Combining high-throughput phenotyping and large-scale QTL mapping dissects the dynamic genetic architecture of maize development by using a RIL population.
Abstract
With increasing demand for novel traits in crop breeding, the plant research community faces the challenge of quantitatively analyzing the structure and function of large numbers of plants. A clear goal of high-throughput phenotyping is to bridge the gap between genomics and phenomics. In this study, we quantified 106 traits from a maize (Zea mays) recombinant inbred line population (n = 167) across 16 developmental stages using the automatic phenotyping platform. Quantitative trait locus (QTL) mapping with a high-density genetic linkage map, including 2,496 recombinant bins, was used to uncover the genetic basis of these complex agronomic traits, and 988 QTLs have been identified for all investigated traits, including three QTL hotspots. Biomass accumulation and final yield were predicted using a combination of dissected traits in the early growth stage. These results reveal the dynamic genetic architecture of maize plant growth and enhance ideotype-based maize breeding and prediction.
Maize (Zea mays) is one of the most widely grown crops worldwide and is not only a staple food for people and animals but also an important industrial material for fuel and many other uses. Maize has served as a model plant with distinct advantages, including its levels of phenotypic and genetic variation (Yan et al., 2011). In the past century, maize yield has increased 8-fold due to the efforts of plant breeders who harnessed genetic variations to breed for improvements in desired traits (Haley, 2011). It is predicted that 9 billion people living on this planet by 2050 will require 70% more food than today’s population and that more than half of the increased demand for cereals will come from maize (Yan et al., 2011). A large gap exists between the current yield increase in global cereal production and the predicted demands for the next few decades. Substantial changes in breeding technologies for agronomic processes and crop improvement will be required (Tester and Langridge, 2010).
With the rapid development of next-generation sequencing and high-density single-nucleotide polymorphism (SNP) genotyping technologies, linkage mapping and genome-wide association studies have been used widely to dissect the genetic architecture of agriculturally important traits in commercial maize (Buckler et al., 2009; Kump et al., 2011; Tian et al., 2011; Li et al., 2013; Wen et al., 2014, 2015). Many genes and variants underlying agriculturally important traits have been discovered in crops (Kesavan et al., 2013; Zuo and Li, 2013; Martinez et al., 2016). However, precision phenotyping still remains a bottleneck (Furbank and Tester, 2011). Traditional phenotyping is usually labor intensive, time consuming, lower throughput, costly, and frequently destructive to plants (Chen et al., 2014) and is far behind the development of other omics studies such as genomics, although efforts have been made to improve phenotyping efficiency (Yang et al., 2013).
Plant phenomics has been defined as the nondestructive and accurate acquisition of high-dimensional phenotypic data on an organism-wide scale across plant development (Houle et al., 2010). Recently, some high-throughput plant phenotyping platforms (Reuzeau et al., 2005; Nagel et al., 2012; Honsdorf et al., 2014) and open-source image-analysis pipelines (Hartmann et al., 2011; Chen et al., 2014; Klukas et al., 2014) were developed to quantify phenotypic traits at the population level for different plant species. High-throughput noninvasive phenotyping also has been adopted successfully to assess the genetics of estimated biomass dynamics in maize (Junker et al., 2015; Muraya et al., 2017). In a previous work, a rice automatic phenotyping platform (RAP) was designed to achieve high-throughput screening of rice (Oryza sativa) plants for genetic studies (Yang et al., 2014). In this study, the RAP was expanded for high-throughput phenotyping of maize plants. The dynamic growth phenotype of a recombinant inbred line (RIL) population containing 167 recombinant individuals was measured from seedling to tasseling stage with 16 time points. A total of 106 different traits were obtained. That, combined with the ultra-high-density linkage map, including 2,496 recombinant bins, allowed us to perform large-scale quantitative trait locus (QTL) mapping. In total, 988 QTLs, including three hotspots, were identified, which has provided useful information for future maize genetic improvement and helps us understand the dynamic genetic architecture of plant development and growth.
RESULTS
A High-Throughput Plant Phenotyping Platform for Maize
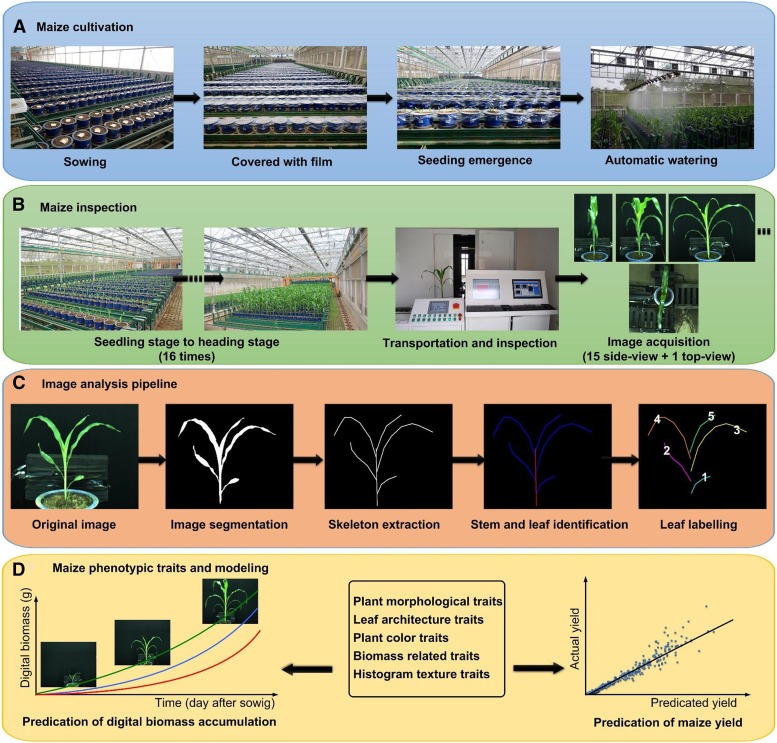
We cultivated the maize RIL population under greenhouse conditions (Fig. 1A) and using the RAP platform phenotyped each individual plant (Fig. 1B) from the seedling to tasseling stage at 16 time points (T1–T16; for the process, see Supplemental Video S1). At each time point, 15 side-view images and one top-view image of each plant were taken at the inspection unit using a color imaging camera (Stingray F-504C; Allied Vision Technologies). Two replicates of the RIL population and the two parents were inspected once every 3 d from seedling to tasseling stage, yielding ∼476 GB of data. With the modified image-analysis pipeline (Fig. 1C), 106 phenotypic traits, including 10 plant morphological traits, 22 leaf architecture traits, one plant color trait, three biomass-related traits, six histogram texture traits, and 64 growth-related traits, were extracted (Fig. 1D). The definitions of these 106 traits are shown in Supplemental Table S1 and Supplemental Note S1. The time costs of plant screening and image analysis for each plant were 45 s and 10 s, respectively. The operating procedure for RAP-Maize is provided in Supplemental Video S1, and details of the image-analysis pipeline are shown in “Materials and Methods” and in Supplemental Figure S1.
