The polyunsaturated fatty acid content of Arabidopsis seeds is determined by both cis-acting variants in FATTY ACID DESATURASE2 and an independent temperature-responsiveness locus on chromosome 2.
Abstract
Plants modify the polyunsaturated fatty acid content of their membrane and storage lipids in order to adapt to changes in temperature. In developing seeds, this response is largely controlled by the activities of the microsomal ω-6 and ω-3 fatty acid desaturases, FAD2 and FAD3. Although temperature regulation of desaturation has been studied at the molecular and biochemical levels, the genetic control of this trait is poorly understood. Here, we have characterized the response of Arabidopsis (Arabidopsis thaliana) seed lipids to variation in ambient temperature and found that heat inhibits both ω-6 and ω-3 desaturation in phosphatidylcholine, leading to a proportional change in triacylglycerol composition. Analysis of the 19 parental accessions of the multiparent advanced generation intercross (MAGIC) population showed that significant natural variation exists in the temperature responsiveness of ω-6 desaturation. A combination of quantitative trait locus (QTL) analysis and genome-wide association studies (GWAS) using the MAGIC population suggests that ω-6 desaturation is largely controlled by cis-acting sequence variants in the FAD2 5′ untranslated region intron that determine the expression level of the gene. However, the temperature responsiveness of ω-6 desaturation is controlled by a separate QTL on chromosome 2. The identity of this locus is unknown, but genome-wide association studies identified potentially causal sequence variants within ∼40 genes in an ∼450-kb region of the QTL.
Temperature is one of the major environmental factors that limit plant distribution. Plants have evolved a range of short-term adaptive responses to cope with temperature variation, including changing their cellular lipid composition (Penfield, 2008). There is typically an inverse relationship between temperature and the polyunsaturated fatty acid (PUFA) content of plant membrane lipids, with lower temperatures leading to an increase in the ratio of PUFAs verses saturated and monounsaturated fatty acids (Willemot et al., 1977; Pearcy, 1978; Slack and Roughan, 1978). The phase transition temperature of lipids is strongly influenced by their degree of acyl group unsaturation, so this response helps to maintain physiologically suitable levels of average lipid order, or fluidity, in membranes (Guschina and Harwood, 2006). A similar response of PUFA content to temperature also was found in the storage lipids of many oilseed species, including major crops such as soybean (Glycine max), oilseed rape (Brassica napus), and sunflower (Helianthus annuus; Canvin, 1965; Harris et al., 1978; Carver et al., 1986). Breeders and seed oil processors consider this effect to be undesirable, since it leads to variation in product quality (Rolletschek et al., 2007; Singer et al., 2016). It may be considered an inevitable consequence of the change in membrane lipid composition, because phosphatidylcholine (PC) is both a substrate for PUFA production (Slack et al., 1978) and a major precursor for triacylglycerol (TAG) biosynthesis in seeds (Bates et al., 2009; Bates and Browse, 2011). However, the temperature-dependent plasticity of storage lipid composition also might have adaptive significance (Sanyal and Linder, 2013). Biogeographic studies suggest that the degree of fatty acid unsaturation in oilseeds has played a role in adaptation to temperature on a microevolutionary and a macroevolutionary scale (Linder, 2000; Sanyal and Linder, 2013).
The enzymes that are primarily responsible for producing PUFAs in seed oils are the microsomal ω-6 and ω-3 fatty acid desaturases FAD2 and FAD3, which act in sequence to convert oleic acid (18:1) to linoleic acid (18:2) and then to α-linolenic acid (18:3), while the fatty acid is esterified to the sn-2 position of PC (Miquel and Browse, 1992; Browse et al., 1993; Lou et al., 2014). Temperature can influence the degree of microsomal fatty acid desaturation indirectly through its effects on substrate availability (Harris and James, 1969; Browse and Slack, 1983; Rolletschek et al., 2007; Li et al., 2015). However, there also is evidence that FAD2 and FAD3 are regulated directly by temperature (Tang et al., 2005; O’Quin et al., 2010). Whether this regulation occurs at the level of gene expression appears to vary depending upon the species, tissue, and gene in question. Several studies have reported a significant up-regulation of FAD2 or FAD3 isogenes at lower temperature (Martz et al., 2006; Kargiotidou et al., 2008; Román et al., 2012; Zhu et al., 2012), while others have reported either down-regulation or no appreciable change (Okuley et al., 1994; Heppard et al., 1996; Horiguchi et al., 2000; Li et al., 2015). In the case of FAD3, evidence also has been provided for translational regulation by temperature (Horiguchi et al., 2000), mediated by the 5′ untranslated region (UTR) of the transcript (Wang and Xu, 2010).
Temperature also is known to affect ω-6 and ω-3 fatty acid desaturation by influencing the rate of FAD2 and FAD3 protein turnover (Covello and Reed, 1996; Dyer et al., 2001; Martínez-Rivas et al., 2003; Tang et al., 2005; O’Quin et al., 2010). In particular, heterologous expression studies in yeast (Saccharomyces cerevisiae) have demonstrated that FAD2 and FAD3 proteins are less stable and, therefore, less abundant at elevated temperatures (Dyer et al., 2001; Sánchez-García et al., 2004; Tang et al., 2005; O’Quin et al., 2010; Khuu et al., 2011). Furthermore, studies have shown that the effect of temperature on FAD2 and FAD3 protein abundance is determined by a combination of cis-acting degradation signals and the ubiquitin-proteasome pathway (Tang et al., 2005; O’Quin et al., 2010). However, it remains to be determined precisely how temperature-dependent regulation of FAD2 and FAD3 turnover is implemented in planta and how the temperature signal is transduced (O’Quin et al., 2010).
A recent study by Sanyal and Linder (2013) has suggested that the fatty acid composition of Arabidopsis (Arabidopsis thaliana) seeds also is affected by temperature and that accessions differ in their response. Therefore, Arabidopsis seeds may provide a convenient experimental system in which to investigate the temperature regulation of microsomal fatty acid desaturation. Many studies have exploited genetic approaches to investigate the function of fatty acid desaturation in plant temperature adaptation (Murata et al., 1992; Miquel et al., 1993; Murakami et al., 2000). However, no studies appear to have used genetic variation in plants to study the underlying regulatory mechanism(s). The aims of this study, therefore, were as follows: (1) to characterize the response of Arabidopsis seed lipid content and composition to temperature across the ambient range (∼10°C–30°C) and (2) to exploit natural phenotypic variation identified within the multiparent advanced generation intercross (MAGIC) recombinant inbred population (Kover et al., 2009; Gan et al., 2011) to map quantitative trait loci (QTLs) for both microsomal fatty acid desaturation and its plasticity in response to temperature.
RESULTS
Temperature Influences the Oil Content of Arabidopsis Seeds
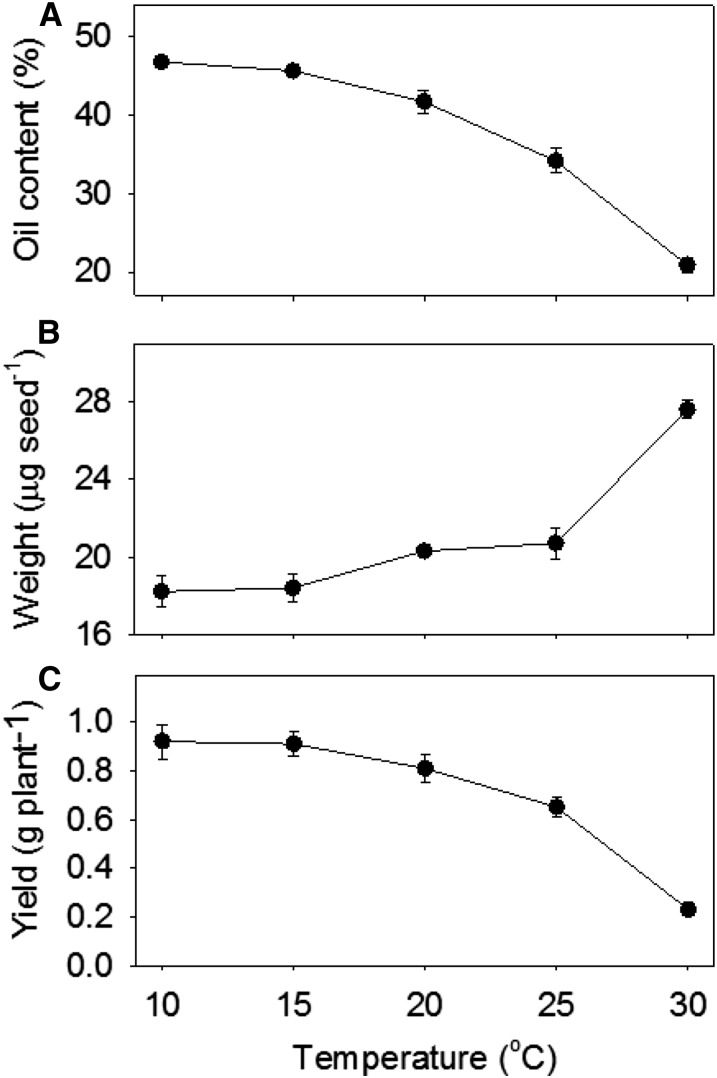
To characterize the effect of maturation temperature on Arabidopsis seed lipids, plants of ecotype Columbia-0 (Col-0) were grown under standard conditions until the onset of flowering and then transferred to a series of cabinets set to constant temperatures of 10°C, 15°C, 20°C, 25°C, and 30°C. At maturity, the seeds were harvested and analyzed. Total oil content, expressed as a percentage of seed weight, was greatest at 10°C and showed a progressive decline as the temperature increased to 30°C (Fig. 1A). A similar relationship between temperature and percentage of oil content has been reported previously in oilseed rape (Canvin, 1965; Zhu et al., 2012). Average seed weight changed relatively little between 10°C and 25°C but increased sharply at 30°C (Fig. 1B; Supplemental Fig. S1), and this coincided with a decrease in seed yield per plant (Fig. 1C). High temperature causes infertility in Arabidopsis (Bac-Molenaar et al., 2015), and we found that 30°C is approaching the higher temperature threshold for Col-0. This may explain the reduction in seed set, which also is known to lead to an increase in seed size (Hughes et al., 2008).
