A salivary endo-β-1,4-glucanase in the rice brown planthopper Nilaparvata lugens facilitates access to the phloem by degrading celluloses in plant cell walls.
Abstract
The brown planthopper (BPH) Nilaparvata lugens is one of the most destructive insect pests on rice (Oryza sativa) in Asia. After landing on plants, BPH rapidly accesses plant phloem and sucks the phloem sap through unknown mechanisms. We discovered a salivary endo-β-1,4-glucanase (NlEG1) that has endoglucanase activity with a maximal activity at pH 6 at 37°C and is secreted into rice plants by BPH. NlEG1 is highly expressed in the salivary glands and midgut. Silencing NlEG1 decreases the capacity of BPH to reach the phloem and reduces its food intake, mass, survival, and fecundity on rice plants. By contrast, NlEG1 silencing had only a small effect on the survival rate of BPH raised on artificial diet. Moreover, NlEG1 secreted by BPH did not elicit the production of the defense-related signal molecules salicylic acid, jasmonic acid, and jasmonoyl-isoleucine in rice, although wounding plus the application of the recombination protein NlEG1 did slightly enhance the levels of jasmonic acid and jasmonoyl-isoleucine in plants compared with the corresponding controls. These data suggest that NlEG1 enables the BPH’s stylet to reach the phloem by degrading celluloses in plant cell walls, thereby functioning as an effector that overcomes the plant cell wall defense in rice.
To protect themselves from attack by herbivores, plants have developed a set of resistance mechanisms, including constitutive and induced defenses (Felton and Tumlinson, 2008; Erb et al., 2012; Stam et al., 2014; Schuman and Baldwin, 2016). Constitutive defenses are physical and chemical defensive traits that plants express regardless of the presence of herbivores. By contrast, induced defenses are activated only when plants are infested by herbivores (Wu and Baldwin, 2010). Defense induction starts with the recognition of specific herbivore-associated molecular patterns and is followed by the activation of a complex signaling network, such as mitogen-activated protein kinase cascades, and jasmonic acid (JA), jasmonoyl-isoleucine (JA-Ile), salicylic acid (SA), and ethylene signaling pathways; the expression of defense-related genes; and the production of defensive chemicals (Erb et al., 2012; Stam et al., 2014; Schuman and Baldwin, 2016).
In response, herbivores have evolved the capacity to suppress and circumvent these plant defenses through the release of effectors (Elzinga and Jander, 2013). Plant cell walls (PCWs), for instance, are thick, rigid structures that consist mainly of a pectin-embedded network of cellulose and hemicellulose (Calderón-Cortés et al., 2012); these structures not only act as physical defenses against herbivores by enhancing the mechanical hardness of plant tissues but also reduce the digestibility of food for herbivores (Santiago et al., 2013), thereby functioning as the first layer of defense against herbivores. Herbivores can secrete salivary PCW-degrading enzymes such as cellulases (consisting of endo-β-1,4-glucanases and β-glucosidases) and pectinases to degrade PCWs (Backus et al., 2012; Calderón-Cortés et al., 2012). Herbivores also can secrete other effectors to overcome plant defenses. C002, for instance, is a salivary protein identified from the salivary glands of the pea aphid Acyrthosiphon pisum; this protein is essential for sustained phloem feeding (Mutti et al., 2008). Expressing C002 from the green peach aphid Myzus persicae in Nicotiana benthamiana increased aphid reproduction on these plants, whereas reducing C002 expression in aphids by plant-mediated RNA interference (RNAi) reduced aphid fecundity (Bos et al., 2010; Pitino et al., 2011). Other salivary proteins, such as Glc oxidase from the corn earworm Helicoverpa zea (Musser et al., 2002), calcium-binding proteins from the vetch aphid Megoura viciae (Will et al., 2007), Mp10 and Mp55 from the green peach aphid (Bos et al., 2010; Elzinga et al., 2014), structural sheath proteins from the grain aphid Sitobion avenae (Abdellatef et al., 2015), Me10 and Me23 from the potato aphid Macrosiphum euphorbiae (Atamian et al., 2013), and Armet from the pea aphid (Wang et al., 2015), also have been found to increase herbivore performance. Thus, herbivore effectors play a central role in overcoming plant defenses and helping the herbivore establish a population on host plants. However, the mechanisms underlying the effector-mediated promotion of herbivore abilities to overcome plant defenses remain mostly unknown (Elzinga and Jander, 2013).
The brown planthopper (BPH) Nilaparvata lugens, one of the most destructive insect pests of the rice (Oryza sativa) plant in Asia, causes substantial losses in rice yield every year (Heong et al., 2014). As a piercing-sucking insect, BPH secretes two primary kinds of saliva during feeding: coagulable and watery. Coagulable saliva forms salivary sheaths around the insect’s stylets that help to stabilize and protect the stylets and may suppress plant defense responses to components of the watery saliva (Miles, 1999; Abdellatef et al., 2015). Watery saliva, which contains a mixture of amino acids, proteins, and digestive enzymes, assists in the digestion of plant material and helps suppress plant defense responses (Miles, 1968, 1999; Harmel et al., 2008; Carolan et al., 2009; Hogenhout and Bos, 2011; Nicholson et al., 2012; Elzinga and Jander, 2013). Thus, BPH saliva plays an important role in BPH feeding. The morphology, transcriptomes, and secreted proteins of BPH salivary glands have been analyzed and identified (Sogawa, 1968; Konishi et al., 2009; Ji et al., 2013). Moreover, several BPH salivary proteins, such as a catalase-like protein, a salivary sheath protein, a salivary glands-specific protein with unknown function, and annexin-like5, have been reported to be secreted into rice and to play an important role in salivary sheath formation and/or BPH feeding (Petrova and Smith, 2014; Huang et al., 2015b, 2016). However, whether other salivary proteins also are involved in BPH feeding and how these proteins regulate BPH feeding remain unclear.
By screening 352 reported genes encoding putative secreted proteins of the salivary gland of BPH (Ji et al., 2013), we identified a BPH gene, NlEG1, which encodes a putative endo-β-1,4-glucanase. Given the role of salivary cellulases in degrading PCWs stated above, we thought that NlEG1 might play a role in BPH feeding by influencing the formation of the salivary sheaths and/or defense in rice. Therefore, we chose NlEG1 and explored its role in rice-BPH interactions. Through a combination of molecular biology and behavioral experiments, we show that NlEG1 is an effector that enables BPH to feed on rice plants and simultaneously circumvents plant defenses.
RESULTS
Isolation and Characterization of NlEG1
Based on the data from BPH salivary gland transcriptomes (Ji et al., 2013), the full-length cDNA of the gene NlEG1 (1,454 bp), including an open reading frame of 1,386 bp, was obtained by reverse transcription (RT)-PCR (Fig. 1; GenBank accession no. KM459012). Its deduced amino acid sequence revealed that NlEG1 encodes a protein of 461 amino acids with a calculated molecular mass of 52.2 kD. The protein possesses an extracellular signal peptide and has no transmembrane domains, suggesting a putative secreted protein. Moreover, two potential O-glycosylation sites and two N-glycosylation sites were identified (Fig. 1). Protein alignment followed by phylogenetic tree analysis revealed that NlEG1 is homologous to insect endo-β-1,4-glucanases and belongs to the glycosyl hydrolase family 9 (GHF 9); this family is characterized by catalytic domains, including a catalytic nucleophile (Asp-77), a probable secondary nucleophile (Asp-80), and a proton acceptor (Glu-435), and two signature motifs (Nakashima et al., 2002; Kim et al., 2008; Willis et al., 2011; Fig. 1; Supplemental Figs. S1 and S2). NlEG1 shares the highest homology (72%) with the endo-β-1,4-glucanase from Isoptera (Zootermopsis nevadensis; KDR16731.1) and Phthiraptera (Pediculus humanus corporis; XP_002426465.1), followed by that from Hemiptera (A. pisum; XP_001944774.2 [71%]) and Hymenoptera (Apis florea; XP_003690676.1 [69%]).
Figure 1.
Nucleotide sequence of NlEG1 and its deduced amino acid sequence. The arrow indicates the signal peptide cleavage site. Predicted N-glycosylation and O-glycosylation sites are underlined in black and highlighted in gray, respectively. The square region denotes the proton-donor catalytic region. The catalytic nucleophile, probable secondary nucleophile, and proton acceptor (Glu) of the conserved catalytic domain for GHF 9 members are represented as a black diamond, a gray diamond, and a black circle, respectively. Two GHF 9 signature motifs are underlined in gray.