Figure 1.
High-throughput maize plant phenotyping. A, Maize plant sowing and maize cultivation. B, The procedure of maize inspection. C, The image-analysis pipeline. D, The extracted maize phenotypic traits and modeling.
Performance Evaluation of RAP-Maize
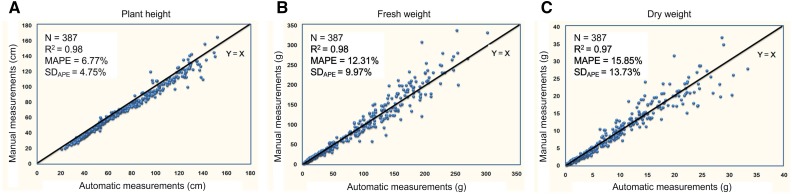
To model the biomass and evaluate the measuring accuracy of RAP-Maize, a subset of the maize association mapping panel (Yang et al., 2011; Li et al., 2013; 387 individuals from 100 genotypes with three to four replicates each [see “Materials and Methods”]) was grown separately and measured for three traits automatically and manually in four stages. In the previous study (Yang et al., 2014), the side-projected area (SA) showed good correlation with manually measured fresh weight and dry weight, with r2 > 0.79. To determine the best model for predicting the fresh weight and dry weight, 10 models (including linear, quadratic, exponential, and power models) were evaluated using the adjusted coefficient of determination (adjusted r2) and the mean absolute percentage error (MAPE) as the index; the statistical details are shown in Supplemental Table S2. The results showed that the power model (modes 9 and 10) had higher r2 and less MAPE for the fresh weight. Finally, mode 9 with one variable maximum side-projected area was selected as the best model for fresh weight modeling, which was one of the reasons why the image with maximum SA was automatically selected for the following image analysis and trait extraction. A similar result was observed with the same model for dry weight (Supplemental Table S3). Scatterplots showed that the r2 was greater than 0.97 between manual versus automatic measurements for plant height (Fig. 2A), fresh weight (Fig. 2B), and dry weight (Fig. 2C). These results demonstrate that automatic measurements are as good as manual measurements but with higher throughput.
Figure 2.
Performance evaluation of RAP-Maize. A, Scatterplot of automatic measurements versus manual measurements for plant height. B, Scatterplot of automatic measurements versus manual measurements for fresh weight. C, Scatterplot of automatic measurements versus manual measurements for dry weight.
Natural Variation of Phenotypic Traits and Heritability
The RIL population manifested high diversity for most of the 106 investigated phenotypic traits at each time point (Supplemental Data S1), ranging from 1.07- to 5.56-fold and from 1.02- to 14.95-fold change at minimum and maximum levels at 16 time points, respectively. For all investigated phenotypic traits, an ∼3-fold change was observed on average at different time points (Table I).
Table I. Range, mean of fold changes, and summary of QTLs for the investigated phenotypic traits identified at 16 time points.
| Time Point | No. of Traitsa | Fold Change (RILs, Mean)b | Fold Change (RILs, Range)b | QTL No.c | No. of QTLs (Mean and Range) | PVE (Mean and Range)d |
|---|---|---|---|---|---|---|
| % | ||||||
| T1 | 29 | 3.06 | 1.10–10.03 | 50 | 1.7 (1–5) | 10.5 (6.8–17.9) |
| T2 | 31 | 3.24 | 1.02–14.95 | 69 | 2.2 (1–5) | 9.3 (6.9–14.7) |
| T3 | 31 | 3.02 | 1.01–12.06 | 59 | 1.9 (1–4) | 9.4 (7.1–16.4) |
| T4 | 30 | 2.67 | 1.02–7.42 | 59 | 2.0 (1–5) | 8.7 (7.0–16.0) |
| T5 | 29 | 2.95 | 1.03–9.31 | 66 | 2.3 (1–6) | 9.4 (6.9–16.0) |
| T6 | 35 | 2.94 | 1.03–9.55 | 82 | 2.3 (1–5) | 9.5 (6.6–19.4) |
| T7 | 36 | 2.97 | 1.04–10.83 | 69 | 1.9 (1–4) | 9.8 (6.4–26.4) |
| T8 | 22 | 2.81 | 1.04–7.34 | 53 | 2.4 (1–5) | 9.9 (6.2–26.6) |
| T9 | 30 | 3.11 | 1.08–10.61 | 65 | 2.2 (1–6) | 9.6 (5.5–24.2) |
| T10 | 23 | 2.83 | 1.09–8.19 | 48 | 2.1 (1–8) | 9.3 (6.8–26.3) |
| T11 | 27 | 2.70 | 1.09–8.00 | 50 | 1.9 (1–6) | 9.7 (6.6–17.6) |
| T12 | 25 | 2.46 | 1.10–5.79 | 48 | 1.9 (1–5) | 9.1 (6.4–15.3) |
| T13 | 22 | 2.58 | 1.08–7.69 | 42 | 1.9 (1–4) | 10.0 (6.2–19.6) |
| T14 | 30 | 2.53 | 1.07–5.56 | 72 | 2.4 (1–6) | 9.7 (6.1–17.7) |
| T15 | 27 | 2.55 | 1.06–6.28 | 56 | 2.7 (1–5) | 10.1 (6.8–19.2) |
| T16 | 27 | 2.87 | 1.06–13.66 | 50 | 1.9 (1–5) | 9.2 (6.7–15.2) |
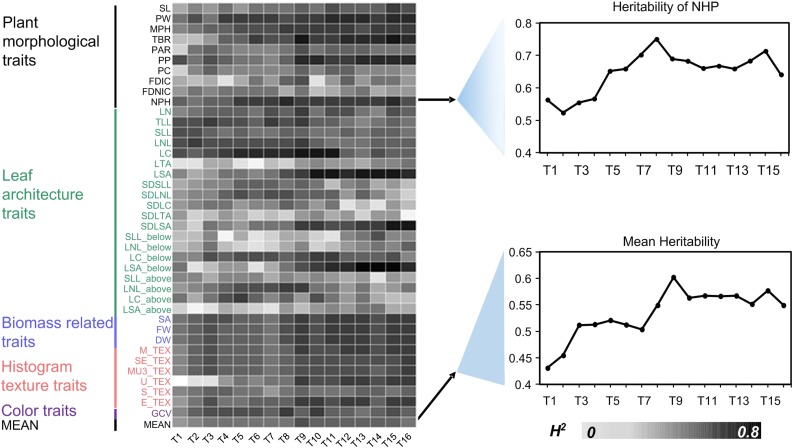
The investigated traits showed greater than 0.5 heritability for most traits at most of the time points (Fig. 3, left), as exemplified by the natural plant height and one of the morphology-related trait digital volumes (Fig. 3, top right). For most traits, heritability estimates were low in the six early developmental stages (i.e. 0.064 for U_TEX [uniformity, one of six histogram texture traits] at the first time point and 0.15 for average leaf tangency angle (LTA) at the sixth time point) and increased with the growth and development of maize in the late developmental stages. Some traits, including LTA, standard deviation of leaf tangency angle (SDLTA), fractal dimension with image cropping (FDIC), fractal dimension without image cropping (FDNIC), and average of leaf straight angle in upper half of plant (LSA_above), have low heritability across all developmental stages (Fig. 3, left), which might be due to the lower genetic variation of these traits. Some traits, including plant perimeter (PP), total projected area / bounding rectangle area ratio (PBR), maximum plant height (MPH), natural plant height (NPH), and average leaf straight angle (LSA), showed higher heritability in late developmental stages, which may due to the genes controlling these traits being expressed primarily in the late stages or environmental effects being larger at early stages, but got averaged over time in later stages (Fig. 3, bottom right).