Figure 1.
Effect of temperature during the seed maturation period on the oil yield of Arabidopsis Col-0. A, Seed oil content. B, Seed weight. C, Seed yield. Values are means ± se of measurements on seeds from five separate plants.
Temperature Influences the Oil Composition of Arabidopsis Seeds
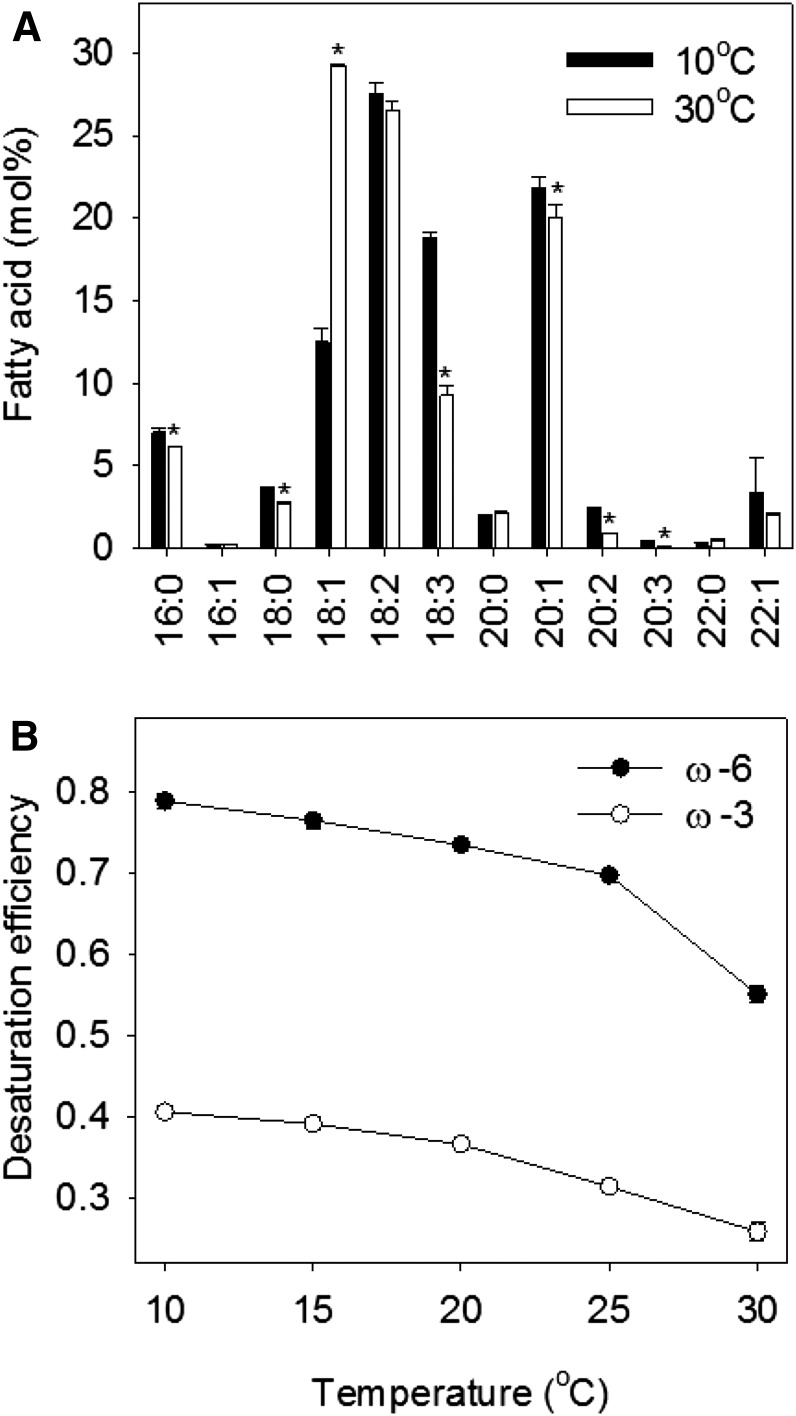
Approximately 94% of the fatty acids in Arabidopsis seeds are found in storage oil (Li-Beisson et al., 2013). Analysis of TAG total fatty acid composition revealed that temperature has a strong effect on the relative quantities of 18:1 and 18:3 in Col-0 seeds. The amount of 18:1 more than doubles on a mol % basis between 10°C and 30°C (becoming the most abundant fatty acid species), whereas the amount of 18:3 is halved (Fig. 2A). By contrast, the quantity of the metabolic intermediate 18:2 varies relatively little (Fig. 2A). These data suggest that higher temperature reduces both ω-6 and ω-3 fatty acid desaturation in Col-0 seeds. To derive values for overall ω-6 and ω-3 desaturation efficiencies (DEs), we calculated the proportion of available substrate that was converted to product(s) in a manner similar to Botella et al. (2016): ω-6 DE = 18:2 + 18:3/18:1 + 18:2 + 18:3 and ω-3 DE = 18:3/18:2 + 18:3. Both ω-6 and ω-3 DEs declined progressively with increasing temperature (Fig. 2B), and linear regression analysis suggests a strong positive relationship (r2 = 0.9368, P = 0.0069). The other fatty acids present in the seed TAG showed much less variation with temperature (Fig. 2A). However, statistically significant differences (P < 0.05) also were detected for palmitic acid (16:0) stearic acid (18:0), and the very-long-chain fatty acids (VLCFAs) eicosenoic acid (20:1), eicosadienoic acid (20:2), and eicosatrienoic acid (20:3; Fig. 2A). The latter two VLCFA species also are minor products of ω-6 and ω-3 desaturation, respectively (Miquel and Browse, 1992; Browse et al., 1993).
Figure 2.
Effect of temperature on the fatty acid composition of TAG in Arabidopsis Col-0 seeds. A, Fatty acid composition at 10°C and 30°C. B, Relationship between ω-6 and ω-3 DEs and temperature. Values are means ± se of measurements on seeds from three separate plants. In A, asterisks denote statistically significant differences (P < 0.05) from 10°C. In B, regression analysis supports a strong positive linear relationship between ω-6 and ω-3 DEs (r2 = 0.9368, P = 0.0069).
