DNA polymerase epsilon plays a key role in replicative stress sensing and signaling.
Abstract
Faithful transmission of the genetic information is essential in all living organisms. DNA replication is therefore a critical step of cell proliferation, because of the potential occurrence of replication errors or DNA damage when progression of a replication fork is hampered causing replicative stress. Like other types of DNA damage, replicative stress activates the DNA damage response, a signaling cascade allowing cell cycle arrest and repair of lesions. The replicative DNA polymerase ε (Pol ε) was shown to activate the S-phase checkpoint in yeast in response to replicative stress, but whether this mechanism functions in multicellular eukaryotes remains unclear. Here, we explored the genetic interaction between Pol ε and the main elements of the DNA damage response in Arabidopsis (Arabidopsis thaliana). We found that mutations affecting the polymerase domain of Pol ε trigger ATR-dependent signaling leading to SOG1 activation, WEE1-dependent cell cycle inhibition, and tolerance to replicative stress induced by hydroxyurea, but result in enhanced sensitivity to a wide range of DNA damaging agents. Using knock-down lines, we also provide evidence for the direct role of Pol ε in replicative stress sensing. Together, our results demonstrate that the role of Pol ε in replicative stress sensing is conserved in plants, and provide, to our knowledge, the first genetic dissection of the downstream signaling events in a multicellular eukaryote.
Faithful duplication of the genome is a key step during cell proliferation in all living-organisms. In eukaryotes, it requires the activity of three replicative polymerases (DNA Pol α, δ, and ε) that are associated to a large protein complex called the “replisome” that encompasses all the core activities required for DNA replication (Kurth and O’Donnell, 2013).
Although it is clear that Pol α is responsible for the synthesis of RNA/DNA primers, the exact roles of Pol δ and ε are still a matter of debate. The most widely accepted view is that Pol δ and ε synthesize the lagging and leading strands, respectively (Pursell et al., 2007; Kunkel and Burgers, 2008). However, according to an alternative model, Pol δ could be the main replicative polymerase, whereas Pol ε would be involved in the repair of replication errors and play a scaffolding role (Johnson et al., 2015). Combination of a collection of mutations with hypomorphic alleles of the three replicative polymerases revealed specialized genetic networks interacting with each polymerase: this observation corroborated the nonoverlapping functions of the three polymerases in yeast as well as the central role of Pol ε at the preinitiation steps of DNA replication (Dubarry et al., 2015).
In animals and yeast, DNA Pol ε consists of four subunits: one large catalytic subunit Pol2 and three accessory subunits Dpb2, 3, and 4. Pol2 and Dpb2 are essential to cell viability, whereas Dpb3 and 4 are dispensable (Pursell and Kunkel, 2008). Pol2 has two functional moieties: the highly conserved N-terminal domain encompassing the polymerase and exonuclease activities, and a C-terminal domain (Tahirov et al., 2009). Surprisingly, only the C-terminal extension is required for cell survival and DNA replication (Kesti et al., 1999), further supporting the notion that Pol ε has an essential scaffolding function, independently from DNA synthesis per se. Furthermore, detailed genetic analysis performed in fission yeast demonstrated that Pol2 is required both for the chromatin loading and the progression of the CMG complex (Handa et al., 2012), a multisubunit complex comprising CDC45, the Mini Chromosome Maintenance heterohexamer, and the Go-Ichi-Ni-San that functions as the replicative helicase, and is connected to Pol2 via Dpb2 (Sengupta et al., 2013).
A number of factors such as DNA lesions, difficult to replicate sequences, collision with the transcription machinery, and others can impede fork progression during the S-phase and cause replicative stress. Stalled forks are fragile structures that can lead to genetic instability; cells have therefore evolved complex sensing mechanisms allowing checkpoint activation in response to replicative stress (Jossen and Bermejo, 2013). Checkpoint activation triggers the expression of multiple genes required for replication fork stabilization, cell cycle arrest, and DNA repair (Friedel et al., 2009; Segurado and Tercero, 2009). In yeast, replicative stress activates the Mec1 kinase (ATR in Animals and Plants), that leads to nucleotide biosynthesis, expression of the DNA repair machinery, and cell cycle arrest (Jossen and Bermejo, 2013). Interestingly, Mec1 activation is mediated via two independent pathways, one triggered by single-stranded DNA (ssDNA) accumulation and the other requiring the C-terminal domain of Pol2a (Navas et al., 1995; Puddu et al., 2011). This sensor role of DNA Pol ε likely involves its ability to interact with the checkpoint protein Rad17 (Post et al., 2003) and the mediator protein Mrc1 (Lou et al., 2008). In addition, in budding yeast, association of the Ctf18-RFC complex with the N terminus of Pol ε was shown to be instrumental for the activation of the S-phase checkpoint, indicating that both domains of the protein can contribute to this sensor role of Pol ε (García-Rodríguez et al., 2015). In Xenopus laevis, POL2A interacts with Claspin (the homolog of Mrc1; Lee et al., 2005), and the essential role of the C terminus is conserved in Drosophila (Suyari et al., 2012). Together, these reports suggest that the dual function of Pol ε catalytic subunit in DNA replication and replicative stress response is conserved in all eukaryotes. However, most of the knowledge regarding the role of POL2A in replicative stress sensing has been obtained in yeast because the lethality of POL2A deficiency has precluded detailed analysis in multicellular organisms.
The genome of Arabidopsis (Arabidopsis thaliana) encompasses two genes encoding the catalytic subunit of Pol ε: POL2A and POL2B, but only POL2A is an essential gene (Ronceret et al., 2005). Over the past 10 years, a number of hypomorphic alleles of POL2A have been isolated (see Supplemental Fig. S1): the early in short days7 (esd7-1) mutant, which harbors a mutated amino acid in the catalytic domain of POL2A close to the junction of N- and C-terminal regions of the protein and shows early flowering as well as overall reduced growth (del Olmo et al., 2010); the abscisic acid oversensitive4 (abo4-1) mutant line, which has a point mutation in the catalytic domain of the polymerase; the abo4-2 mutant (with a T-DNA insertion; Supplemental Fig. S1), which displays enhanced homologous recombination in somatic cells and constitutive activation of DNA repair genes (Yin et al., 2009); and the tilted1 (til1-4) mutant, which displays prolonged cell cycle during embryo development (Jenik et al., 2005). By contrast, disruption of POL2B has no visible effect on plant development, although esd7-1 pol2b double mutants show more severe growth defects than esd7-1 single mutants, providing evidence for some level of redundancy between the two genes (del Olmo et al., 2010).
Defects observed in POL2A hypomorphic mutants suggest that the role of DNA Pol ε in replicative stress sensing is also conserved in plants. In plants as in other eukaryotes, the Ataxia Telangiectasia Mutated (ATM, also called “Tel1” in yeast) and ATM- and Rad3-related (ATR, also called “Mec1” in yeast) are the two main kinases involved in the response to double-strand breaks (DSB) and replicative stress, respectively (Yoshiyama et al., 2013). In plants, signals from these two pathways converge toward the SOG1 transcription factor that can activate cell cycle inhibitors as well as DNA repair genes (Yoshiyama et al., 2013). We demonstrated previously that overexpression of the DPB2 subunit of Pol ε activates the DNA damage response (DDR; Pedroza-García et al., 2016) via both the ATM and the ATR pathways. However, in yeast and animals, the catalytic subunit POL2 rather than its accessory subunits is thought to be directly involved in replicative stress sensing (Lee et al., 2005; Puddu et al., 2011). In this work, we took advantage of the viability of Arabidopsis hypomorphic mutant lines to investigate the role of POL2A in replicative stress sensing and to genetically test its interaction with the main players of DDR that are conserved in all eukaryotes (Yoshiyama et al., 2013). Our results indicate that plant POL2A functions upstream of the DDR Kinase ATR to activate replication stress response, providing evidence for the conservation of its key role in genome stability in multicellular eukaryotes.