Figure 3.
Phenotypic trait heritability during 16 growth stages. The heat map shows broad-sense heritability (H2) of the investigated phenotypic traits over 16 time points (left) as exemplified by the digital volume for natural plant height (top right) and for mean H2 (bottom right). Trait identifiers are given as in the heat map and colored according to their classification as indicated: black, plant morphological traits; green, leaf architecture traits; blue, biomass-related traits; pink, histogram texture feature; purple, color trait. MEAN, H2 of the mean value for all traits.
QTL Analysis
In this study, 167 of the genotyped RILs were used for QTL mapping for all the investigated phenotypic traits at each time point. In total, 42 to 82 QTLs were identified at each time point (Table I). The number of QTLs for each trait ranged from one to eight, with a mean of 1.7 to 2.7 across the 16 time points. In total, 938 QTLs were identified for 42 investigated phenotypic traits across 16 time points of maize growth. The percentage of phenotypic variation that each QTL could explain ranged from 5.5% to 26.6%, with a mean ranging from 8.7% to 10.5% at the 16 time points, respectively (Table I). For the growth rate-related traits dry weight and fresh weight, across 16 time points, a total of 50 QTLs were detected. The number of QTLs for each growth rate-related trait ranged from one to four, with a mean of 1.5 to 1.9. The percentage of phenotypic variation that each QTL could explain ranged from 7.3% to 17.8%, with a mean ranging from 8.4% to 10% for these 64 growth rate-related traits (Supplemental Table S4). For these 988 QTLs, the mapping resolution (QTL support interval) ranged from 0.3 to 10.8 centimorgan (cM), with a mean of 2.7 cM (∼3.9 Mb; Supplemental Fig. S2). Furthermore, these QTLs can be categorized into 152 nonredundant QTLs (Supplemental Data S2), which may be due to the highly positive or negative correlations between paired traits (Supplemental Fig. S3).
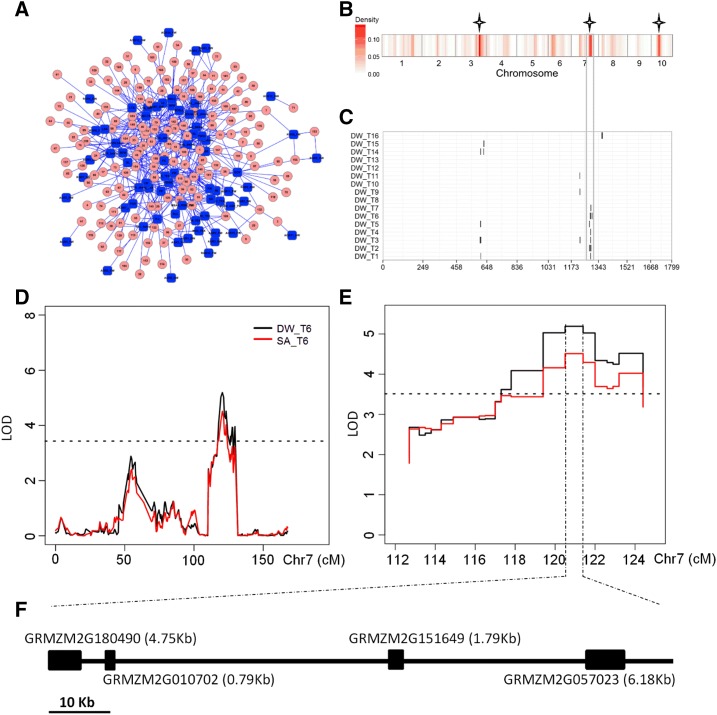
Detailed information, including location, peak marker, additive effect, QTL support interval, and explained phenotypic variance (r2), of each QTL for each trait is shown in Supplemental Data S2. A trait-locus network, including all traits across 16 development stages and their corresponding significant loci, shows the complex relationships among traits and detected loci. Some obvious connection nodes were found that could correspond to factors regulating maize growth (Fig 4A). QTL distribution across chromosomes (Supplemental Fig. S4) was not random, and three QTL hotspots were observed across the maize genome, on chromosomes 3, 7, and 10 (Fig. 4B). Moreover, the following observations were made. (1) A single QTL affecting a particular trait mapped at several time points of development; for example, a QTL affecting dry weight located at 120.5 cM on chromosome 7 was detected at seven time points (Fig. 4C) and a QTL for total leaf length per plant (TLL) on chromosome 2 was detected at 10 time points (Supplemental Figure S4). This implies that the gene affecting these traits may have expressed in early stages. (2) There was some overlap between QTLs affecting dry weight and SA (Fig. 4, D and E), since high correlations were observed between the two traits (Supplemental Table S3), indicating that SA can replace dry weight in QTL analysis in maize. (3) At a particular stage, most traits were controlled by a number of QTLs with minor to moderate effects (Supplemental Fig. S4; Supplemental Data S2).