Storage Oil Composition Reflects That of PC in Arabidopsis Seeds
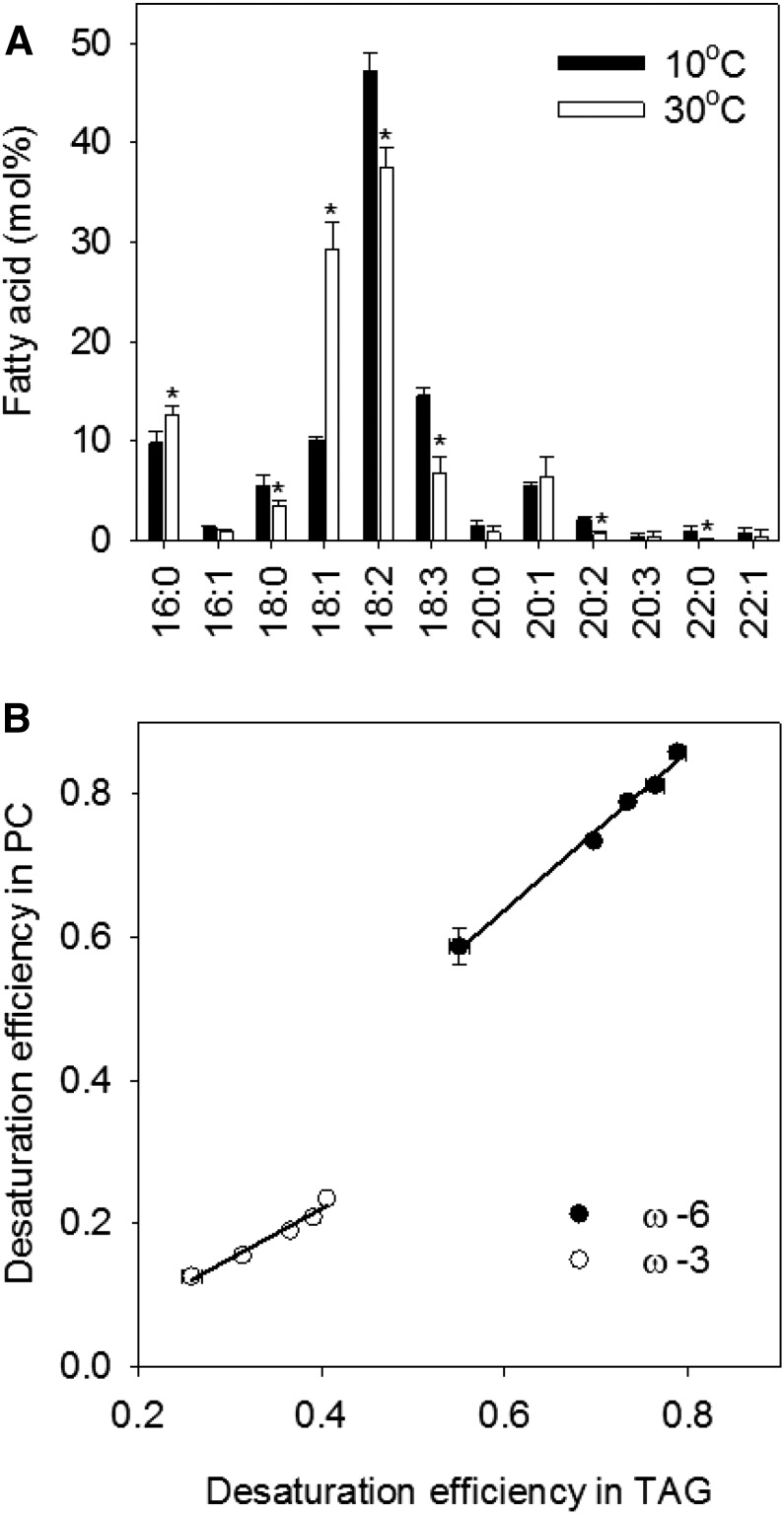
Temperature modifies the fatty acid composition of seed oil through its effect on PC (Slack and Roughan, 1978), which is the site of ω-6 and ω-3 fatty acid desaturation (Slack et al., 1978) and also a direct precursor for TAG biosynthesis in Arabidopsis (Bates and Browse, 2011). Analyzing the fatty acid composition of PC confirmed that 18:1 content also doubles on a mol % basis between 10°C and 30°C, while 18:3 content halves (Fig. 3A). Furthermore, linear regression analysis shows that ω-6 and ω-3 DEs for TAG and PC exhibit a strong positive correlation across the temperature range (Fig. 3B): r2 = 0.992 and 0.9687 and P = 0.0003 and 0.0024 for ω-6 and ω-3 DEs, respectively. TAG and PC molecular species composition in seeds matured at 10°C and 30°C also clearly reflected the difference in total fatty acid composition. There was a general shift in the number of double bonds in C52, C54, C56, and C58 species of TAG and in C34 and C36 species of PC (Supplemental Fig. S2).
Figure 3.
Relationship between the effect of temperature on the fatty acid composition of PC and TAG in Arabidopsis Col-0 seeds. A, Fatty acid composition of PC at 10°C and 30°C. B, Relationship between ω-6 and ω-3 DEs in PC and TAG. Values are means ± se of measurements on seeds from three separate plants. In A, asterisks denote statistically significant differences (P < 0.05) from 10°C. In B, data are plotted for 10°C, 15°C, 20°C, 25°C, and 30°C. The lines were fitted by regression. r2 = 0.992 and 0.9687 and P = 0.0003 and 0.0024 for ω-6 and ω-3 DEs, respectively.
Natural Variation Exists in the Response of Seed Oil Composition to Temperature
Kover et al. (2009) previously developed a MAGIC mapping population in Arabidopsis that is derived from 19 founder accessions and consists of more than 500 recombinant inbred lines (RILs). The founders were selected based on their wide geographical distribution (e.g. latitude ranging from ∼29° to ∼60°) and also include the reference ecotype Col-0. To investigate whether the MAGIC population would contain significant variation in the response of seed lipids to temperature, the 19 founder accessions were vernalized, grown under standard conditions until the onset of flowering, and then transferred to cabinets set to constant temperatures of 15°C and 25°C. Analyzing the fatty acid composition of the seed TAG revealed that, in all 19 accessions, ω-6 DEs were significantly lower (P < 0.05) at 25°C than at 15°C (Fig. 4A). Furthermore, the extent of this difference (∆DE) also varied by more than 4-fold (Fig. 4A). In all 19 accessions, ω-3 DEs also were significantly lower (P < 0.05) at 25°C than at 15°C, but the variation was only ∼2-fold (Fig. 4B). Can-0 displayed the largest response in ω-6 DE between 15°C and 25°C (0.23), and this accession originates from near Las Palmas in the Canary Islands, where the mean temperature in the spring is ∼17°C. Wil-2 displayed the smallest response (0.054), and this accession originates from near Vilnius in Lithuania, where the mean temperature in the spring is ∼6°C. Linear regression analysis (Supplemental Fig. S3) suggests that there is a significant positive relationship between spring mean temperature in the location where the 19 founder accessions were originally collected and ω-6 ∆DE (r2 = 0.6010, P = 0.0003). However, no significant correlation was present for ω-3 ∆DE (r2 = 0.1208, P = 0.1398). Linear regression analysis (Fig. 4C) also revealed that the relationship between ω-6 and ω-3 ∆DEs across all 19 accessions is relatively weak (r2 = 0.2280, P = 0.0387). These data suggest that there is substantial genetic variation in the temperature responsiveness of ω-6 DE within the MAGIC founders and that this trait also might have adaptive significance (Sanyal and Linder 2013), given that accessions from cooler climates appear to respond less strongly to heat.
Figure 4.
Effect of temperature on ω-6 and ω-3 DEs measured in TAG from seeds of the 19 founder accessions of the MAGIC population. A, ω-6 DE. B, ω-3 DE. C, Association between the change in ω-6 and ω-3 DEs. Values are means ± se of measurements on seeds from three separate plants of each genotype. Wil-2, Wilna-2; Wu-0, Wurzburg-0; Tsu-0, Tsushima-0; Kn-0, Kaunas-0; Ws-0, Wassilewskija-0; Rsch-4, Rschew-4; Po-0, Poppelsdorf-0; Bur-0, Burren-0; Ler-0, Landsberg erecta; Zu-0, Zurich-0; No-0, Nossen-0; Hi-0, Hilversum-0; Mt-0, Martuba-0; Edi-0, Edinburgh-0; Oy-0, Oystese-0; Sf-2, San Feliu-2; Col-0, Columbia-0; Ct-1, Catania-1; Can-0, Canary Islands-0. In C, the dotted lines mark the 95% confidence interval (CI).
Variation in FAD2 Controls ω-6 Desaturation in Seeds
We previously analyzed the fatty acid composition of seed from ∼450 RILs of the MAGIC population that were vernalized and grown under standard conditions (∼23°C, 16 h of light/∼18°C, 8 h of dark) in the glasshouse (Bryant et al., 2016). To determine whether this population can be used to map QTLs that control microsomal ω-6 fatty acid desaturation, we calculated DE values for our existing data set. Across the recombinant inbred population, ω-6 DE ranged from 0.65 to 0.84 and the estimated broad sense heritability was 0.87. The mean value for each RIL was used for both multiple QTL modeling (Kover et al., 2009) and genome-wide association studies (GWAS), exploiting genomic resources and software tools developed by Richard Mott and colleagues (Kover et al., 2009; Gan et al., 2011; Imprialou et al., 2016). QTL analysis resulted in the detection of a single major QTL for ω-6 DE (genome-wide P < 0.001), situated on chromosome 3 at ∼4.1 Mb (Table I). The estimated 90% CI for this QTL is ∼0.6 ± 0.3 Mb, based on the −log10 ANOVA P value score of ∼30 (Kover et al., 2009).
Table I. QTL analysis of the effect of temperature on ω-6 fatty acid desaturation in seeds from the Arabidopsis MAGIC population.