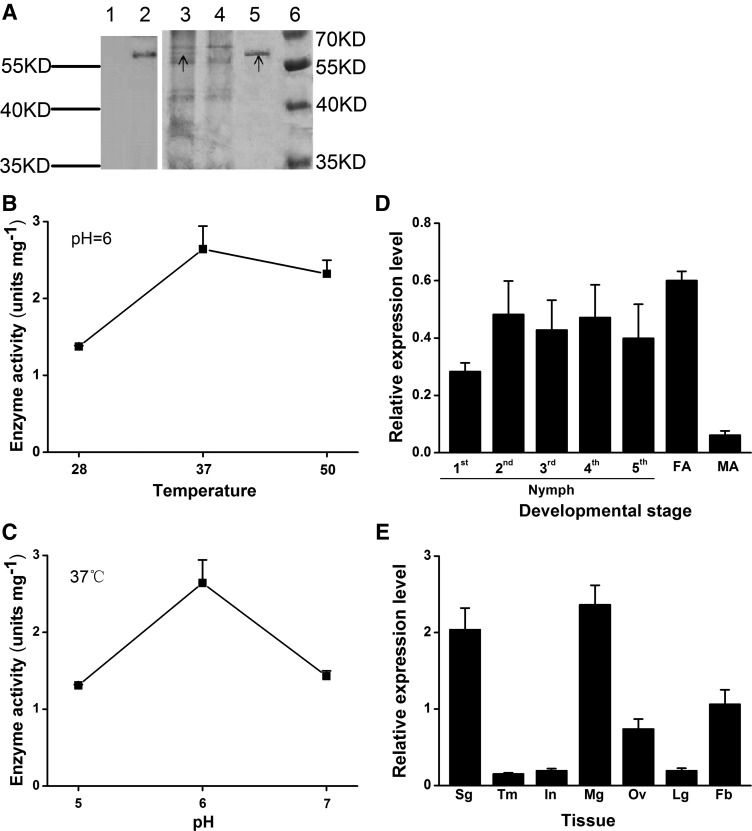
To confirm that NlEG1 has endoglucanase activity, the recombination protein NlEG1 was produced in Pichia pastoris (Fig. 2A). The mass of the recombination protein NlEG1 was about 60 kD (Fig. 2A). Enzyme activity assays demonstrated that NlEG1 acted hydrolytically on carboxymethyl cellulose (CMC) and showed the highest activity at pH 6 at 37°C (Fig. 2, B and C). Moreover, NlEG1 also acted hydrolytically on filter paper and cellulose from rice plants (0.55 and 0.79 units mg−1, respectively, at pH 6 at 37°C) but had no activity on crystalline cellulose (Avicel), curdlan, laminarin, and xylan. The catalytic activity of NlEG1 against CMC showed a Km of 6.72 mg mL−1 with a Vmax of 9.91 units mg−1. Quantitative real-time (qRT)-PCR analysis revealed that NlEG1 was expressed in most life stages of BPH (Fig. 2D) and was highly expressed in the salivary gland, midgut, fat body, and ovary (Fig. 2E).
Figure 2.
Molecular characterization of NlEG1. A, Expression and purification of NlEG1. Samples for western-blot (lanes 1 and 2) and SDS-PAGE (lanes 3–6) analyses were as follows: concentrated supernatant from P. pastoris with the empty vector pPICZɑ A (lanes 1 and 4; control); concentrated supernatant from P. pastoris with the recombinant vector NlEG1:pPICZɑ A (lanes 2 and 3); purified recombinant protein NlEG1 (lane 5); and protein maker (lane 6). The black arrows represent the target band. Rabbit anti-NlEG1 polyclonal antibodies were used to develop the western blot. B and C, Mean enzyme activity + se (n = 3) of the purified recombinant protein NlEG1 on CMC at different temperatures at pH 6 (B) and at 37°C at different pH levels (C). D and E, Mean transcript levels + se (n = 3) of NlEG1 in whole bodies at various developmental stages (D) and in different tissues (E). FA, Female adults; MA, male adults; Sg, salivary gland; Tm, thorax muscle; In, integument; Mg, midgut; Ov, ovary; Lg, leg; Fb, fat body.
NlEG1 Is Secreted into Rice during BPH Feeding
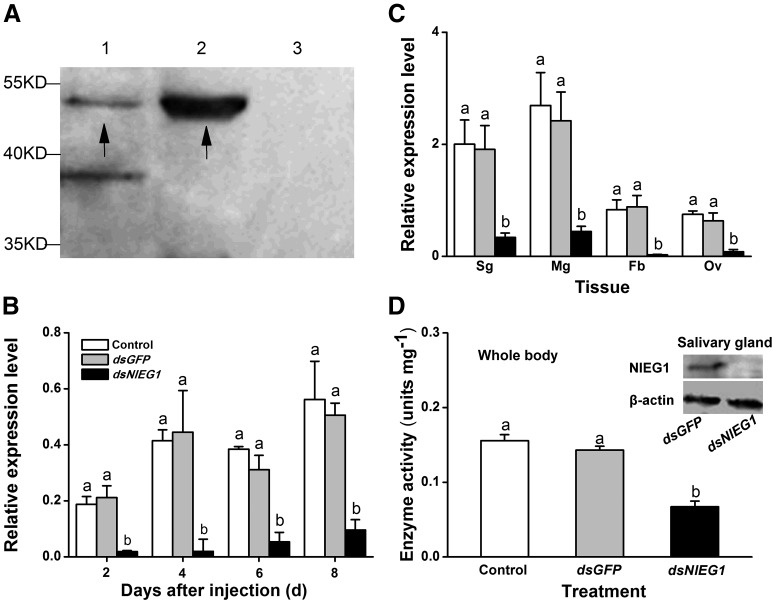
To explore whether BPH excretes NlEG1 through salivation during feeding, rice stems were infested individually with 200 fourth- and fifth-instar nymphs for 24 h, after which the outer three leaf sheaths were harvested and the proteins were extracted. Western-blot analysis was performed using polyclonal anti-NlEG1 rabbit antibodies. As shown in Figure 3A (lane 2), a band of about 50 kD was detected in plants attacked by BPH. The same band also was detected in extracts from BPH salivary glands (Fig. 3A, lane 1). On the other hand, the band of NlEG1 was not detected in control plants (noninfested plants; Fig. 3A, lane 3). These results indicate that NlEG1 is transferred from BPH salivary glands to the plant during feeding. A band of about 38 kD also was detected in extracts of the salivary glands (Fig. 3A, lane 1) but could not be detected in the plant extracts either before or after BPH feeding (Fig. 3A, lanes 2 and 3). It appears that the rabbit used for raising antibodies against the protein NlEG1 polypeptide also contained antibodies that matched another salivary gland protein of 38 kD. These results show that NlEG1 accumulates in rice plants after BPH feeding and, therefore, is likely excreted into the plant by the herbivore.
Figure 3.
NlEG1 in rice and its silencing efficiency by RNAi. A, Detection of protein NlEG1 in rice infested by BPH nymphs. Protein samples for western-blot analysis were as follows: extract from the salivary glands (lane 1) and extracts from rice plants that were infested by nymphs (lane 2) or kept noninfested (lane 3). The black arrows represent the target band. B and C, Mean transcript levels + se (n = 3) of NlEG1 in whole bodies on different days (B) and in different tissues of newly emerged brachypterous female adults 3 d (C) after they (fifth-instar nymphs) had been injected with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP) or kept noninjected (control). Sg, Salivary gland; Mg, midgut; Ov, ovary; Fb, fat body. D, Mean endo-β-1,4-glucanase activities + se (n = 4) in the whole body 3 d after the insects received the same treatments as above. The inset shows western-blot analysis for NlEG1 in the salivary glands of BPH that received the same treatments as above. Letters indicate significant differences among different treatments (P < 0.05, Duncan’s multiple range test).
