The transcription factor MYB29 integrates various hormonal, growth, and stress signals with mitochondrial retrograde signaling in Arabidopsis.
Abstract
Plants sense and integrate a variety of signals from the environment through different interacting signal transduction pathways that involve hormones and signaling molecules. Using ALTERNATIVE OXIDASE1a (AOX1a) gene expression as a model system of retrograde or stress signaling between mitochondria and the nucleus, MYB DOMAIN PROTEIN29 (MYB29) was identified as a negative regulator (regulator of alternative oxidase1a 7 [rao7] mutant) in a genetic screen of Arabidopsis (Arabidopsis thaliana). rao7/myb29 mutants have increased levels of AOX1a transcript and protein compared to wild type after induction with antimycin A. A variety of genes previously associated with the mitochondrial stress response also display enhanced transcript abundance, indicating that RAO7/MYB29 negatively regulates mitochondrial stress responses in general. Meta-analysis of hormone-responsive marker genes and identification of downstream transcription factor networks revealed that MYB29 functions in the complex interplay of ethylene, jasmonic acid, salicylic acid, and reactive oxygen species signaling by regulating the expression of various ETHYLENE RESPONSE FACTOR and WRKY transcription factors. Despite an enhanced induction of mitochondrial stress response genes, rao7/myb29 mutants displayed an increased sensitivity to combined moderate light and drought stress. These results uncover interactions between mitochondrial retrograde signaling and the regulation of glucosinolate biosynthesis, both regulated by RAO7/MYB29. This common regulator can explain why perturbation of the mitochondrial function leads to transcriptomic responses overlapping with responses to biotic stress.
Plants are exposed to a variety and combination of abiotic and biotic stresses on a daily to seasonal basis that can negatively affect growth. To survive and reproduce, plants must sense and cope with a multitude of environmental cues and mount an integrated defense response (Suzuki et al., 2014), which is different from, and cannot be accurately predicted from, the analysis of single-stress responses (Rizhsky et al., 2004; Mittler, 2006). The combined response is coordinated via various signaling pathways that converge or interact at different levels. Plant hormones play a central role in mediating and integrating these responses, with the well-characterized example of abscisic acid (ABA) that is required for acclimation to combined salt and heat stresses (Suzuki et al., 2016). The responses can be direct, often referred to as anterograde signaling, in which the signal is transmitted straight to the nucleus without any impact on organellar functions. In addition, environmental conditions may affect the organelles (mitochondria or plastids) and, subsequently, elicit a signal transduction pathway to the nucleus, often referred to as retrograde signaling.
When plants are exposed to multiple adverse conditions, they might respond by compromising or superseding the response to another condition. The crosstalk between salicylic acid (SA) and ethylene (ET)/jasmonic acid (JA) signaling pathways, both involved in biotic stress responses, not only fine-tunes plant responses to different pathogens, but also allows the prioritization of one pathway above another under multiple biotic stimuli (Pieterse et al., 2012). In Arabidopsis (Arabidopsis thaliana), treatment with the bacterial elicitor flagellin 22 (flg22) that mimics some biotic stresses strongly suppresses flavonol biosynthesis by UV-B stress (abiotic stress; Schenke et al., 2011), a mechanism believed to allow plants to prioritize secondary metabolism in favor of pathogen defense (Schenke et al., 2014). A comprehensive analysis of stress combinations has revealed potentially negative or positive interactions (i.e. enhanced damage or cross-protection, respectively, due to more than one stress condition; Mittler, 2006). Drought or heat stresses have been shown to decrease resistance to some biotic stresses, such as high-temperature suppression of the host resistance against Tobacco mosaic virus (Király et al., 2008). Transcriptome responses to sequential stress treatments have shown that significant first-class signatures could still be detected in subsequent stress responses, although the transcriptome signature of the second stress applied dominated (Coolen et al., 2016). A number of interactions of abiotic and biotic responses have been analyzed, and the underlying plant hormones, transcription factors (TFs), and kinase signaling pathways that mediate the interactions between these pathways are beginning to be elucidated (Mittler, 2006; Atkinson and Urwin, 2012; Prasch et al., 2015).
The nucleus-encoded mitochondrial ALTERNATIVE OXIDASE (AOX) is widely used as a model to study mitochondrial retrograde signaling (MRS; Rhoads and Subbaiah, 2007; Vanlerberghe, 2013; Ng et al., 2014). AOX is induced in a variety of plants by a broad range of adverse conditions, ranging from nutrient limitation, drought, high/low temperature to attack by various biotic pests (Vanlerberghe, 2013). This wide induction array indicates that different pathways can trigger AOX; for instance, AOX is known to be induced by reactive oxygen species (ROS)-dependent and ROS-independent pathways (Gray et al., 2004). However, the crosstalk between these pathways, with a few exceptions, is not elucidated at the molecular level. AOX1a has been shown to be regulated in Arabidopsis by SUCROSE-NONFERMENTING-RELATED PROTEIN KINASE CATALYTIC SUBUNIT α (KIN10) and CYCLIN-DEPENDENT KINASE E;1 (CDKE;1; Ng et al., 2013a) kinases and by ABSCISIC ACID INSENSITIVE4 (Giraud et al., 2009), NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON13 (ANAC013) and ANAC017 (De Clercq et al., 2013; Ng et al., 2013b), and different WRKY TFs (Van Aken et al., 2013). Furthermore, whereas some hormones are known to induce or trigger AOX, such as ET and SA, respectively (Rhoads and McIntosh, 1992; Norman et al., 2004; Ederli et al., 2006), others, such as auxin, have been shown to be antagonistic to the induction of AOX and MRS in general (Ivanova et al., 2014). In a number of studies, the overall changes in the transcriptome upon mitochondrial perturbation in Arabidopsis display most prominent overlaps with biotic stress responses (Clifton et al., 2006; Schwarzländer et al., 2012; Umbach et al., 2012; Cvetkovska and Vanlerbergh, 2013), suggesting a mechanistic link between biotic stress responses and the transcriptional regulation of AOX1a.
While the molecular components involved in regulating AOX expression are being uncovered, the role of AOX in plant defense/stress responses still has to be defined. Arabidopsis plants with reduced amounts of AOX1a, the predominantly inducible AOX gene in Arabidopsis (Clifton et al., 2005), displayed a small, but noticeable, cold-altered phenotype (Fiorani et al., 2005). During a combined drought and moderate light stress, Arabidopsis aox1a mutants had a more sensitive phenotype, but not during drought or moderate light treatment alone (Giraud et al., 2008). AOX also has been reported to be induced upon a variety of biotic challenges (Vanlerberghe, 2013), including treatment with flg22 (Van Aken et al., 2013). Separate challenge of Nicotiana attenuata with multiple biotic factors suggested that AOX plays a role in resistance to cell piercing-sucking insects, but not against the rapid feeding herbivore Manduca sexta (Zhang et al., 2012). A role for AOX in producing a superoxide burst that promotes, without being absolutely necessary, a hypersensitive response has been found after infection with Pseudomonas syringae (Cvetkovska and Vanlerberghe, 2013). The role of AOX in virus threats has been reported frequently, but these results should be interpreted carefully because the use of inhibitors to induce AOX may have secondary effects, such as the production of ROS or reactive nitrogen species that can trigger defense responses (Vanlerberghe, 2013). An overall theme that emerges from studies investigating the role of AOX triggered by biotic or abiotic stimuli is that AOX and the mitochondrial electron transport chain, in general, are key players in generating ROS/reactive nitrogen species signals that are important mediators of stress signaling (Vanlerberghe, 2013; Ng et al., 2014).
A forward-genetic approach with the AOX1a promoter driving the expression of a firefly luciferase (LUC) successfully identified both positive and negative regulators of AOX1a in Arabidopsis (Ng et al., 2014). Here, we discovered the TF V-myb myeloblastosis viral oncogene homolog29 (MYB DOMAIN PROTEIN29 [MYB29]) as a negative regulator of AOX1a expression upon the inhibition of electron flow through the cytochrome electron transport chain. Previously, MYB29 had been characterized as a regulator of Met-derived aliphatic glucosinolate biosynthesis in Arabidopsis, along with other MYB-type TFs (Sønderby et al., 2010), which are involved in the defense against biotic feeding pests (Fahey et al., 2001) and abiotic stress (Martínez-Ballesta et al., 2015). The role of MYB29 as a negative regulator of AOX1a reveals a hitherto unknown interaction between mitochondrial retrograde/stress signaling and the regulation of aliphatic glucosinolate levels. A model is presented for how this interaction may be mediated to integrate and optimize AOX1a expression.