Approximately 450 RILs from the population were grown in the glasshouse (GH) or in controlled-environment chambers at 15°C and 25°C. The DEs were derived from the total fatty acid composition of the mature seeds, and QTL analysis was performed using the HAPPY R package (Kover et al., 2009). logPmax = −log10 ANOVA P value and 90% CI = estimated 90% CI. QTLs with a genome-wide P < 0.01 are shown.
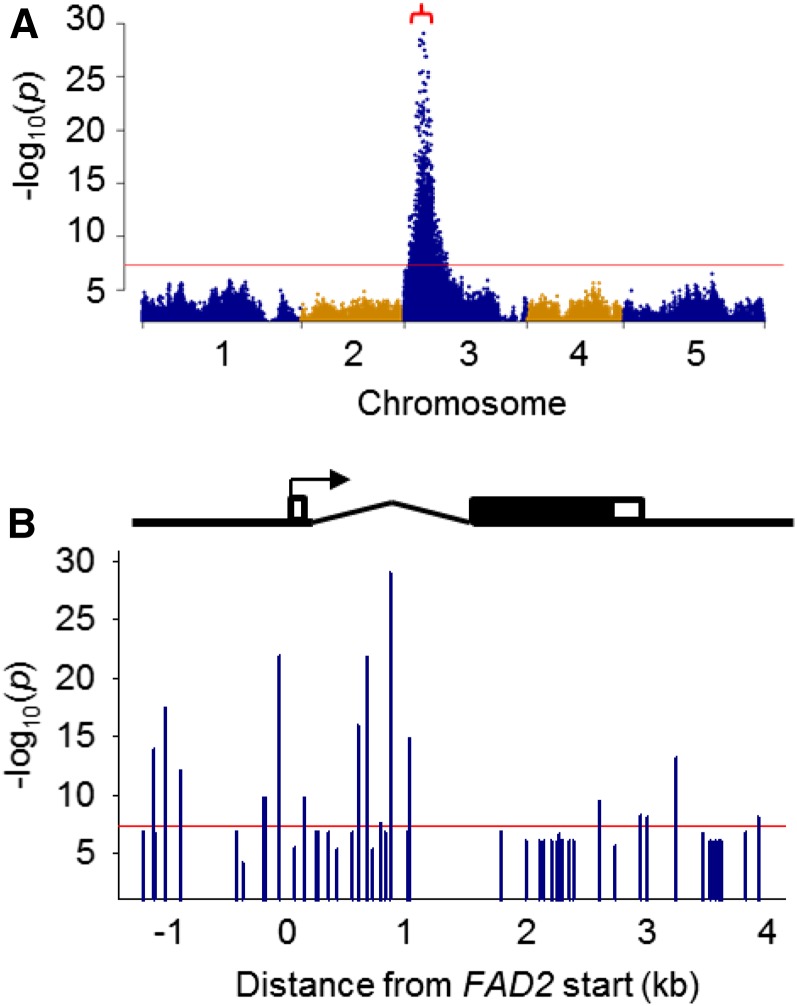
GWAS also was performed using all ∼3 million individual sequence variants in the imputed genomes of the RILs (Imprialou et al., 2016). Polymorphisms with the highest –log10(P) scores lay within the 90% CI for the ω-6 DE QTL present on chromosome 3 (Fig. 5A). All the polymorphisms above the genome-wide significance threshold within the QTL 90% CI were ranked by –log10(P) score, and genes that mapped within 1 kb upstream or downstream were identified and listed (Supplemental Data S1). The highest ranked polymorphism (P1) for ω-6 DE [–log10(P) score of 29] is a single-nucleotide substitution (T/G) situated at 3,862,256 bp on chromosome 3. P1 can explain ∼29% of the phenotypic variation in the trait and lies in the intron within the 5′ UTR of the FAD2 gene (At3g12120; Fig. 5B; Table II). The population contains more than 50 sequence variants within 1 kb of FAD2, and six additional polymorphisms within the intron and promoter also had a –log10(P) score greater than 10 (Fig. 5B; Table II). Arabidopsis contains a single FAD2 gene that is responsible for greater than 95% of ω-6 fatty acid desaturation in the seed (Okuley et al., 1994). Therefore, it is probable that one or more of these associate polymorphisms at the FAD2 locus cause the phenotypic variation observed in ω-6 DE.
Figure 5.
Manhattan plots showing the association of greater than 3M individual sequence variants with ω-6 DE in seeds from ∼450 RILs of the MAGIC population that were grown in the glasshouse. A, Whole-genome scan. B, Closeup of the FAD2 locus including the region 1 kb upstream and downstream of the start and finish of transcription. GWAS were performed using the magic_src_v4.0.tar.gz package. In A, the 90% CI for the corresponding QTL is bracketed, and the red line marks the genome-wide significance threshold. In B, the arrow marks the start and direction of transcription for FAD2, the kinked line indicates the intron, and the white and black bars are UTRs and exon, respectively.
Table II. Sequence polymorphisms within the region of the FAD2 locus on chromosome 3 that are strongly associated with ω-6 DEs.
Approximately 450 RILs from the MAGIC population were grown in the glasshouse (GH) or in controlled-environment chambers at 15°C and 25°C. The ω-6 DEs were derived from the total fatty acid composition of the mature seeds, and GWAS were performed using the magic_src_v4.0.tar.gz package. Polymorphisms are marked relative to the start of transcription in Col-0 (Kim et al., 2006).
| Position | –log10(P), GH | –log10(P), 15°C | –log10(P), 25°C | Location Versus FAD2 Start Site | Polymorphism Versus Col-0 |
|---|---|---|---|---|---|
| 3,863,947 |
17.40 |
13.57 |
6.90 |
−925 (promoter) |
A/T |
| 3,863,832 |
12.05 |
9.16 |
6.22 |
−810 (promoter) |
−AAT |
| 3,863,088 |
21.91 |
10.76 |
6.21 |
−66 (promoter) |
−TC to −TCTCTC |
| 3,862,499 |
15.96 |
12.24 |
9.18 |
+523 (5′ UTR intron) |
C/A or +A to +AAAA |
| 3,862,428 |
21.83 |
15.27 |
7.68 |
+594 (5′ UTR intron) |
A/G |
| 3,862,256 |
29.03 |
15.63 |
13.23 |
+766 (5′ UTR intron) |
T/G (P1) |
| 3,862,109 | 14.82 | 11.08 | 10.71 | +913 (5′ UTR intron) | T/C |
Variation in FAD2 Determines the Expression Level of the Gene
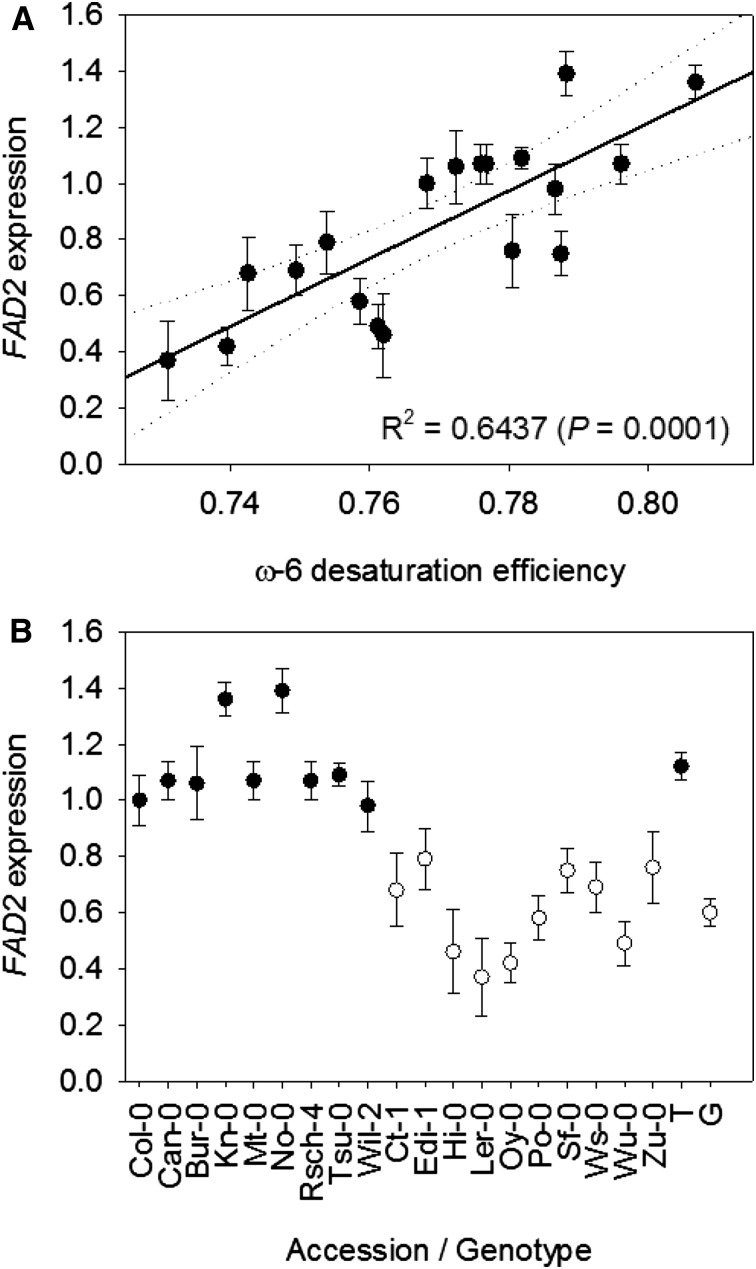
Although the MAGIC population contains ∼3 million sequence variants, there are no nonsynonymous substitutions in FAD2 (Gan et al., 2011). As a result, the polymorphisms at this locus that are associated with ω-6 DE most likely act by altering the expression level of the gene. Gan et al. (2011) previously showed that transcript abundance data for the 19 founder accessions is sufficient to identify potential cis-acting polymorphisms associated with expression (i.e. cis-eQTL). Therefore, we used quantitative PCR to measure FAD2 transcript abundance in seeds from the 19 founders that were grown alongside the RIL population in the glasshouse (Bryant et al., 2016), and we found that there is a significant positive correlation between FAD2 expression level and ω-6 DE (r2 = 0.6437, P = 0.0001; Fig. 6A). Furthermore, we also tested whether FAD2 expression was associated with P1, the single-nucleotide polymorphism identified previously in the intron of the 5′ UTR (Table II). We found that there was a highly significant relationship (P < 0.0001), with P1 variant T associated with high expression and variant G associated with low expression (Fig. 6B).