Silencing NlEG1 Impairs BPH Feeding and Fecundity
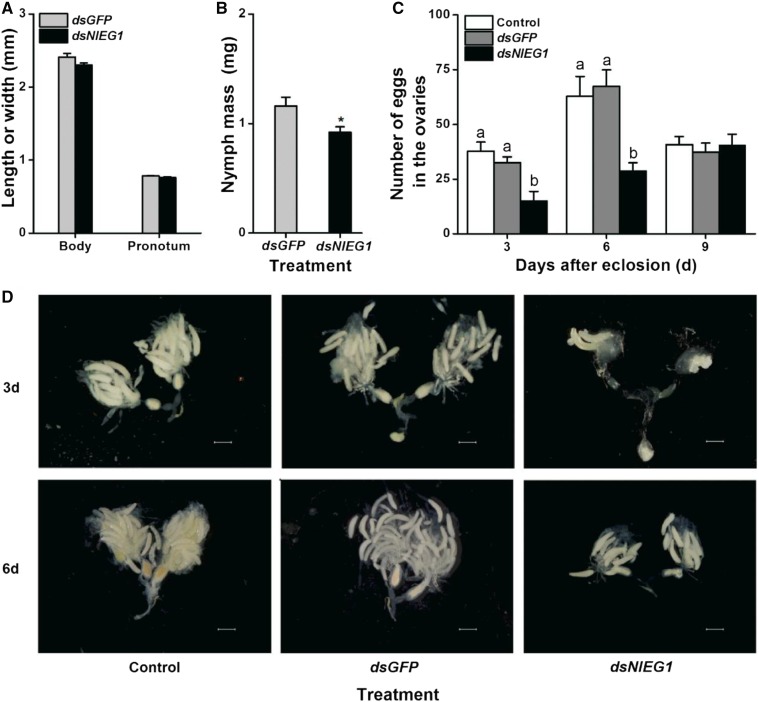
To explore the function of NlEG1, we used RNAi as described by Liu et al. (2010). Injection with double-stranded RNA (dsRNA) of NlEG1 decreased the transcript levels of NlEG1 in the whole body, salivary gland, midgut, fat body, and ovary of BPH by 81% to 95% over a period of 6 d (2–8 d post injection; Fig. 3, B and C). Silencing also reduced the abundance of the protein in the salivary glands and the enzyme activity of NlEG1 (Fig. 3D). Silencing NlEG1 did not influence the body length and pronotum width of BPH nymphs (Fig. 4A; Supplemental Fig. S3). However, the mass of NlEG1-silenced nymphs and the number of eggs in the ovaries of NlEG1-silenced female adults were reduced by about 21% and 54% to 57%, respectively, although the eggs were normal (Fig. 4, B–D).
Figure 4.
Growth phenotypes of BPH nymphs that were injected with dsRNA of NlEG1 or GFP or kept noninjected. A and B, Mean body length and pronotum width + se (n = 11; A) as well as individual mass + se (n = 5; B) of BPH nymphs 6 d after they had been injected with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP). The asterisk indicates a significant difference between treatments (P < 0.05, Student’s t test). C, Mean number of eggs + se (n = 11–15) in the ovary of a female adult that had been injected with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP) or kept noninjected (control) at the fifth-instar nymph stage. Letters indicate significant differences among different treatments (P < 0.05, Duncan’s multiple range test). D, Photographs of ovaries of female adults at 3 and 6 d after eclosion that received the same treatments as in C, showing that knocking down NlEG1 reduces the number of eggs in the ovaries of female adults but does not result in deformities. Bars = 500 µm.
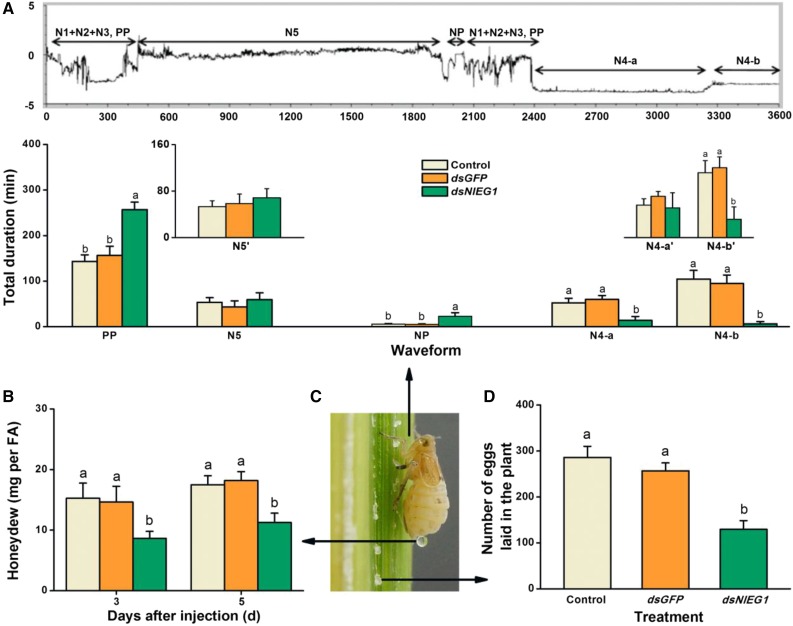
To study the effect of NlEG1 on BPH feeding, we used the electrical penetration graph (EPG) technique, a powerful method used to profile the feeding behavior of piercing-sucking insects (Seo et al., 2009; Cao et al., 2013). Five main phases of feeding can be distinguished by EPG: nonpenetration, the pathway phase (including penetration initiation, salivation and stylet movement, and extracellular activity near the phloem), intracellular activity in the phloem, phloem sap ingestion, and the xylem phase. Representative EPG traces from BPH, including the different phases, are shown in Figure 5A (top). NlEG1 silencing significantly increased the nonpenetration (Fig. 5A, NP) and the pathway phase (Fig. 5A, PP) times. On the other hand, the phloem intracellular activity and ingestion phases were reduced significantly (Fig. 5A, N4-a and N4-b). Eleven of the 15 tested NlEG1-silenced individuals did not reach the phloem during the 6-h trial period, whereas all control individuals did. The few silenced individuals that reached the phloem still spent significantly less time ingesting phloem sap than did controls (Fig. 5A, N4-b′). Silencing NlEG1 also significantly and consistently reduced the amount of secreted honeydew, which is an indicator of the amount of food intake (Fig. 5B). Moreover, in accord with the result on the number of eggs in the ovaries, the number of eggs laid by NlEG1-silenced female adults was decreased by 50% (Fig. 5D). Collectively, these results show that NlEG1 is required for cell wall penetration and feeding as well as fecundity.
Figure 5.
Knocking down NlEG1 reduces the feeding and fecundity of female BPH adults. A, Overall typical view of EPG waveforms generated by the feeding behavior of BPH on rice (top) and mean duration + se (n = 15–19) at different feeding phases of female adults (bottom) that had been injected with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP) or kept noninjected (control) at the fifth-instar nymph stage. NP, Nonpenetration; PP, pathway phase (N1 + N2 + N3), including penetration initiation (N1), salivation and stylet movement (N2), and extracellular activity near the phloem (N3); N4-a, intracellular activity in the phloem region; N4-b, phloem sap ingestion; N5, xylem phase. N4-a′, N4-b′, and N5′ indicate mean duration + se (n = 4–19) excluding the numerical value of zero in N4-a, N4-b, and N5, respectively. EPGs were recorded for 6 h per insect. B, Mean amount of honeydew per day + se (n = 15) secreted by a female BPH adult that received the same treatments as above. FA, Female adult. C, A gravid female adult of BPH on rice, showing its feeding, excreted honeydew, and laying eggs. D, Mean number of eggs + se (n = 11–15) laid by a female adult on plants that received the same treatments as above. Letters indicate significant differences among different treatments (P < 0.05, Duncan’s multiple range test).