RESULTS
POL2A Hypomorphic Mutants Are Tolerant to Replicative Stress Induced by Hydroxyurea and Display Constitutive Activation of the DNA Damage Response
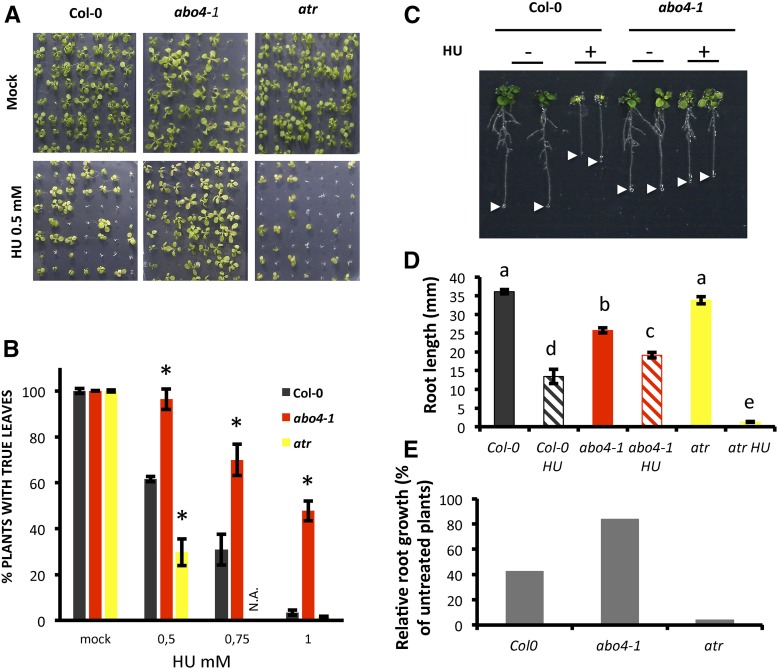
To investigate the role of the catalytic subunit of Pol ε in the activation of the DDR, we initially analyzed the abo4-1 mutant (Yin et al., 2009). This mutant harbors a point mutation leading to a Gly-534 to Arg change affecting a highly conserved amino acid in the catalytic domain of the protein (Supplemental Fig. S1). These mutants are hypersensitive to the replicative polymerase inhibitor aphidicolin, providing evidence for the impairment of Pol ε activity (Pedroza-García et al., 2016), and have been reported to display impaired cell cycle progression and activation of DNA repair genes, suggesting that replicative stress might be constitutively activated. To test this hypothesis, we first asked whether S-phase progression was impaired in abo4-1 mutants as we reported previously for DPB2 overexpressing lines (Pedroza-García et al., 2016). Indeed, flow cytometry analysis of flower buds nuclei (Supplemental Fig. S2) and cumulative EdU incorporation assay (Supplemental Table S1) revealed that S-phase is prolonged in abo4-1 mutants, and the total cell cycle length is increased by almost 70%. However, by contrast with DPB2 overexpressors, abo4-1 mutants are hypersensitive to a wide range of genotoxic stresses [(Yin et al., 2009) and this study Supplemental Fig. S3], such as the DSB-inducing agents Mitomycin C or zeocin, and UV-B irradiation. These drugs not only directly damage DNA but also can produce replication-blocking lesions, and the hypersensitivity of abo4-1 to genotoxins might thus result either from defects in response to DNA damage or in failure to activate the appropriate response upon fork blockage. To explore the role of POL2A in the replicative stress response without using DNA damaging agents, abo4-1 mutants were challenged with hydroxyurea (HU), an inhibitor of ribonucleotide reductase (RNR) that induces stalling of replication forks by depleting cellular deoxyribonucleotide pools. Treatment with low doses of HU thus induces fork stalling without creating DNA damage directly, although it can result in DSB formation as a consequence of fork collapse (Singh and Xu, 2016). As shown in Figure 1, abo4-1 was more tolerant to replication fork stalling than the wild type.
Figure 1.
The abo4-1 mutant shows increased tolerance to HU-induced replicative stress. A and B, Wild-type (Col-0) and abo4-1 mutant seedlings were germinated on HU-supplemented medium and plants with true leaves were counted after 12 d. The atr mutant was used as a hypersensitive control. In , values are average ± se of three biological replicates. Asterisks indicate statistically relevant differences with respect to the wild type in the same conditions (Student t test, P < 0.05). C to E, Wild type (Col-0) and abo4-1 mutant seedlings were grown for 4 d on half-strength MS and transferred to HU-supplemented medium (1 mM) for 9 d to monitor root growth. C, By contrast with wild-type plants, root length was almost unchanged by HU exposure in the abo4-1 mutant; arrowheads mark the position of the root tip. D, Average root length was measured after 9 d on HU; at least 20 plantlets were used for each treatment; values are average ± se. Different letters indicate significantly different values (Student t test, P < 0.05). Data are representative of two independent experiments. E, The relative growth of each genotype after 9 d on HU was calculated compared to untreated plants of the same genotype.
Previous studies have shown that a number of genes involved in the DDR are constitutively activated in POL2A-deficient mutants (Yin et al., 2009), which may account for their improved tolerance to HU. To obtain a global view of this response genomewide, the transcriptome of the abo4-1 line was compared to that of wild-type plants by RNA sequencing. The abo4-1 line showed that 218 genes were significantly induced while 154 were repressed (absolute fold change ≥1.5 P value ≤0.01; Supplemental Table S2); despite discrepancies between the sets of genes identified as differentially expressed that likely result from differences in growth conditions or data analysis, we observed significant overlap with previous RNA-seq analysis of this mutant [(Han et al., 2015), Supplemental Fig. S4A; χ2 P value less than 2.2e-16]. As expected, gene-ontology analysis of significantly up-regulated genes revealed overrepresentation of DNA metabolic process, response to DNA damage, and cell cycle (Supplemental Fig. S4B). Among these were several genes involved in DNA replication. Three genes encoding the ssDNA binding proteins RPA1C, D, and E as well as RAD17 were up-regulated, indicating that abo4-1 mutants are subjected to constitutive replicative stress. In addition, different ATR- downstream targets such as the WEE1 kinase, that participates in the inactivation of cyclin-dependent kinases (CDKs), or SMR7, a plant-specific CDKs inhibitor, were activated together with B-type CDKs (CDKB1;1, CDKB1;2, and CDKB2;1) and B-type cyclin (CYCB1;1, CYCB1;4, CYCB2;1, and CYCB2;4), consistent with previous reports (Yin et al., 2009). Finally, expression of genes involved in DNA repair was also induced in the abo4-1 mutant line (Supplemental Table S3).
This analysis confirms that constitutive DNA replicative stress results in the activation of cell cycle checkpoints in the abo4-1 line.
The ATR-WEE1-Dependent Checkpoint Is Required for the Viability of POL2A Mutants
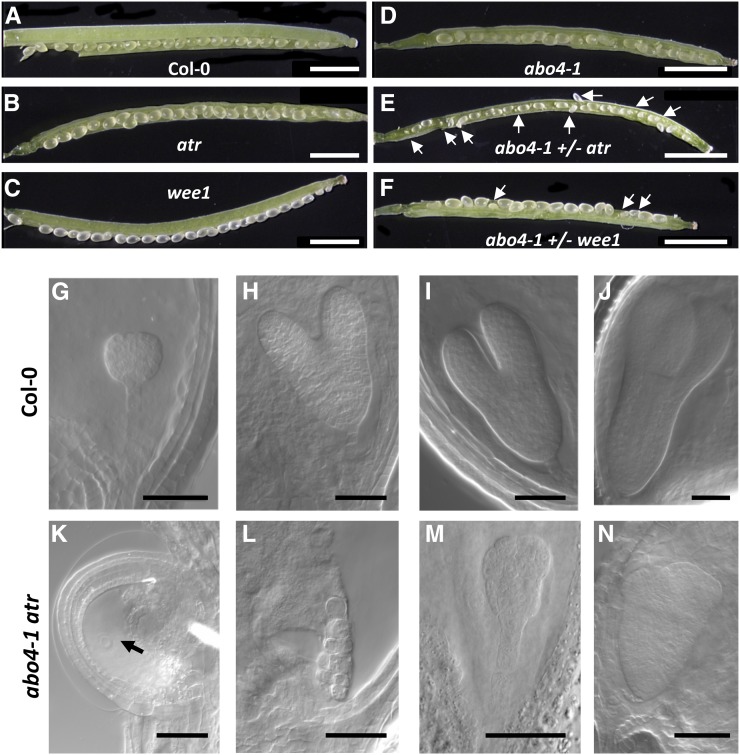
We next used a genetic approach to determine which DDR pathways are activated in abo4-1. We first crossed abo4-1 with the atr and wee1 mutants that are deficient for replicative stress response (Hu et al., 2015). Double mutants could never be recovered, and siliques of sesquimutants contained approximately 1/4 of aborted seeds (Fig. 2, A–F). Closer observation of embryo development in abo4-1 atr/+ and abo4-1 wee1/+ sesquimutants showed that 1/4 of embryos stopped development at various stages and displayed aberrant division patterning (Fig. 2, G–N). Similar defects in embryo development have already been described in T-DNA insertion homozygous mutants for pol2a, and also when wild-type embryos were exposed to aphidicolin, an inhibitor of replicative polymerases (Jenik et al., 2005). These results thus suggest that ATR and WEE1 are required during embryo development in abo4-1 for cell proliferation progress despite replicative stress.
Figure 2.
ATR and WEE1 are required for abo4-1 mutant viability. A to F, Open siliques of wild type (A), atr (B), wee1 (C), abo4-1 (D) mutants, and atr/+ abo4-1 (E) and wee1/+ abo4-1 (F) sesquimutants. Arrows point to aborted seeds. Bar = 2 mm for all panels. G to N, Embryo development in wild type (G to J) and wee1/+ abo4-1 sesquimutants. G, Globular stage. H, Late heart stage. I and J, Early and late torpedo stage. In the siliques of wee1/+ abo4-1 mutants, approximately 3/4 of embryos undergo normal development as in the wild type, abo4-1, or wee1 single mutants. However, 1/4 of embryos stop development at various stages and show aberrant division patterning. K, Arrested embryo just after fertilization; the arrow points to the single nucleus of the endosperm. L and M, Embryos at the globular stage with abnormal cell organization. N, Embryo at the late torpedo stage with misshapen cotyledons. Bar = 50 µm for all panels.