Figure 6.
FAD2 expression analysis in the 19 founder accessions of the MAGIC population. Relationships between expression and ω-6 DE (A) and expression and genotype at P1 (3,862,256 bp on chromosome 3; B) are shown. Quantitative PCR was performed on RNA from seeds of five separate plants of each genotype, and values are normalized to Col-0 and expressed as means ± se. In A, the dotted lines mark the 95% CI. In B, black circles represent accessions that have nucleotide variant T and white circles represent accessions that have nucleotide variant G. The averages shown on the left are significantly different (P < 0.0001).
Variation at an Unknown Locus Controls the Response of ω-6 Desaturation to Temperature
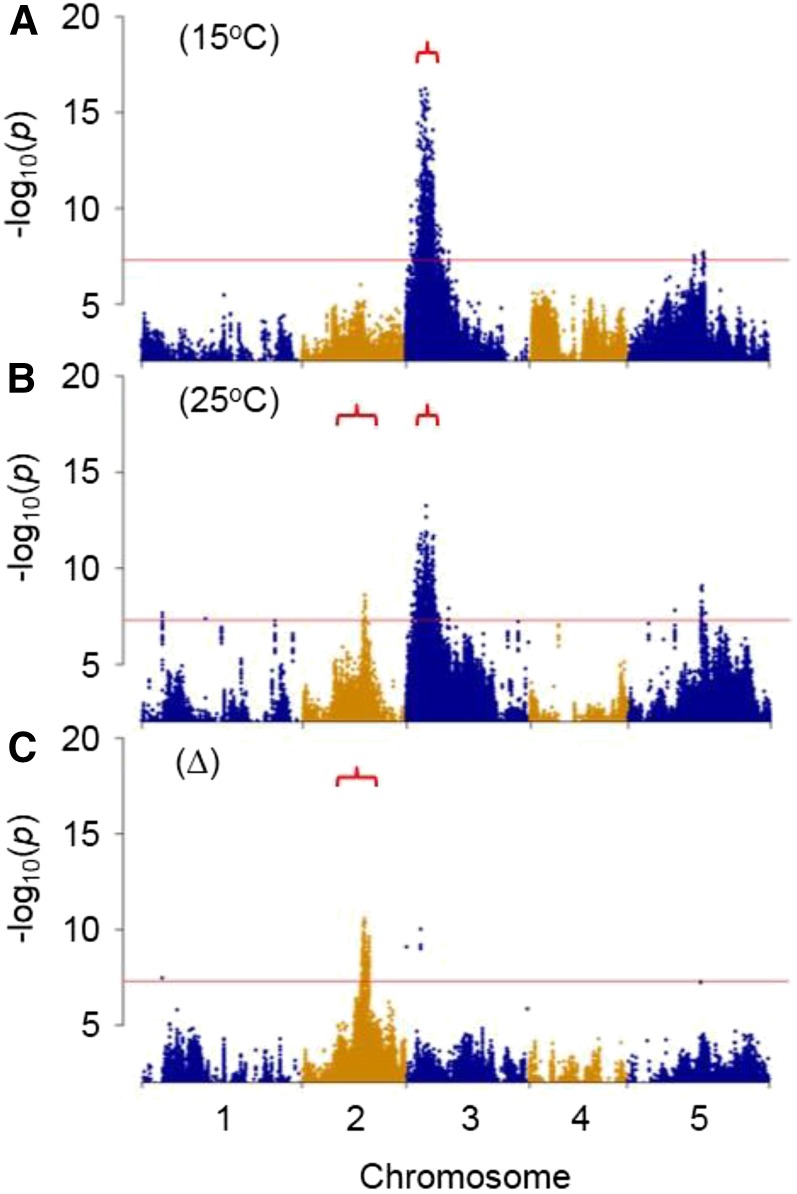
To investigate the genetic basis of natural variation in the response of seed fatty acid composition to temperature, ∼450 RILs from the MAGIC population were vernalized, grown under standard conditions until the onset of flowering, and then transferred to controlled-environment chambers set to constant temperatures of 15°C and 25°C. The fatty acid composition of the mature seeds was determined, and ω-6 DE and ΔDE values were calculated. These data were then analyzed using both multiple QTL modeling and GWAS (Kover et al., 2009; Imprialou et al., 2016). One major and one minor QTL were detected for ω-6 DE at 15°C (P < 0.001; Table I). The major QTL is situated on chromosome 3 in the same region as FAD2 (∼3.5 ± 0.4 Mb), while the minor QTL is on chromosome 5 at ∼7.7 ± 1.5 Mb. At 25°C, a major QTL for ω-6 DE also was detected on chromosome 3 near FAD2 (∼3.3 ± 0.8 Mb), while a more minor QTL was detected on chromosome 2 at ∼11.8 ± 1.4 Mb. When the ΔDE was examined, no QTL was detected on chromosome 3, near the location of FAD2, and instead a single QTL was present on chromosome 2 at ∼11.8 ± 1 Mb. This QTL is in the same approximate location as the minor QTL found for ω-6 DE at 25°C (Table I). QTL analysis, therefore, suggests a relatively simple genetic architecture for ω-6 DE. The trait appears to be controlled primarily by variation at a single locus on chromosome 3, but the response of ω-6 DE to temperature is determined by variation at another locus on chromosome 2. Furthermore, variation at this locus also might exert a stronger influence on ω-6 DE at higher temperature if it is the same QTL as that detected in this region at 25°C.
GWAS revealed that individual polymorphisms with the largest –log10(P) scores also coincided with the location of the major QTL on chromosome 3 for ω-6 DE at both 15°C and 25°C (Fig. 7, A and B). The polymorphisms were ranked, and the P1 variant situated at 3,862,256 bp on chromosome 3 within the 5′ UTR intron of FAD2 was third at 25°C and first at 15°C (Table II). The other six associated polymorphisms, which were identified previously within 1 kb of FAD2 in the glasshouse experiment, also had a comparatively high –log10(P) score at both 25°C and 15°C (Table II). Therefore, variation at the FAD2 locus appears to determine ω-6 DE at both 15°C and 25°C. However, the responsiveness of ω-6 DE to the change in temperature is determined by a separate QTL on chromosome 2 (Table I). GWAS performed on this trait also identified highly associated polymorphisms located within the 90% CI of the QTL (Fig. 5C). All the polymorphisms within this region that were above the threshold for genome-wide significance were ranked by –log10(P) score, and genes that map within 1 kb upstream or downstream were identified and listed (Supplemental Data S1). In total, ∼240 genes were found to contain one or more of these polymorphisms. However, the highest scoring polymorphisms lie within a smaller set of ∼40 genes (Supplemental Table S1) that are all located in an ∼450-kb region of chromosome 3 (11,735,058–12,189,131 bp). The function of these genes was investigated by searching relevant databases such as ARALIP (Li-Beisson et al., 2013), The Arabidopsis Information Resource (https://www.arabidopsis.org/), and KnetMiner (Hassani-Pak et al., 2016). Two of the genes (ATP-BINDING CASSETTE G3 [ABCG3] and DEPHOSPHO-COENZYME A KINASE [CoAE]) could play a metabolic role in lipid biosynthesis (Kupke et al., 2003; Li-Beisson et al., 2013). However, many of the other genes on the list are known or predicted to be involved in gene regulation or posttranslational protein modification. Therefore, these genes also are candidates for involvement in the temperature regulation of ω-6 DE and may potentially target FAD2 function.