Figure 4.
Chromosomal distribution of identified QTLs. A, Network of QTLs associated with the traits obtained from RAP-Maize. Blue nodes represent the 42 traits, and pink nodes represent all identified significant loci from QTL mapping. B, Density of QTLs of all investigated phenotypic traits across the genome. The window size is 10 cM. Three QTL hotspots are marked with stars. Detailed information for all detected QTLs is shown in Supplemental Data S2. C, Chromosomal distribution of dry weight (DW) QTLs identified across 16 time points. QTL regions (represented by the confidence interval for each QTL assigned as 1 − log of odds [LOD] drop of the peak) across the maize genome responsible for dry weight are shown as black solid boxes. QTL hot regions for dry weight on the chromosome are marked with the gray hollow box. D, LOD curves of QTL mapping for SA and dry weight around the chromosome 7 QTLs at the sixth time point. E, LOD values of the bins at the peak of the QTL interval shown as a function of their genetic positions. F, Candidate genes (GRMZM2G180490, GRMZM2G010702, GRMZM2G151649, and GRMZM2G057023) located in the peak bin. Other candidates genes located in the two bins next to the peak bin are listed in Supplemental Table S9.
Recently, large-scale metabolic trait QTL mapping was also performed in the same RIL population in different tissues including leaf (Wen et al., 2015). A common QTL hotspot was observed on chromosome 10 (Supplemental Fig. S5) in the metabolic traits QTL mapping and in this study, indicating that this genome region may not only affect the variation of most metabolic traits but also control maize growth traits.
Prediction of Digital Biomass Accumulation
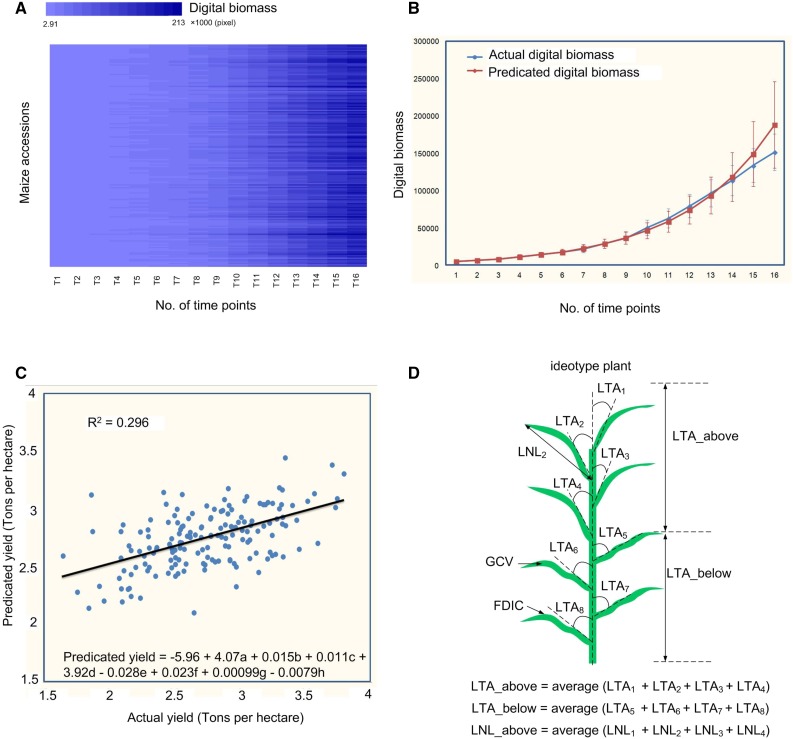
It would be very helpful for maize breeding if we could use the phenotypic data obtained in the early growth stage to predict the final biomass or yield. The digital biomass (SA) of the 387 individuals was calculated and showed good correlation with the manually measured fresh weight and dry weight (Supplemental Tables S2 and S3) and, thus, can be used to represent the biomass. After the digital biomass of the RIL population at 16 different time points was obtained (Fig. 5A), we tested the six models (linear, power, exponential, logarithm, quadratic, and logistic models) for their ability to predict digital biomass in the early growth stage. The digital biomass measurements at 16 time points were divided into a training set and a testing set. For example, if the number of time points for the training set was six (T1–T6), the corresponding number of time points for the testing set was 10 (T7–T16). The results were evaluated by comparison of r2, MAPE, and standard deviation of absolute percentage error (SDAPE) values. As shown in Supplemental Figure S6, when the number of time points for the training set is large enough (such as 11), the prediction results of the power, exponential, quadratic, and logistic models were all satisfactory (the r2 was 0.96 and the MAPE and SDAPE were both below 30%). However, when the number of time points for the training set was six, only the exponential model with the testing set showed good prediction ability (the r2 was 0.96 and the MAPE and SDAPE were both below 20%). The comparison of actual digital biomass and predicted digital biomass is shown in Figure 5B and indicates that, from the seedling stage to the tasseling stage, the exponential model had better prediction ability for digital biomass accumulation, even in the early growth stage (Supplemental Fig. S6; Supplemental Table S5).
Figure 5.
Predication of maize digital biomass accumulation and yield. A, Heat map showing the difference of digital biomass accumulation with different maize RILs. B, Comparison of actual digital biomass (blue line) and predicted digital biomass (red line; using digital biomass of the first six time points to predict the digital biomass of the remaining 10 time points). Error bars represent the se of the dry weight of 167 samples at each time point. C, Scatterplot showing the relationship between the actual grain yield and the predicted yield with the predicted formula; a, b, c, d, e, f, g, and h represent FDIC_1, LTA_above_1, GCV_8, SDLC_8, LTA_above_9, LTA_below_9, LNL_above_16, and LSA_below_16, respectively. FDIC_1 and LNL_above_16 are leaf morphological traits; LTA_above_1, SDLC_8, LTA_above_9, LTA_below_9, and LSA_below_16 are leaf angle traits; GCV_8 is a plant color trait. The black line is the fitting line, and the standardized coefficients are shown in Supplemental Table S7. D, Predicted ideotype maize plant based on the associated traits in early stages with higher GCV, FDIC, LNL_above, and LTA_below and lower LTA_above.