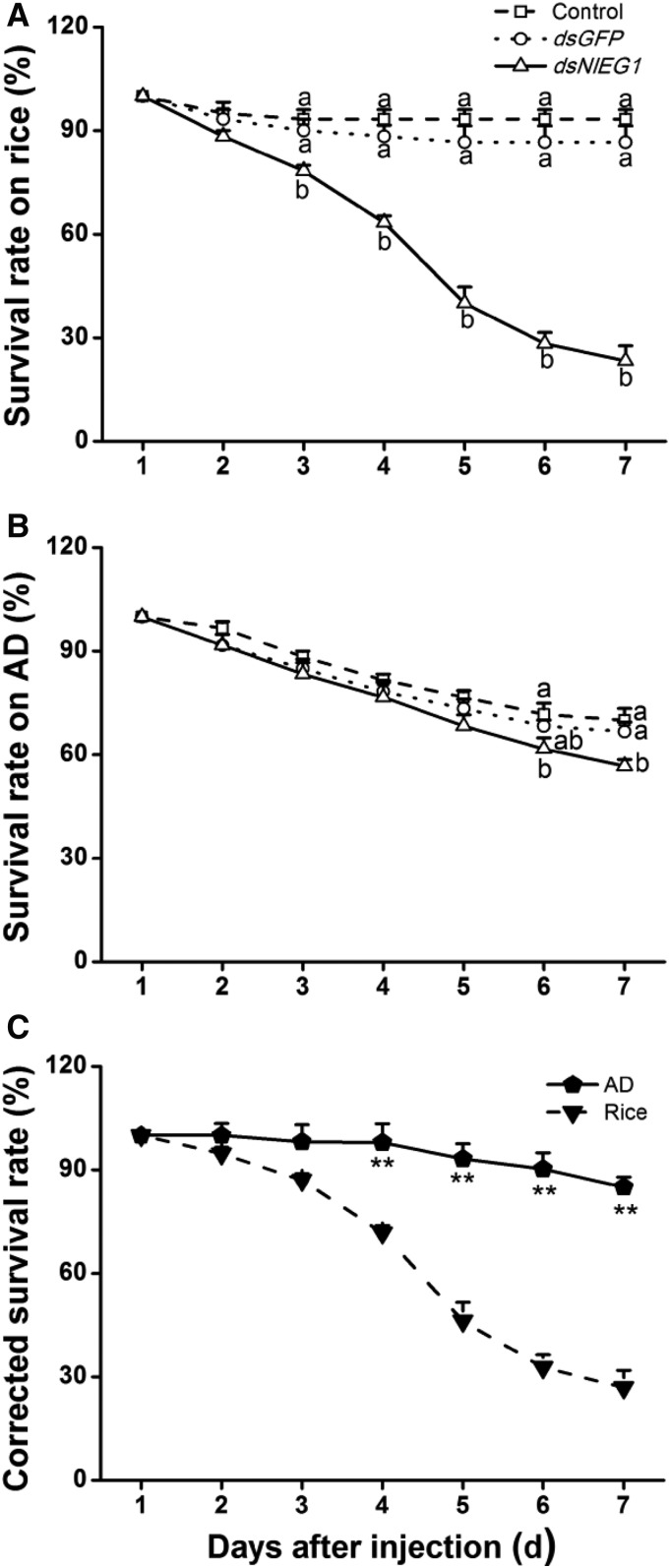
NlEG1 Is Indispensable for BPH Survival on Rice
To further test whether the presence of NlEG1 influences BPH survival rates on rice and whether this influence is related to the effect of NlEG1 on the ability of BPH to reach the phloem, we compared the performance of BPH nymphs on different food matrices. NlEG1 silencing generally reduced the survival rate (Fig. 6), and the effect was most pronounced on rice plants: 3 d after dsRNA injection, the survival rate of BPH on rice dropped significantly, and it was 23% at 7 d postinjection (Fig. 6A). In contrast, the survival rate of BPH nymphs with silenced NlEG1 was much higher in insects raised on artificial diet than in those raised on rice; moreover, the survival rate of BPH nymphs with knocked down NlEG1 raised on artificial diet was reduced only slightly compared with that of control BPH nymphs 7 d after the start of the experiment (Fig. 6B). Compared with that of BPH nymphs with silenced NlEG1 raised on artificial diet, the corrected survival rate of BPH nymphs with silenced NlEG1 raised on rice was significantly lower 4 to 7 d after injection (Fig. 6C). These results demonstrate that NlEG1 contributes to the survival rate of BPH nymphs raised on rice.
Figure 6.
Knocking down NlEG1 decreases survival rates among BPH nymphs. A and B, Mean survival rates + se (n = 4) of BPH nymphs that had been injected with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP) or kept noninjected (control) at the third-instar nymph stage, feeding on rice (A) or artificial diet (AD; B). Letters indicate significant differences among different treatments (P < 0.05, Duncan’s multiple range test). C, Mean corrected survival rates + se (n = 4) of BPH nymphs with injected NlEG1 dsRNA, using BPH nymphs with injected GFP dsRNA as controls, feeding on rice or artificial diet. Asterisks indicate significant differences between treatments (P < 0.01, Student’s t test).
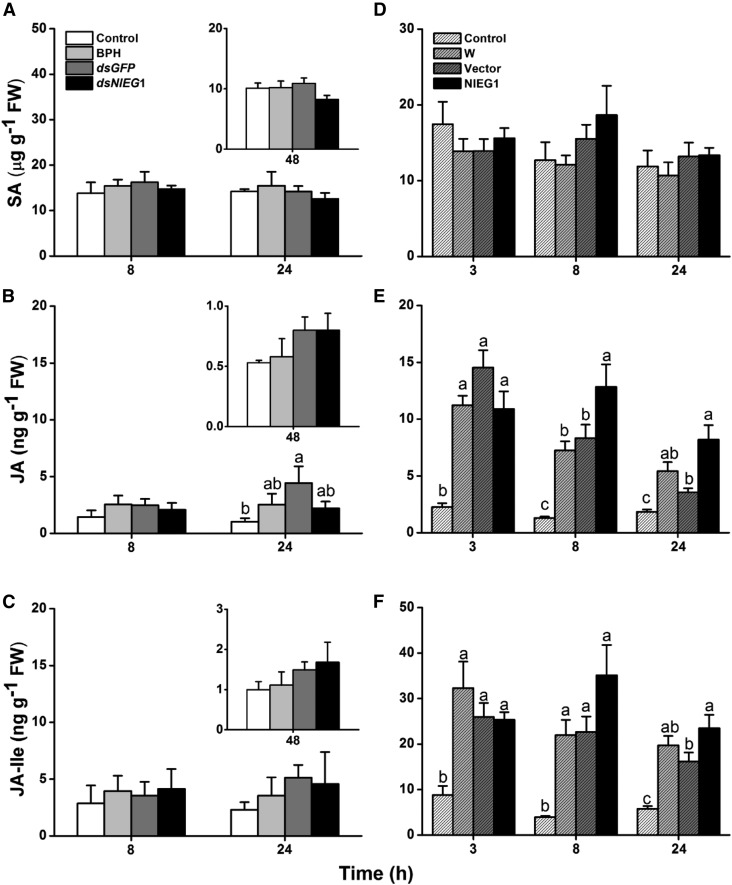
NlEG1 Secreted by BPH Does Not Induce Defense Responses in Rice
Plant hormones, namely, JA, JA-Ile, and SA, play major roles in rice defense against herbivores (Zhou et al., 2009; Lu et al., 2011, 2015). To determine if the salivary protein NlEG1 influences the production of these phytohormones and thus modulates defense responses in rice, we investigated the levels of SA, JA, and JA-Ile in rice after the plant was either infested by fifth-instar BPH nymphs whose ability to produce NlEG1 had been silenced or treated with the recombination protein NlEG1. The results showed that BPH nymph feeding did not induce, or very weakly induced, the production of SA, JA, and JA-Ile from 8 to 48 h after feeding (Fig. 7, A–C). Silencing NlEG1 did not alter the effect of BPH nymph feeding on SA, JA, and JA-Ile levels in plants: levels of these signals in all three treatments (feeding by nymphs, by nymphs with injected dsRNA of GFP, or by nymphs with knocked down NlEG1) and in controls were similar (Fig. 7, A–C). However, compared with plants that were treated with wounding plus the purified elution products from the empty vector, plants treated with wounding plus the application of the recombination protein NlEG1 showed slightly higher levels of JA (8 and 24 h after treatment) and JA-Ile (24 h after treatment) but not SA (Fig. 7, D–F). These findings indicate that the NlEG1 secreted by BPH nymphs during feeding did not elicit JA- and JA-Ile-mediated defense responses in rice, although the recombination protein NlEG1 did slightly induce the production of these signals in plants that had been mechanically wounded.
Figure 7.
NlEG1 secreted by BPH does not affect the levels of SA, JA, and JA-Ile in rice. A to C, Mean levels + se (n = 5) of SA (A), JA (B), and JA-Ile (C) in rice plants at 8 and 24 h after they were kept nonmanipulated (control) or were infested individually by 25 fifth-instar nymphs, which had been injected 3 d earlier with dsRNA of NlEG1 (dsNlEG1) or GFP (dsGFP) or kept noninjected (BPH). Insets show mean levels + se (n = 5) of SA, JA, and JA-Ile in rice plants 48 h after they received the same treatments as stated above. This experiment was not done at the same time as the above experiment. FW, Fresh weight. D to F, Mean levels + se (n = 5) of SA (D), JA (E), and JA-Ile (F) in rice plants at 3, 8, and 24 h after they were kept nonmanipulated (control) or were treated individually with wounding plus 20 μL of the purified recombinant protein NlEG1 (NlEG1), the purified products of the empty vector (Vector), or water (W). Letters indicate significant differences among different treatments (P < 0.05, Duncan’s multiple range test).