Figure 7.
Manhattan plots showing the association of greater than 3M individual sequence variants with ω-6 DE in seeds from ∼450 RILs of the MAGIC population grown at 15°C (A) and 25°C (B) and the difference in DE values (∆) between the two temperatures (C). GWAS were performed using the magic_src_v4.0.tar.gz package. The 90% CIs for corresponding QTLs are bracketed, and the red lines mark the genome-wide significance thresholds.
The Genetic Architecture of ω-3 Desaturation in Arabidopsis Seeds Is More Complex
Within the 19 MAGIC founder accessions, there was relatively little variation in ω-3 ∆DE compared with ω-6 ∆DE (Fig. 4B). Nevertheless, we also performed a genome-wide analysis of this trait using the MAGIC population (Supplemental Table S2; Supplemental Fig S4). These analyses suggested that ω-3 DE has a more complex genetic architecture than ω-6 DE. In our glasshouse experiment (Bryant et al., 2016), QTLs were detected on chromosomes 2, 3, and 5 at 15.8 ± 2, 4.6 ± 2.8, and 19.1 ± 2.6 Mb, respectively (Supplemental Table S2). The QTL on chromosome 2 coincides with the location of FAD3 (At2g29980), and GWAS revealed that two of the fifth highest ranked polymorphisms in this region lie within 1 kb of this gene [12,782,970 and 12,783,194 bp; −log10(P) = 8.384]. As with FAD2, Arabidopsis contains a single FAD3 gene, and this is responsible for nearly all ω-3 fatty acid desaturation in the seed (Browse et al., 1993). Therefore, allelic variation at this locus could potentially explain some of the phenotypic variation in ω-3 DE. When ω-3 ∆DE was analyzed, a single QTL with a relatively low level of significance was identified on chromosome 2 at ∼13.7 ± 3 Mb (Supplemental Table S2). The 90% CI for this QTL is quite large and encompasses the location of both FAD3 and the QTL for ω-6 ∆DE, but using GWAS, no individual sequence variant was detected with a −log10(P) value above the genome-wide significance threshold (Supplemental Fig. S4). Therefore, the genetic basis for variation in the plastic response of ω-3 DE to temperature in Arabidopsis seeds is harder to define than that of ω-6 DE using the MAGIC population, and this question requires an alternative approach.
DISCUSSION
In this study, we show that Arabidopsis can be used as a genetic model to study the plastic response of storage oil content and composition to variation in ambient temperature during seed maturation. The response of Arabidopsis is similar to that of several major oilseed crops such as soybean, oilseed rape, and sunflower (Canvin, 1965; Harris et al., 1978; Carver et al., 1986). The genetic basis of this plasticity is poorly understood, but it is of relevance to breeders and seed oil processors because variation in temperature can adversely affect both oil yield and quality in the field (Rolletschek et al., 2007; Zhu et al., 2012; Singer et al., 2016). The most obvious effect of temperature on Arabidopsis seed lipid composition is to alter the PUFA content of the TAG. Between 10°C to 30°C, the 18:1 content of Col-0 seed TAG doubles (on a mol % basis) and 18:3 content halves, while 18:2 changes relatively little. This pattern is consistent with a progressive reduction in both ω-6 and ω-3 DE. The response in Col-0 also appears to become stronger at higher temperature, particularly for ω-3 DE. Temperature modifies the PUFA content of seed TAG through its effect on PC (Slack and Roughan, 1978), which is the main substrate for ω-6 and ω-3 fatty acid desaturation in seeds (Slack et al., 1978) and also a direct precursor for TAG biosynthesis (Bates et al., 2009; Bates and Browse, 2011). Accordingly, we observed a very strong positive correlation between PC and TAG for both ω-6 and ω-3 DEs in Col-0 seeds within the range of 10°C to 30°C. Arabidopsis seeds accumulate the VLCFA 20:1 in addition to PUFAs, and 18:1 is a substrate for both fatty acid elongation and desaturation (Li-Beisson et al., 2013). Disruption of ω-6 desaturation leads to an increase in 20:1 content in Col-0 seeds (Okuley et al., 1994). However, we found that 20:1 content actually decreases slightly between 10°C and 30°C. These data suggest that fatty acid elongation and/or 20:1 incorporation into TAG also may be inhibited as temperature increases.
Analysis of the 19 founder accessions of the Arabidopsis MAGIC population (Kover at al., 2009) revealed that natural variation exists in the plastic response of both ω-6 and ω-3 DEs to a change in temperature from 15°C and 25°C. However, there is only a relatively weak positive correlation between ω-6 and ω-3 ∆DEs, suggesting that they are not entirely under the same genetic control. Within the 19 accessions, the variation in the plastic response also was much larger for ω-6 ∆DE (greater than 4-fold) than for ω-3 ∆DE (∼2-fold). The MAGIC founders were originally selected based on their wide geographical distribution and originate from substantially different latitudes, ranging from ∼29° to ∼60° (Kover at al., 2009). Arabidopsis usually flowers in the spring. Linear regression analysis suggested that there is a significant positive relationship between spring mean temperature in the location where the accessions were first collected and ω-6 ∆DE, but no significant correlation was detected for ω-3 ∆DE. Therefore, our data suggest that the plastic response of ω-6 fatty acid desaturation (and, therefore, PUFA content) to temperature in seed may be subject to natural selection. However, ideally, a larger survey of accessions would be required to test this hypothesis more rigorously. Sanyal and Linder (2013) previously analyzed 84 Arabidopsis accessions and also reported that there is variation in the plastic response, but they did not relate the size of this response to climatic conditions where the accessions originated.
Whether plasticity in seed TAG composition is under direct selection also is not clear, since the composition of the major membrane phospholipid PC also changes with temperature. The PUFA content of Arabidopsis leaf lipids also responds to temperature (Falcone et al., 2004). However, the change reported in leaf PC composition between 10°C and 30°C is very modest (Li et al., 2015) compared with our data in seeds. Sanyal and Linder (2013) also reported that there is no significant relationship between the temperature response of fatty acid composition in seed TAG and leaf phospholipids across 84 Arabidopsis accessions. Interpreting the effect of temperature on leaf lipid composition is complicated by the existence of parallel glycerolipid biosynthetic pathways in the chloroplast and endoplasmic reticulum, which utilize different ω-6 and ω-3 desaturases (Li-Beisson et al., 2013). Li et al. (2015) recently presented data that suggest that temperature alters the balance of metabolic flux between these compartments, which contributes to changes in lipid composition. However, in both Arabidopsis seeds and roots, the glycerolipid biosynthetic pathway in the endoplasmic reticulum is predominant (Li-Beisson et al., 2013). Analysis of root fatty acid composition in the 19 MAGIC founders revealed that ω-3 DE changes substantially between 10°C and 30°C in this tissue, while the change in ω-6 DE is very small (Supplemental Fig. S5). Therefore, the high degree of plasticity in ω-6 DE we observed in Arabidopsis seeds appears to be rather specific to this tissue.
Analysis of seeds from the MAGIC founder accessions suggested that the recombinant inbred population may have utility for mapping QTLs associated with ω-6 ∆DE in this tissue. The MAGIC population developed by Kover et al. (2009) is a powerful tool for genetic trait dissection and can be used for both QTL analysis and GWAS (Kover et al., 2009; Gan et al., 2011; Imprialou et al., 2016). Using ω-6 DE values from ∼450 RILs that we grew previously under standard glasshouse conditions (Bryant et al., 2016), we found that the trait is under relatively simple genetic control. A single major QTL was detected on chromosome 3 at 4.1 ± 0.3 Mb that maps precisely to the location of FAD2. Previous studies using biparental populations also identified a QTL in this region (Hobbs et al., 2004; O’Neill et al., 2012; Sanyal and Linder, 2012). Of the ∼3 million sequence variants present in the imputed genomes of the population (Imprialou et al., 2016), the one that is most strongly associated with the trait [–log10(P) score of ∼ 29] is P1 situated in the intron of the FAD2 5′ UTR, and six other sequence variants with a –log10(P) score greater than 10 also lie either within the 5′ UTR intron or in the ∼1-kb promoter region of this gene. The population contains no nonsynonymous mutations in FAD2; therefore, these cis-acting sequence variants are most likely to control the expression of the gene (Gan et al., 2011). In support of this hypothesis, we found that there is a statistically significant association between the FAD2 expression level in seeds of the 19 founder accessions and their genotype for P1. Interestingly, recent GWAS experiments performed on the seed fatty acid composition of 391 Arabidopsis accessions found that the same sequence variant (P1) also was the most strongly associated with 18:1, 18:2, 20:2, and total PUFA levels (Branham et al., 2016a) as well as with the oil melting point (Branham et al., 2016b). Furthermore, when we surveyed the 1,135 resequenced Arabidopsis accessions (1001 Genomes Consortium, 2016), we found that there is a significant relationship between the latitude of their site of collection and the genotype at P1 (T = 49.26° ± 0.35°, n = 458 and G = 47.47° ± 0.28°, n = 569; P = 0.000076).