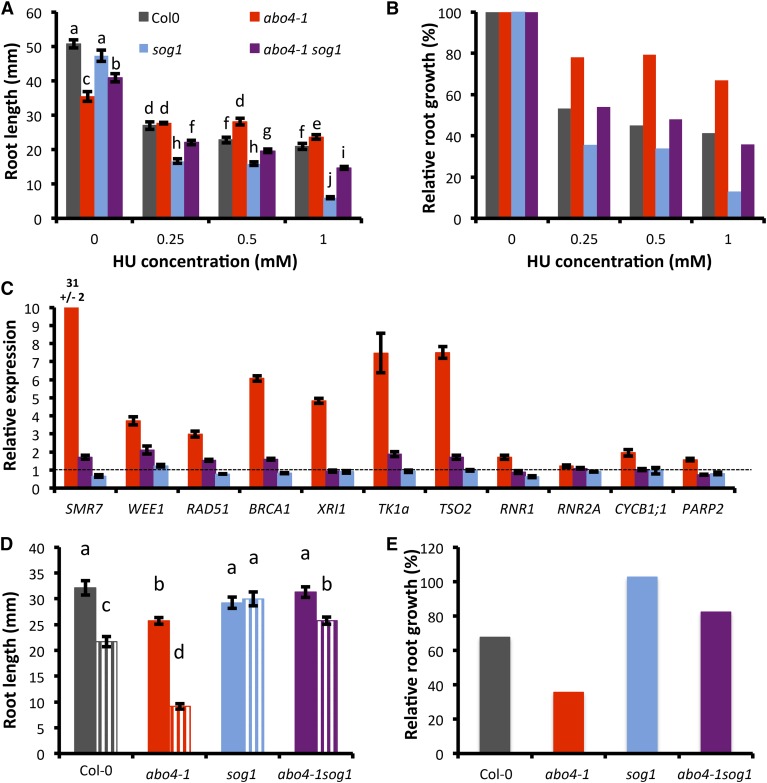
Part of the response to replicative stress mediated by ATR depends on the SOG1 transcription factor that acts independently of WEE1 (Hu et al., 2015). We therefore asked whether SOG1 also contributed to the checkpoint activation observed in abo4-1 mutants. abo4-1 sog1 mutants were viable, albeit smaller than abo4-1 single mutants (Supplemental Fig. S5), indicating that SOG1 activity is required to sustain growth in abo4-1, but not for embryo development.
We next asked whether the tolerance to replicative stress observed in abo4-1 plants required SOG1 activation. As previously demonstrated in Hu et al. (2015), sog1 was hypersensitive to HU. By contrast, abo4-1 sog1 mutants behaved like wild-type plants on medium supplemented with 0.25 mm or 0.5 mm of HU and thus displayed an intermediate phenotype between the two parental lines (Fig. 3, A and B, and Supplemental Fig. S6, A and B). In addition, we performed qRT-PCR on 11 genes that were up-regulated in abo4-1 seedlings according to the RNA-seq data (Supplemental Table S3). These genes are representative of different mechanisms such as cell cycle regulation (CYCB1;1, WEE1, and SMR7), DNA repair genes (RAD51, BCRA1, XRI1, and PARP2), and nucleotide synthesis genes (TK1a, TSO2, and RNR1). Up-regulation of some DDR genes was lost in abo4-1 sog1 mutants while others were still up-regulated, albeit to a lower extent than in abo4-1 single mutants (Fig. 3C), consistent with the notion that Pol ε deficiency activates the replication stress response via SOG1-dependent and -independent pathways.
Figure 3.
The checkpoint activated by the abo4-1 mutation is partially dependent on SOG1. A and B, HU sensitivity in wild type (Col-0), abo4-1, sog1, and abo4-1 sog1 mutants. Plantlets were grown on half-strength MS for 4 d and transferred to control medium or HU-supplemented medium for 9 d. A, Root length; values are average ± se obtained on at least 15 plantlets. B, Relative root growth; values are expressed as percentage of length on MS medium. C, qRT-PCR analysis of the expression of selected genes in abo4-1, sog1, and abo4-1 sog1 mutants; values are average ± sd. D and E, Zeocin sensitivity in wild-type (Col-0), abo4-1, sog1, and abo4-1 sog1 mutants. Plantlets were grown on half-strength MS for 4 d and transferred to control medium (full bars) or zeocin-supplemented medium (10 µM, dashed bars) for 9 d. D, Root length; values are average ± se obtained on at least 15 plantlets. In A and D, different letters indicate significantly different values (Student t test, P < 0.05). For all panels, data are representative of at least two independent experiments.
Finally, we tested the contribution of SOG1 to the sensitivity of abo4-1 mutants to DSB-inducing agents. As shown in Figure 3, D and E, and Supplemental Figure S6, the abo4-1 sog1 double mutant was tolerant to zeocin, an intercalating agent that induces the formation of DSBs, like the sog1 mutant, suggesting that the sensitivity of abo4-1 mutant to DBSs requires SOG1 activity.
The Checkpoint Activated by POL2A Deficiency Is ATM-independent
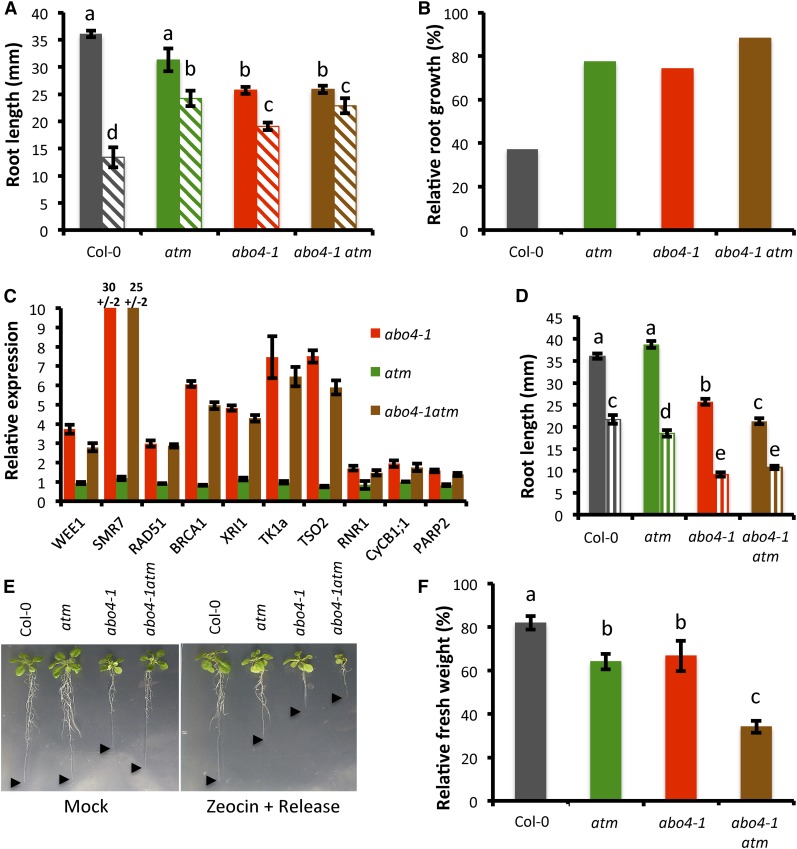
Response to DSBs is activated by the ATM kinase (Culligan et al., 2006). We therefore asked whether the abo4-1 mutation also activated the ATM pathway: abo4-1 atm double mutants could be recovered, indicating that the ATM pathway is dispensable for abo4-1 survival. To determine whether constitutive activation of DDR in abo4-1 involves ATM activity, we treated atm and abo4-1 atm seedlings with HU. Intriguingly, atm seedlings also displayed HU tolerance compared to wild-type plants (Fig. 4, A and B), suggesting that HU affects root growth at least partly via ATM activation, likely due to DSB formation after fork collapse. HU tolerance was the same in abo4-1 atm mutants as in abo4-1 single mutants (Fig. 4, A and B). Consistently, expression of DDR genes was induced to the same level in abo4-1 and abo4-1 atm mutants compared to wild-type plants, indicating that constitutive activation of the replicative stress response in abo4-1 does not require ATM (Fig. 4C).
Figure 4.
The checkpoint activated by the abo4-1 mutation is ATM-independent. A and B, HU sensitivity in wild type (Col-0), abo4-1, atm, and abo4-1 atm mutants. Plantlets were grown on half-strength MS for 4 d and transferred to control medium (full bars) or HU-supplemented medium (1 mM, dashed bars) for 9 d. A, Root length; values are average ± se obtained on at least 15 plantlets. B, Relative root growth; values are expressed as percentage of length on MS medium. C, qRT-PCR analysis of the expression of selected genes in abo4-1, atm, and abo4-1 atm mutants; values are average relative expression compared to the wild type ± sd. D, Zeocin sensitivity in wild type (Col-0), abo4-1, atm, and abo4-1 atm mutants. Plantlets were grown on half-strength MS for 4 d and transferred to control medium (full bars) or zeocin-supplemented medium (10 µM, dashed bars) for 9 d. D, Root length; values are average ± se obtained on at least 15 plantlets. E, Representative picture of plantlets grown on half-strength medium (mock), or grown on MS supplemented with zeocin (10 µM) for 6 d and allowed to recover for another 6 d. Arrowheads mark the position of the root tip. F, Relative fresh weight of plantlets after recovery. Values are average ± se from six replicates. In A, D, and F, different letters indicate statistically relevant differences (Student t test, P < 0.05).