RESULTS
Identification of MYB29 as a Negative Regulator of AOX1a Expression
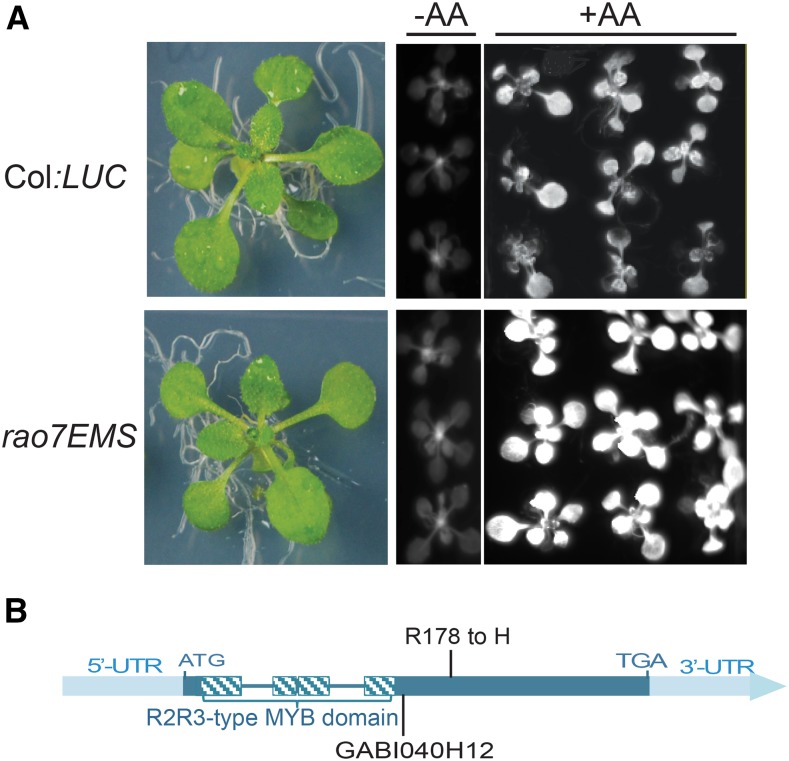
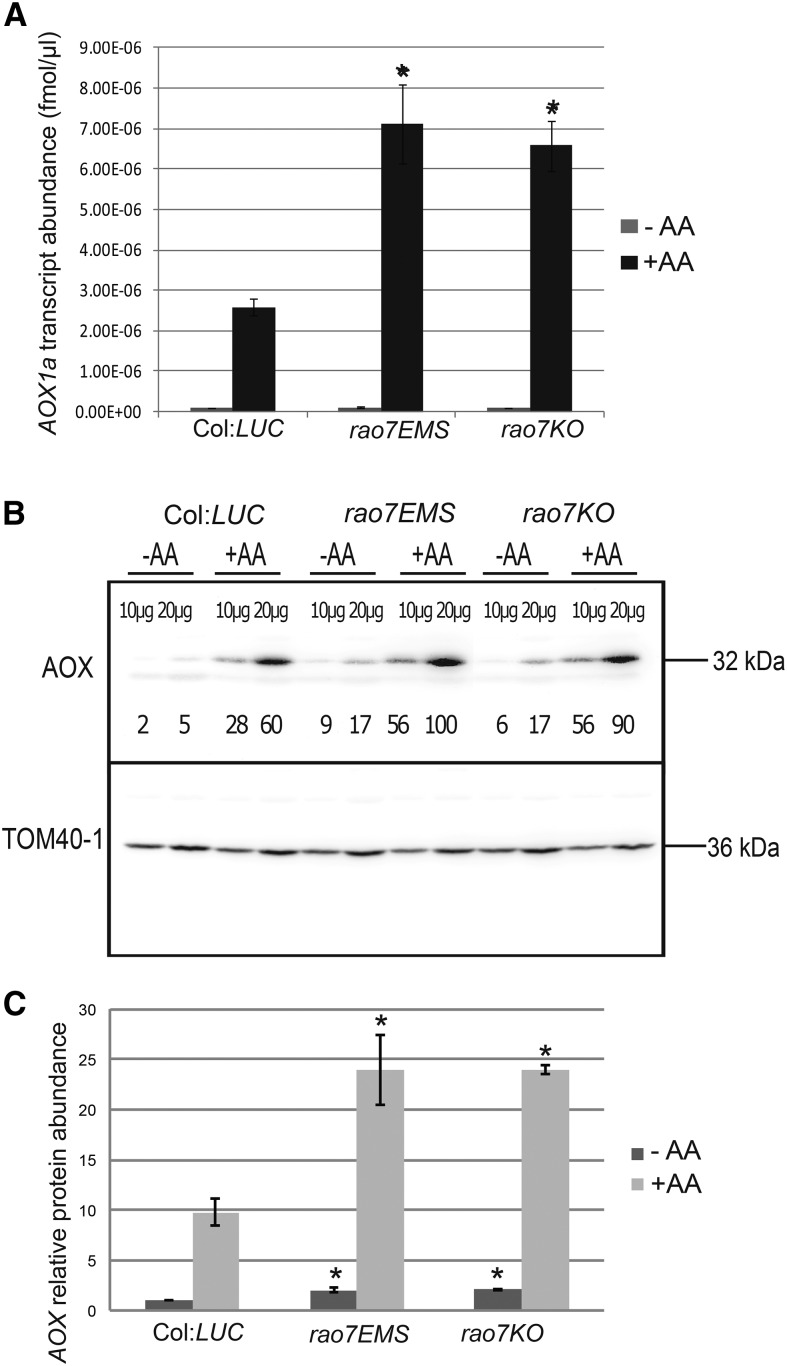
Using the AOX1a promoter driving the expression of LUC, we previously identified CDKE;1/REGULATOR OF ALTERNATIVE OXIDASE1a 1 (RAO1; Ng et al., 2013a), ANAC017/RAO2 (Ng et al., 2013b), and four proteins involved in auxin metabolism/transport (RAO3, RAO4, RAO5, and RAO6; Ivanova et al., 2014) as regulators of AOX1a expression in Arabidopsis. By means of the same approach to characterize these regulators, application of antimycin A (AA), and visualization of the luminescence signal 6 h later, another mutation was identified that did not map to any of the previously identified loci. This mutant displayed a higher level of luminescence than the wild-type control (Columbia[Col]:LUC) (Fig. 1A). Map-based cloning combined with next-generation mapping as outlined previously (Ng et al., 2013a, 2014) revealed a mutation, resulting in an Arg-to-His modification confirmed by Sanger sequencing, at locus At5g07690 that encodes the R2R3-type MYB domain-containing TF MYB29 (Stracke et al., 2001; Fig. 1B). The mutant was designated regulator of alternative oxidase1a 7 ethyl methanesulfonate (rao7EMS). The phenotype of the rao7EMS mutant could be rescued by complementation with constructs containing the wild-type MYB29-coding sequence that restored the wild-type LUC induction in response to AA treatment (Supplemental Fig. S1). These results confirm that the rao7 phenotype results from a mutation in MYB29. To prove that the luminescence signal was an accurate proxy for AOX1a transcript and protein abundance, quantitative reverse transcription (qRT)-PCR and western-blot analyses were carried out in both the ethyl methanesulfonate (EMS) line (rao7EMS) and in a T-DNA knockout for MYB29 (rao7KO; Fig. 1B; Supplemental Fig. S2). Transcript and protein abundances for AOX1a were ∼2-fold greater than those of the wild-type control under AA stress conditions (Fig. 2), in agreement with the higher luminescence observed (Fig. 1A). Thus, RAO7/MYB29 was concluded to be a negative regulator of the stress-dependent AOX1a induction. Under unstressed conditions (mock treatment), the AOX1a transcript abundance was similar between the wild type and rao7/myb29 mutants, whereas the AOX protein levels were ∼2-fold higher in both mutants than those in the wild type (Fig. 2C). However, under the unstressed growth conditions used, the levels of AOX1a transcript and AOX protein were low.
Figure 1.
Identification of RAO7/MYB29 as a regulator of Arabidopsis AOX1a. A, Fourteen-day-old seedlings (left) and luminescence images of Col:LUC and the rao7EMS mutant before (middle) and after (right) treatment with 50 µm antimycin A (AA). Col:LUC plants were generated from Col-0 plants transformed with a construct containing a firefly luciferase reporter gene driven by the AOX1a promoter (P-AOX1a:LUC), as described previously (Ng et al., 2013a). LUC activity was visualized in a NightOWL bioluminescence imager. B, MYB29 gene model. The positions of the rao7EMS mutation and the T-DNA insertion of a GABI line (GABI040H12; Supplemental Fig. S2) are indicated. UTR, Untranslated region.
Figure 2.
AOX1a transcript and protein abundance in rao7EMS and rao7KO mutant lines. Two-week-old seedlings were sprayed with water (−AA) or 50 µm AA (+AA) and harvested 3 h (transcript abundance) or 6 h (protein abundance) after treatment. A, qRT-PCR analysis of the transcript abundance of AOX1a. Bars represent average expression from three biological repeats ± se. B, Quantified AOX protein abundance. AOX values were normalized against the corresponding TRANSLOCASE OF THE OUTER MITOCHONDRIAL MEMBRANE40-1 (TOM40-1) as a loading control for mitochondrial protein abundance. Relative protein abundance, below each blot, is expressed as a percentage of the highest value of the set. C, AOX protein quantifications (averages ± se) from three biological replicate experiments. Asterisks indicate significant differences (P < 0.05) compared with Col:LUC as indicated by Student’s t test.
RAO7/MYB29 Mutation Alters Plant Stress Tolerance
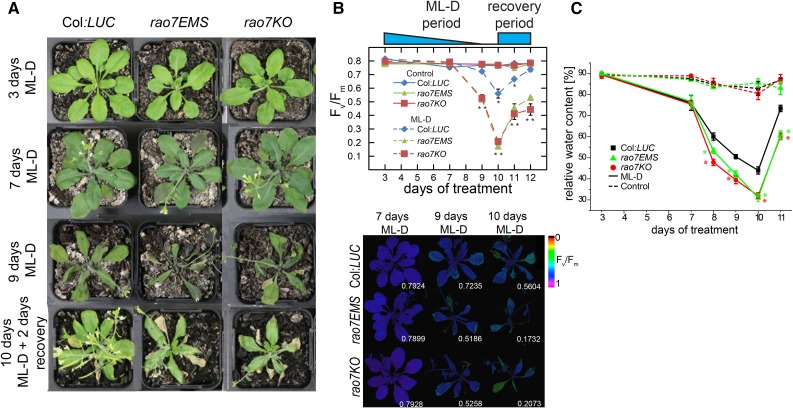
Mutation of RAO7/MYB29 accentuates the induction of AOX1a transcript and protein levels following stress treatment. Previously, aox1a mutants were found to be more sensitive to combined moderate light and drought stress treatments (Giraud et al., 2008). Therefore, we assessed whether the RAO7/MYB29 mutation also affected the plant tolerance against this condition. Under unstressed conditions (well watered at 120 µmol m−2 s−1 light intensity), the phenotypes of the rao7EMS and rao7KO mutants did not differ noticeably from that of wild-type seedlings and plants, except for the significantly increased rosette area of mature (from 5 weeks old) rao7EMS plants, earlier stem and inflorescence emergence, and earlier flowering (stages 5.10 and 6.00; Supplemental Fig. S3; Boyes et al., 2001). However, under combined moderate light intensity (500 µmol m−2 s−1) and drought stress treatment, both rao7EMS and myb29KO displayed more severe stress phenotypes than the wild type (Fig. 3A), as also indicated by photochemical efficiency reduction (Fig. 3B). The relative water content between rao7 mutants and Col:LUC did not differ under unstressed conditions but was 15% to 30% reduced in the rao7 mutants compared with Col:LUC after 8 d of moderate light intensity and drought stress treatment (Fig. 3C).
Figure 3.
Physiological analysis of combined drought and moderate-light response in rao7 mutants. A, Col:LUC wild-type and rao7EMS and rao7KO mutant plants grown under a sufficient watering regime and normal light conditions (120 µmol m−2 s−1) for 3 weeks before water was withheld and plants placed at moderate light intensities (500 µmol m−2 s−1) for 9 d (ML-D). Plants were rewatered under a normal light regime on day 10 and reobserved after 2 d (recovery). B, Top, maximal photosystem II (PSII) quantum efficiency (Fv/Fm; Baker, 2008) of Col:LUC, rao7EMS, and rao7KO plants under ML-D conditions before and after recovery. Bars represent averages of nine leaves from three plants ± se. Blue shapes indicate soil water content upon water retention (triangle) and the rewatering stage (box). Asterisks indicate significant differences compared with well-watered Col:LUC as indicated by Student’s t test (*, P < 0.05 and **, P < 0.01). Bottom, false-color images of Fv/Fm from ML-D-treated Col:LUC, rao7EMS, and rao7KO plants. Average values are displayed in the bottom right corner of each image. C, Relative water content of Col:LUC and rao7 mutant plants grown under normal or ML-D conditions. Averages ± se of three plants are shown, and asterisks indicate significant (P < 0.05) differences compared with Col:LUC (Student’s t test).
Genome-Wide Transcriptional Responses to Mitochondrial Perturbation Affected by the RAO7/MYB29 Mutation
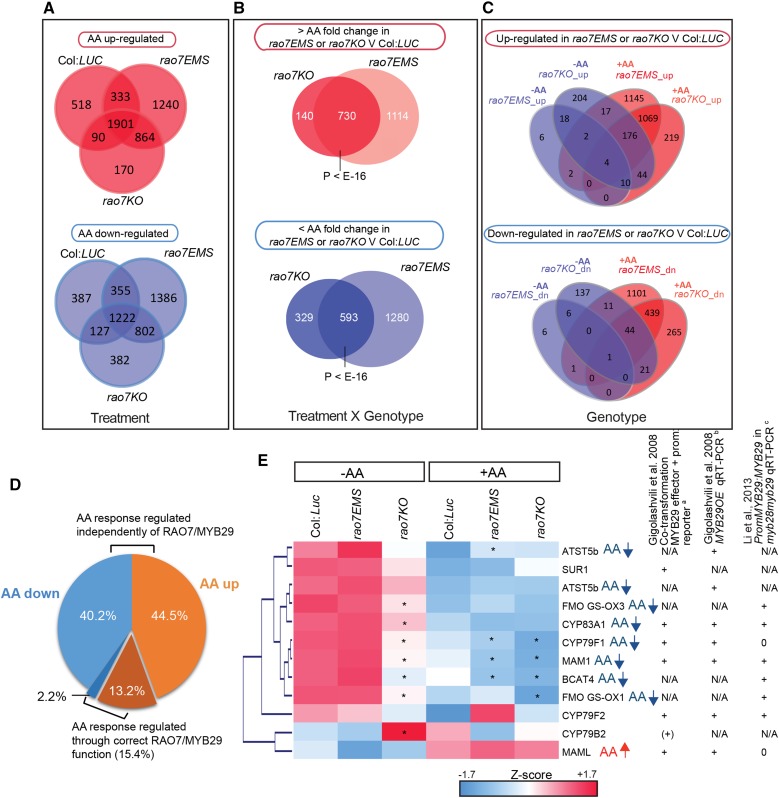
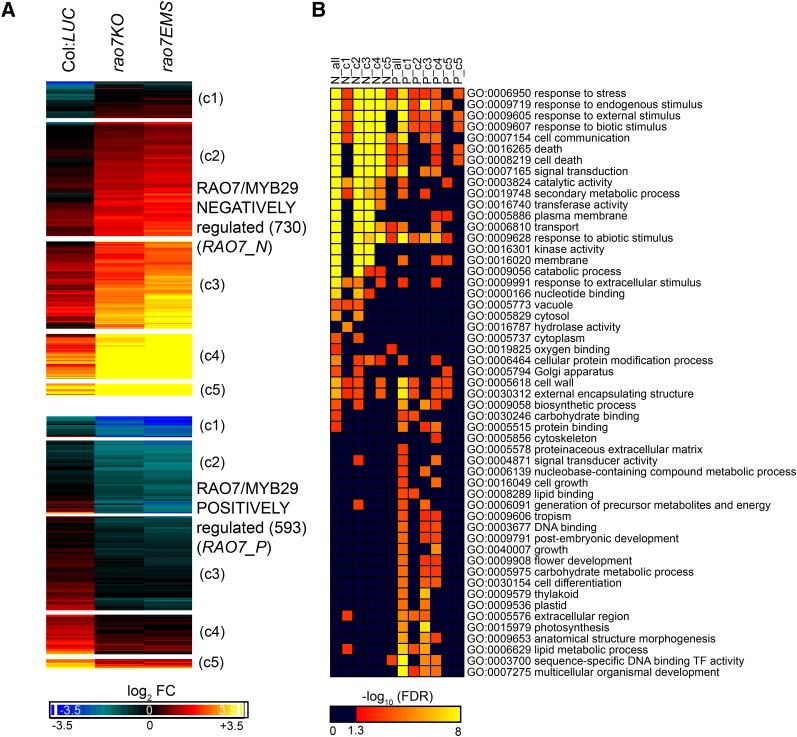
In a yeast (Saccharomyces cerevisiae) one-hybrid assay spanning the 1.85-kb upstream region of the translational start site, no binding of RAO7/MYB29 to any region of the AOX1a promoter could be detected (Supplemental Fig. S4). To assess whether and how the RAO7 mutation affects the genome-wide transcriptional responses to AA treatment, RNA sequencing (RNA-Seq) analysis was performed on wild-type (Col:LUC), rao7EMS, and rao7KO plants treated with AA for 3 h. Comparison of AA responses between the three genotypes revealed that MYB29 plays a role in regulating partly, but clearly not completely, the AA response (Fig. 4A). To gain insight into the gene expression changes induced by the RAO7/MYB29 mutation, we identified transcripts for which the AA-induced fold change differed significantly (two-way ANOVA [treatment × genotype interaction] with false discovery rate [FDR] < 0.05 and greater than 1.25-fold change; see “Materials and Methods”) between rao7EMS or rao7KO and Col:LUC. The overlap between the rao7EMS and rao7KO positively and negatively affected transcript levels was 730 (hypergeometric [HG] P < E-16) and 593 (HG P < E-16), respectively (Fig. 4B). Hereafter, these transcripts will be referred to as RAO7/MYB29 negatively (increased AA fold change in both rao7 mutants [RAO7_N]) and MYB29/RAO7 positively (decreased AA fold change in both rao7 mutants [RAO7_P]) regulated transcripts (Supplemental Table S1). Compared with the AA stress-responsive transcriptome in wild-type plants (Col:LUC), MYB29 loss of function affected the correct expression of ∼15% of AA-responsive genes (Fig. 4D). rao7EMS affected the AA-responsive transcriptome more drastically than rao7KO, with 1,114 genes with positively and 1,280 genes with negatively affected AA fold change specifically in the rao7EMS mutant (further referred to as RAO7-EMS_N and RAO7-EMS_P, respectively; Fig. 4B; Supplemental Table S1), implying a difference between the effects of the two types of mutations in the rao7 mutant lines. However, this situation is reversed under unstressed conditions. Between rao7KO and Col:LUC, 695 genes are differentially regulated (FDR < 0.05 and greater than 1.5-fold change), whereas in the rao7EMS mutant compared with Col:LUC, only 56 transcripts were altered, among which 41 were similarly regulated in both mutants (Fig. 4C).