The 5′ UTR intron is a conserved feature of FAD2 genes (Kim et al., 2006; Lozinsky et al., 2014). Kim et al. (2006) have shown that the 5′ UTR introns of both sesame (Sesamum indicum) and Arabidopsis FAD2 mediate the enhancement of gene expression. The Arabidopsis FAD2 5′ UTR intron contains several putative cis-elements, including an E box (CANNTG) that is situated only 3 bp away from P1 (Supplemental Fig. S6; Kim et al., 2006). Kim et al. (2007) identified a basic region/helix-loop-helix transcription factor that transactivates FAD2 expression in sesame by binding both E box and G box elements in the promoter. Therefore, it is possible that a related transcription factor also enhances FAD2 expression in Arabidopsis by interacting with the E box situated near P1 and that this sequence variant might influence DNA binding. Several other polymorphisms within the FAD2 promoter and 5′ UTR intron that are associated with ω-6 DE also collocate with putative cis-elements (Supplemental Fig. S6; Kim et al., 2006). Further work will be required to determine which polymorphisms are causal and to establish the precise mechanism of action.
When we grew the MAGIC population at two temperatures (15°C and 25°C) and performed genetic analysis on seed ω-6 DE, we also identified a major QTL overlying FAD2, and the same sequence variants within the 5′ UTR exon and promoter region of this gene (including P1) were closely associated with the trait. However, when the plastic response to temperature (ω-6 ∆DE) was analyzed, no QTL was found in the region of FAD2. Instead, a single major QTL was detected on chromosome 2 at ∼11.8 ± 1 Mb. A minor QTL for ω-6 DE also was detected at this position at 25°C but not at 15°C. These data suggest that the cis-acting sequence variants identified in FAD2 control ω-6 DE independently of temperature and that a separate QTL on chromosome 2 controls the temperature response of ω-6 DE. Control of ω-6 DE also appears to be shared between this QTL and FAD2 at 25°C but not at 15°C, indicating that variation at the locus may primarily affect the trait at higher temperature. There are two models that could explain how the QTL on chromosome 2 controls ω-6 ∆DE. The underlying gene could either regulate FAD2 directly (Tang et al., 2005) or it could affect desaturation indirectly by influencing the availability of substrate (Harris and James, 1969; Browse and Slack, 1983; Rolletschek et al., 2007; Li et al., 2015).
Using GWAS, it was possible to identify individual sequence variants within the region of the ω-6 ∆DE QTL that are most strongly associated with the trait. The decay in linkage disequilibrium is more than 10-fold slower in the MAGIC population than in a panel of Arabidopsis accessions (Kover et al., 2009); therefore, a substantial number of polymorphisms were identified within the QTL 90% CI (Supplemental Data S1). However, virtually every sequence variant that exists in the MAGIC population was scored, including those that must be causal. The sequence variants with the highest –log10(P) values lie within ∼40 genes (Supplemental Table S1) that are all located within an ∼450-kb region of chromosome 2 (11,735,058–12,189,131 bp). Among these genes, ABCG3 and CoAE could play a metabolic role in influencing substrate provision for ω-6 fatty acid desaturation (Kupke et al., 2003; Li-Beisson et al., 2013). The substrate of ABCG3 is not known, but CoAE encodes the last step in CoA biosynthesis (Kupke et al., 2003), and although CoA is not directly required for ω-6 fatty acid desaturation, it is an essential cofactor for de novo fatty acid and glycerolipid biosynthesis (Li-Beisson et al., 2013).
On the short list of candidate genes for the ω-6 ∆DE QTL, there are also several with regulatory functions, such as HISTONE DEACETYLASE2D (HD2D), ARGONAUTE5, subunits of two protein kinases (cyclin-dependent kinase1 and 5′-AMP-activated protein kinase β-2), two proteases, a putative E3 ligase, several transcription factors, RNA-binding proteins, several proteins of unknown function, and the microRNA MIR172 (Supplemental Table S1). Transcriptional regulators within this list could either act directly on FAD2 or function within the upstream signal transduction pathway that links ω-6 DE to temperature. Although FAD2 expression is not cold inducible in Arabidopsis leaves (Okuley et al., 1994; Li et al., 2015), direct action cannot be discounted entirely. Zhu et al. (2012) reported that FAD2 expression is enhanced by low temperatures in the developing seeds of oilseed rape. Interestingly, Wang et al. (2016) recently reported that Arabidopsis mutants in a number of histone deacetylases exhibit changes in seed fatty acid composition (ω-6 and ω-3 DEs) and that H3K9/14 acetylation of FAD3 influences its expression level (Wang et al., 2016). DNA acetylation in Arabidopsis has been implicated in heat stress-responsive gene expression (Hu et al., 2015). Therefore, although an hd2d mutant has yet to be analyzed, it is conceivable that this gene might play a role in controlling ω-6 ∆DE. The expression level of MIR172A also has been shown to respond to temperature, and this microRNA targets several APETALA2 domain transcription factors involved in temperature-sensitive processes such as flowering (May et al., 2013). There also is evidence that FAD2 is phosphorylated in soybean (Tang et al., 2005) and Arabidopsis (Durek et al., 2010) and that elevated temperature promotes the degradation of FAD2 isoforms via the ubiquitin-proteasome pathway (Tang et al., 2005). Therefore, protein kinases, proteases, and ubiquitin-proteasome pathway components also are potential candidates for regulators of ω-6 ∆DE. Further work will be required to determine which gene is responsible for the ω-6 ∆DE QTL identified in this study and to fully characterize its mode of action.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type (Col-0) Arabidopsis (Arabidopsis thaliana) seeds, the MAGIC population parental accessions, and RILs (N782242) were obtained from the European Arabidopsis Stock Centre (University of Nottingham). Col-0 seeds were routinely surface sterilized, plated on agar plates containing one-half-strength Murashige and Skoog salts (Sigma-Aldrich), and imbibed in the dark for 4 d at 4°C. The plates were then placed in a chamber set to standard growth conditions (16 h of light, 22°C/8 h of dark, 18°C, photosynthetic photon flux density = 250 μmol m−2 s−1, and 70% relative humidity). After 5 d, seedlings were transplanted to moist Levington F2 compost in 7-cm2 pots and returned to standard conditions. For the MAGIC parental accessions and RILs, seeds were sown on moist Levington F2 compost in P40 trays and vernalized for 6 weeks at 4°C before being placed in standard growth conditions. At the onset of flowering, about five plants of each genotype were transferred to controlled-environment chambers set to the same photoperiod, photosynthetic photon flux density, and relative humidity but different constant temperatures ranging from 10°C to 30°C. Plants in 7-cm2 pots were bagged to retain all seeds as described previously (van Erp et al., 2014). At maturity, seeds were harvested from either individual plants in 7-cm2 pots or pools of approximately five plants for each genotype in P40 trays and used for further analysis.
Seed Lipid Analysis
Seed oil and moisture contents were measured by low-resolution time domain NMR spectroscopy using a Minispec MQ20 device (Bruker) fitted with a robotic sample-handling system (Rohasys) as described previously (van Erp et al., 2014). The total fatty acid composition of seeds was measured by gas chromatography analysis after combined digestion and fatty acid methyl ester formation using the method described by Browse et al. (1986). For PC and TAG analysis, lipids were extracted from seeds by homogenization in chloroform:methanol:formic acid (1:1:0.1, v/v) followed by phase partitioning with 1 m KCl/0.2 m H3PO4 (Dörmann et al., 1995). The lipids were applied to silica gel 60A plates (Merck) and separated by thin-layer chromatography using hexane:diethyl ether:acetic acid (70:30:1, v/v) as the mobile phase. Lipids were visualized under UV light having been sprayed with primulin solution (0.01% [w/v] in 60% [v/v] acetone). PC and TAG were identified by cochromatography with commercial standards (Sigma-Aldrich), and their fatty acid composition was determined by gas chromatography analysis (Browse et al., 1986). DEs were derived from fatty acid profiles in a manner similar to that of Botella et al. (2016) using the following calculations: ω-6 DE = 18:2 + 18:3/18:1 + 18:2 + 18:3 and ω-3 DE = 18:3/18:2 + 18:3. Lipid extracts also were analyzed by electrospray-tandem mass spectrometry using an ABSiex 4000 QTRAP system as described previously (Mendes et al., 2013).