We next asked if ATM and POL2A act in the same or in parallel pathways in the DSBs response. If POL2A was required for proper activation of DSB repair genes upstream of ATM, failure to activate these genes could account for the enhanced sensitivity of abo4-1 mutants to DSB inducing agents. To test this hypothesis, we monitored the transcriptional response to zeocin-induced DSBs in abo4-1 (Supplemental Fig. S7). As shown before, expression of all tested genes was increased in control conditions. Zeocin treatment induced further induction of target gene expression in all cases, except when the basal expression was already comparable to the expression observed in wild-type plants treated with zeocin. The sensitivity of abo4-1 mutants to DNA damaging agents can therefore not be attributed to a reduced expression level of DDR genes in response to DSB. These results suggest that POL2A and ATM function in parallel in DSB response. To corroborate this finding, we monitored the sensitivity of single and double mutants to zeocin. Prolonged exposure to zeocin had a similar effect on root growth in abo4-1 and atm abo4-1 (Fig. 4D). However, when the seedlings were allowed to recover for 6 d after a 6-d treatment with zeocin, abo4-1 atm seedlings displayed reduced growth compared to abo4-1 single mutant, and their fresh weight was dramatically decreased (Fig. 4, E and F), indicating that the atm and abo4-1 mutations have additive effects on tolerance to DSBs, leading to enhanced sensitivity in the double mutant compared to parental lines.
Contrasting Outcomes of Distinct POL2A Mutations on DDR
Mutations in different regions of the POL2 protein led to diverse defects in other eukaryotes (Henninger and Pursell, 2014; Rayner et al., 2016): mutations affecting the exonuclease activity mainly lead to a decrease in replication fidelity, whereas mutations affecting the processivity of the protein or its C terminus could be expected to impact S-phase progression or replicative stress response. We therefore tested whether other hypomorphic alleles of POL2A could lead to constitutive activation of DNA stress response and tolerance to HU. The abo4-2 mutant harbors a T-DNA insertion in the 12th intron [(Yin et al., 2009); Supplemental Fig. S1], but this mutation is not lethal. By contrast, pol2a null mutants arrest early during embryogenesis (Ronceret et al., 2005), suggesting that some POL2A protein or at least a portion of it accumulates in abo4-2. We therefore characterized POL2A expression in this mutant into more detail. We observed that the 5′ and 3′ portions of the cDNA situated on each side of the T-DNA insertion accumulate at almost wild-type levels (Supplemental Fig. S8A). In addition, RT-PCR analysis using primers located on each side of the insertion revealed the accumulation of low levels of wild-type mRNA and additional splicing variants corresponding to the elimination of exon 12 or exons 12 and 13 (amino acids 427 to 481 or 427 to 540, respectively); these splicing variants do not generate a frameshift, and thus could allow the production of a modified protein lacking conserved amino acids of the active site (Supplemental Fig. S8B). This mutant thus likely accumulates different isoforms of the POL2A protein and possibly its N- or C-terminal domain on its own. In addition, abo4-2 displays defects in cell-cycle regulation like the abo4-1 mutant (Supplemental Fig. S9, and Supplemental Table S1).
As observed in abo4-1, the abo4-2 mutant also displays tolerance to HU and hypersensitivity to zeocin (Supplemental Fig. S10, A and B) and shows constitutive expression of genes involved in DDR (Supplemental Fig. S10C). To determine whether the downstream signaling pathways activated in abo4-2 and abo4-1 were identical, we crossed abo4-2 with atr, atm, and sog1 mutants: only abo4-2 sog1 and abo4-2 atm double mutants were viable. As in abo4-1, transcriptional activation of DDR genes was largely SOG1-dependent but ATM-independent (Supplemental Fig. S10C), and activation of DDR genes in response to DSB was not impaired (Supplemental Fig. S7). However, although the sog1 mutation induced a further reduction of rosette growth in the abo4-1 background, the abo4-2 sog1 mutant displayed improved development compared to the abo4-2 single mutant (Supplemental Figs. S5 and S10A), indicating that the abo4-1 and abo4-2 mutations have different consequences on DDR activation. To gain further insight into these differences, we tested the sensitivity of abo4-2, abo4-2 sog1, and abo4-2 atm mutants to HU and zeocin. As for abo4-1, the HU-tolerant phenotype of abo4-2 was lost in the sog1 background. Unexpectedly, the atm mutation also reduced HU tolerance in abo4-2, providing evidence for the activation of different signaling pathways in the two mutants.
To complete our study, we also analyzed the esd7-1 mutant that harbors a point mutation leading to a substitution of Gly by Arg at position 992, situated at the extremity of the catalytic domain [Supplemental Fig. S1; (del Olmo et al., 2010)]. Like abo4-1 and abo4-2, this mutant was tolerant to HU (Supplemental Fig. S10A), but hypersensitive to a variety of genotoxic stresses (data not shown), further corroborating that incorporation of modified POL2A at the fork leads to replicative stress. Interestingly, the til1-4 mutation that affects the exonuclease domain of the POL2A protein [Supplemental Fig. S1; (Jenik et al., 2005)] did not confer tolerance to replicative stress (Supplemental Fi. S11A). On the contrary, this mutant was hypersensitive to HU even though the cell cycle length was reported to be longer during embryo development (Jenik et al., 2005). This difference in terms of response to HU treatment correlated with an overall lower expression level of DDR genes in til1-4 compared to the abo4-1 or esd7-1 mutants (Supplemental Fig. S11B). Finally, we tested the contribution of the POL2B gene to the activation of the DDR observed in pol2a hypomorphic mutants. As previously observed, esd7-1 pol2b double mutants were smaller than esd7-1 single mutants. However, they showed similar HU tolerance, indicating that POL2B function has no major contribution in the activation of the replicative stress response (Supplemental Fig. S11).
As stated above, DNA Pol ε plays a dual role at the replication fork because it performs both a catalytic and a scaffolding function, and defects described in hypomorphic POL2A mutants could either be direct consequences of the signaling role of DNA Pol ε or indirect effects of replicative stress. To discriminate between these two hypotheses, we generated POL2A RNA interference lines (POL2A-RNAi). If the replicative stress observed in POL2A hypomorphic mutants is a consequence of defects in fork stabilization and reflects the scaffolding role of POL2A, POL2A down-regulation would be expected to lead to activation of replicative stress and tolerance to HU like accumulation of a partially inactive form of the protein. By contrast, if POL2A itself is required for replicative stress signaling, POL2A knock-down would fail to activate the replicative stress response, and the plants should then become hypersensitive to HU like the atr and sog1 mutants. POL2A-RNAi plants display a range of developmental alterations such as reduced size, and partial sterility (Supplemental Fig. S12A). These features are shared with POL2A hypomorphic mutants. However, only POL2A-RNAi lines with a mild phenotype reached a similar size, as the wild type showed a sufficiently stable phenotype over generations to be used for further analysis (Supplemental Fig. S12B). These lines showed an approximately 2-fold reduction in POL2A mRNA accumulation (Supplemental Fig. S12C). Although POL2A-RNAi lines showed a slight increase in S-phase length, flow cytometry revealed no obvious accumulation of cells in S-phase (Supplemental Table S1 and Supplemental Fig. S9).
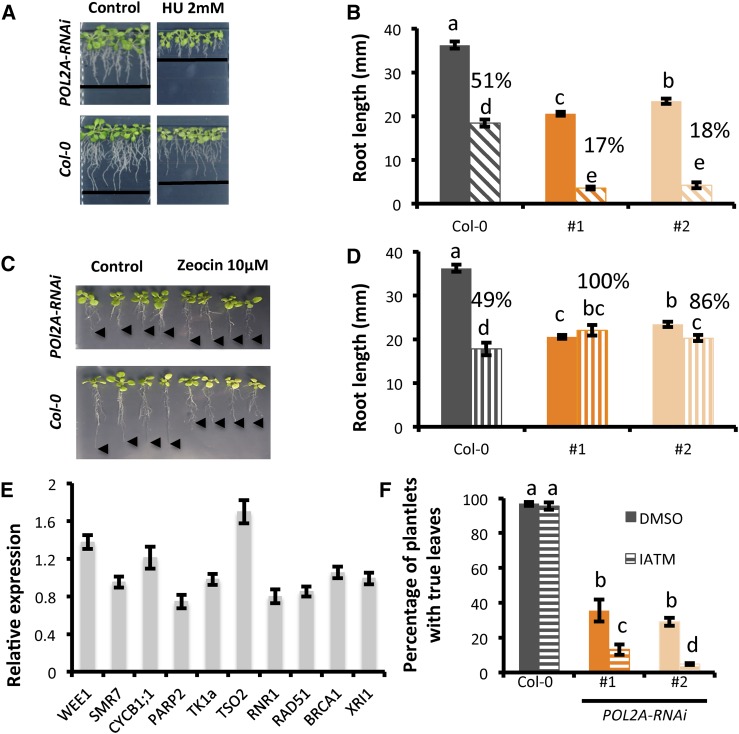
By contrast to abo4-1 and abo4-2, POL2-RNAi seedlings were hypersensitive to HU (Fig. 5, A and B), and they were not affected by zeocin exposure (Fig. 5, C and D). Consistently, POL2A-RNAi did not display constitutive up-regulation of DDR genes (Fig. 5E), suggesting that adequate levels of POL2A are essential for transcriptional activation triggered by replication stress checkpoint.
Figure 5.