Figure 4.
Changes in the transcriptome of rao7EMS and rao7KO under normal and AA-treated cellular conditions. A, Venn diagrams comparing the number of up- and down-regulated genes after AA treatment in Col:LUC, rao7EMS, and rao7KO (FDR < 0.05 and greater than 1.5-fold change). B, Identification of the number of genes for which the AA response (AA-induced fold change) is significantly affected in rao7EMS and/or rao7KO (two-way ANOVA [treatment × genotype interaction] FDR < 0.05 and greater than 1.25-fold change). C, Overview of genes differentially expressed in rao7EMS or rao7KO compared with Col:LUC under normal cellular conditions (mock treatment [−AA]) and AA stress conditions (FDR < 0.05 and greater than 1.5-fold change). D, Proportion of AA up- and down-regulated genes through the MYB29 function. E, Expression analysis of known potential MYB29 target genes involved in aliphatic glucosinolate biosynthesis in the transcriptome study. MYB29 targets were obtained from the literature (Gigolashvili et al., 2008; Li et al., 2013), and their expression was analyzed with the transcriptome data from Col:LUC, rao7EMS, and rao7KO grown under normal cellular conditions (−AA) or treated with AA for 3 h (+AA). For heat-map visualization, the z scores of log2-transformed reads per kilobase per million mapped read values were hierarchically clustered based on Euclidian distance with Genesis 1.6.0 (Sturn et al., 2002). Significant differences between rao7EMS and rao7KO mutants relative to Col:LUC are indicated with asterisks (FDR < 0.05). Genes regulated (FDR < 0.05) by AA treatment in the wild type (Col:LUC) are indicated with arrows. Evidence from the literature about genes transcriptionally targeted by MYB29 is indicated. Gigolashvili et al. (2008) used acotransformation of MYB29 effector and target gene promoter:reporter constructs and bqRT-PCR analysis of MYB29 overexpression lines (MYB29OE), whereas cLi et al. (2013) used qRT-PCR analysis on the myb28myb29 mutants containing the PromoterMYB29:MYB29 construct. +, Induced target gene/promoter; 0, unaffected target gene/promoter; N/A, data not available.
Previous studies had identified potential MYB29 target genes involved in aliphatic glucosinolate biosynthesis: METHYLTHIOALKYLMALATE SYNTHASE1 (MAM1), MAM-LIKE (MAML), CYTOCHROME P450 79B1 (CYP79B1), CYP79F1, CYP79F2, CYP83A1, FLAVIN-MONOOXYGENASE GLUCOSINOLATE S-OXYGENASE1 (FMO GS-OX1), FMO GS-OX3, BRANCHED-CHAIN AMINOTRANSFERASE4 (BCAT4), ARABIDOPSIS SULFOTRANSFERASE5B (ATST5B), ATST5C, and SUPERROOT1 (SUR1; Gigolashvili et al., 2008; Li et al., 2013), of which six transcripts were decreased and one was increased in rao7KO compared with Col:LUC plants under unstressed conditions (Fig. 4E, −AA). However, all transcripts displayed similar expression levels in rao7EMS compared with the wild type under unstressed conditions (Fig. 4E, −AA). The six rao7KO-repressed genes were all down-regulated upon AA treatment in the wild-type situation (Col:LUC; FDR < 0.05), and for four of them, the expression remained lower in rao7KO than in Col:LUC (Fig. 4E, +AA). Interestingly, three genes also were repressed in rao7EMS compared with Col:LUC under the AA stress conditions. As the transcriptome of rao7EMS is very similar to that of wild type under unstressed conditions, but clearly altered in its response to AA, the effect of the Arg178His amino acid change in rao7EMS is largely specific during stress conditions.
RAO7/MYB29 Mutation Interacts with Growth, Hormone, and Stress Signaling in Response to Mitochondrial Perturbations
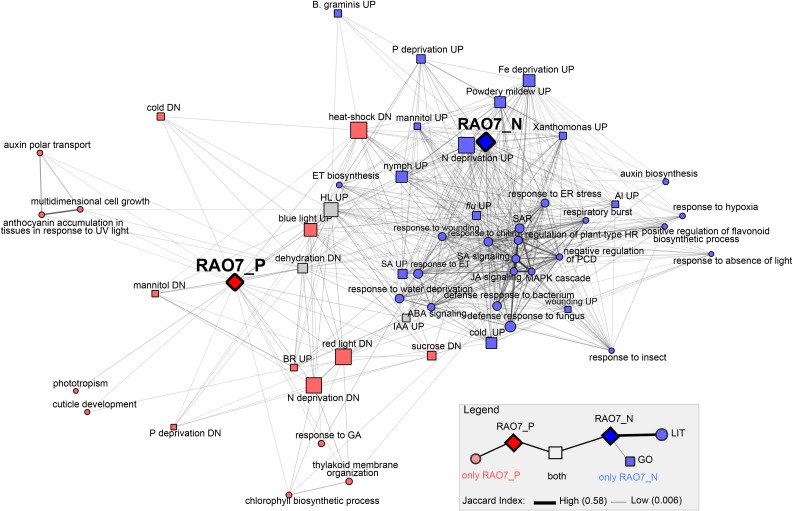
As the rao7EMS and rao7KO mutants respond in a similar manner to AA treatment, the commonly affected part of the transcriptome was functionally analyzed (transcripts positively [RAO7_P] or negatively [RAO7_N] regulated by RAO7/MYB29 in both mutants; Figs. 5 and 6; Supplemental Fig. S5). In addition to the complete RAO7_P and RAO7_N gene sets, RAO7_P and RAO7_N subgroups based on similarity in their expression profiles also were examined (Fig. 5A). A functional enrichment analysis with the Gene Ontology (GO) and the broad parent GO Slim categories revealed that the MYB29 positively regulated transcriptome (RAO7_P) was enriched with genes related to growth and developmental processes, including energy and lipid metabolism (Fig. 5B; Supplemental Fig. S5). In contrast, RAO7_N genes are involved in responses to stress, cell death, secondary metabolism, and signal transduction (Fig. 5B; Supplemental Fig. S5). In addition to GO analysis, the Plant Gene Set Enrichment Analysis (PlantGSEA; Yi et al., 2013) tool was searched for differentially expressed (DE) genes from stress or hormone treatment studies, and 41 lists of stress- or hormone-responsive genes were found that overlapped significantly with RAO7_P and/or RAO7_N (see “Materials and Methods”). By means of the gene lists from the PlantGSEA tool and from 49 nonredundant GO Biological Process (BP) terms identified from the above analysis (Supplemental Fig. S5), and the RAO7_P and RAO7_N genes as input, all pairwise overlaps were calculated and visualized in a network (Fig. 6; Supplemental Table S2; see “Materials and Methods”). Most gene lists were connected specifically to either RAO7_P (Fig. 6, red nodes) or RAO7_N (Fig. 6, blue nodes). Again, RAO7_N genes were predominantly stress up-regulated genes, with overlapping gene sets responding to biotic factors, cold, singlet oxygen-induced cell death (fluorescent mutant), water and nutrient deprivation, and endoplasmic reticulum (ER) stress. RAO7_N genes also were involved in processes of mitogen-activated protein kinase (MAPK) signaling, cell death and hypersensitive response, and ET and auxin biosynthesis. In contrast, RAO7_P genes were involved in fundamental cellular processes, such as cell size regulation, polar auxin transport, thylakoid membrane organization, and phototropism, and include genes regulated by light and down-regulated by several stress factors, such as temperature, osmotic stress, and nutrient deprivation. Also interesting is the effect of RAO7 on various hormone signaling processes, such as ABA, ET, JA, and SA signaling, which are negatively regulated (RAO7_N), and, in contrast, on the brassinosteroid (BR) and gibberellic acid (GA) responses, which are positively regulated (RAO7_P), as well as on auxin responses that are both positively and negatively regulated.
Figure 5.
Transcriptional response to AA regulated through the RAO7/MYB29 function. A, Heat-map representation of the expression of genes for which the AA response is either negatively or positively regulated through the RAO7/MYB29 function. Genes were classified as negatively (RAO7_N) or positively (RAO7_P) regulated by RAO7/MYB29 when their AA fold change was increased or decreased in both rao7EMS and rao7KO mutants compared to the wild type (Col:LUC), respectively. Genes were classified in clusters (c1, c2, c3, c4, and c5) according to their expression characteristics in response to AA in the different genotypes by means of K-means clustering. Colors represent log2 fold change (FC) of AA treatment compared with mock treatment, with blue and red/yellow representing transcripts that are down-regulated and up-regulated by AA treatment, respectively. B, GO Slim enrichment analysis of the RAO7_N and RAO7_P genes and their respective coexpression clusters. Color codes represent the negative logarithm (base 10) of the FDR-adjusted P value. Significantly (FDR < 0.05) enriched GO terms are indicated in red-yellow.
Figure 6.