DISCUSSION
Our experiments demonstrate that NlEG1 acts as an herbivore effector that enables BPH to overcome the defense of PCWs of rice plants. NlEG1 has endo-β-1,4-glucanase activity but no exoglucanase activity (Fig. 2, B and C) and is injected into rice during BPH feeding (Fig. 3A). Knocking down NlEG1, which significantly reduced the transcript and protein levels as well as the enzyme activity of NlEG1 (Fig. 3, B–D), caused BPH to spend more time in nonpenetration and the pathway phase and less time feeding on phloem (Fig. 5A); this, in turn, decreased the amount of food intake, nymph mass, survival rate, and fecundity of BPH fed on rice (Figs. 4–6). By contrast, NlEG1 silencing did not affect the early ability of BPH to feed on artificial diet without cellulose and rigid PCWs (i.e. for the first 6 d). These results suggest that NlEG1 aids phloem access through cell wall penetration by degrading PCW celluloses.
Endo-β-1,4-glucanases play an important role in degrading PCWs by randomly cleaving amorphous sites of cellulose chains. Thus far, endogenous endo-β-1,4-glucanases have been reported in 16 insect orders; most belong to the GHF 9 and are expressed mainly in the salivary glands and midguts (Calderón-Cortés et al., 2012). NlEG1 also is classified as a GHF 9 protein and is most highly expressed in the midgut and salivary glands of BPH (Fig. 2E; Supplemental Fig. S1), an expression pattern that fits its biological functions well. Interestingly, NlEG1 also was found to be highly expressed in the fat bodies and ovaries of BPH (Fig. 2E), with highest expression levels in female adults (Fig. 2D). This suggests that NlEG1 may have other biological functions. NlEG1 produced in the ovaries, for instance, may be secreted into the plants by the ovipositor of the female adult to soften plant tissues and assist egg deposition; such was found to be the case in Deraeocoris nebulosus, which uses salivary pectinases to soften plant materials before oviposition (Boyd et al., 2002). Our silencing experiments also illustrate that the gene is directly required for egg production in the ovaries (Fig. 4, B and C). NlEG1 expression in the fat body might be related to the detoxification of plant defense chemicals, as has been reported for some PCW-degrading enzymes in insects (Calderón-Cortés et al., 2012). Further research will be necessary to elucidate these roles in NlEG1.
Using the P. pastoris expression system, we obtained the purified recombinant NlEG1 with an estimated molecular mass of about 60 kD by SDS-PAGE (Fig. 2A); this observed mass was about 10 kD greater than the predicted mass of mature NlEG1, 49.6 kD. This inconsistency can be explained by the 36 additional amino acids from the expression vector (which caused the mass of the recombinant protein to increase to 53.9 kD) and the mobility retardation due to the His tag consisting of six His residues in the recombinant NlEG1. That the molecular mass of a His tag fusion protein determined by SDS-PAGE is greater than expected has been reported by many researchers (Niu and Guiltinan, 1994; Qiu et al., 2010; Yin et al., 2013; Wang et al., 2016); and the His tag is known to decrease the mobility of a His tag fusion protein (Tang et al., 2000). In a specific enzyme activity assay, we found that NlEG1 acted hydrolytically on CMC, filter paper, and rice cellulose but not on crystalline cellulose, Glc polymers with β-1,3 or β-1,6 linkages, and Xyl polymers, suggesting a similar substrate specificity of NlEG1 to insect GHF 9 endo-β-1,4-glucanases, such as rCfEG5 and rCfEG3a from Coptotermes formosanus (Zhang et al., 2009, 2011). Using CMC as a substrate, the Km of NlEG1 (6.72 mg mL−1) was similar to the values of other purified GHF 9 enzymes from termites, such as rCfEG5 (2 mg mL−1), rCfEG3a (4.67 mg mL−1), and RsEG (15 mg mL−1) from Reticulitermes speratus; however, the Vmax of NlEG1 (9.91 units mg−1) was much lower than the Vmax of the three enzymes stated as above (548, 590, and 89 units mg−1, respectively; Ni et al., 2010; Zhang et al., 2011). This difference may reflect the various efficiency levels among insect species in degrading celluloses (Oppert et al., 2010; Zhang et al., 2011). As a piercing-sucking herbivore, BPH might not have to possess cellulase activity at levels comparable to those of wood-feeding herbivores, such as termites. We also investigated the optimal temperature and pH condition of NlEG1. The result showed that the pH optimum of NlEG1 was 6, consistent with the pH optimum of cellulases reported in most insects that had an optimal pH between 4 and 6 (Tokuda et al., 1997; Willis et al., 2011). The optimal temperature of NlEG1 was 37°C, at the lower limit of the range of the optimal temperature (37°C–65°C) reported in insect endo-β-1,4-glucanases (Tokuda et al., 1997). This difference may reflect the adaptation of NlEG1 to the environmental conditions in which BPH lives.
It has been reported that cellulases and/or their degraded cell wall fragments, such as oligosaccharides, can induce plant defense responses (Martinez et al., 2001). Similarly, here, we found that wounding plus the application of the recombination protein NlEG1 elicited slightly higher levels of JA and JA-Ile in rice than did wounding plus the addition of the purified elution products from the empty vector (Fig. 7, D–F). However, levels of JA, JA-Ile, and SA in plants infested by BPH nymphs whose NlEG1 had been knocked down were similar to the levels in plants infested by control BPH nymphs and in control plants (Fig. 7, A–C), suggesting that NlEG1 secreted by BPH did not induce these signal-mediated defense responses in rice. This discrepancy is probably related to the feeding behavior of BPH, namely, its use of a stylet to penetrate intercellular spaces, allowing it to suck phloem sap (Seo et al., 2009); this behavior causes little tissue damage and circumvents plant defenses. Moreover, the salivary sheath and some components of the watery saliva secreted and/or formed during the piercing-sucking insect feeding also can reduce the chance that plants will produce defensive responses (Miles, 1968, 1999). These reasons also may explain why BPH nymph feeding had little induction on the production of JA, JA-Ile, and SA in rice (Fig. 7, A–C). Whether NlEG1 elicits the biosynthesis of other defense-related signals, such as ethylene, and thus also acts as a plant defense elicitor, needs to be elucidated. However, our experiments here show that the benefit of NlEG1 as an effector outweighs its potential costs as an elicitor.
CONCLUSION
In summary, our study shows that NlEG1, a salivary endo-β-1,4-glucanase of BPH, is an effector that enables BPH to feed on rice by degrading PCW celluloses and, simultaneously, to circumvent JA- and JA-Ile-mediated defense responses in rice. This finding reveals the molecular basis of how piercing-sucking insects overcome PCW-based resistance traits in plants and provides a plausible mechanism that helps to explain the extraordinary success and impact of BPH as a rice pest since the Green Revolution started in the 1960s; that revolution resulted in extensive plantations of semidwarf rice varieties whose GA pathways had been impaired, varieties that have lower levels of lignin and cellulose than those with normal GA pathways (Okuno et al., 2014; Huang et al., 2015a).
MATERIALS AND METHODS
Plant Growth and Insect Rearing
Mudgo, a rice (Oryza sativa) variety containing the resistance gene Bph1, was used for experiments. Plants were grown as described by Lu et al. (2011), and 35- to 40-d-old plants individually planted in 500-mL hydroponic plastic pots were used. Colonies of BPH were originally provided by the Chinese National Rice Research Institute and maintained on Mudgo at 27°C ± 1°C and 70% ± 10% relative humidity under a 14/10-h light/dark photoperiod.
Cloning, Sequence Alignment, and Phylogenetic Analysis of NlEG1
The full-length cDNA of NlEG1 was obtained by RT-PCR from total RNA isolated from salivary glands of adult BPH females. The primers (Supplemental Table S1) were designed based on the consensus sequence of endo-β-1,4-glucanase genes in insects and transcriptome data of BPH salivary glands and whole bodies (Xue et al., 2010; Ji et al., 2013). PCR-amplified fragments were cloned into the pMD19-T vector (TaKaRa) and sequenced. The NlEG1 sequence was translated and analyzed with the Compute pI/MW tool to predict the pI and molecular mass of the predicted protein (http://expasy.org/tools). NetOGlyc 4.0 (http://www.cbs.dtu.dk/services/NetOGlyc/) and NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc) were used to predict O- and N-glycosylation sites, respectively. Predictions of the signal peptide and transmembrane domain were made using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), respectively. Amino acid sequences of insect endo-β-1,4-glucanase sequences downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) were aligned using ClustalX2. Phylogenetic relationships were determined using MEGA 5.0 with the neighbor-joining method.