Proper levels of POL2A is required for checkpoint activation in DDR. A and B, Wild-type (Col-0) and POL2A-RNAi seedlings were grown for 4 d on half-strength MS and transferred to HU-supplemented medium (1 mM) for 9 d. POL2A-RNAi lines were hypersensitive to this drug: lines indicate the extremity of roots (A). After 9 d, root length was measured on plants kept on control medium (full bars) or on HU-supplemented medium (dashed bars; B). Values above the bar indicate the relative root growth compared to the respective untreated control. C and D, Wild-type (Col-0) and POL2A-RNAi seedlings were grown for 4 d on half-strength MS and transferred to zeocin-supplemented medium (10 µM) for 9 d. POL2-RNAi lines were unaffected by this drug (C). Arrowheads mark the position of the root tip. After 9 d, root length was measured on plants kept on control medium (full bars) or on zeocin-supplemented medium (dashed bars; D). E, qRT-PCR quantification of selected genes in POL2A-RNAi seedlings. Values are average ± sd compared to the wild type. F, POL2A-RNAi plantlets are hypersensitive to IATM (Ku55933). Plants were germinated on MS medium containing DMSO or IATM (10 µM). After 10 d, the percentage of plants with true leaves was monitored. Germination and development are severely affected in POL2A-RNAi lines, and the proportion of plants with true leaves was therefore reduced compared to the wild type on control medium. However, this reduction was even more pronounced in the presence of IATM, whereas this compound had no effect on wild-type plants. In B, D, and E, values are average ± se of data obtained on at least 15 plantlets. Different letters indicate statistically relevant differences (Student t test, P < 0.05). All data are representative of at least two biological replicates.
Failure to activate suitable response upon replication stress can lead to fork collapse and thus generate DSBs that in consequence trigger ATM activation. To determine whether ATM activity is essential to POL2-RNAi plants survival, we tested the effect of a specific inhibitor of ATM activity [IATM, Ku55933; (Amiard et al., 2011)], and found that POL2A-RNAi lines are hypersensitive to this drug (Fig. 5F), supporting the notion that ATM pathway activation is required for plant survival when POL2A accumulation is decreased.
In summary, these results suggest that the presence of mutated POL2A in the replication fork affects cell cycle progression, leading to an increase in S-phase length due to checkpoint activation that confers tolerance to HU, whereas lowered concentration of POL2A prevents proper checkpoint activation in response to replicative stress.
Role of Pol ε Catalytic Subunit during Arabidopsis Reproductive Development
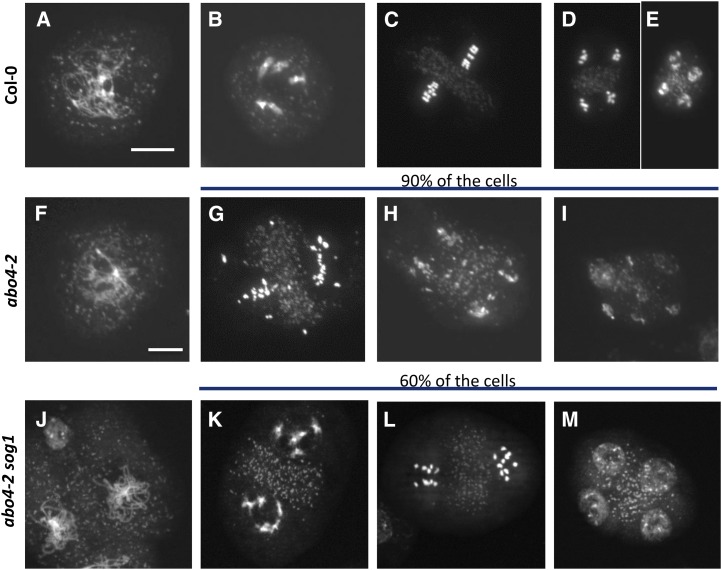
Recently, we have shown that increased accumulation of the Pol ε accessory subunit DPB2 led to the activation of a cell cycle checkpoint during premeiotic DNA replication and induced SOG1-dependent DNA fragmentation (Pedroza-García et al., 2016). Furthermore, Huang et al. (2015) reported that the til4-1 and abo4-2 alleles of POL2A display meiotic DNA fragmentation, although they attributed these defects to impaired DSB repair. We observed that the fertility of abo4-2 sog1 mutants was improved compared to abo4-2, suggesting that meiotic defects are at least partly due to SOG1 function. To clarify whether POL2A also participates in a premeiotic checkpoint, we analyzed meiosis progression in abo4-2 and in abo4-2 sog1 mutants. In our hands, DNA fragmentation was observed in 89% of meiocytes (n = 123) in the abo4-2 mutant (Fig. 6). This proportion was lowered to 40% in abo4-2 sog1 mutants (n = 93), confirming the hypothesis that in pol2a hypomorphic mutants, the SOG1-dependent checkpoint is activated in response to defects during premeiotic DNA replication.
Figure 6.
abo4-2 mutants show SOG1-dependent meiotic fragmentation. Meiosis progression in the wild type (A to E), abo4-2 mutant (F to I), and abo4-2 sog1 mutant. In the wild type, after early prophase (A), bivalents were formed (B), homologous chromosomes segregated during division I, and sister chromatids segregate during division II (C, metaphase; D, anaphase) to form tetrads (E). In the abo4-2 mutant, early prophase was normal (F), but bivalents were never observed. Instead, in 90% of the cells, extensive DNA fragmentation was observed both during the first (G) and the second division (H), leading to the formation of polyads (I). In abo4-2 sog1 mutants, 40% of the cells still showed DNA fragmentation (J), but 60% of meiocytes were wild-type-like (K, end of division I; L, anaphase of division II; M, tetrad). Bar = 10 µm for all panels.
Surprisingly, the fertility of abo4-1 atm and abo4-2 atm double mutants was modified compared to parental lines. Indeed, abo4-1 atm plants showed improved fertility compared to atm (Supplemental Fig. S13, A and B), suggesting that constitutive activation of the DDR in abo4-1 might partially rescue the meiotic defects of atm (García et al., 2003). We also observed that the fertility of abo4-2 atm mutants was improved compared to abo4-2: the number of seeds per silique was comparable to what was observed in atm (Supplemental Fig. S13, C and D). Previous studies have shown that atm mutants are partially deficient for repair of meiotic DSBs (García et al., 2003), as described for abo4-2. However, the rescue of abo4-2 sterility by the atm mutation suggests that the SOG1-dependent checkpoint triggered by premeiotic replication defects requires ATM activity.
Taken together, our results indicate that POL2A plays a role in replicative stress checkpoint activation both in somatic and in reproductive tissues, but that the signaling events differ between the different cell types.
DISCUSSION
Arabidopsis POL2A Participates in Cell Cycle Checkpoint Activation and Fork Stabilization
DNA Pol ε is required not only for DNA synthesis per se during DNA replication, but also for sensing of replication stress. This dual role is well established in yeast, and likely conserved in animals, but detailed investigation has been hampered by the lethality of mutants deficient for its catalytic subunit. In this work, we took advantage of hypomorphic alleles encompassing partially defective versions of the Pol ε catalytic subunit POL2A [abo4-1, abo4-2 (Yin et al., 2009), esd7-1 (del Olmo et al., 2010), and til1-4 (Jenik et al., 2005)] available in Arabidopsis to explore its contribution in checkpoint activation upon replicative stress. Although they are hypersensitive to DNA damaging agents [(Yin et al., 2009); and this study], abo4-1, abo4-2, and esd7-1 mutant alleles display specific tolerance to HU-induced replicative stress. This phenotype is likely due to basal activation of the replication stress checkpoint, as evidenced by the prolonged S-phase and constitutive expression of DDR genes observed in the mutants. Indeed, abo4-1 seedlings displayed up-regulation of genes encoding proteins required for replicative stress response such as the ssDNA sensors RAD 17 and RPA (Heitzeberg et al., 2004; Aklilu et al., 2014) or B-type CDKs and Cyclins that promote G2 arrest (Cools et al., 2011), and are specifically involved in DSB repair (Weimer et al., 2016).
One of the key elements during replication stress response is the activation of nucleotide biosynthesis by the RNR that in yeast has been shown to depend on Pol ε (Navas et al., 1995, 1996; Zhao and Rothstein, 2002). In addition to the de novo pathway involving the RNR, plants like all eukaryotes with the exception of yeast also rely on a salvage pathway comprising Thymidine Kinase 1 (Boldt and Zrenner, 2003), and these two pathways have redundant functions in DDR (Wang and Liu, 2006; Roa et al., 2009; Chen et al., 2010; Pedroza-García et al., 2015). Elements of both pathways are up-regulated in abo4 mutants, which may account for their tolerance to HU. Interestingly, mutation of the exonuclease domain of POL2A in the til1-4 mutant did not lead to improved HU tolerance, possibly because lower fidelity of Pol ε does not impact fork progression as severely as mutations affecting the polymerase active site. Consistently, the increase in cell cycle length observed in abo4-1 and abo4-2 was much more severe than what was described in ti1-4 (Jenik et al., 2005), and up-regulation of DDR genes was also less pronounced in til1-4. Together, these observations suggest that the presence of a mutated variant of Pol ε with reduced polymerase activity gums up DNA replication, leading to fork stalling and activation of the DDR (Fig. 7A).