Correlation of the MYB29-regulated gene sets with functional annotations. The MYB29 positively and negatively regulated genes (RAO7_P and RAO7_N) were used for gene set enrichment analysis with GO BP (FDR < 3.00E-05; see Supplemental Fig. S5) and DE genes (UP, up-regulated; DN, down-regulated) after hormone or stress treatments, the latter obtained from the literature through the PlantGSEA tool (FDR < 3.00E-03; Yi et al., 2013). Afterward, all gene lists were compared with each other in a pairwise manner. Only gene list pairs with significant overlap (FDR < E-03) and containing at least 10% of genes from either or both gene sets were used for network construction, and only gene sets directly connected to RAO7_P or RAO7_N were retained. Edge thickness and distance in the network correspond to the Jaccard index of similarity. Node shape indicates the type of gene list (squares, GO BP; circles, DE gene lists from the literature [LIT]). Node color refers to whether nodes are connected only with RAO7_P (red), only with RAO7_N (blue), or with both (gray). HL, high light; HR, hypersensitive response; IAA, indole-3-acetic acid; PCD, programmed cell death; SAR, systemic acquired resistance.
As the transcriptomes of rao7EMS and Col:LUC are very similar under unstressed conditions and rao7EMS exhibits AA-specific responses that are not observed in stressed rao7KO, we also analyzed the specific transcriptional response of rao7EMS (RAO7-EMS_P and RAO7-EMS_N; Supplemental Figs. S6–S8). Like the RAO7_P and RAO7_N genes, the RAO7-EMS_P genes were enriched for functions related to growth and development, whereas the RAO7-EMS_N genes were associated with stress responses (Supplemental Figs. S6 and S7). Comparison of the four gene lists (RAO7_P, RAO7_N, RAO7-EMS_P, and RAO7-EMS_N) in a functional correlation network revealed that seven terms related to light response, down-regulation by nutrient deficiency or sugar, or thylakoid membrane organization were commonly enriched between the RAO7_P and RAO7-EMS_P gene lists (Supplemental Table S2, red), whereas 25 functions were shared between the RAO7_N and RAO7-EMS_N lists (Supplemental Table S2, blue) and were related mainly to responses to biotic stress factors, including up-regulation by pathogens, insects, or wounding, ET, JA, and SA signaling, and ET biosynthesis, but also up-regulation upon abiotic stress factors and ABA signaling (Supplemental Fig. S8). Thus, RAO7/MYB29 seems to positively regulate growth and to negatively affect stress responses, potentially through interference at the hormonal crosstalk level.
Downstream Transcriptional Regulatory Network Analysis Reveals MYB29 Direct Targets and Intermediate TFs
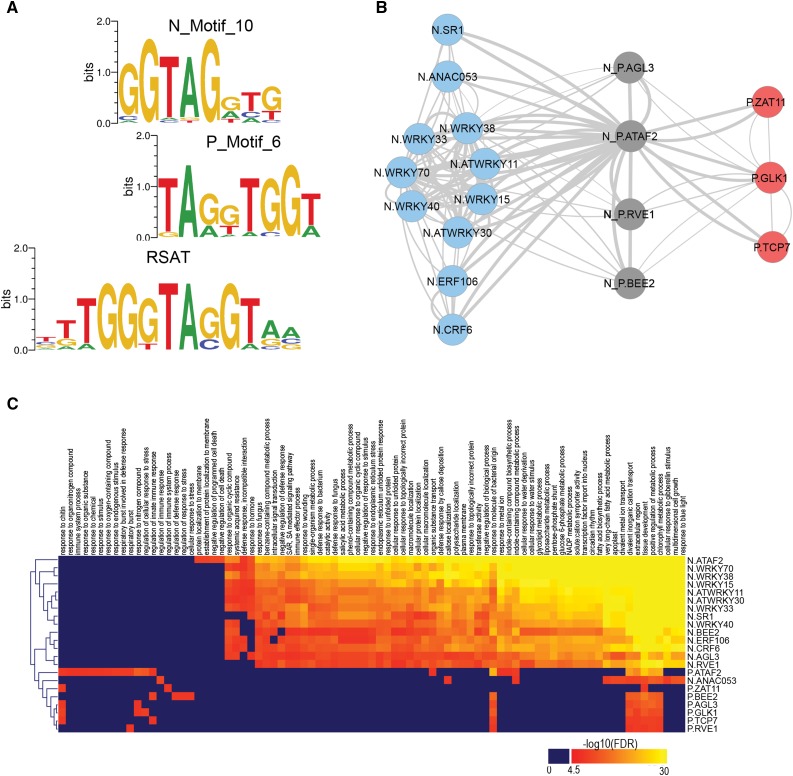
To unravel the transcriptional regulatory networks downstream of MYB29, we analyzed together DE genes, cis-regulatory elements, and GO enrichment. For the identification of direct MYB29 target genes, RAO7_P and RAO7_N gene promoters were searched for de novo enriched motifs (see “Materials and Methods”; Supplemental Fig. S9). The DNA-binding site of MYB29 as well as its closest homologs (phylogenetic subgroup S12 within the R2R3-MYB family; Dubos et al., 2010) had not, to our knowledge, been experimentally determined. P_Motif_6 and N_Motif_10 (Fig. 7A) were retained as putative MYB29-binding sites, because they resemble the consensus binding sites of R2R3-MYB family members that are homologous to MYB29. More specifically, MYB29 homologs have the binding sites GGTAGGT[AG] (MYB3; subgroup S4; Dubos et al., 2010), [AG]GT[AT]GGT[AG] (ATMYB4; subgroup S4), [GA][GT]TAGGT[AG] (MYB111; subgroup S7), or GGTAGGTGG (MYB15; subgroup S2; Franco-Zorrilla et al., 2014; Weirauch et al., 2014). To gain further insight into the MYB29 DNA-binding characteristics, we searched the promoters that are potentially targeted by MYB29 (MAM1, MAML, CYP79F1, CYP79F2, CYP79B2, CYP83A1, FMO-GS OX1, FMO-GS OX3, BCAT4, ATST5b, ATST5c, and SUR1; Gigolashvili et al., 2008; Li et al., 2013) for common sequence motifs with the Regulatory Sequence Analysis Tools (RSAT) oligo-analysis tool (van Helden et al., 1998) and identified TGGGTAGGT that resembled both P_Motif_6 and N_Motif_10 (Fig. 7A). Genome-wide mapping of these motifs revealed that the P_Motif_6 is present in 25% of the promoters of the MYB29 positively regulated genes (HG P = 2.4E-06) but is also, to a minor extent, enriched in the MYB29 negatively regulated genes (frequency of 20% and HG P = 0.036). Similarly, the N_Motif_10 occurs in 35% of the MYB29 negatively regulated genes (HG P = 2.51E-10) and in 28% of the MYB29 positively regulated genes (HG P = 0.046). In total, 584 (∼44%) RAO7/MYB29-affected genes had at least one P_Motif_6 or N_Motif_10 MYB29-binding site.
Figure 7.
Transcriptional regulatory network downstream of MYB29. A, Identification of potential MYB29-binding sites (P_Motif_6 and N_Motif_10) by de novo motif analysis of RAO7_P and RAO7_N gene set promoters with the Amadeus software (Linhart et al., 2008) and comparison with a de novo discovered motif in 12 known MYB29 target genes from Figure 4E by means of the RSAT (van Helden et al., 1998). Sequence logos were made with WebLogo 2.8.2 (P_Motif_6 and N_Motif_10) and WebLogo 3.5.0 (RSAT; Crooks et al., 2004). B, Identification of intermediate regulators of MYB29-dependent genes. RAO7_P and RAO7_N gene promoters were searched for enriched binding sites for 671 TFs of Arabidopsis. Only TFs containing the P_Motif_6 or N_Motif_10 MYB29-binding site and with MYB29-dependent expression were retained, of which 18 intermediate TFs were predicted to regulate RAO7_P genes (red), RAO7_N genes (blue), or both (gray) downstream of MYB29. TFs were compared based on the overlap of their target genes in a network, with edges representing pairwise overlap of more than 50%. C, Functional analysis of intermediate TFs based on GO enrichment analysis of their target genes. Color codes represent the negative algorithm (base 10) of the FDR-adjusted P values, with significantly enriched GO terms (FDR < E-4.5) indicated in red-yellow.
The occurrence of putative MYB29-binding sites in less than half of the DE genes, combined with a significant GO enrichment for sequence-specific DNA-binding TF activity in the RAO7_P genes and nucleotide binding in the RAO7_N genes (Fig. 5B), indicated that many of the RAO7/MYB29-affected genes might be regulated indirectly through TFs present in RAO7_P or RAO7_N. To identify intermediate regulators, we devised a novel and unbiased regulator prediction strategy based on known binding sites for Arabidopsis TFs. After integrating and mapping 744 motifs, known to bind 671 TFs, we performed a systematic motif enrichment analysis on the set of RAO7_P and RAO7_N genes. By retaining only TFs that are part of the RAO7_N or RAO7_P DE genes and that contain the P_Motif_6 or N_Motif_10 MYB29-binding site, we identified 18 potential intermediate TFs, of which 11 were specific for RAO7_N (Fig. 7B, blue nodes), three were specific for RAO7_P (Fig. 7B, red nodes), and four were common for both gene sets (Fig. 7B, gray nodes; Supplemental Table S3). We defined the functional TF similarity based on the target gene overlaps and visualized them in a network with edges representing a pairwise overlap of more than 50% (Fig. 7B). TFs from the same family (i.e. WRKY and ETHYLENE RESPONSE FACTOR [ERF] family TFs) clustered together, as expected from the similarity in their binding sites. In addition, the function of the respective TFs was further analyzed and compared based on the GO functional annotations of their predicted target genes (Fig. 7C). Various GO terms (FDR < E-4.5) were identified, ranging from functions in developmental processes to defense reactions to stress. Interestingly, the predicted transcriptional regulators of RAO7_N and those of RAO7_P clustered together.
For the RAO7_N gene set, seven WRKYs (ATWRKY11, WRKY15, ATWRKY30, WRKY33, WRKY38, WRKY40, and WRKY70), two ERFs (ERF106 and CYTOKININ RESPONSE FACTOR6), one NAC (ANAC053), and one calmodulin-binding TF, SIGNAL RESPONSIVE1 (SR1), were found with functions in biotic, oxidative, dehydration/salt stress, cold responses, and/or senescence (Zheng et al., 2006; Galon et al., 2008; Jiang and Deyholos, 2009; Besseau et al., 2012; Birkenbihl et al., 2012; Lee et al., 2012; Vanderauwera et al., 2012; Kim et al., 2013; Scarpeci et al., 2013; Jiang et al., 2016). Interestingly, both SR1 and WRKY40 regulate glucosinolate levels (Laluk et al., 2012; Schön et al., 2013). A strong overrepresentation of WRKY-binding sites also was obvious from the de novo motif analyses, with WRKY-like binding sites (W-boxes) as most significantly enriched motifs in RAO7_N (N_Motif_1 [HG P < E-16] and N_Motif_2 [HG P < E-16]; Supplemental Fig. S9). Interestingly, ATWRKY11, WRKY33, WRKY38, WRKY40, and WRKY70 all act at the interface of the antagonistic crosstalk between SA and JA/ET of defense responses to biotrophic pathogens, and necrotrophic pathogens and insects, respectively (Journot-Catalino et al., 2006; Li et al., 2006; Kim et al., 2008; Birkenbihl et al., 2012; Liu et al., 2015). In contrast, intermediate TFs specifically of the RAO7_P gene set (GOLDEN2-LIKE1 [GLK1], TEOSINTE BRANCHED1, CYCLOIDEA AND PCF FAMILY7 [TCP7], and ZINC FINGER OF ARABIDOPSIS THALIANA11) or shared between RAO7_P and RAO7_N (AGAMOUS-LIKE3 [AGL3], ATAF2, BRASSINOSTEROID ENHANCED EXPRESSION1 [BEE2], and REVEILLE1 [RVE1]) mainly have functions in various vegetative and reproductive developmental processes or chloroplast development, regulated, among others, through brassinosteroid or auxin (Friedrichsen et al., 2002; Ditta et al., 2004; Rawat et al., 2009; Waters et al., 2009; Huh et al., 2012; Aguílar-Martínez and Sinha, 2013; Liu et al., 2014; Peng et al., 2015; Xu et al., 2015). These results are consistent with the de novo motif analyses: MADS box-like binding site (N_Motif_13; P = 2.8E-14) for AGL3, RVE1-like binding site (P_Motif_12; P = 1.0E-12; Franco-Zorrilla et al., 2014) for RVE1, GLK1-like binding site (P_Motif_12; P = 1.0E-12; Franco-Zorrilla et al., 2014) for GLK1, G-box/basic helix-loop-helix-like (bHLH) binding site (P_Motif_2; P < E-16) for BEE1, and TCP type I-like binding site (P_Motif_1; P < E-16; Kosugi and Ohashi, 2002) for TCP7.