A Number of Novel Traits Could Be Used as Indicators for Final Yield Prediction
It would be important for plant breeding if we could use the measured traits, especially the traits measured in early development stages, to predict the final grain yield. To test whether this is possible, the variance explained for grain yield with different traits in different stages was evaluated (Supplemental Table S6). Up to 54.6% of the phenotypic variance of grain yield could be explained by combining 16 traits across all 16 time points. If only eight phenotypic traits at four time points (T1, T8, T9, and T16) were used, 29.6% of the grain yield variance was explained (Fig. 5C). Based on the values of coefficients in the selected model for grain yield (Supplemental Table S7), we established an ideotype maize plant (Fig. 5D). The grain yield had the following features. (1) A positive correlation with the leaf morphological traits (FDIC_1; r = 0.261) and the leaf angle in the upper half of the plant (LTA_above_1; r = 0.133). This implies that a more wavy-shaped leaf may maximize the area receiving light in the seedling stage. (2) A positive correlation with the leaf angle in the lower half of the plant (LTA_below_9; r = 0.156) and a negative correlation with the leaf angle in the upper half of the plant (LTA_above_9; r = −0.152). This distribution of leaf architecture can maximize the light-harvesting area, thus increasing photosynthetic efficiency. (3) A positive correlation with the SDLC_8 (sd of leaf curvature per plant; r = 0.171); higher SDLC indicates higher variability of leaf angles within a plant. (4) A positive correlation with GCV_8 (r = 0.114); higher GCV means more dark green leaves, which may be related to greater chlorophyll content. (5) A positive correlation with leaf length in the upper half of the plant (LNL_above_16; r = 0.192). These results were consistent with the smart canopy concept for the maize plant, which promises to maximize the potential for light harvesting per unit of land area (Ort et al., 2015). Interestingly, a QTL hotspot was identified on chromosome 10 for SDLC, which overlapped with a metabolic trait QTL (Wen et al., 2015; Supplemental Figs. S4 and S5). Cloning of this QTL may be helpful for understanding plant architecture regulation and the associations with grain yield.
DISCUSSION
Due to the limitations of traditional phenotyping, which is labor intensive, time consuming, low throughput, and costly (Chen et al., 2014), most previous QTL studies focused on a limited number of traits, usually at the mature stage (Yan et al., 2003; Osman et al., 2013; Zhang et al., 2013). Plant growth is a dynamic process, and the timing of end-point measurement will greatly influence the outcome of mapping (El-Lithy et al., 2004). In this study, using a modified image-analysis pipeline, the RAP was expanded for use in a maize RIL population for high-throughput quantification of multiple traditional and novel features at 16 development stages accurately (r2 > 0.97) and efficiently (45 s was needed for screening and image analysis per plant). Both traditional traits (i.e. plant height) and many novel features (i.e. FDIC, GCV, and LTA) could be investigated nondestructively at multiple time points. RAP-Maize provides a good opportunity to study the dynamic development process in maize and to better understand the underlying genetic mechanisms.
By combining the high-throughput maize phenotyping platform and the high-density linkage map, 988 QTLs were identified that provided the opportunity to investigate QTL distribution at the genome-wide level (Supplemental Data S2; Supplemental Fig. S4). The following observations were made. (1) QTLs were different at different stages, indicating that the plant growth regulation mechanism changes over time. (2) Most traits were controlled by a large number of QTLs, consistent with the quantitative nature of these traits as well as with previously studied agronomic traits (Buckler et al., 2009; Xiao et al., 2016). (3) QTL distribution across chromosomes was not random (Fig. 4B; Supplemental Fig. S4), and three QTL hotspots were identified (Fig. 4B), including one hotspot located in the genomic region identified previously for a metabolic trait QTL (Wen et al., 2015), which could be the key node for regulating plant growth. In total, 53 candidate genes within the first three peak bins (91.96–95.21 Mb) of this hotspot were identified (Supplemental Table S8), and great effort was still required to find the casual gene(s). Another QTL hotspot located on chromosome 7 associated with dry weight was identified across seven time points (Fig. 4, C–E). In total, 28 candidate genes were located in the first three peak bins (161.62–162.16 Mb; Supplemental Table S9), and only four candidate genes (GRMZM2G180490, GRMZM2G010702, GRMZM2G151649, and GRMZM2G057023) were located within the peak bin (161.95–162.04 Mb) of the QTL (Fig. 4F). GRMZM2G180490 encodes an adenylyl-sulfate kinase; GRMZM2G010702 has an unknown function; GRMZM2G151649, the homolog of AT3G01400.1, encodes an armadillo (ARM) repeat superfamily protein, which is involved in the ubiquitination pathway regulating the development of seed size in soybean (Glycine max; Xie et al., 2014); GRMZM2G057023 encodes an interferon-related developmental regulator and is expressed highly in leaf (Supplemental Fig. S7), and it might be the most likely candidate gene affecting dry weight.
Furthermore, for the biomass, significant loci detected here did not overlap with other studies (Barrière et al., 2010; Riedelsheimer et al., 2012; Rincent et al., 2014; Muraya et al., 2017), and only one QTL located in the hotspot on chromosome 7 coincided with the fresh shoot biomass and metabolite QTL identified previously (Wen et al., 2015). Cloning this QTL hotspot should help to explain an underlying mechanism of plant growth and metabolic regulation. In a recent study, an automated noninvasive phenotyping method was also used to monitor the plant sizes of 252 diverse maize inbred lines by focusing on biomass at 11 different developmental time points, and 12 main-effect marker-trait associations were identified (Muraya et al., 2017). Association mapping in maize is a very powerful tool to identify genes with high resolution if millions of molecular markers and a large population size are used (Yan et al., 2011; Liu et al., 2016). Only a few marker-trait associations were identified for biomass in the recent study (Muraya et al., 2016), since the marker density was low and the sample size was median. We also compared the merits and demerits of high-throughput phenotyping technology in different plant species, especially in crops (Supplemental Table S10). Our platform has obvious advantages for detecting trait/QTL number; more importantly, we detected many novel traits undetected previously that can be used for yield and biomass prediction. However, this was indoor shoot-based phenotyping and needs to expand to the field or field plot level.