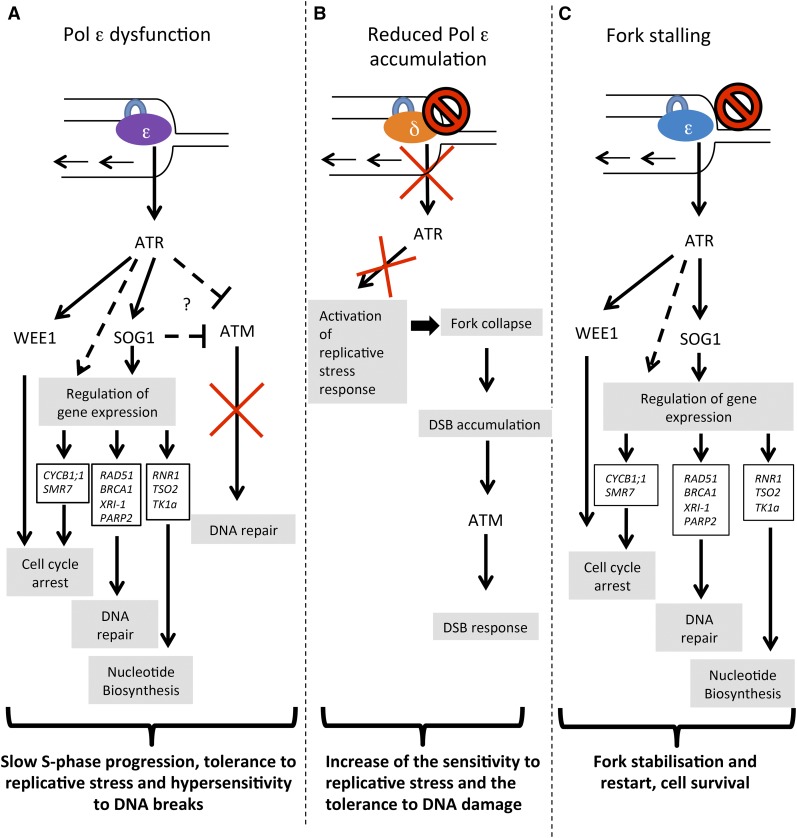
Figure 7.
Model for Pol ε function in plant DDR. A, In the wild type, Pol ε catalytic subunit POL2A is involved in replication stress sensing; this leads to ATR-dependent activation of the WEE1 and SOG1 pathways, allowing the expression of genes involved in cell cycle arrest, DNA repair, and nucleotide biosynthesis, ultimately leading to fork stabilization and completion of DNA replication and cell survival. B, In pol2A mutants with point mutations affecting POL2A activity, the abnormal Pol ε subunit likely gums up replication, leading to constitutive replication stress and activating ATR. The WEE1 branch of the downstream pathway is essential to plant survival, whereas the SOG1 branch of the pathway is dispensable, but confers tolerance to replicative stress. SOG1 activation may also negatively regulate ATM signaling leading to enhanced sensitivity to DNA damaging agents. C, When accumulation of POL2A is reduced, Pol δ likely replaces it and replicates both DNA strands. In the absence of Pol ε, replicative stress signaling is not properly activated, which may lead to fork collapse and DNA lesions that can in turn activate ATM signaling and promote tolerance to DSB-inducing agents. In all panels, dashed arrows indicate putative pathways that remain to be molecularly identified.
To further explore the role of Pol ε in replicative stress response, we generated knock-down lines expressing lower levels of POL2A mRNA. These lines were viable, and the less severe ones reached a similar size to wild-type plants, either because Pol δ can synthetize both DNA strands when Pol ε accumulation is reduced as has been hypothesized in other organisms (Pursell and Kunkel, 2008; Johnson et al., 2015), or because residual expression of Pol ε is still sufficient to allow proper S-phase progression. By contrast with pol2a hypomorphic mutants, POL2A knock-down lines did not display constitutive activation of the replicative stress checkpoint but were hypersensitive to HU, further supporting the direct involvement of plant Pol ε in replicative stress sensing (Fig. 7B). In line with this conclusion, it is worth noting that up-regulation of DDR genes was less pronounced in abo4-2 than in abo4-1, possibly because accumulation of POL2A is reduced in abo4-2 due to the T-DNA insertion. Nevertheless, the hypersensitivity of POL2A-RNAi lines to HU might also be a consequence of defects in fork stabilization due to decreased POL2A accumulation. Indeed, Pol ε plays an essential scaffolding role to stabilize stalled forks (Lou et al., 2008; Pursell and Kunkel, 2008; Henninger and Pursell, 2014), independent of checkpoint activation (Branzei and Foiani, 2009). Alternative mechanisms have been described to stabilize the replisome when Pol ε is limiting (Mejia-Ramirez et al., 2015), but whether they are conserved in plants and could be operating in POL2A-RNAi lines remains to be established.
Signaling Downstream of POL2A
Replicative stress sensing depends on the ATR kinase that activates WEE1 and SOG1 via two independent pathways (Hu et al., 2015), but does not involve ATM (Culligan et al., 2006). Consistently, the viability of abo4 mutants does not require ATM, but depends on components of the replication stress checkpoint: ATR and WEE1. In Schizosaccharomyces pombe, deletion of the POL2 catalytic domain led to tolerance to HU and hypersensitivity to MMS, and survival of this mutant was strictly dependent on Rad3 (ATR) and Polδ (Feng and D’Urso, 2001), because ATR is required to stabilize the association of Pol ε with stalled forks (Pursell and Kunkel, 2008). The lethality of abo4 atr double mutants thus indicates that the mechanisms described in yeast are conserved in multicellular organisms. Interestingly, abo4 sog1 mutants are viable, suggesting that Pol ε deficiency can activate both branches of the replicative stress response as described in Pedroza-García et al. (2016), and demonstrates that only the ATR-WEE1 branch of the pathway is required for embryo development in the presence of replicative stress.
Unexpectedly, despite the constitutive activation of DDR genes, pol2A hypomorphic mutants were hypersensitive to DNA damaging agents. Induction of DDR genes upon zeocin exposure was not impaired in these mutants, suggesting that their hypersensitivity to DNA damage either reflects the direct involvement of Pol ε in DNA repair (Pursell and Kunkel, 2008), or defects in the DDR that were not detected in this study. Indeed, the sensitivity of abo4 mutants to DSBs was SOG1-dependent, indicating that POL2A deficiency simultaneously triggers the expression of DDR genes and hampers proper activation of the ATM-dependent pathway (Fig. 7A). In addition, POL2A-RNAi lines are tolerant to zeocin. This observation suggests that POL2A is not required for DSB repair, and that bypass mechanisms possibly involving translesion synthesis polymerases compensate Pol ε down-regulation. The activation of translesion synthesis polymerases in response to DNA damage has been shown to require ATM (Curtis and Hays, 2011), consistent with the hypersensitivity of POL2A-RNAi lines to the ATM inhibitor. Interestingly, sog1 mutants are hypersensitive to HU and tolerant to zeocin, and the sog1 mutation was epistatic on the abo4 mutations for these responses. All together, these results indicate (1) that abo4 functions upstream of SOG1 to confer tolerance to HU and (2) that the sensitivity of abo4 mutants to zeocin requires SOG1 activity, suggesting that activation of this DDR pathway interferes with proper DSB response even though it leads to the activation of DNA repair genes. In yeast, the Mec1 (ATR) pathway has been shown to attenuate Tel1 (ATM) signaling (Clerici et al., 2014): constitutive activation of SOG1 via ATR may thus prevent proper response to DSBs in Arabidopsis and account for the sensitivity of POL2A hypomorphic mutants to DNA damaging agents (Fig. 7A).
The antagonism between the ATR and ATM pathways may explain the puzzling observation that the sog1 mutation had opposite effects on the growth of the two abo4 alleles studied here. Indeed, one can postulate that the abo4-1 mutation triggers mainly the ATR branch of the DDR, whereas the abo4-2 mutation also activates the ATM branch of the pathways, possibly because this particular mutated allele affects not only POL2A activity but also its accumulation, leading to endogenous DNA damage as a consequence of fork collapse. In this model, loss of SOG1 in the abo4-1 background would enhance the growth defects, because SOG1 is required for plant survival upon replicative stress (Hu et al., 2015). By contrast, in abo4-2 mutants, loss of SOG1 may allow more effective activation of the ATM pathway and thus partly rescue growth defects. Consistently, HU resistance was lost in the abo4-2 atm mutant but not in abo4-1 atm, suggesting that the abo4-2 mutant may be more prone to fork collapse.