Gene regulatory networks also were predicted downstream of RAO7-EMS by means of a workflow similar to that for RAO7. Genes that were specifically altered by RAO7-EMS and not by KO mutation (RAO7-EMS_P and RAO7-EMS_N) were searched also for de novo enriched motifs in their promoters, but no R2R3-MYB-like binding sites were found (Supplemental Fig. S10). Therefore, to predict intermediate TFs, the previously defined P_Motif_6 and N_Motif_10 were used as potential MYB29-binding sites. For RAO7-EMS_P, no intermediate TFs were detected that contain a MYB29-binding site and regulate gene expression downstream of MYB29-EMS, but for the RAO7-EMS_N genes, ATWRKY18 and WRKY53 were predicted as intermediate regulators (Supplemental Table S3). Similar to what was observed for the RAO7 intermediate TFs, ATWRKY18 and WRKY53 are involved in antagonistic SA - JA/ET crosstalk and in glucosinolate homeostasis (Miao and Zentgraf, 2007; Murray et al., 2007; Schön et al., 2013).
To further validate the intermediate TF network that regulates RAO7/MYB29-affected genes, we gathered published microarray studies on loss- and/or gain-of-function mutants for ATAF2, ATWRKY11, WRKY15, WRKY33, WRKY40, and SR1 (Delessert et al., 2005; Journot-Catalino et al., 2006; Birkenbihl et al., 2012; Vanderauwera et al., 2012; Van Aken et al., 2013; Prasad et al., 2016). For each TF, DE genes were split into groups of up- and down-regulated genes under control and stress conditions. For all TFs, except ATAF2, a significant overlap was observed between the RAO7/MYB29-dependent genes containing the TF-binding site and at least one of the DE gene sets from the TF perturbation (Supplemental Table S4). The overlap was most significant with DE genes negatively regulated by the TF in the microarray studies and to a lesser extent or not with the positively regulated DE genes. This result is consistent with the observation that many WRKYs can act either as repressor or as bifunctional activator/repressor, depending on the sequences surrounding the W-boxes (Miao et al., 2004; Chen et al., 2010; Vanderauwera et al., 2012; Liu et al., 2015). WRKY33-bound genes in chromatin immunoprecipitation experiments also overlapped significantly with the predicted targets of WRKY33 downstream of RAO7/MYB29 (HG P < E-16; Liu et al., 2015). Comparison with WRKY33-bound targets that were differentially regulated by WRKY33 also revealed a larger overlap with the WRKY33-repressed genes than with the WRKY33-induced genes. However, these results of negative regulation do not explain the direction of the expression changes observed in the rao7 mutants (i.e. the TFs [ATWRKY11, WRKY15, WRKY33, WRKY40, and SR1] and their predicted target genes downstream of RAO7 are both up-regulated in the rao7 mutants). Therefore, these expression changes are probably the consequence of the concerted action of multiple TFs on promoters and/or various heterodimer combinations of activators and repressors, as described previously for WRKY TFs (Chen et al., 2010; Bakshi and Oelmüller, 2014).
DISCUSSION
MYB29 Is a Negative Regulator of the Mitochondrial Stress Response
We demonstrated that RAO7/MYB29 is a negative regulator of the mitochondrial stress response using AOX1a, an MRS marker in Arabidopsis. Transcripts of 16 of 26 mitochondrial genes that had been defined previously as responsive under a number of different stress treatments were identified as negatively regulated by MYB29 (Van Aken et al., 2009). Included in this list of 16 genes are not only AOX1d and ALTERNATIVE NAD(P)H DEHYDROGENASE genes but also the mitochondrial membrane protein 66 (OM66), involved in regulating cell death (Zhang et al., 2014). Furthermore, of seven transcripts defined as general mitochondrial stress markers (Schwarzländer et al., 2012), six were up-regulated in the rao7/myb29 mutants treated with AA, including ANAC013, shown to be a core regulator of the mitochondrial stress response (De Clercq et al., 2013).
MYB29, together with MYB28 and MYB76, is a transcriptional regulator of the biosynthesis of glucosinolates (Hirai et al., 2007; Beekwilder et al., 2008; Sønderby et al., 2010), which had not been connected previously to retrograde signaling. These secondary metabolites occur mainly in the Brassicales order (Mithen et al., 2010) and are correlated with plant defense against herbivorous insects and pathogens. After tissue damage, vacuolar glucosinolates are released and hydrolyzed into toxic compounds (Barth and Jander, 2006). Glucosinolates also accumulate during various abiotic stress conditions, such as salinity, drought, temperature, and light (Del Carmen Martínez-Ballesta et al., 2013), and play a role in ABA and methyl jasmonate signaling of stomatal closure via an unknown mechanism (Zhao et al., 2008; Islam et al., 2009). rao7 mutants have an increased sensitivity and exhibited a reduced leaf water content during combined light and drought stress (Fig. 3), but it is unclear whether this feature is due to altered glucosinolate levels affecting stomatal closure.
Remarkably, the transcriptomes of the rao7EMS and rao7KO mutants were not identical. In particular, the rao7EMS mutant had a more drastically perturbed transcriptome during AA treatment (Fig. 4). A possible explanation might be that, whereas probably no MYB29 is produced in rao7KO, the single amino acid change (Arg178His in rao7EMS that resides outside the R2R3-MYB DNA-binding domain (Fig. 1B) might produce a MYB29 protein that can still bind to DNA, but without its full regulatory potential or activity. A phylogenetic analysis identified that Arg178 is conserved in a Brassicaceae-specific clade, including MYB28 and MYB76 (Supplemental Fig. S11). A compensatory mechanism in the rao7KO mutant through functional redundancy with MYB28/MYB76 binding to the free MYB29-binding sites might be the reason for the more severe stress phenotype in rao7EMS. For example, a compensatory up-regulation of MYB28 has been shown to exist in the myb29/rao7KO mutant (Sønderby et al., 2010). However, we cannot exclude the possibility of any remaining EMS mutations in rao7EMS underlying rao7EMS-specific phenotypes and, therefore, have focused the main analyses and conclusions on the shared rao7EMS/rao7KO transcriptomic effects that are unlikely to be the results of any side mutations (rao7EMS) or transformation artifacts (rao7KO). An analysis of transcripts of previously defined MYB29 targets involved in glucosinolate biosynthesis revealed that, specifically under unstressed conditions, rao7EMS plants displayed wild-type–like levels, in contrast with strongly perturbed levels in the rao7KO mutant (Fig. 4E). The dissimilarity between rao7EMS and rao7KO under unstressed conditions also was displayed at the genome-wide transcriptome, because rao7KO affected approximately 700 transcripts, whereas only 56 transcripts were altered in rao7EMS. These observations indicate that RAO7-EMS nearly specifically affects the stress-responsive transcriptome. Altogether, our findings support and are in agreement with a specific/non-redundant function for MYB29 (Sønderby et al., 2010), because its impact on the mitochondrial stress response is clearly significant.
MYB29 did not bind to the AOX1a promoter in yeast one-hybrid assays. Also, the predicted MYB29-binding sites (Fig. 7A) could not be found in the AOX1a promoter or in most other MRS marker genes (Schwarzländer et al., 2012; De Clercq et al., 2013), indicating that MYB29 indirectly controls mitochondrial retrograde gene expression. However, MYB TFs often cooperate in dynamic complexes with bHLH TFs for DNA binding (Pireyre and Burow, 2015), the absence of which in the yeast system could have prevented the DNA binding of MYB29. Indeed, MYB29 has been shown to interact physically with the bHLH TF MYC2 to regulate glucosinolate biosynthesis (Schweizer et al., 2013). The lack of evidence for the direct binding of MYB29 to the AOX1a promoter, together with the lack of potential MYB-binding sites in more than half of the promoters of transcripts deregulated in both rao7EMS and rao7KO mutants in response to mitochondrial stress, made us postulate and detect that MYB29 affects part of the MRS through a network of downstream TFs (Fig. 7B). Among these TFs, WRKY40 has been demonstrated to be a negative regulator of AOX1a and OM66 under AA treatment (Van Aken et al., 2013). In addition, ANAC053 was shown previously to be a positive regulator by binding a cis-element in the AOX1a promoter, redundant with other NAC family members, such as ANAC013 and ANAC017 (De Clercq et al., 2013; Van Aken et al., 2016).
MYB29 Interacts with ET Signaling through ERF TFs
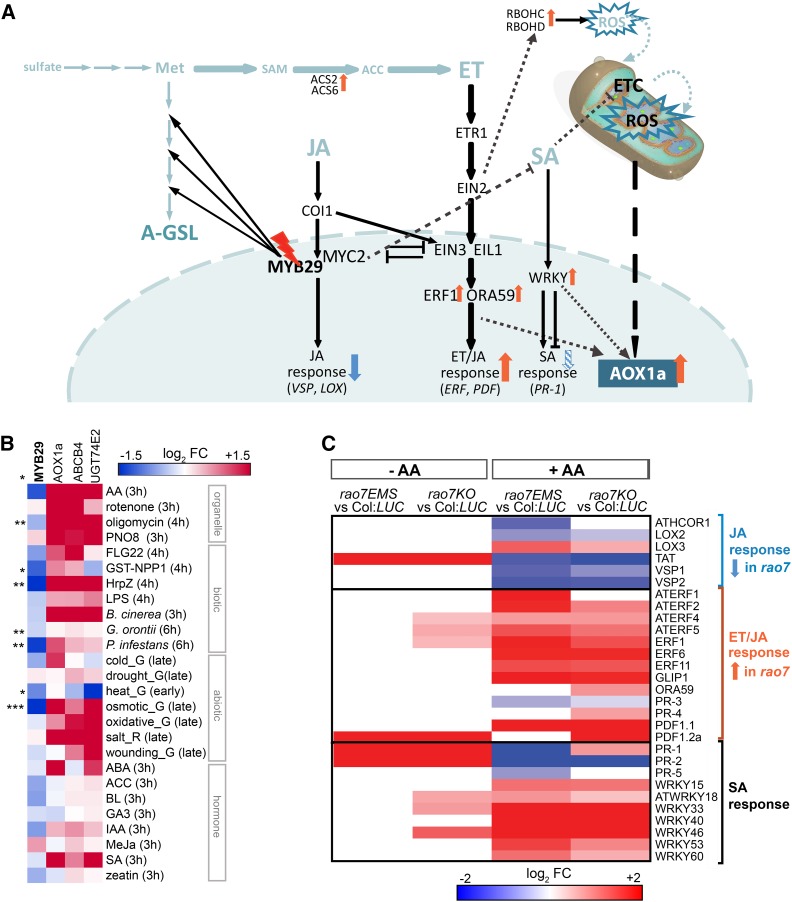
MYB29 expression was assessed under various stress, hormone, and chemical treatments, indicating that it is repressed by several stresses, including pathogens, osmotic, and mitochondrial stresses (Fig. 8B). Thus, MYB29 appears to be induced specifically by wounding and JA (Hirai et al., 2007; Gigolashvili et al., 2008) and probably functions downstream of the JA signaling pathway, as shown for its interaction partner MYC2 (Schweizer et al., 2013). JA signaling is strongly connected with ET signaling, both synergistically and antagonistically (Pieterse et al., 2012). The synergistic pathway (ET/JA pathway) mediates defense against necrotrophic pathogens, whereas JA and ET antagonistically regulate resistance against insects (the latter positively mediated by the JA pathway). This negative crosstalk is mediated by downstream TFs: ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1; ET/JA pathway) and MYC2 (JA pathway) reciprocally repress each other’s transcriptional functions (Song et al., 2014). AOX expression in response to various stresses has been shown independently to be mediated through ET signaling. Salt, ozone, metal, and pathogen induction of AOX was abolished in mutants of ETHYLENE RECEPTOR1 (ETR1) and/or EIN2 proteins that act upstream of EIN3/EIL1 in ET signal transduction (Simons et al., 1999; Tuominen et al., 2004; Wang et al., 2010; Keunen et al., 2015). It is unclear how the AOX transcription is regulated downstream from the ET signaling pathway, but dysfunctional MYC2 due to the absence of the functional MYB29 in the rao7 mutants could derepress EIN3/EIL1 and activate downstream ET transcriptional responses. An analysis of marker genes responsive to ET and/or JA indicated that crosstalk between ET and JA is perturbed in the rao7 mutants specifically under AA stress conditions: most markers of the ET and JA synergistic signaling (ET/JA) pathway are induced, whereas several markers of the antagonistic JA signaling pathway are repressed in the mutants under AA stress (Fig. 8C). Most evident was the negative regulation by MYB29 (up-regulated in both rao7 mutants) of ERF family TFs that are transcriptionally induced by ET (ERF1, ATERF1, ATERF2, ATERF4, ATERF5, ATERF6, ATERF11, and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF 59 [ORA59]) and mediate ET signaling responses downstream of EIN3/EIL1 by binding the GCC-box element in the promoters of ET-responsive genes (Solano et al., 1998; Fujimoto et al., 2000; Zarei et al., 2011; Dubois et al., 2015). Consistently, a GCC-box-like motif (N_Motif_8; P = 2.3E-12) as well as an ET response element (N_Motif_12; P = 1.6E-14) are overrepresented among the MYB29 negatively regulated genes (Supplemental Fig. S9). In addition, an analysis of the expression of all 122 ERF genes revealed that other ERF family members from subfamilies VIII and IX (Nakano et al., 2006) also are transcriptionally down-regulated by MYB29 (up-regulated in rao7EMS and/or rao7KO; Supplemental Table S5). This observation hints at the involvement of repressed ET signaling/responses in the negative regulation of AOX1a by RAO7/MYB29 (Fig. 8A). In addition, ERF106, predicted as an intermediate regulator of RAO7_N genes, has been found to be involved in the degradation of oxylipins that antagonize ET signaling during oxidative stresses (Walper et al., 2016). Moreover, ET is mandatory for the stress-induced accumulation of ROS, and ET-regulated AOX expression also is dependent on ET-mediated ROS production through the up-regulation of certain NADP (NADPH) oxidases (Jakubowicz et al., 2010; Jiang et al., 2013; Keunen et al., 2015), among which the respiratory burst oxidase homologs RBOHC and RBOHD also are up-regulated in both rao7EMS and rao7KO mutants specifically under AA stress, but the RBOHF expression is not modified (Fig. 8A; Supplemental Table S1).
