11: 224–231 [DOI] [PubMed] [Google Scholar]
- Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JGB, McCourt P, Samuel MA (2013) ABI3 controls embryo degreening through Mendel’s I locus. Proc Natl Acad Sci USA 110: E3888–E3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AM. (2012) The complex regulation of senescence. Crit Rev Plant Sci 31: 124–147 [Google Scholar]
- Gao S, Gao J, Zhu X, Song Y, Li Z, Ren G, Zhou X, Kuai B (2016) ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol Plant 9: 1272–1285 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2011) AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol 156: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk ADJ, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Aoki S, Ando E, Kitatsuji A, Watanabe A, Ohnishi M, Takahashi K, Inoue S, Nakamichi N, Tamada Y, et al. (2015) A flowering integrator, SOC1, affects stomatal opening in Arabidopsis thaliana. Plant Cell Physiol 56: 640–649 [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254 [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I (2008) SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J 55: 832–843 [DOI] [PubMed] [Google Scholar]
- Li S, Gao J, Yao L, Ren G, Zhu X, Gao S, Qiu K, Zhou X, Kuai B (2016) The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep 35: 1729–1741 [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao Y, Liu X, Peng J, Guo H, Luo J (2014) LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res 42: D1200–D1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H (2009) Regulation of floral patterning by flowering time genes. Dev Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23: 3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Noodén LD, Penney JP (2001) Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). J Exp Bot 52: 2151–2159 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci Rep 6: 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, et al. (2015) EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Zhou Q, Wu S, Zhang Y, Zhang L, Huang J, Sun Z, Kuai B (2010) Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. J Integr Plant Biol 52: 496–504 [DOI] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C (2013) Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol 162: 1992–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM (1996) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC (2012) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM (2010) Cytosolic pyruvate,orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J 62: 641–652 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD (1998) DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res 26: 5277–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Li Z, Yang L, Xie Z, Chen J, Zhang W, Liu T, Gao S, Gao J, Zhu Y, et al. (2016) NON-YELLOWING2 (NYE2), a close paralog of NYE1, plays a positive role in chlorophyll degradation in Arabidopsis. Mol Plant 9: 624–627 [DOI] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XR, Xie XL, Xia XJ, Yu JQ, Ferguson IB, Giovannoni JJ, Chen KS (2016) Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J 86: 403–412 [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan SS (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B (2015) Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J 84: 597–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.