RNA Preparation and qRT-PCR
Total RNA was extracted from the following materials: (1) whole bodies of BPH at different developmental stages (from first- to fifth-instar nymphs and newly emerged brachypterous male and female adults) and (2) specific tissue of BPH: salivary glands, thorax muscles, integuments, midguts, ovaries, legs, and fat bodies that had been dissected from newly emerged brachypterous female adults. Total RNA was isolated using the SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions. One microgram of each total RNA sample was reverse transcribed using the PrimeScript RT-PCR Kit (TaKaRa). qRT-PCR was performed on the CFX96 Real-Time System (Bio-Rad) using SsoFast probes supermix (Bio-Rad). A linear standard curve, threshold cycle number versus log (designated transcript level), was constructed using a series dilution of a specific cDNA standard, and the relative levels of the transcript of the target gene in all unknown samples were determined according to the standard curve. A BPH actin gene (GenBank accession no. EU179848) was used as an internal standard to normalize cDNA concentrations. The primers and probes used for qRT-PCR for all tested genes are provided in Supplemental Table S1. Three to four independent biological replicates were analyzed in each experiment.
Expression of NlEG1 in Pichia pastoris
The full-length open reading frame of NlEG1 was PCR amplified using a pair of primers (Supplemental Table S1) and cloned into the P. pastoris expression vector pPICZα A (Invitrogen). The recombinant vector NlEG1:pPICZɑ A and empty vector pPICZɑ A (as a control) were transformed into P. pastoris strain KM71 (Invitrogen). Expression was induced by adding 100% methanol to a final concentration of 1% (v/v). The expressed products from the empty vector and the recombinant vector were purified using nickel-nitrilotriacetic acid agarose columns and following the instructions in the Ni-NTA Superflow Cartridge Handbook (Qiagen), and the purified products were concentrated with a YM-10 Microcon centrifugal filter device (Millipore) to remove imidazole. The final purified concentrated products from P. pastoris cells with the empty vector and recombinant vector were mixed with 2× SDS loading buffer, separated by SDS-PAGE on a 12% (w/v) gradient gel, and stained with 0.025% (w/v) Coomassie Blue R-250 in water. The predicted mass of the mature recombinant protein NlEG1, including six C-terminal His tags, is 53.9 kD.
Polyclonal Antibody Preparation and Western-Blot Analysis
Based on the Optimum Antigen design tool, a polypeptide (WRGDSSLNDRGLKGC) of NlEG1 was selected as the antigen to produce the rabbit polyclonal antibodies, and the polyclonal antibodies were purified by GenScript. The following protein samples used for western-blot analysis were prepared. (1) Proteins extracted from salivary glands of BPH. The salivary glands of 100 newly emerged adult females were collected and homogenized in 1 mL of phosphate-buffered saline. The extract was centrifuged at 12,000g for 5 min at 4°C, and the supernatant was collected as samples. (2) Proteins from rice leaf sheaths, infested by BPH or not. Rice stems were confined individually within glass cylinders (diameter, 4 cm; height, 8 cm; with 48 small holes, diameter, 0.8 mm) in which 200 fourth- or fifth-instar nymphs were released, and 24 h later, the herbivore was removed. Plants in empty glass cylinders were used as controls. For each rice stem, the outer three leaf sheaths were harvested, and the entire leaf sheaths (0.9 g) from three rice stems were merged and homogenized in 4 mL of phosphate-buffered saline in liquid nitrogen. The extract was centrifuged at 12,000g for 5 min at 4°C, and the supernatant was collected and concentrated to 200 µL using a YM-3 Microcon centrifugal filter device (Millipore). SDS-PAGE loading buffer (2×) was added to samples, and these samples were then subjected to SDS-PAGE on a 12% (w/v) gradient gel and transferred onto a polyvinylidene difluoride membrane. Nonspecific binding sites were blocked with 5% (w/v) instant nonfat dry milk, and membranes were incubated with the purified polyclonal antibody. The antigen-antibody complexes were visualized with horseradish peroxidase-conjugated goat anti-rabbit IgG (Multisciences) at a dilution of 1:5,000 at 37°C for 1 h followed by extensive washing for 30 min with frequent changes of Tris-buffered saline plus Tween 20 and detected by FluorChem FC2 (Alpha Innotech).
Endo-β-1,4-Glucanase Activity Assay
The endo-β-1,4-glucanase activity of the protein samples was determined, using CMC as a substrate, by a reducing sugar-releasing assay as described previously with some modifications (Li et al., 1998). Briefly, 400 μL of 1% (w/v) CMC-Na solution in 50 mm sodium acetate buffer (pH 6) mixed with 15 μL of the sample was incubated at certain temperatures (see below) for 30 min. We first used this method to investigate the optimal pH and temperature condition of NlEG1 produced from P. pastoris. For the optimum temperature, the enzyme activity was measured at 28°C, 37°C, and 50°C at pH 6, and for the optimum pH, the activity was determined at 37°C at pH 5, 6, and 7 (adjusted by 50 mm sodium acetate buffer solution). Based on these experimental data, we found that NlEG1 has its highest activity at 37°C at pH 6. Thus, we investigated the enzyme activity of the proteins extracted from newly emerged adult females 3 d after injection at 37°C at pH 6. For the extraction of BPH proteins, newly emerged BPH female adults were homogenized in 50 mm sodium acetate buffer (pH 6) on ice, and then the solution was centrifuged at 12,000 rpm for 20 min at 4°C. The supernatant (proteins) was collected as samples. Endo-β-1,4-glucanase activity was defined as units per mg of protein, where 1 unit of enzyme activity was defined as the amount of enzyme that produced 1 µmol of reducing sugars (Glc equivalent) per min. The concentration of the proteins was measured in triplicate using the Bio-Rad Bradford Protein Assay.
Specific Enzymatic Activity and Kinetics of NlEG1
Using the reducing sugar assay, the specific enzyme activity of NlEG1 was measured. Six substrates were used in the assay, including Whatman No. 1 filter paper (Whatman), cellulose extracted from rice plants using the method described by Rosa et al. (2012), Avicel (PH101-type crystalline cellulose; Sigma-Aldrich), curdlan (β-1,3-glucan; Sigma-Aldrich), laminarin (β-1,3:β-1,6-glucan; Sigma-Aldrich), and xylan (poly β-1,4-xylopyranose; Sigma-Aldrich). An aliquot of 400 μL of 1% (w/v) substrate solution in 50 mm sodium acetate buffer (pH 6) was mixed with 10 μL of NlEG1 (500 ng), and then the mixture was incubated for 1 h at 37°C.
The enzyme kinetics of NlEG1 was measured using serial concentrations of CMC (from 1 to 10 mg mL−1) in 50 mm sodium acetate buffer (pH 6). An aliquot of 20 μL of NlEG1 (1,000 ng) was added into 400 μL of CMC solution, and then the mixture was incubated for 5 min at 37°C. Lineweaver-Burk plots were drawn using Microsoft Excel 2010, and Km and Vmax were determined.
RNAi Experiment
A 215-bp fragment of NlEG1 and a 657-bp fragment of control gene GFP were amplified by RT-PCR with primers including a T7 promoter sequence (Supplemental Table S1). The PCR products were used to synthesize dsRNA in vitro using the MEGAscript RNAi kit (Ambion). Third- or fifth-instar nymphs (for details, see descriptions of different experiments) were injected as described previously (Liu et al., 2010). Each nymph was injected with about 0.1 μg of dsRNA of NlEG1 or GFP (control) or not injected (control). To determine the efficiency of gene silencing after dsRNA injection, the levels of NlEG1 transcripts in the whole body, salivary gland, midgut, ovary, and fat body of the insect that had been injected with NlEG1 or GFP dsRNA, or not injected, were investigated at 2, 4, 6, and 8 d after injection.
BPH Bioassays
To measure survival rates of BPH, third-instar nymphs injected with NlEG1 or GFP dsRNA, or kept noninjected, were allowed to feed on rice plants or artificial diet. The treated insects were first kept on rice seedlings at 27°C ± 1°C with 70% ± 10% relative humidity and a 14/10-h (light/dark) photoperiod to recover for 1 d, and then the healthy ones were used for the following bioassay. Stems of rice plants (one plant per pot) were confined individually within glass cylinders as stated above into which 15 third-instar BPH nymphs were released. In the artificial diet experiment, 15 third-instar BPH nymphs were introduced into individual feeding chambers (9 cm long and 2 cm in diameter) as described previously (Fu et al., 2001). The number of surviving BPH nymphs in each cylinder or feeding chamber was recorded every day. The survival rate of each BPH treatment and the corrected survival rates of BPH nymphs with injected NlEG1 dsRNA, using BPH nymphs with injected GFP dsRNA as controls, on rice or artificial diet were calculated. The experiment was repeated four times.