Figure 8.
Model for the proposed function of RAO7/MYB29 in the regulation of AOX1a. A, Diagram showing the proposed mechanisms of AOX1a regulation induced by the RAO7 mutation. Upon the RAO7 mutation, the synthesis of aliphatic glucosinolates (A-GSL) from Met is down-regulated; as a result, other biochemical pathways that use the limiting Met and S-adenosyl Met (SAM) as substrate, such as the biosynthesis of ET (Sauter et al., 2013), are probably promoted. Concomitantly, genes encoding aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) that catalyze the conversion of SAM to the ET biosynthesis intermediate ACC are up-regulated in the rao7 mutants. ET signaling is mandatory for stress-induced AOX expression that is impaired in the etr1 and ein2 ET signaling mutants. ET works both synergistically and antagonistically with the JA signaling pathway. Downstream TFs (ERF1 and ORA59) and marker genes (ERF and PDF) of the synergistic ET/JA pathway are up-regulated in the rao7 mutants (see below). In contrast, antagonistic JA response markers (VEGETATIVE STORAGE PROTEIN [VSP] and LIPOXYGENASE [LOX]) are down-regulated in the rao7 mutants, indicating that MYB29 alters the crosstalk between ET and JA, with an impact on AOX transcription likely as a consequence. This negative crosstalk is mediated at the level of downstream TFs (MYC2 for JA and EIN3 and EIL1 for ET/JA), inhibiting each other’s functions (Song et al., 2014). MYC2 is known to interact with and work in a transcriptional complex with MYB29 downstream of the JA signaling (Schweizer et al., 2013). Therefore, MYB29 mutation might relieve the repression of ET signaling by JA (MYC2). In addition, genes encoding RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) proteins that produce apoplastic ROS and mediate AOX expression downstream of the ET signaling (Keunen et al., 2015) also are up-regulated in the rao7 mutants. JA signaling also interacts with SA signaling, mainly antagonistically, and MYC2 can down-regulate SA biosynthesis (Zheng et al., 2012). WRKY TFs active at the interface of the SA versus ET/JA crosstalk were identified to function downstream of MYB29. Various WRKYs function in positive or negative regulation of AOX1a expression (Van Aken et al., 2013). Furthermore, SA itself is known to induce mitochondrial signaling and AOX expression through perturbations of the electron transport chain (ETC) and subsequent mitochondrial ROS production. Arrows indicate changes observed in the rao7 mutants, with orange and blue arrows representing increased and decreased levels of expression compared to the wild type, respectively. Dashed lines mark indirect or uncertain regulations/effects. COI1, CORONATINE INSENSITIVE1. B, Expression characteristics of MYB29 in response to various stress and hormone treatments compared with those of AOX1a, ATP-BINDING CASSETTE B4 (ABCB4), and URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE 74E2 (UGT74E2) genes that are negatively regulated through MYB29. Expression data were obtained from the AtGenExpress global stress expression data set (Kilian et al., 2007) and the AtGenExpress hormone and chemical treatment data set (Goda et al., 2008) with Genevestigator (Hruz et al., 2008) and from the public microarray database Genevestigator (Hruz et al., 2008; www.genevestigator.com). Significant differences of MYB29 expression between stress or hormone and control treatments are indicated with asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Student’s t test). The heat map was generated with Genesis 1.6.0 (Sturn et al., 2002). BL, Brassinolide; flg22, flagellin 22; G, green tissue; GST-NPP1, GLUTATHIONE S-TRANSFERASE-NECROSIS-INDUCING PHYTOPHTHORA PROTEIN1; Hrpz, hairpin Z; LPS, lipopolysaccharide; MeJa, methyl jasmonate; R, root tissue. C, Meta-analysis of the expression of marker genes responsive to ET, JA, and SA in the rao7 mutants grown under nonstress (−AA) conditions or AA-induced stress (+AA). The color code represents log2 fold change (FC) in the rao7 mutants compared with the wild type, with red and blue representing transcripts with increased and decreased expression, respectively, in the mutant versus the wild type (FDR < 0.05). ATHCOR1, ARABIDOPSIS THALIANA CORONATINE-INDUCED PROTEIN1; GLIP1, GDSL LIPASE1; TAT, TYROSINE AMINOTRANSFERASE.
Another indication that ET signaling/responses are altered is the early-flowering phenotype in rao7EMS plants under normal growth conditions (Supplemental Fig. S3B). Alternatively, the RAO7/MYB29 mutation could interfere with ET biosynthesis through the availability of Met, the aliphatic glucosinolate precursor (Gigolashvili et al., 2008; Sauter et al., 2013; Fig. 8A), in accordance with the enrichment of genes involved in ET biosynthesis among the MYB29 negatively regulated genes, including the aminocyclopropane-1-carboxylic acid synthase genes ACS2 and ACS6 (Fig. 6; Supplemental Figs. S5, S7, and S8) that are mandatory for the stress-induced accumulation of AOX1a (Keunen et al., 2015).
MYB29 Interacts with SA Signaling through WRKY TFs
TF network analysis predicted various WRKY TFs downstream of MYB29 (Fig. 7B). The WRKY TF superfamily is a major regulator of SA signaling in plant defense responses (Vidhyasekaran, 2015). SA has been shown previously to affect AOX and other MRS gene expression and to mediate part of its responses through mitochondrial ROS production (Gleason et al., 2011; Berkowitz et al., 2016). The SA marker genes PATHOGENESIS-RELATED1 (PR-1), PR-2, and PR-5 all were repressed in the rao7EMS mutant, whereas 16 SA-inducible WRKY TF-encoding genes (WRKY6, ATWRKY11, WRKY15, WRKY18, WRKY25, WRKY26, ATWRKY30, WRKY33, WRKY39, WRKY40, WRKY41, WRKY46, WRKY51, WRKY53, WRKY60, and WRKY63; Dong et al., 2003), of which some are negative or positive regulators of PR gene expression, were up-regulated in both rao7EMS and rao7KO mutants, implying a complex regulation of SA responses downstream of RAO7/MYB29 (Fig. 8C; Supplemental Table S6; Li et al., 2004, 2006; Xu et al., 2006; Murray et al., 2007; Kim et al., 2008; Birkenbihl et al., 2012; Hu et al., 2012; Jiang et al., 2016). Interestingly, seven WRKY TFs that either negatively (ATWRKY11, ATWRKY18, WRKY33, WRKY38, and WRKY40) or positively (WRKY53 and WRKY70) regulate SA responses were predicted to be intermediate regulators of the MYB29 negatively regulated genes (Fig. 7; Supplemental Table S3). These WRKYs work at the interface of antagonistic interactions between SA and JA signaling and, concomitantly, have altered signaling through the JA and/or ET pathways. For example, WRKY33 stimulated JA and ET/JA marker genes (ATERF2, ORA59, PLANT DEFENSIN1.1 [PDF1.1], PDF1.2a, and LIPOXYGENASE2) through inhibition of the SA-mediated repression of JA signaling (Birkenbihl et al., 2012). Thus, these results imply an altered ET, JA, and SA signaling crosstalk induced by MYB29 mutation through WRKY TFs (Fig. 8A). Moreover, the myc2 mutant was shown previously to have elevated SA biosynthesis and responses through the modulation of ANAC019, ANAC055, and ANAC072, which regulate SA biosynthesis and metabolism genes (Zheng et al., 2012). Overall, MYB29 together with MYC2 acts in the complex interplay of ET, JA, and SA signaling that is largely mediated at the downstream transcriptional level (Koornneef and Pieterse, 2008; Song et al., 2014), more specifically by the modulation of ERF and WRKY TF levels.
MYB29, a Molecular Link between Retrograde, Growth, and Stress Signaling
RAO7/MYB29 mutation also affected the levels of transcripts related to growth, developmental processes, and energy metabolism, in addition to stress responses, albeit regulated in an opposite direction (Figs. 5 and 6; Supplemental Figs. S6 and S8). Adverse growth conditions compromise photosynthesis and respiration and deplete the plant’s energy resources. As a result, plants need to adapt their energy metabolism and balance vegetative and reproductive growth in response to stresses. An important sensor and integrator of energy depletion signals during stress is KIN10 (Baena-González et al., 2007). Interestingly, RAO7 coregulated genes also are coregulated by KIN10 and by sugar availability (Supplemental Fig. S12, A–F; Gonzali et al., 2006). KIN10 was shown previously to integrate energy sensing with mitochondrial signaling through its physical interaction with a cyclin-dependent kinase, CDKE;1, in the regulation of AOX1a (Ng et al., 2013a). Interestingly, ET also was previously linked to growth regulation upon stress sensing by regulation of the CDKA activity and the cell cycle (Skirycz et al., 2011). Thus, MYB29 and CDKE;1 probably integrate a wide variety of cellular signals to control stress responses, partially through KIN10 and ET signaling. Thus, whereas AOX in general is induced by various treatments, it now appears that it is also repressed at the transcriptional level by different stress signals. Auxin is considered a negative regulator, but this is hypothesized to be linked to growth optimization and stress response turning off. Here, a TF that regulates responses to biotic stimuli represses the induction of AOX. Thus, although AOX has been linked to biotic stress responses, it is clear that important regulators of aliphatic glucosinolates involved in biotic stress responses act to repress AOX, hence allowing the prioritization of different stress response pathways. Alternatively, the repression of AOX during biotic stress may function to amplify mitochondrial ROS signaling that, in turn, amplifies biotic stress defense responses.
MATERIALS AND METHODS
Plant Material and Forward Genetic Screening
The cloning of the 2-kb AOX1a upstream promoter region and the generation of the Arabidopsis (Arabidopsis thaliana) Col:LUC line were described previously (Ng et al., 2013a). EMS mutagenesis, stress treatments, screening, genetic mapping, gene identification, and verifications were done as described previously (Ng et al., 2013a). The EMS mutant (M2) was homozygous for the mutation and was backcrossed to Col:LUC twice to reduce the number of noncausal mutations (Jander et al., 2003).
The T-DNA insertion line for MYB29 (GABI_040H12) was obtained from the European Arabidopsis Stock Centre. The T-DNA insertion homozygous lines were confirmed by PCR with gene-specific primers (LP and RP) and a T-DNA-specific primer (LB; Supplemental Table S7). The location of the T-DNA insert was confirmed by sequencing. From RNA-Seq data, GABI_040H12 has 20% residual MYB29 mRNA levels compared with the wild type.
rao7EMS mutant plants were complemented with the full-length coding sequence of MYB29 that was PCR amplified from Columbia-0 cDNA and cloned into the binary vector pK7WG2 (Karimi et al., 2002) under the control of the constitutive cauliflower mosaic virus 35S promoter. This binary vector was transformed into rao7EMS mutant plants with Agrobacterium tumefaciens. T1 transformants were screened on kanamycin, and the presence of the transgene was confirmed by PCR analysis. LUC assays were conducted on the heterozygous T2 complemented plants.