Predicting crop yield using simple phenotypic indicators available early in development would greatly aid maize breeding. Molecular markers have been used widely in breeding diagnostics and are especially efficient for traits controlled by major genes (Collard and Mackill, 2008). In this study, we found that a few indicators in the early growth stage of maize could be used to predict the final grain yield. About 30% of the variance could be explained using only eight phenotypic traits at four time points. This is an impressive result, given that the grain yield data came from seven different field environments in different years and only a few measured traits from seedling to tasseling stage were used. These findings provide useful clues for ideotype-based maize breeding by optimization of leaf and plant architecture, such as (1) smaller to bigger leaf angles from top to ground; (2) more wave-like and dark green leaf; and (3) longer leaf, especially around and up ears. We simplified the model as shown in Figure 5D. More importantly, most of the mentioned traits are very difficult to measure by the traditional method and now can be manipulated automatically in the early stage of maize growth. The smart canopy for maize is an integrated concept that still needs to be tested and proven in the field. Combining the current field phenotyping platforms, such as the aerial sensing platform (Berni et al., 2009), ground-base field phenotyping at the plot level (Andradesanchez et al., 2013), and the movable imaging chamber in the field (Busemeyer et al., 2013), the robust image-analysis pipeline also could be transferred to the field (Berni et al., 2009; Andradesanchez et al., 2013; Busemeyer et al., 2013) for high-throughput phenotyping. In summary, combining the high-throughput phenotyping technology and large-scale QTL analysis not only greatly expanded our knowledge of the maize dynamic development process but also provided a new strategy for breeders to optimize plant architecture toward ideotype breeding in maize.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Experiment Design
In this study, a maize (Zea mays) RIL population (Chander et al., 2008a, 2008b; Wen et al., 2015) with its parents (B73 and BY804) were planted in the RAP (Yang et al., 2014) with two replications. All the maize RILs were screened at 16 different developmental time points (once every 3 d starting from 22 to 67 d after sowing; e.g. T1–T16 represent 22, 25, 28, 31, 34, 37, 40, 43, 46, 49, 52, 55, 58, 61, 64, and 67 d after sowing, respectively); the sowing date and inspection dates are provided in Supplemental Table S11. The growth conditions were as follows. Fertilizing was carried out at sowing, V5, and V9 stage (60 kg of water + 370.68 g of carbamide + 330.76 g of potassium dihydrogen phosphate + 94.24 g of potassium chloride, to be fully dissolved with 150 mL of liquid fertilizer for each plant per time). A subset of the association mapping population (Yang et al., 2011; Li et al., 2013), including 100 diversity inbred lines, was randomly selected and sown on the same day in a phenotypic platform with four replications, screened using the RAP, and measured manually at 36, 48, 60, and 70 d after sowing (Supplemental Table S11). Destructively measured traits were obtained for biomass modeling, and the correlation between manual measurement and automatic measurement was calculated. Growth movies of the parents and four selected recombinant lines are shown in Supplemental Videos S2 to S7.
The RIL population was also planted in Henan, Hubei, Chongqing, Yunnan, and Hainan provinces, China, during 2011 and 2012. In the seven environments, at least five good open-pollinated ears were harvested from each row for measuring ear weight and cob weight. Yield data (ear weight minus cob weight) for each line was recorded. Best linear unbiased prediction (BLUP) was obtained by fitting the mixed liner model in the R package lem4 (R Development Core Team, 2013) for estimation of the breeding value of each line across all environments, and the BLUP values were then combined to reduce the prediction bias caused by the unbalanced data. Finally, the BLUP data of grain yield across seven environments and all investigated phenotypic traits obtained across 16 time points were put into the grain yield prediction model. The experimental design is shown in Supplemental Figure S8.
Image Analysis and Trait Extraction
Maize image analysis was carried out as shown in Supplemental Figure S1. First, the image with the maximum plant width was selected automatically from 15 different angles (Supplemental Fig. S1A); then, the excess green vegetation index (E × G) component was extracted, and the OTSU method (Ohtsu, 1979) was applied to acquire the binary image (Supplemental Fig. S1B). Second, a region growing algorithm was used to obtain the whole-plant binary image (Supplemental Fig. S1C). Using the binary image, plant morphological traits and projected area were calculated. Moreover, the color trait and histogram texture traits were computed by matching the binary image and original color image. Then, the parallel thinning algorithm was performed to create the skeleton image (Supplemental Fig. S1D), and the Hough transformation was applied to distinguish the leaf skeleton from the stem skeleton (Supplemental Fig. S1E). With this information, the stem length and total leaf were identified. Finally, each leaf skeleton was identified and labeled (Supplemental Fig. S1F), and the traits for each leaf were calculated, including leaf angle, leaf length, and leaf curvature. With biomass (fresh weights and dry weights) obtained at different time points, growth-related traits were calculated. Detailed information for trait definitions is shown in Supplemental Table S1, and details of trait extraction are described in Supplemental Note S1. The software interface and the source code of the image-analysis pipeline are given in Supplemental Figures S9 and S10. In addition, the LabVIEW programs and dynamic link library enrolled in the maize image-analysis pipeline are listed in Supplemental Table S12. All the source code and software applications, including LabVIEW programs, dynamic link library, and cpp documents, can be downloaded using the link http://plantphenomics.hzau.edu.cn/checkiflogin_en.action (username, UserPP; password, 20170108pp).
Biomass Modeling and Digital Biomass Accumulation Modeling
To determine the best model for measuring fresh weight and dry weight, 10 models (including linear, quadratic, exponential, and power models) were evaluated using the adjusted coefficient of determination (adjusted r2) and the MAPE. Statistical analysis (mainly the linear stepwise regression) for biomass modeling was implemented with SPSS software (Statistical Product and Service Solutions, version 13.0). After the predicted fresh weights and dry weights at 16 different time points were obtained, maize plant growth was modeled with six models: linear, power, exponential, logarithm, quadratic, and logistic. In order to test the prediction ability of the different models in the early growth stage, the fresh weight values at 16 time points were divided into two parts: a training set and a testing set. The results of trait fitting were evaluated by comparison of r2, MAPE, and SDAPE values. The statistical analyses of the six models (linear, power, exponential, logarithm, quadratic, and logistic) for maize plant growth were implemented with LabVIEW 8.6 (National Instruments) and MATLAB 2011 (The Mathworks).
Grain Yield Predication Using Plant Phenotypic Traits in the Early Growth Stage
To evaluate the variance explained for the maize grain yield in the early growth stages, linear stepwise regression was used with maize plant phenotypic traits. The entry value of use probability of F in the stepping method criterion was 0.05, and the removal value of use probability of F was 0.1. The variable was added into the model if the F value was less than the entry value; however, the variable was removed if the F value was higher than the removal value. The linear stepwise regression analysis for grain yield was implemented with SPSS software.