Pol ε and Meiosis
Recently, we reported that overexpression of the Pol ε accessory subunit DPB2 results in SPO11-independent DNA fragmentation during meiosis, and showed that this process required SOG1 activity, suggesting that the observed fragmentation is the consequence of an active process triggered by defects occurring during premeiotic replication (Pedroza-García et al., 2016). By contrast, Huang et al. (2015) recently described SPO11-dependent DNA fragmentation in pol2a mutants, and proposed that they were due to defects in DNA repair (). However, abo4-1 mutants that are also hypersensitive to genotoxic stress do not display meiotic defects, suggesting that impairment of the POL2 activity per se does not trigger DNA fragmentation. Furthermore, we showed that SOG1-deficiency partially rescued the meiotic defects of abo4-2 mutants [pol2a-1 in Huang et al. (2015)]. Together, our results provide evidence for the involvement of POL2A in the premeiotic checkpoint previously described, although residual DNA fragmentation observed in abo4-2 sog1 mutant likely results from defects in the repair of programmed DSBs. Furthermore, the finding that the atm mutation partially rescues the fertility of abo4-2 mutants suggests that the meiotic checkpoint activated by Pol ε deficiency involves ATM signaling. One possible model would thus be that inactivation of Pol ε triggers fork collapse, thereby generating DSBs and ATM-dependent SOG1 activation. Consistently, the SOG1-dependent fragmentation phenotype was observed in abo4-2 mutants that accumulate reduced levels of full-length POL2A but not in abo4-1 mutants, where production of the full-length protein is unchanged.
Intriguingly, the abo4-1 mutation also improved the fertility of the atm mutant. Both in budding and in fission yeast, replicative stress induced by stalled forks inhibits the formation of DSBs (Subramanian and Hochwagen, 2014); it is therefore possible that the constitutive activation of replicative stress in the abo4-1 mutant leads to the formation of fewer DSBs, thereby alleviating the subsequent repair defects caused by the atm mutations. Further investigation of meiosis progression in double mutants should help clarify this point. Together, our results further demonstrate that premeiotic DNA replication is a critical step for gamete formation, and that defects occurring during this phase activate a cell death program that requires the SOG1 transcription factor.
Concluding Remarks
Overall, this work has shed light on the diverse roles of plant POL2 in DDR activation, as summarized on Figure 7. We show that the role of Pol ε in S-phase checkpoint activation is a universal mechanism operating similarly in a multicellular organism as in yeast. Mutations affecting Pol ε can lead to a rare autosomal recessive disease (Pachlopnik Schmid et al., 2012), and have been associated with various types of cancer (Rayner et al., 2016). Intriguingly, the consequences of Pol ε deficiency are much less dramatic in plants, possibly because of the plasticity of their development that allows replacement of damaged cells by neighboring ones in meristems (Heyman et al., 2014). Because many mutants deficient for DDR are viable in plants but lethal in animal systems, future studies in Arabidopsis could reveal mechanisms that have not been elucidated in other multicellular eukaryotes and could be translated into Mammalian cells to further elucidate the association of Pol ε deficiency with tumorigenesis.
MATERIALS AND METHODS
Cloning Procedures
Transgenic POL2ARNAi lines were generated after Agrobacterium-mediated transformation of Col plants with the plasmid CATMA1a07250, which harbors a fragment of 155 bp corresponding to the nucleotides 3368 to 3522 of the coding sequence of ESD7/POL2A cDNA, cloned in sense and anti-sense orientation in the pAgrikola vector (Hilson et al., 2004), a Gateway destination vector based closely on the Hellsgate 12 vector. Several transgenic independent plants were selected in medium containing phosphinothricin at 10 mg/mL and later were established as homozygous lines. All of them displayed lower levels of expression of POL2A mRNA in comparison to nontransformed control plants, and no modification of POL2B expression.
Plant Material and Growth Conditions
Seeds were surface-sterilized by treatment with bayrochlore for 20 min, then washed and imbibed in sterile-water for 2 to 4 d at 4°C to obtain homogeneous germination. Seeds were sown on commercially available 0.5× Murashige and Skoog (MS) medium (Basalt Salt Mixture M0221; Duchefa) with the appropriate antibiotic if needed and solidified with 0.8% agar (Phyto-Agar HP696; Kalys), and grown in a long d (16 h light, 8 h night, 21°C) growth chamber. After 2 weeks, the plants were transferred to soil in a glasshouse under short-d conditions (8 h light 20°C, 16 h night at 18°C) for 2 weeks before being transferred to long-d conditions.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from seedlings with the RNeasy MiniPrep kit (Qiagen, according to the manufacturer's instructions. First strand cDNA was synthesized from 2 µg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. 1/25th of the synthesized cDNA was mixed with 100 nM of each primer and LightCycler 480 Sybr Green I master mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals acquired with a LightCycler 480 detection system (Roche Applied Science). The specificity of amplification products was determined by melting curves. PP2AA3 was used as internal control for signals normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates relative expression level of the selected genes with algorithms based on ΔΔCt method. Data were from triplicates and are representative of at least two biological replicates. The sequence of primers used in this study is provided in Supplemental Table S4.
Transcriptome Studies
Three independent biological replicates were produced. For each biological repetition and each point, RNA samples were obtained by pooling RNAs from more than 200 plants. Whole plantlets were collected on plants at 1.04 developmental growth stages (Boyes et al., 2001), cultivated in vitro under long-d conditions. Total RNA was extracted as described above. RNA-seq experiment was carried out at the POPS Transcriptomic Platform, Institute of Plant Sciences - Paris-Saclay in Orsay, France. PolyA RNA was purified using the Dynabeads mRNA direct micro kit (Ambion). The sequencing libraries were constructed with the Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) and the sequencing spheres were prepared with the Ion PI Template OT2 200 Kit v3 (Life Technologies) before sequencing on an Ion Proton using the Ion PI Sequencing 200 Kit v3 (Thermo Fisher Scientific) and Ion PI v2 chips (Life Technologies) with 520 run flows.
RNA-seq Bioinformatic Treatment and Analysis
To allow comparisons, each RNA-Seq sample followed the same pipeline from trimming to count of transcript abundance as follows. Read preprocessing criteria included trimming library adapters and performing quality control checks using the Torrent suite (Version 4.2.1; ThermoFisher Scientific) with default settings. The reads corresponding to rRNAs were identified by mapping rRNAs on Arabidopsis (Arabidopsis thaliana) using Bowtie, Version 2 [http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; with –local option; Langmead and Salzberg (2012)]and removed. The same software was used to align the remaining reads against the Arabidopsis transcriptome [33,602 mRNA from TAIR 10; Lamesch et al. (2012)] without ambiguous hits (multihits are removed). According to these rules, approximately 75% of the initial reads aligned to transcripts for each sample. Genes that do not have at least 1 read after a counts-per-million normalization in at least three samples among the six were discarded. The differential analysis has been performed by using a likelihood ratio test in a negative binomial generalized linear model where the dispersion is estimated by the method proposed in the software edgeR (Bioconductor) and where a biological replicate effect was taken into account. A gene was declared differentially expressed if its raw P value adjusted by the Benjamini-Hochberg procedure to control the FDR is ≤0.01 and its absolute fold change is ≥1.5. Analyses were performed with the software R (Version 3.1.0) and the edgeR package (version 3.6.8; Bioconductor).
Light and Fluorescence Microscopy
Fresh siliques were opened under a stereo-microscope (SVII; Carl Zeiss) and images were captured with a color CCD camera (Power HAD; Sony).
For meiotic analyses, flower buds were fixed in ethanol: Acetic Acid (3:1). 4∼,6-Diamidino-2-phenylindole staining of meiotic chromosomes was performed according to the method of Ross et al. (1996). Slides were observed on an epi-fluorescence videomicroscope (SVII; Darl Zeiss), and images were captured with a color CCD camera (Power HAD; Sony).
Observations were done with a wide field fluorescence microscope (AxioImager Z.2; Carl Zeiss) fitted with a metal halide lamp and the appropriate shifted free filter sets for imaging DAPI dye (Cat. no. 49; Carl Zeiss). Images were acquired with a cooled CCD camera (AxioCam 506 monochrome; Carl Zeiss) operated using Zen Blue software (Carl Zeiss).
Cell cycle length analysis was performed as described in Pedroza-García et al. (2016).
Flow Cytometry
For flow cytometric nuclei analysis, tissues were chopped with a razor blade in 1 mL of Gif nuclei-isolation buffer [45 mm MgCl2, 30 mm sodium citrate, 60 mm MOPS, 1% (w/v) polyvinylpyrrolidone 10,000, pH 7.2] containing 0.1% (w/v) Triton X-100, supplemented with 5 mm sodium metabisulphite and RNAse (5 U/mL). Propidium iodide was added to the filtered supernatants to a final concentration of 50 µg/mL. Endoreduplication levels of 5000 to 10,000 stained nuclei were determined using a Cyflow SL3 flow cytometer (Partec-Sysmex) with a 532-nm solid-state laser (30 mW) excitation and an emission collected after a 590-nm long-pass filter. For cell cycle analysis, we used the algorithm available in the FloMax software (flomax.software.informer.com).
Statistical Analysis
All statistical analysis was performed using the R software (https://www.r-project.org/).
Data Deposition
RNAseq data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE71002) and at CATdb (http://urgv.evry.inra.fr/CATdb/; Project: NGS2014_10_Epsilon) according to the “Minimum Information About a Microarray Experiment” standards.
Accession Numbers
Accession numbers of genes mentioned in this work are as follows: POL2A (AT1G08260), POL2B (AT2G27120), RNR1 (AT2G21790), TSO2, (AT3G27060), SMR7 (AT3G27630), PARP2 (AT2G31320), TK1a (AT3G07800), CYCB1-1 (AT4G37490), XRI-1 (AT5G48720), WEE1 (AT1G02970), BRCA1 (AT4G21070), RNR2A (AT3G23580), UBC21 (AT5G25760), RAD51 (AT5G20850), ATM (AT3G48190), ATR (AT5G40820), SOG1 (AT1G25580), PP2AA3 (AT1G13320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Positions of the mutations in POL2A used in this study.