Plant Growth Conditions, Stress Treatments, and LUC Assays
Fourteen-day-old seedlings were grown on Gamborg’s B5 medium (PhytoTechnology) supplemented with 3% (w/v) Suc and 0.75% (w/v) agar at 22°C with 16 h of light (120 µmol m−2 s−1)/8 h of darkness. Stress treatments were carried out by spraying the plants with 50 µm AA or water (mock) for 6 h. After treatments, plants were sprayed with 2.5 mm luciferin (GoldBio) and incubated in the dark for 30 min, and luminescence was measured with a NightOWL bioluminescence imaging system (Berthold Technologies) and analyzed with Image Lab software (version 5, build 18; Bio-Rad Laboratories).
For drought and moderate-light treatments, plants were grown in soil at 22°C with 16 h of light (120 µmol m−2 s−1)/8 h of darkness with watering every 3 d, referred as normal conditions. After 21 d, plants were not watered for 3 d and then transferred to 500 µmol m−2 s−1 light without subsequent watering, referred to drought and moderate-light stress conditions. Progressive drought and moderate-light responsiveness was monitored for 9 d. Plants were rewatered and placed under the normal light regime on day 10. For the determination of relative water contents, whole rosettes were weighed directly after excision from the plant (FW, fresh weight), after submersion in water for 4 h (TW, turgid weight), or after dehydration at 80°C overnight (DW, dry weight) as described previously (Giraud et al., 2008). Relative water content was calculated as (FW − DW)/(TW − DW) × 100 (%).
Phenotypic Analysis
Detailed growth and developmental phenotypic analyses of Col:LUC and mutant lines were carried out according to Boyes et al. (2001). At least 15 plants per genotype were used for analysis.
qRT-PCR
Green tissue from 14-d-old Col:LUC, rao7EMS, and rao7KO (GABI_040H12) seedlings treated with 50 µm AA or water as a mock treatment were harvested in biological triplicates at 3 h after treatment. RNA isolation, cDNA synthesis, and qRT-PCR were done as described previously (Giraud et al., 2008).
Mitochondrial Isolation and Immunoblotting
Mitochondria were isolated from 2-week-old seedlings 6 h after treatment with 50 µm AA (Murcha and Whelan, 2015). Immunodetection was carried out as described previously (Wang et al., 2012) with antibodies to AOX (Elthon et al., 1989) and TOM40-1 (Carrie et al., 2009). To ensure the linearity of detection, two dilutions of mitochondria were loaded. The band intensity was quantitated with Quantity One imaging software (Bio-Rad). The intensity of the cross-reacting bands probed with the antibody to AOX was adjusted to the intensity of TOM40-1 that was used as a loading control. The pixel density was set to 100 for the highest band detected (i.e. rao7EMS treated with AA) and that of the remaining bands relative to it.
Yeast One-Hybrid Assays
The coding sequence of MYB29 was amplified from Arabidopsis cDNA and cloned into the pGADT7-rec2 prey vector (Clontech) with EcoRI and BamHI restriction digestion and T4 ligation. In total, 25 AOX1a promoter fragments spanning the 1.85-kb upstream region were cloned into the pHIS2 bait vector with EcoRI and SacI restriction sites. Each fragment was ∼100 bp long and overlapped with the previous and next fragment for 25 bp (for primer sequences, see Supplemental Table S7). Yeast transformations and yeast one-hybrid assays were performed as described previously (Van Aken et al., 2013).
Measurement of Photosynthetic Parameters
Chlorophyll fluorescence emission of 21-d-old seedlings exposed to normal growth conditions or moderate-light and drought conditions for 3, 7, 9, and 10 d and after 1 and 2 d of recovery was measured with a pulse amplitude modulation fluorometer (PAM-101) and analyzed with ImagingWin software (Walz). After 10 min of dark acclimation, Fv/Fm was determined as (Fm − F0)/Fm, where Fm is the maximum photosystem II (PSII) fluorescence in the dark-adapted state and F0 is the initial (minimum) PSII fluorescence in the dark-adapted state (Baker, 2008). For each line, data were averaged over three biological repeats with three leaves from individual plants per replicate.
RNA-Seq Data Analysis
The analysis was performed with the R software package edgeR (version 3.10.5; Robinson et al., 2010; R Core Team, 2014). Prior to our analysis, reads mapped to rRNA, pre-tRNA, small nuclear/nucleolar RNA, and other RNA (The Arabidopsis Information Resource 10 annotation) were omitted. Only genes with a minimum of five read counts in at least three samples were retained for the analysis; in our analysis, i.e. 19,889 genes. Trimmed mean of M-values (TMM) normalization (Robinson and Oshlack, 2010) was applied by means of the calcNormFactors function. Variability in the data set was assessed with MDSplot. All three biological replicates clustered together, except the second biological replicate of the mock-treated EMS mutant. Thus, TMM normalization was repeated without this sample. Trended negative binomial dispersion parameters were estimated with the default Cox-Reid method based on a model with main effects of treatment, genotype and replicate, and an interaction term between treatment and genotype with the estimateGLMTrendedDisp function, followed by the estimation of the empirical Bayes dispersion for each gene. A negative binomial regression model was then used to model the overdispersed counts for each gene separately, with fixed values for the dispersion parameter as outlined by McCarthy et al. (2012) and as implemented in the function glmFit using the model described above. A likelihood ratio test (LRT) was applied to compare this model with a model without replicate to assess possible replicate (batch) effects. No genes were found with a significant batch effect (FDR < 0.05, Benjamini-Hochberg (BH) adjustment), as was expected by the MDS plot. The estimate of the dispersions and the fitting of the model were repeated with now only the main effects of genotype and treatment and their interaction. The significance of the interaction term was assessed with the LRT comparing the full model with the main effects model. At an FDR level of 0.05, 3,894 genes were found with a significant interaction. To test user-defined hypotheses, the model was reparameterized. The factors were combined to one factor with six levels (three genotypes × two treatments), and a no-intercept single-factor model was fitted to the data. With this design, dispersions were reestimated and the model was refit. Contrasts of interest were the difference between treatments for each genotype and the difference between genotypes for each treatment. The treatment contrast in each genotype was contrasted with those of other genotypes to assess the interaction between genotype and treatment. Significance was assessed with the LRT, and, as before, FDR adjustments of P values were applied. All edgeR functions were applied with default values. RNA-Seq data were deposited to the National Center for Biotechnology Information Sequence Read Archive database under project identifier PRJNA342199.
Gene Functional Enrichment Analysis
First, for each of the four RAO7/MYB29-regulated gene sets (RAO7_P, RAO7_N, RAO7-EMS_P, and RAO7-EMS_N), genes were clustered by means of the centroid-based K-means clustering (Hartigan and Wong, 1979) as follows. The total within-group sum of squares was calculated by increasing the cluster size from k = 2 to 20 (Supplemental Fig. S13). Based on this analysis and from visual inspection of the clustering results in heat maps, we determined an appropriate number of clusters as k = 5 (RAO7_N) or k = 6 (RAO7_P, RAO7-EMS_P, and RAO7-EMS_N). For the K-means clustering, the kmeans function in R was used with 25 random sets of centers. Clusters with less than 25 genes were omitted for further analysis. GO enrichment analysis was done on the complete RAO7-regulated gene sets and on their respective clusters using the HG distribution. P values were corrected for multiple testing by means of the BH FDR correction. GO annotations were obtained from The Arabidopsis Information Resource (March 2015), and all annotations were extended to include all parental GO terms to compute fold enrichments (ratio of frequency in the test set over frequency in the genome). Nonredundant GO results refer to enriched GO terms discarding parental GO terms. Results were visualized in heat maps by means of the Partek Genomics Suite software, version 6.6 (www.partek.com), for gene expression or Genesis 1.6.0 (Sturn et al., 2002) for GO.
For the network construction of enriched gene sets, MYB29 positively and negatively regulated gene sets (RAO7_P, RAO7_N, RAO7-EMS_P, and RAO7-EMS_N) were searched against the PlantGSEA database for enrichment of DE gene sets from stress or hormone treatments (FDR < 3.00E-03; Yi et al., 2013). In addition, nonredundant GO BP terms from the above GO enrichment analysis (FDR < 3.00E-05) were included. Closely resembling PlantGSEA and/or GO BP terms as well as GO general biological processes were removed manually prior to analyses, resulting in a set of 90 enriched gene sets. All pairwise overlaps and Jaccard indices (intersection of two sets divided by union) were calculated between the MYB29 regulated genes and enriched gene sets and, in addition, between the enriched gene sets mutually. The P value of an overlap size equal to or greater than observed was calculated with a cumulative HG test in R for gene sets sharing at least one gene. Afterward, the BH adjustment was used for multiple testing correction. Only pairwise overlaps were retained with an FDR value lower than E-03 and containing at least 10% of genes from either or both gene sets. Only gene sets (nodes) showing interaction with MYB29 were retained, and their pairwise overlaps were loaded into Cytoscape (version 3.1.0; Shannon et al., 2003). The edge-weighted spring-embedded option was used for layout with the Jaccard index as edge weight. Furthermore, edge visualization was augmented by correlating edge transparency and width with the Jaccard index and setting a minimum baseline for visibility. Node color and shape were in accordance with the MYB29 relation (MYB29 negative regulation, blue; MYB29 positive regulation, red; both, black), whereas the node size was correlated with the total number of genes in the gene set.
Promoter Analysis
De novo motif examination was performed on intergenic regions 1 kb upstream from the translational start site with Amadeus and default settings and motif length 8 (Linhart et al., 2008). Motif logos were created with WebLogo and with 8-mers as input and without the small sample correction option (Crooks et al., 2004). De novo motifs were compared with known plant cis-regulatory elements in the Similarity, Tree-building, and Alignment of DNA Motifs and Profiles database (Mahony and Benos, 2007).
To identify intermediate TF regulators, first, known TF-binding sites were mapped as positional count matrices on the 2-kb upstream intergenic sequence for all genes using matrix-scan with a P value cutoff of less than 1E-05 (Thomas-Chollier et al., 2008). All Arabidopsis TF-binding sites from the CIS-BP database (Weirauch et al., 2014) were integrated together with positional count matrices from AthaMap (Steffens et al., 2004). In total, 671 TFs could be associated with one or more DNA motifs (744 motifs with known binding TFs). Motif enrichment was determined with the HG distribution and BH FDR correction. Only significantly (P < 1E-03) enriched motifs were retained, and TFs with binding sites present in at least 10% of either of the RAO7 DE gene lists were considered as candidate regulators.
Accession Numbers
Sequence data can be found in the TAIR database under Locus identifiers (Arabidopsis Genome Initiative codes) displayed in Supplemental Table S8.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Complementation of rao7EMS with wild-type MYB29 restoring normal AA induction of the P-AOX1a:LUC-driven luminescence.
Supplemental Figure S2. Confirmation of the T-DNA knockout line for the gene encoding RAO7/MYB29.
Supplemental Figure S3. Phenotypic analysis of rao7 mutants under nonstress conditions.
Supplemental Figure S4. Yeast one-hybrid analysis for MYB29 binding to the AOX1a 1.85-kb promoter.
Supplemental Figure S5. GO enrichment analysis of genes regulated by RAO7/MYB29.
Supplemental Figure S6. Transcriptional response to AA regulated through the RAO7/MYB29-EMS function.
Supplemental Figure S7. GO enrichment analysis of genes regulated by RAO7/MYB29-EMS.
Supplemental Figure S8. Correlation of the MYB29-EMS-regulated gene sets with functional annotations.
Supplemental Figure S9. De novo promoter motif discovery in RAO7/MYB29-regulated genes.
Supplemental Figure S10. De novo promoter motif discovery in RAO7/MYB29-EMS-regulated genes.
Supplemental Figure S11. Sequence conservation of potential MYB29 homologs.
Supplemental Figure S12. Comparison of genes regulated by RAO7, KIN10, and Suc.
Supplemental Figure S13. Plot of the total within-group sum of squares against the number of clusters in K-means solutions.
Supplemental Table S1. Summary of RNA-Seq data.