Heritability Analysis
Heritability (H2) was calculated for each trait as follows:
where σ2G is the genotypic variance, σ2e is the error variance, and r is the number of replications. The estimates of σ2G and σ2e were analyzed by AVOVA using the lmer function in the lme4 package in the R environment (R Development Core Team, 2013; version 3.1.3; http://www.r-project.org/).
Genotype, Linkage Map Construction, and QTL Mapping
The genotypic data for the RIL population obtained from a former study (Wen et al., 2015) showed that a linkage map was 1,790.2 cM in length, including 2,496 recombinant bins, 0.72 cM per bin on average. Details about the map construction and its description were reported in previous studies (Wen et al., 2015; Pan et al., 2016). In summary, the RIL population was genotyped using the Illumina MaizeSNP50 BeadChip, which contains 56,110 SNPs (Ganal et al., 2011). SNPs with both missing rate and heterozygosity of less than 10% were used to construct the genetic linkage map, which contains 2,496 recombinant bins. In this study, 167 of the genotyped RILs were used for QTL mapping using the seedling emergence and growth phenotypes. QTL analysis was performed by the composite interval mapping method (Zeng, 1994) using the software Windows QTL Cartographer version 2.5 (Wang and Zeng, 2007) for each investigated phenotypic trait and growth-related traits at 16 time points. Walking step was set to 0.5 cM, and zmap (model 6) with a 10-cM window was used. The bins or genetic blocks (a genomic region in which no recombination exists) were clearly defined, and a uniform LOD value was assigned for each bin. To determine the LOD threshold, 25 trait-time point combinations from all 736 trait-time point combinations were selected randomly for permutation tests with 500 times (P = 0.05). The results indicated that the LOD threshold ranged from 3.23 to 3.65, with a mean of 3.42. To simplify it, a threshold of LOD ≥ 3.5 was used to establish the presence of a QTL for all the traits. A QTL support interval was defined as the 1 − LOD drop position ranging from the QTL peak. All QTLs with overlapping QTL support intervals were categorized as nonredundant QTLs. Possible candidate genes were identified within the QTL hotspots on chromosomes 7 and 10 based on the filtered working gene list of the maize genome downloaded from MaizeGDB (http://www.maizegdb.org). Candidate genes were annotated according to InterProScan (http://www.ebi.ac.uk/interpro/scan.html).
Construction of the Trait-Locus Network
The nodes of the trait-locus network contain all traits across 16 development stages and their corresponding significant loci with the threshold value of LOD ≥ 3.5. All traits were labeled blue, and all loci were labeled pink (Fig. 4A). The network was visualized using the software Cytoscape version 2.6.3 (Shannon et al., 2003).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Maize image analysis and trait extraction.
Supplemental Figure S2. Distribution of QTL mapping resolution.
Supplemental Figure S3. Correlation coefficients between paired traits for 42 traits investigated at 16 time points.
Supplemental Figure S4. Chromosomal distribution of identified QTLs with 42 primary phenotypic traits and 64 growth-related traits.
Supplemental Figure S5. Comparison of heat maps for QTL density between metabolic and investigated phenotypic traits in the BY804/B73 recombination population.
Supplemental Figure S6. Predication ability comparison of six models for digital biomass accumulation.
Supplemental Figure S7. The RNA sequencing gene atlas for four genes (GRMZM2G180490, GRMZM2G010702, GRMZM2G151649, and GRMZM2G057023).
Supplemental Figure S8. Experimental design.
Supplemental Figure S9. The image-analysis interface designed in this study.
Supplemental Figure S10. Flow chart of the program.
Supplemental Table S1. Classification and abbreviations for 106 traits.
Supplemental Table S2. Statistical summary of the 10 developed models for fresh weight estimation.
Supplemental Table S3. Statistical summary of the 10 developed models for dry weight estimation.
Supplemental Table S4. Summary of QTLs for growth rate-related traits identified at 16 time points.
Supplemental Table S5. Statistical summary of the six developed models for digital biomass accumulation.
Supplemental Table S6. Detecting the phenotypic traits significantly associated with yield and calculating the percentage of the phenotypic variance explanation.
Supplemental Table S7. Statistical details of coefficients in a selected model for yield.
Supplemental Table S8. Candidate genes and their annotations located in the first three peak bins in the QTL hotspot located on chromosome 10.
Supplemental Table S9. Candidate genes and their annotations located in the first three peak bins in the QTL hotspot located on chromosome 7.
Supplemental Table S10. Comparison of published work for the combination of high-throughput phenotyping and QTL/genome-wide association study analysis.
Supplemental Table S11. Experimental schedule of maize plant phenotyping.
Supplemental Table S12. Detailed information of Subvi and the dynamic link library used in this study.
Supplemental Data S1. Variation of investigated phenotypic traits in the BY804/B73 population and two parents at different stages.
Supplemental Data S2. QTL information summary of all traits across 16 time points.
Supplemental Video S1. Operating procedure for RAP-Maize.
Supplemental Video S2. Growth movie of the parent B73.
Supplemental Video S3. Growth movie of the parent BY804.
Supplemental Video S4. Growth movie of the recombinant line BB048.
Supplemental Video S5. Growth movie of the recombinant line BB054.
Supplemental Video S6. Growth movie of the recombinant line BB078.
Supplemental Video S7. Growth movie of the recombinant line BB096.
Supplemental Notes S1. Definitions of the features.
Supplementary Material
Glossary
- SNP
single-nucleotide polymorphism
- RAP
rice automatic phenotyping platform
- RIL
recombinant inbred line
- QTL
quantitative trait locus
- SA
side-projected area
- MAPE
mean absolute percentage error
- cM
centimorgan
- BLUP
Best linear unbiased prediction
- LOD
log of the odds
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0100103), the National Program on High Technology Development (grant no. 2013AA102403), the National Natural Science Foundation of China (grant nos. 31600287 and 31525017), the Hubei Provincial Natural Science Foundation of China (grant no. 2015CFB529), and the Fundamental Research Funds for the Central Universities.
Articles can be viewed without a subscription.