To investigate the influence of NlEG1 knockdown on the growth phenotype of BPH nymphs, third-instar nymphs injected with NlEG1 or GFP dsRNA were allowed to feed on rice plants. Six days later, the body length and pronotum width of BPH nymphs (using the MshotDigital Imaging System) as well as nymph mass (to an accuracy of 0.1 mg) were measured. Photographs of nymphs also were taken with a light stereomicroscope (Olympus SZX7). The measurement for body length and pronotum width was repeated 11 times. The mass measurement was replicated five times; each time, the total mass of 20 nymphs was measured and the average individual mass was calculated.
To assess the effect of the knockdown of NlEG1 on BPH feeding, a brachypterous female adult at 3 d (newly emerged) and 5 d after the injection of NlEG1 or GFP dsRNA, or no injection (fifth-instar nymphs were injected), was placed into a small Parafilm bag (6 × 5 cm), which was then fixed on the stem of a rice plant. The amount of honeydew excreted by a female adult was weighed (to an accuracy of 0.1 mg) at 24 h after the start of the experiment. The experiment was replicated 15 times.
The effect of NlEG1 knockdown on the fecundity of BPH female adults also was investigated. Stems of rice plants (one plant per pot) were confined individually within glass cylinders into which were released one newly emerged BPH female adult, 2 d after the injection of NlEG1 or GFP dsRNA or no injection (fifth-instar nymphs were injected), and one newly emerged BPH male adult without treatment. Eleven days later, the insect was removed and the number of eggs laid by female adults in each rice plant was counted with a microscope. The number of eggs in the ovary of each female adult from the three treatment groups, at 3, 6, and 9 d after eclosion, also was counted with a microscope. The experiment was repeated 11 to 15 times.
EPG Recording of BPH Feeding Behavior
The feeding behavior of BPH was recorded on a direct-current EPG system (Wageningen Agricultural University). The method was the same as that described by Cao et al. (2013). All experiments were carried out at 26°C ± 1°C and 70% ± 10% relative humidity under continuous light conditions. The feeding behavior of individual newly emerged adult females, 4 d after the injection of NlEG1 or GFP dsRNA, or not injected (fifth-instar nymphs were injected), on rice was monitored for 6 h. For each treatment (group), 15 to 19 replications were recorded. The signals recorded were analyzed using PROBE version 3.4 software (Wageningen Agricultural University). The output signals from EPG recordings were classified into five typical waveforms associated with the stylet penetration behavior of BPH (Seo et al., 2009; Cao et al., 2013), including NP for nonpenetration, PP (N1 + N2 + N3) for the pathway phase (including penetration initiation, salivation and stylet movement, and extracellular activity near the phloem), N4-a for intracellular activity in the phloem, N4-b for phloem sap ingestion, and N5 for the xylem phase (Fig. 5A). The durations of each sequential waveform event for each insect were measured, and the average waveform duration per insect (in minutes) for each waveform was calculated for each treatment (Cao et al., 2013). Another variable used in this experiment was the duration of N4-a′, N4-b′, and N5′, meaning the duration excluding the numerical value of zero in N4-a, N4-b, and N5, respectively.
JA, JA-Ile, and SA Analysis
Potted plants (one per pot) were randomly assigned to the following treatments. (1) Infestation by different BPH nymph groups. Plant stems were confined individually in glass cylinders into which 25 fifth-instar nymphs that had been injected with NlEG1 or GFP dsRNA, or kept noninjected for 3 d, were released. (2) NlEG1 treatment. Plants were pierced individually 200 times on the lower part of the stems (about 2 cm) with a #00 insect pin and treated with 20 µL of the recombinant protein NlEG1 (32.7 ng µL−1), the purified products of the empty vector, or water, or they were kept nonmanipulated (control). The outer three leaf sheaths of stems were harvested at different time points (for details, see Fig. 7) after the start of the treatment. Samples were ground in liquid nitrogen, and SA, JA, and JA-Ile were extracted with ethyl acetate spiked with labeled internal standards ([2D4]SA, [2D6]JA, and [2D6]JA-Ile) and then analyzed with HPLC-mass spectrometry/mass spectrometry following the method described by Lu et al. (2015).
Data Analysis
Differences in BPH performance, expression levels of the gene, and JA, JA-Ile, and SA levels between treatments were determined by ANOVA (Student’s t test for comparing two treatments). All tests were carried out with Statistica (SAS Institute; http://www.sas.com/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KM459012 (NlEG1) and EU179848 (Nlactin) .
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein alignment of GHF 9 enzymes from insects.
Supplemental Figure S2. Phylogenetic tree for amino acid sequences of NlEG1 and reported insect endogenous endo-β-1,4-glucanases.
Supplemental Figure S3. Growth phenotypes of BPH nymphs 6 d after they had been injected with either GFP or NlEG1 dsRNA.
Supplemental Table S1. Primers and probes used for qRT-PCR and PCR.
Supplementary Material
Acknowledgments
We thank Jian Xue for providing the BPH transcriptome data; Meng Ye, Tingting Cao, Zhen Zhang, Qiyao Chai, and Meng Zou for invaluable assistance with the experiments; Ian T. Baldwin and Matthias Erb for insightful discussions; and Matthias Erb and Emily Wheeler for editorial assistance.
Glossary
- JA
jasmonic acid
- JA-Ile
jasmonoyl-isoleucine
- SA
salicylic acid
- PCW
plant cell wall
- BPH
brown planthopper
- RT
reverse transcription
- CMC
carboxymethyl cellulose
- qRT
quantitative real-time
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- EPG
electrical penetration graph
Footnotes
This study was supported by Projects of International Cooperation and Exchanges of the National Natural Science Foundation of China (grant no. 31520103912), the Special Fund for Agro-Scientific Research in the Public Interest (grant no. 201403030), the Special Fund for Agro-Scientific Research in the Public Interest of Zhejiang (grant no. 2014C22004), and the earmarked fund for the China Agriculture Research System (grant no. CARS-01-21).
Articles can be viewed without a subscription.
References
- Abdellatef E, Will T, Koch A, Imani J, Vilcinskas A, Kogel KH (2015) Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol J 13: 849–857 [DOI] [PubMed] [Google Scholar]
- Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I (2013) In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol Plant Microbe Interact 26: 67–74 [DOI] [PubMed] [Google Scholar]
- Backus EA, Andrews KB, Shugart HJ, Greve LC, Labavitch JM, Alhaddad H (2012) Salivary enzymes are injected into xylem by the glassy-winged sharpshooter, a vector of Xylella fastidiosa. J Insect Physiol 58: 949–959 [DOI] [PubMed] [Google Scholar]
- Bos JI, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DW, Cohen AC, Alverson DR (2002) Digestive enzymes and stylet morphology of Deraeocoris nebulosus (Hemiptera: Miridae), a predacious plant bug. Ann Entomol Soc Am 95: 395–401 [Google Scholar]
- Calderón-Cortés N, Quesada M, Watanabe H, Cano-Camacho H, Oyama K (2012) Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu Rev Ecol Evol Syst 43: 45–71 [Google Scholar]
- Cao TT, Lü J, Lou YG, Cheng JA (2013) Feeding-induced interactions between two rice planthoppers, Nilaparvata lugens and Sogatella furcifera (Hemiptera: Delphacidae): effects on feeding and honeydew excretion. Environ Entomol 42: 1281–1291 [DOI] [PubMed] [Google Scholar]
- Carolan JC, Fitzroy CI, Ashton PD, Douglas AE, Wilkinson TL (2009) The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9: 2457–2467 [DOI] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga DA, Jander G (2013) The role of protein effectors in plant-aphid interactions. Curr Opin Plant Biol 16: 451–456 [DOI] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Tumlinson JH (2008) Plant-insect dialogs: complex interactions at the plant-insect interface. Curr Opin Plant Biol 11: 457–463 [DOI] [PubMed] [Google Scholar]
- Fu Q, Zhang Z, Hu C, Lai F, Sun Z (2001) A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl Entomol Zool (Jpn) 36: 111–116 [Google Scholar]
- Harmel N, Létocart E, Cherqui A, Giordanengo P, Mazzucchelli G, Guillonneau F, De Pauw E, Haubruge E, Francis F (2008) Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect Mol Biol 17: 165–174 [DOI] [PubMed] [Google Scholar]