Supplemental Table S2. Overview of GO terms and transcriptome studies used in the functional correlation network of RAO7/MYB29-regulated genes.
Supplemental Table S3. Overview of identified intermediate TFs and their predicted target genes.
Supplemental Table S4. Validation of the predicted target genes of the intermediate TFs with public transcriptome studies.
Supplemental Table S5. Expression of the ERF family genes in rao7 mutants.
Supplemental Table S6. Expression of SA-responsive WRKY genes in the rao7 mutants.
Supplemental Table S7. List of primers used.
Supplemental Table S8. Locus identifiers (Arabidopsis Genome Initiative codes) for genes analyzed in this study.
Supplementary Material
Acknowledgments
We thank Yan Wang and Jasmien Vercruysse for help with the data analysis and Martine De Cock for help in preparing the article.
Glossary
- ABA
abscisic acid
- SA
salicylic acid
- ET
ethylene
- JA
jasmonic acid
- TF
transcription factor
- MRS
mitochondrial retrograde signaling
- ROS
reactive oxygen species
- AA
antimycin A
- EMS
ethyl methanesulfonate
- qRT
quantitative reverse transcription
- RNA-Seq
RNA sequencing
- FDR
false discovery rate
- HG
hypergeometric
- GO
Gene Ontology
- PlantGSEA
Plant Gene Set Enrichment Analysis
- DE
differentially expressed
- BP
Biological Process
- LRT
likelihood ratio test
Footnotes
This work was supported by the Australian Research Council Centre of Excellence Program (grant no. CE140100008), the Interuniversity Attraction Poles Program (grant no. IUAP P7/29) initiated by the Belgian Science Policy Office, the Ghent University Multidisciplinary Research Partnership Biotechnology for a Sustainable Economy (grant no. 01MRB510W), the Agency for Innovation by Science and Technology (IWT) in Flanders (predoctoral fellowship to J.V.d.V and D.V.), the Australian Research Council APD (fellowship and grant no. DP110102868 to O.V.A.), the Ghent University Special Research Fund (grant no. 01J11311 to P.W.), and the Research Foundation-Flanders (postdoctoral fellowship no. 12N2415N and travel grant no. V450215N to I.D.C.).
References
- Aguilar-Martínez JA, Sinha N (2013) Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R (2014) WRKY transcription factors: jack of many trades in plants. Plant Signal Behav 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, et al. (2008) The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3: e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, De Clercq I, Van Breusegem F, Whelan J (2016) Interaction between hormonal and mitochondrial signalling during growth, development and in plant defence responses. Plant Cell Environ 39: 1127–1139 [DOI] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Coolen S, Proietti S, Hickman R, Davila Olivas NH, Huang P-P, Van Verk MC, Van Pelt JA, Wittenberg AHJ, De Vos M, Prins M, et al. (2016) Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J 86: 249–267 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ 36: 721–732 [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43: 745–757 [DOI] [PubMed] [Google Scholar]
- del Carmen Martínez-Ballesta M, Moreno DA, Carvajal M (2013) The physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int J Mol Sci 14: 11607–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Claeys H, Van Vlierberghe K, Matsui M, Inzé D (2015) The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol 169: 166–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Ederli L, Morettini R, Borgogni A, Wasternack C, Miersch O, Reale L, Ferranti F, Tosti N, Pasqualini S (2006) Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiol 142: 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 59: 237. [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Müller C, Flügge U-I (2008) HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol 177: 627–642 [DOI] [PubMed] [Google Scholar]
- Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan Y-F, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LHM, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P (2006) Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res 119: 115–123 [DOI] [PubMed] [Google Scholar]
- Gray GR, Maxwell DP, Villarimo AR, McIntosh L (2004) Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep 23: 497–503 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hartigan JA, Wong MA (1979) Algorithm AS 136: a k-means clustering algorithm. J R Stat Soc Ser C 28: 100–108 [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D (2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185-186: 288–297 [DOI] [PubMed] [Google Scholar]
- Huh SU, Lee S-B, Kim HH, Paek K-H (2012) ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol Cells 34: 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Tani C, Watanabe-Sugimoto M, Uraji M, Jahan MS, Masuda C, Nakamura Y, Mori IC, Murata Y (2009) Myrosinases, TGG1 and TGG2, redundantly function in ABA and MeJA signaling in Arabidopsis guard cells. Plant Cell Physiol 50: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee J-H, Zhang B, Van Aken O, Radomiljac JD, van der Merwe M, Yi K, et al. (2014) A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative oxidase1a expression in Arabidopsis. Plant Physiol 165: 1233–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowicz M, Gałgańska H, Nowak W, Sadowski J (2010) Exogenously induced expression of ethylene biosynthesis, ethylene perception, phospholipase D, and Rboh-oxidase genes in broccoli seedlings. J Exp Bot 61: 3475–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, Last RL (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol 131: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Belfield EJ, Cao Y, Smith JAC, Harberd NP (2013) An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25: 3535–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C-H, Huang Z-Y, Xie P, Gu C, Li K, Wang D-C, Yu Y-Y, Fan Z-H, Wang C-J, Wang Y-P, et al. (2016) Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J Exp Bot 67: 157–174 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keunen E, Schellingen K, Van Der Straeten D, Remans T, Colpaert J, Vangronsveld J, Cuypers A (2015) ALTERNATIVE OXIDASE1a modulates the oxidative challenge during moderate Cd exposure in Arabidopsis thaliana leaves. J Exp Bot 66: 2967–2977 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim K-C, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park S, Gilmour SJ, Thomashow MF (2013) Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75: 364–376 [DOI] [PubMed] [Google Scholar]
- Király L, Hafez YM, Fodor J, Király Z (2008) Suppression of tobacco mosaic virus-induced hypersensitive-type necrotization in tobacco at high temperature is associated with downregulation of NADPH oxidase and superoxide and stimulation of dehydroascorbate reductase. J Gen Virol 89: 799–808 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Laluk K, Prasad KVSK, Savchenko T, Celesnik H, Dehesh K, Levy M, Mitchell-Olds T, Reddy ASN (2012) The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol 53: 2008–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee H-J, Park C-M (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sawada Y, Hirai A, Sato M, Kuwahara A, Yan X, Hirai MY (2013) Novel insights into the function of Arabidopsis R2R3-MYB transcription factors regulating aliphatic glucosinolate biosynthesis. Plant Cell Physiol 54: 1335–1344 [DOI] [PubMed] [Google Scholar]
- Linhart C, Halperin Y, Shamir R (2008) Transcription factor and microRNA motif discovery: the Amadeus platform and a compendium of metazoan target sets. Genome Res 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kracher B, Ziegler J, Birkenbihl RP, Somssich IE (2015) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 4: e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-M, An J, Han HJ, Kim SH, Lim CO, Yun D-J, Chung WS (2014) ZAT11, a zinc finger transcription factor, is a negative regulator of nickel ion tolerance in Arabidopsis. Plant Cell Rep 33: 2015–2021 [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV (2007) STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35: W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ballesta M, Moreno-Fernández DA, Castejón D, Ochando C, Morandini PA, Carvajal M (2015) The impact of the absence of aliphatic glucosinolates on water transport under salt stress in Arabidopsis thaliana. Front Plant Sci 6: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R, Bennett R, Marquez J (2010) Glucosinolate biochemical diversity and innovation in the Brassicales. Phytochemistry 71: 2074–2086 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Whelan J (2015) Isolation of intact mitochondria from the model plant species Arabidopsis thaliana and Oryza sativa. Methods Mol Biol 1305: 1–12 [DOI] [PubMed] [Google Scholar]
- Murray SL, Ingle RA, Petersen LN, Denby KJ (2007) Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant-Microbe Interact 20: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J (2014) Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol Plant 7: 1075–1093 [DOI] [PubMed] [Google Scholar]
- Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA, et al. (2013a) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288: 3449–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013b) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C, Howell KA, Millar AH, Whelan JM, Day DA (2004) Salicylic acid is an uncoupler and inhibitor of mitochondrial electron transport. Plant Physiol 134: 492–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zhao J, Neff MM (2015) ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142: 4129–4138 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pireyre M, Burow M (2015) Regulation of MYB and bHLH transcription factors: a glance at the protein level. Mol Plant 8: 378–388 [DOI] [PubMed] [Google Scholar]
- Prasad KVSK, Abdel-Hameed AAE, Xing D, Reddy ASN (2016) Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress. Sci Rep 6: 27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasch CM, Ott KV, Bauer H, Ache P, Hedrich R, Sonnewald U (2015) β-amylase1 mutant Arabidopsis plants show improved drought tolerance due to reduced starch breakdown in guard cells. J Exp Bot 66: 6059–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, Zhao Y, Ljung K, Harmer SL (2009) REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci USA 106: 16883–16888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, McIntosh L (1992) Salicylic acid regulation of respiration in higher plants: alternative oxidase expression. Plant Cell 4: 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Subbaiah CC (2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M (2013) Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451: 145–154 [DOI] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Mueller-Roeber B, Valle EM (2013) Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol 83: 265–277 [DOI] [PubMed] [Google Scholar]
- Schenke D, Böttcher C, Scheel D (2011) Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ 34: 1849–1864 [DOI] [PubMed] [Google Scholar]
- Schenke D, Cai D, Scheel D (2014) Suppression of UV-B stress responses by flg22 is regulated at the chromatin level via histone modification. Plant Cell Environ 37: 1716–1721 [DOI] [PubMed] [Google Scholar]
- Schön M, Töller A, Diezel C, Roth C, Westphal L, Wiermer M, Somssich IE (2013) Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol Plant-Microbe Interact 26: 758–767 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, König A-C, Sweetlove LJ, Finkemeier I (2012) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P (2013) Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BH, Millenaar FF, Mulder L, Van Loon LC, Lambers H (1999) Enhanced expression and activation of the alternative oxidase during infection of Arabidopsis with Pseudomonas syringae pv tomato. Plant Physiol 120: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo SD, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA (2010) A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol 153: 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R (2004) AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res 32: D368–D372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Bassil E, Hamilton JS, Inupakutika MA, Zandalinas SI, Tripathy D, Luo Y, Dion E, Fukui G, Kumazaki A, et al. (2016) ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 11: e0147625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203: 32–43 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohée S, van Helden J (2008) RSAT: regulatory sequence analysis tools. Nucleic Acids Res 36: W119–W127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Overmyer K, Keinänen M, Kollist H, Kangasjärvi J (2004) Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J 39: 59–69 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, Ruckle ME, McIntosh L, Hock JJ, Bingham S, White SJ, George RM, Subbaiah CC, et al. (2012) Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS ONE 7: e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, Whelan J (2016) Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol 171: 2150–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Vandenbroucke K, Inzé A, van de Cotte B, Mühlenbock P, De Rycke R, Naouar N, Van Gaever T, Van Montagu MCE, Van Breusegem F (2012) AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 109: 20113–20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, André B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasekaran P. (2015) Salicylic acid signaling in plant innate immunity. In Vidhyasekaran P, ed, Plant Hormone Signaling Systems in Plant Innate Immunity. Signaling and Communication in Plant Series, Vol. 2 Springer, Dordrecht, The Netherlands, pp 27–122 [Google Scholar]
- Walper E, Weiste C, Mueller MJ, Hamberg M, Dröge-Laser W (2016) Screen identifying Arabidopsis transcription factors involved in the response to 9-lipoxygenase-derived oxylipins. PLoS ONE 11: e0153216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang J, Bi Y (2010) Induction of alternative respiratory pathway involves nitric oxide, hydrogen peroxide and ethylene under salt stress. Plant Signal Behav 5: 1636–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Guo H, Zhang D, Chen D, Jiang Z, Lin R (2015) REVEILLE1 promotes NADPH:protochlorophyllide oxidoreductase A expression and seedling greening in Arabidopsis. Photosynth Res 126: 331–340 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z (2013) PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res 41: W98–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2. promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Van Aken O, Thatcher L, De Clercq I, Duncan O, Law SR, Murcha MW, van der Merwe M, Seifi HS, Carrie C, et al. (2014) The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J 80: 709–727 [DOI] [PubMed] [Google Scholar]
- Zhang L, Oh Y, Li H, Baldwin IT, Galis I (2012) Alternative oxidase in resistance to biotic stresses: Nicotiana attenuata AOX contributes to resistance to a pathogen and a piercing-sucking insect but not Manduca sexta larvae. Plant Physiol 160: 1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM (2008) Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Spivey NW, Zeng W, Liu P-P, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.