Supplemental Figure S2. Cell cycle regulation is altered in abo4-1 mutants.
Supplemental Figure S3. The abo4-1 mutant is hypersensitive to genotoxic stress.
Supplemental Figure S4. Gene-ontology analysis of significantly induced genes in abo4-1 seedlings.
Supplemental Figure S5. The sog1 mutation significantly reduces vegetative growth of the abo4-1 mutant, but partially rescues the abo4-2 mutant.
Supplemental Figure S6. The SOG1 transcription factor is partly responsible for the tolerance of abo4-1 mutants to HU and their sensitivity to zeocin.
Supplemental Figure S7. Transcriptional response to zeocin in abo4 mutants.
Supplemental Figure S8. The T-DNA insertion in the abo4-2 mutant leads to production of different variants of the POL2A mRNA.
Supplemental Figure S9. Partial inactivation and down-regulation of POL2A have contrasting effects on cell cycle regulation.
Supplemental Figure S10. The abo4-2 mutation confers HU tolerance that is partly dependent on SOG1.
Supplemental Figure S11. Different mutations in POL2A confer tolerance or hypersensitivity to replicative stress.
Supplemental Figure S12. Down-regulation of POL2A affects plant growth and fertility.
Supplemental Figure S13. Genetic interactions between Pol e and DDR genes during reproductive development.
Supplemental Table S1. Cell cycle is drastically modified in abo4 mutants, but only mildly in POL2A-RNAi lines.
Supplemental Table S2. List of significantly misregulated genes in the abo4-1 mutant.
Supplemental Table S3. List of genes involved in cell cycle, DNA repair, mitosis, or meiosis that are up-regulated in abo4-1.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Lieven De Veylder (VIB, Gent) and Patricia Kannouche (IGR, Villejuif), for helpful discussions about this work. We thank J. Drouin-Wahbi (IPS2) for assistance with plant growth and material preparation. This work has benefited from the core facilities of Imagerie-Gif (http://www.i2bc.paris-saclay.fr), a member of IBiSA (http://www.ibisa.net).
References
- Aklilu BB, Soderquist RS, Culligan KM (2014) Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication. Nucleic Acids Res 42: 3104–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Depeiges A, Allain E, White CI, Gallego ME (2011) Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell 23: 4254–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt R, Zrenner R (2003) Purine and pyrimidine biosynthesis in higher plants. Physiol Plant 117: 297–304 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2009) The checkpoint response to replication stress. DNA Repair (Amst) 8: 1038–1046 [DOI] [PubMed] [Google Scholar]
- Chen Y-L, Eriksson S, Chang Z-F (2010) Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J Biol Chem 285: 27327–27335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Trovesi C, Galbiati A, Lucchini G, Longhese MP (2014) Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J 33: 198–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, van den Daele H, van Isterdael G, Schnittger A, De Veylder L (2011) The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Curtis MJ, Hays JB (2011) Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair (Amst) 10: 1272–1281 [DOI] [PubMed] [Google Scholar]
- del Olmo I, López-González L, Martín-Trillo MM, Martínez-Zapater JM, Piñeiro M, Jarillo JA (2010) EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J 61: 623–636 [DOI] [PubMed] [Google Scholar]
- Dubarry M, Lawless C, Banks AP, Cockell S, Lydall D (2015) Genetic networks required to coordinate chromosome replication by DNA polymerases α, δ, and ε in Saccharomyces cerevisiae. G3 (Bethesda) 5: 2187–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, D’Urso G (2001) Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol 21: 4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- García V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez LJ, De Piccoli G, Marchesi V, Jones RC, Edmondson RD, Labib K (2015) A conserved Polϵ binding module in Ctf18-RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res 43: 8830–8838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-F, Huang H-W, Li L, Cai T, Chen S, He X-J (2015) The cytosolic iron-sulfur cluster assembly protein MMS19 regulates transcriptional gene silencing, DNA repair, and flowering time in Arabidopsis. PLoS One 10: e0129137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa T, Kanke M, Takahashi TS, Nakagawa T, Masukata H (2012) DNA polymerization-independent functions of DNA polymerase epsilon in assembly and progression of the replisome in fission yeast. Mol Biol Cell 23: 3240–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzeberg F, Chen I-P, Hartung F, Orel N, Angelis KJ, Puchta H (2004) The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J 38: 954–968 [DOI] [PubMed] [Google Scholar]
- Henninger EE, Pursell ZF (2014) DNA polymerase epsilon and its roles in genome stability. IUBMB Life 66: 339–351 [DOI] [PubMed] [Google Scholar]
- Heyman J, Kumpf RP, De Veylder L (2014) A quiescent path to plant longevity. Trends Cell Biol 24: 443–448 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, Chardakov V, Cognet-Holliger C, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Cools T, Kalhorzadeh P, Heyman J, De Veylder L (2015) Deficiency of the Arabidopsis helicase RTEL1 triggers a SOG1-dependent replication checkpoint in response to DNA cross-links. Plant Cell 27: 149–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Cheng Z, Wang C, Hong Y, Su H, Wang J, Copenhaver GP, Ma H, Wang Y (2015) Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc Natl Acad Sci USA 112: 12534–12539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Jurkuta REJ, Barton MK (2005) Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17: 3362–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Klassen R, Prakash L, Prakash S (2015) A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell 59: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossen R, Bermejo R (2013) The DNA damage checkpoint response to replication stress: a game of forks. Front Genet 4: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C (1999) DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 3: 679–685 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, O’Donnell M (2013) New insights into replisome fluidity during chromosome replication. Trends Biochem Sci 38: 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, García-Hernandez M, Karthikeyan AS, Lee CH, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gold DA, Shevchenko A, Shevchenko A, Dunphy WG (2005) Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol Biol Cell 16: 5269–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL (2008) Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Ramirez E, Limbo O, Langerak P, Russell P (2015) Critical function of γH2A in S-Phase. PLoS Genet 11: e1005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas TA, Sanchez Y, Elledge SJ (1996) RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev 10: 2632–2643 [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39 [DOI] [PubMed] [Google Scholar]
- Pachlopnik Schmid J, Lemoine R, Nehme N, Cormier-Daire V, Revy P, Debeurme F, Debré M, Nitschke P, Bole-Feysot C, Legeai-Mallet L, Lim A, de Villartay JP, et al. (2012) Polymerase ε1 mutation in a human syndrome with facial dysmorphism, immunodeficiency, livedo, and short stature (“FILS syndrome”). J Exp Med 209: 2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza-García JA, Domenichini S, Mazubert C, Bourge M, White C, Hudik E, Bounon R, Tariq Z, Delannoy E, del Olmo I, Piñeiro M, Jarillo JA, et al. (2016) Role of the polymerase epsilon sub-unit DPB2 in DNA replication, cell cycle regulation and DNA damage response in Arabidopsis. Nucleic Acids Res 44: 7251–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza-García JA, Nájera-Martínez M, de la Paz Sanchez M, Plasencia J (2015) Arabidopsis thaliana thymidine kinase 1a is ubiquitously expressed during development and contributes to confer tolerance to genotoxic stress. Plant Mol Biol 87: 303–315 [DOI] [PubMed] [Google Scholar]
- Post SM, Tomkinson AE, Lee EY-HP (2003) The human checkpoint Rad protein Rad17 is chromatin-associated throughout the cell cycle, localizes to DNA replication sites, and interacts with DNA polymerase ε. Nucleic Acids Res 31: 5568–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M (2011) Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase epsilon. PLoS Genet 7: e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA (2007) Yeast DNA polymerase ε participates in leading-strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Kunkel TA (2008) DNA polymerase epsilon: a polymerase of unusual size (and complexity). Prog Nucleic Acid Res Mol Biol 82: 101–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN (2016) A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer 16: 71–81 [DOI] [PubMed] [Google Scholar]
- Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME (2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A, Guilleminot J, Lincker F, Gadea-Vacas J, Delorme V, Bechtold N, Pelletier G, Delseny M, Chabouté ME, Devic M (2005) Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J 44: 223–236 [DOI] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Segurado M, Tercero JA (2009) The S-phase checkpoint: targeting the replication fork. Biol Cell 101: 617–627 [DOI] [PubMed] [Google Scholar]
- Sengupta S, van Deursen F, de Piccoli G, Labib K (2013) Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol 23: 543–552 [DOI] [PubMed] [Google Scholar]
- Singh A, Xu Y-J (2016) The cell killing mechanisms of hydroxyurea. Genes (Basel) 7: E99 10.3390/genes7110099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, Hochwagen A (2014) The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6: a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyari O, Kawai M, Ida H, Yoshida H, Sakaguchi K, Yamaguchi M (2012) Differential requirement for the N-terminal catalytic domain of the DNA polymerase epsilon p255 subunit in the mitotic cell cycle and the endocycle. Gene 495: 104–114 [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV (2009) Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol ε and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z (2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, van Damme D, Sugimoto K, Koncz C, et al. (2016) The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Zhang X, Liu J, Wang Y, He J, Yang T, Hong X, Yang Q, Gong Z (2009) Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Sakaguchi K, Kimura S (2013) DNA damage response in plants: conserved and variable response compared to animals. Biology (Basel) 2: 1338–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R (2002) The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA 99: 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.