References
- Andradesanchez P, Gore MA, Heun JT, Thorp KR, Carmosilva AE, French AN, Salvucci ME, White JW (2013) Development and evaluation of a field-based high-throughput phenotyping platform. Funct Plant Biol 41: 68–79 [DOI] [PubMed] [Google Scholar]
- Barrière Y, Méchin V, Denoue D, Bauland C, Laborde J (2010) QTL for yield, earliness, and cell wall quality traits in topcross experiments of the F838 × F286 early maize RIL progeny. Crop Sci 50: 1761–1772 [Google Scholar]
- Berni J, Zarco-Tejada PJ, Suarez L, Fereres E (2009) Thermal and narrowband multispectral remote sensing for vegetation monitoring from an unmanned aerial vehicle. IEEE Trans Geosci Remote Sens 47: 722–738 [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Busemeyer L, Ruckelshausen A, Möller K, Melchinger AE, Alheit KV, Maurer HP, Hahn V, Weissmann EA, Reif JC, Würschum T (2013) Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci Rep 3: 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chander S, Guo Y, Yang X, Yan J, Zhang Y, Song T, Li J (2008a) Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and the candidate gene approach. Mol Breed 22: 353–365 [Google Scholar]
- Chander S, Guo YQ, Yang XH, Zhang J, Lu XQ, Yan JB, Song TM, Rocheford TR, Li JS (2008b) Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor Appl Genet 116: 223–233 [DOI] [PubMed] [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy ME, Clerkx EJ, Ruys GJ, Koornneef M, Vreugdenhil D (2004) Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol 135: 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Tester M (2011) Phenomics: technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner EM, Hansen M, Joets J, et al. (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. (2011) A cornucopia of maize genes. Nat Genet 43: 87–88 [DOI] [PubMed] [Google Scholar]
- Hartmann A, Czauderna T, Hoffmann R, Stein N, Schreiber F (2011) HTPheno: an image analysis pipeline for high-throughput plant phenotyping. BMC Bioinformatics 12: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsdorf N, March TJ, Berger B, Tester M, Pillen K (2014) High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 9: e97047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D, Govindaraju DR, Omholt S (2010) Phenomics: the next challenge. Nat Rev Genet 11: 855–866 [DOI] [PubMed] [Google Scholar]
- Junker A, Muraya MM, Weigelt-Fischer K, Arana-Ceballos F, Klukas C, Melchinger AE, Meyer RC, Riewe D, Altmann T (2015) Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Front Plant Sci 5: 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan M, Song JT, Seo HS (2013) Seed size: a priority trait in cereal crops. Physiol Plant 147: 113–120 [DOI] [PubMed] [Google Scholar]
- Klukas C, Chen D, Pape JM (2014) Integrated analysis platform: an open-source information system for high-throughput plant phenotyping. Plant Physiol 165: 506–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, et al. (2011) Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet 43: 163–168 [DOI] [PubMed] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, et al. (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45: 43–50 [DOI] [PubMed] [Google Scholar]
- Liu H, Luo X, Niu L, Xiao Y, Chen L, Liu J, Wang X, Jin M, Li W, Zhang Q, et al. (2016) Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol Plant (in press) doi/10.1016/j.molp.2016.06.016 [DOI] [PubMed] [Google Scholar]
- Martinez AK, Soriano JM, Tuberosa R, Koumproglou R, Jahrmann T, Salvi S (2016) Yield QTLome distribution correlates with gene density in maize. Plant Sci 242: 300–309 [DOI] [PubMed] [Google Scholar]
- Muraya MM, Chu J, Zhao Y, Junker A, Klukas C, Reif JC, Altmann T (2017) Genetic variation of growth dynamics in maize (Zea mays L.) revealed through automated non-invasive phenotyping. Plant J (in press) doi/10.1111/tpj.13390 [DOI] [PubMed] [Google Scholar]
- Nagel KA, Putz A, Gilmer F, Heinz K, Fischbach A, Pfeifer J, Faget M, Blossfeld S, Ernst M, Dimaki C, et al. (2012) GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol 39: 891–904 [DOI] [PubMed] [Google Scholar]
- Ohtsu N. (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9: 62–66 [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman KA, Tang B, Wang Y, Chen J, Yu F, Li L, Han X, Zhang Z, Yan J, Zheng Y, et al. (2013) Dynamic QTL analysis and candidate gene mapping for waterlogging tolerance at maize seedling stage. PLoS ONE 8: e79305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Li L, Yang X, Tong H, Xu S, Li Z, Li W, Muehlbauer GJ, Li J, Yan J (2016) Genome-wide recombination dynamics are associated with phenotypic variation in maize. New Phytol 210: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Reuzeau C, Pen J, Frankard V, de Wolf J, Peerbolte R, Broekaert W, van Camp W (2005) TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Mol Plant Breed 3: 753–759 [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA 109: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincent R, Nicolas S, Bouchet S, Altmann T, Brunel D, Revilla P, Malvar RA, Moreno-Gonzalez J, Campo L, Melchinger AE, et al. (2014) Dent and Flint maize diversity panels reveal important genetic potential for increasing biomass production. Theor Appl Genet 127: 2313–2331 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327: 818–822 [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Wang SBC, Zeng Z (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, et al. (2015) Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell 27: 1839–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Tong H, Yang X, Xu S, Pan Q, Qiao F, Raihan MS, Luo Y, Liu H, Zhang X, et al. (2016) Genome-wide dissection of the maize ear genetic architecture using multiple populations. New Phytol 210: 1095–1106 [DOI] [PubMed] [Google Scholar]
- Xie FT, Niu Y, Zhang J, Bu SH, Zhang HZ, Geng QC, Feng JY, Zhang YM (2014) Fine mapping of quantitative trait loci for seed size traits in soybean. Mol Breed 34: 2165–2178 [Google Scholar]
- Yan J, Tang H, Huang Y, Shi Y, Li J, Zheng Y (2003) Dynamic analysis of QTL for plant height at different developmental stages in maize (Zea mays L.). Chin Sci Bull 48: 2601–2607 [Google Scholar]
- Yan J, Warburton M, Crouch J (2011) Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci 51: 433–449 [Google Scholar]
- Yang W, Duan L, Chen G, Xiong L, Liu Q (2013) Plant phenomics and high-throughput phenotyping: accelerating rice functional genomics using multidisciplinary technologies. Curr Opin Plant Biol 16: 180–187 [DOI] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, et al. (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5: 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Li J, Yan J (2011) Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed 28: 511–526 [Google Scholar]
- Zeng ZB. (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu Z, Cui Z, Hu Y, Wang B, Tang J (2013) Genetic analysis of grain filling rate using conditional QTL mapping in maize. PLoS ONE 8: e56344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Li J (2013) Molecular dissection of complex agronomic traits of rice: a team effort by Chinese scientists in recent years. Natl Sci Rev 1: 253–276 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.