- Heong KL, Cheng J, Escalada MM (2014) Rice Planthoppers: Ecology, Management, Socio Economics and Policy. Zhejiang University and Springer, Hangzhou, China [Google Scholar]
- Hogenhout SA, Bos JI (2011) Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol 14: 422–428 [DOI] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, et al. (2015a) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HJ, Liu CW, Cai YF, Zhang MZ, Bao YY, Zhang CX (2015b) A salivary sheath protein essential for the interaction of the brown planthopper with rice plants. Insect Biochem Mol Biol 66: 77–87 [DOI] [PubMed] [Google Scholar]
- Huang HJ, Liu CW, Huang XH, Zhou X, Zhuo JC, Zhang CX, Bao YY (2016) Screening and functional analyses of Nilaparvata lugens salivary proteome. J Proteome Res 15: 1883–1896 [DOI] [PubMed] [Google Scholar]
- Ji R, Yu H, Fu Q, Chen H, Ye W, Li S, Lou Y (2013) Comparative transcriptome analysis of salivary glands of two populations of rice brown planthopper, Nilaparvata lugens, that differ in virulence. PLoS ONE 8: e79612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Choo YM, Lee KS, Hong SJ, Seol KY, Je YH, Sohn HD, Jin BR (2008) Molecular cloning and characterization of a glycosyl hydrolase family 9 cellulase distributed throughout the digestive tract of the cricket Teleogryllus emma. Comp Biochem Physiol B Biochem Mol Biol 150: 368–376 [DOI] [PubMed] [Google Scholar]
- Konishi H, Noda H, Tamura Y, Hattori M (2009) Proteomic analysis of the salivary glands of the rice brown planthopper, Nilaparvata lugens (Stal) (Homoptera: Delphacidae). Appl Entomol Zool (Jpn) 44: 525–534 [Google Scholar]
- Li X, Lin W, Gao P, Chen F (1998) Endoglucanase S, a novel endocellulase exhibiting exoglucanase activity from a newly isolated Streptomyces sp. LX. J Appl Microbiol 85: 347–356 [Google Scholar]
- Liu S, Ding Z, Zhang C, Yang B, Liu Z (2010) Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol 40: 666–671 [DOI] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y (2011) An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J 68: 583–596 [DOI] [PubMed] [Google Scholar]
- Lu J, Robert CAM, Riemann M, Cosme M, Mène-Saffrané L, Massana J, Stout MJ, Lou Y, Gershenzon J, Erb M (2015) Induced jasmonate signaling leads to contrasting effects on root damage and herbivore performance. Plant Physiol 167: 1100–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C, Blanc F, Le Claire E, Besnard O, Nicole M, Baccou JC (2001) Salicylic acid and ethylene pathways are differentially activated in melon cotyledons by active or heat-denatured cellulase from Trichoderma longibrachiatum. Plant Physiol 127: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles P. (1968) Insect secretions in plants. Annu Rev Phytopathol 6: 137–164 [Google Scholar]
- Miles PW. (1999) Aphid saliva. Biol Rev Camb Philos Soc 74: 41–85 [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW (2002) Herbivory: caterpillar saliva beats plant defences. Nature 416: 599–600 [DOI] [PubMed] [Google Scholar]
- Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen MS, Park Y, Dittmer N, Marshall J, Reese JC, et al. (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci USA 105: 9965–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Watanabe H, Saitoh H, Tokuda G, Azuma JI (2002) Dual cellulose-digesting system of the wood-feeding termite, Coptotermes formosanus Shiraki. Insect Biochem Mol Biol 32: 777–784 [DOI] [PubMed] [Google Scholar]
- Ni J, Takehara M, Watanabe H (2010) Identification of activity related amino acid mutations of a GH9 termite cellulase. Bioresour Technol 101: 6438–6443 [DOI] [PubMed] [Google Scholar]
- Nicholson SJ, Hartson SD, Puterka GJ (2012) Proteomic analysis of secreted saliva from Russian wheat aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J Proteomics 75: 2252–2268 [DOI] [PubMed] [Google Scholar]
- Niu X, Guiltinan MJ (1994) DNA binding specificity of the wheat bZIP protein EmBP-1. Nucleic Acids Res 22: 4969–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno A, Hirano K, Asano K, Takase W, Masuda R, Morinaka Y, Ueguchi-Tanaka M, Kitano H, Matsuoka M (2014) New approach to increasing rice lodging resistance and biomass yield through the use of high gibberellin producing varieties. PLoS ONE 9: e86870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppert C, Klingeman WE, Willis JD, Oppert B, Jurat-Fuentes JL (2010) Prospecting for cellulolytic activity in insect digestive fluids. Comp Biochem Physiol B Biochem Mol Biol 155: 145–154 [DOI] [PubMed] [Google Scholar]
- Petrova A, Smith CM (2014) Immunodetection of a brown planthopper (Nilaparvata lugens Stål) salivary catalase-like protein into tissues of rice, Oryza sativa. Insect Mol Biol 23: 13–25 [DOI] [PubMed] [Google Scholar]
- Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE 6: e25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Wang Y, Wang J, Zhang F, Ma J (2010) Expression of biologically active recombinant antifreeze protein His-MpAFP149 from the desert beetle (Microdera punctipennis dzungarica) in Escherichia coli. Mol Biol Rep 37: 1725–1732 [DOI] [PubMed] [Google Scholar]
- Rosa SML, Rehman N, Miranda MIGD, Nachtigall SMB, Bica CID (2012) Chlorine-free extraction of cellulose from rice husk and whisker isolation. Carbohydr Polym 87: 1131–1138 [Google Scholar]
- Santiago R, Barros-Rios J, Malvar RA (2013) Impact of cell wall composition on maize resistance to pests and diseases. Int J Mol Sci 14: 6960–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman MC, Baldwin IT (2016) The layers of plant responses to insect herbivores. Annu Rev Entomol 61: 373–394 [DOI] [PubMed] [Google Scholar]
- Seo BY, Kwon YH, Jung JK, Kim GH (2009) Electrical penetration graphic waveforms in relation to the actual positions of the stylet tips of Nilaparvata lugens in rice tissue. J Asia Pac Entomol 12: 89–95 [Google Scholar]
- Sogawa K. (1968) Studies on the salivary glands of rice plant leafhopper. III. Salivary phenolase. Appl Entomol Zool (Jpn) 3: 13–25 [Google Scholar]
- Stam JM, Kroes A, Li Y, Gols R, van Loon JJ, Poelman EH, Dicke M (2014) Plant interactions with multiple insect herbivores: from community to genes. Annu Rev Plant Biol 65: 689–713 [DOI] [PubMed] [Google Scholar]
- Tang W, Zhang J, Wang Z, Hong M (2000) The cause of deviation made in determining the molecular weight of His-tag fusion proteins by SDS-PAGE. Acta Phytophysiol Sin 26: 64–68 [Google Scholar]
- Tokuda G, Watanabe H, Matsumoto T, Noda H (1997) Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): distribution of cellulases and properties of endo-β-1,4-glucanase. Zoolog Sci 14: 83–93 [DOI] [PubMed] [Google Scholar]
- Wang W, Dai H, Zhang Y, Chandrasekar R, Luo L, Hiromasa Y, Sheng C, Peng G, Chen S, Tomich JM, et al. (2015) Armet is an effector protein mediating aphid-plant interactions. FASEB J 29: 2032–2045 [DOI] [PubMed] [Google Scholar]
- Wang W, Li M, Lin H, Wang J, Mao X (2016) The Vibrio parahaemolyticus-infecting bacteriophage qdvp001: genome sequence and endolysin with a modular structure. Arch Virol 161: 2645–2652 [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJ (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104: 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JD, Oppert B, Oppert C, Klingeman WE, Jurat-Fuentes JL (2011) Identification, cloning, and expression of a GHF9 cellulase from Tribolium castaneum (Coleoptera: Tenebrionidae). J Insect Physiol 57: 300–306 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Xue J, Bao YY, Li BL, Cheng YB, Peng ZY, Liu H, Xu HJ, Zhu ZR, Lou YG, Cheng JA, et al. (2010) Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS ONE 5: e14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang Y, Yan M, Zhang X, Pan H, Xu X, Han Z (2013) Molecular cloning, polyclonal antibody preparation, and characterization of a functional iron-related transcription factor IRO2 from Malus xiaojinensis. Plant Physiol Biochem 67: 63–70 [DOI] [PubMed] [Google Scholar]
- Zhang D, Lax AR, Bland JM, Allen AB (2011) Characterization of a new endogenous endo-β-1,4-glucanase of Formosan subterranean termite (Coptotermes formosanus). Insect Biochem Mol Biol 41: 211–218 [DOI] [PubMed] [Google Scholar]
- Zhang D, Lax AR, Raina AK, Bland JM (2009) Differential cellulolytic activity of native-form and C-terminal tagged-form cellulase derived from Coptotermes formosanus and expressed in E. coli. Insect Biochem Mol Biol 39: 516–522 [DOI] [PubMed] [Google Scholar]
- Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, Lou Y (2009) Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J 60: 638–648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.