Genetic Analysis
QTL mapping with the MAGIC population was performed as described by Kover et al. (2009) using the HAPPY R package available from http://archive.is/mus.well.ox.ac.uk. GWAS were performed using the magic_src_v4.0.tar.gz package, which is available from the same site and includes detailed instructions. In brief, the program reconstruction first generates imputed genomes for the RILs with a mosaic break-point accuracy of greater than 2 kb using polymorphism calls from low-coverage sequence and 1.2 million biallelic variants from the full genomes of the 19 founder accessions (Imprialou et al., 2016). The program genome_scan then performs association mapping using all ∼3 million individual sequence variants imputed from the founders. Specific sequence variants in the genomes of the 19 MAGIC founder lines were visualized using the Rätsch laboratory GBrowse tool (http://gbrowse.inf.ethz.ch/gb/gbrowse/thaliana-19magic).
Gene Expression Analysis
DNase-treated RNA was purified from seeds using the RNeasy kit from Qiagen with modifications described previously (Mendes et al., 2013). Single-stranded cDNA synthesis was carried out using SuperScript II RNase H− reverse transcriptase from Invitrogen. Real-time PCR was carried out in a MyiQ single-color real-time PCR detection system (Bio-Rad) using qPCR Mastermix Plus from Eurogentec. Data were analyzed with Bio-Rad iQ5, Optical System Software, version 2.0. The primer pairs used for real-time PCR were FAD2_F (5′-CGGAAACCGACACCACAAA-3′) plus FAD2_R (5′-TTCAGATCTCCCACCGAGAAA-3′) and 18S_F (5′-TCCTAGTAAGCGCGAGTCATC-3′) plus 18S_R (5′-CGAACACTTCACCGGATCAT-3′).
Statistical Analyses
The number of replicates (n) and the se are shown for all measurements. Either unpaired Student’s t test or one-way ANOVA was used to assess the differences between genotypes. For ANOVA, following significant (P < 0.05) F test results, means of interest were compared using the appropriate lsd value at the 5% (P = 0.05) level of significance on the corresponding degrees of freedom. Linear regression analysis was performed using the function in SigmaPlot version 13.0 (Systat Software).
Accession Numbers
Sequence data for the genes described in this article can be found in the GenBank/EMBL data libraries using the Arabidopsis gene identifiers At3g12120 (FAD2) and At2g29980 (FAD3).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effect of temperature on Arabidopsis Col-0 seed morphology.
Supplemental Figure S2. Analysis of TAG and PC molecular species composition in Col-0 seeds matured at 10°C and 30°C.
Supplemental Figure S3. Relationship between spring mean temperature and the responsiveness of desaturation in seeds of the MAGIC parental accessions.
Supplemental Figure S4. Manhattan plots showing the association of individual sequence variants with ω-3 DE.
Supplemental Figure S5. Effect of temperature on fatty acid composition of Arabidopsis roots.
Supplemental Figure S6. Sequence of the 5′ region of FAD2 in Arabidopsis showing the positions of sequence variants that are strongly associated with ω-6 DE.
Supplemental Table S1. Short list of genes containing one or more sequence polymorphisms with a −log10(P) score greater than 10 for association with ω-6 ∆DE in the MAGIC population.
Supplemental Table S2. QTL analysis of the effect of temperature on ω-3 fatty acid desaturation in seeds from the Arabidopsis MAGIC population.
Supplemental Data S1. GWAS data for associations within ω-6 DE QTL CIs.
Supplementary Material
Acknowledgments
We thank Dr. Sue Welham for assistance with statistical analysis; Dr. Ana Mendes, Dr. Christian Craddock, Dr. Nicola Adams, Eve Shaw, Osama Butt, Jerome Dussard-McFarlane, and Daniel Tomkins for assistance in harvesting, cleaning seed, and performing fatty acid analysis for the glasshouse experiment; and the horticultural staff at the University of Warwick and Rothamsted Research for assistance with plant growth.
Glossary
- PUFA
polyunsaturated fatty acid
- PC
phosphatidylcholine
- TAG
triacylglycerol
- UTR
untranslated region
- MAGIC
multiparent advanced generation intercross
- QTL
quantitative trait locus
- Col-0
Columbia-0
- DE
desaturation efficiency
- VLCFA
very-long-chain fatty acid
- RIL
recombinant inbred line
- GWAS
genome-wide association studies
- CI
confidence interval
Footnotes
This work was supported by the U.K. Biotechnology and Biological Sciences Research Council (grant nos. BBS/E/C/00005207 and BB/E022197/1).
Articles can be viewed without a subscription.
References
- 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar JA, Fradin EF, Becker FF, Rienstra JA, van der Schoot J, Vreugdenhil D, Keurentjes JJ (2015) Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27: 1857–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella C, Sautron E, Boudiere L, Michaud M, Dubots E, Yamaryo-Botté Y, Albrieux C, Marechal E, Block MA, Jouhet J (2016) ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol 170: 1300–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham SE, Wright SJ, Reba A, Linder CR (2016a) Genome-wide association study of Arabidopsis thaliana identifies determinants of natural variation in seed oil composition. J Hered 107: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham SE, Wright SJ, Reba A, Morrison GD, Linder CR (2016b) Genome-wide association study in Arabidopsis thaliana of natural variation in seed oil melting point: a widespread adaptive trait in plants. J Hered 107: 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, McConn M, James D Jr, Miquel M (1993) Mutants of Arabidopsis deficient in the synthesis of α-linolenate: biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J Biol Chem 268: 16345–16351 [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Browse J, Slack R (1983) The effects of temperature and oxygen on the rates of fatty acid synthesis and oleate desaturation in safflower (Carthamus tinctorius) seed. Biochim Biophys Acta 753: 145–152 [Google Scholar]
- Bryant FM, Munoz-Azcarate O, Kelly AA, Beaudoin F, Kurup S, Eastmond PJ (2016) ACYL-ACYL CARRIER PROTEIN DESATURASE2 and 3 are responsible for making omega-7 fatty acids in the Arabidopsis aleurone. Plant Physiol 172: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin DT. (1965) The effect of temperature on the oil content and fatty acid composition of the oils from several oilseed crops. Can J Bot 43: 63–69 [Google Scholar]
- Carver BF, Burton JW, Carter TE Jr, Wilson RF (1986) Response to environmental variation of soybean lines selected for altered unsaturated fatty acid composition. Crop Sci 26: 1176–1180 [Google Scholar]
- Covello PS, Reed DW (1996) Functional expression of the extraplastidial Arabidopsis thaliana oleate desaturase gene (FAD2) in Saccharomyces cerevisiae. Plant Physiol 111: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Chapital DC, Cary JW, Pepperman AB (2001) Chilling-sensitive, post-transcriptional regulation of a plant fatty acid desaturase expressed in yeast. Biochem Biophys Res Commun 282: 1019–1025 [DOI] [PubMed] [Google Scholar]
- Falcone DL, Ogas JP, Somerville CR (2004) Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL (2006) Mechanisms of temperature adaptation in poikilotherms. FEBS Lett 580: 5477–5483 [DOI] [PubMed] [Google Scholar]
- Harris HC, McWilliam JR, Mason WK (1978) Influence of temperature on oil content and composition of sunflower seed. Aust J Agric Res 29: 1203–1212 [Google Scholar]
- Harris P, James AT (1969) The effect of low temperatures on fatty acid biosynthesis in plants. Biochem J 112: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani-Pak K, Castellote M, Esch M, Hindle M, Lysenko A, Taubert J, Rawlings C (2016) Developing integrated crop knowledge networks to advance candidate gene discovery. Appl Transl Genomics 11: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppard EP, Kinney AJ, Stecca KL, Miao GH (1996) Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol 110: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ (2004) Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol 136: 3341–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Fuse T, Kawakami N, Kodama H, Iba K (2000) Temperature-dependent translational regulation of the ER omega-3 fatty acid desaturase gene in wheat root tips. Plant J 24: 805–813 [DOI] [PubMed] [Google Scholar]
- Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, et al. (2015) Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J 84: 1178–1191 [DOI] [PubMed] [Google Scholar]
- Hughes R, Spielman M, Schruff MC, Larson TR, Graham IA, Scott RJ (2008) Yield assessment of integument-led seed growth following targeted repair of auxin response factor 2. Plant Biotechnol J 6: 758–769 [DOI] [PubMed] [Google Scholar]
- Imprialou M, Kahles A, Steffen JB, Osborne EJ, Gan X, Lempe J, Bhomra A, Belfield EJ, Visscher A, Greenhalgh R, et al. (2016) Genomic rearrangements considered as quantitative traits. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargiotidou A, Deli D, Galanopoulou D, Tsaftaris A, Farmaki T (2008) Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J Exp Bot 59: 2043–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuu N, Gidda S, Shockey JM, Dyer JM, Mullen RT (2011) The N termini of Brassica and tung omega-3 fatty acid desaturases mediate proteasome-dependent protein degradation in plant cells. Plant Signal Behav 6: 422–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kim H, Shin JS, Chung CH, Ohlrogge JB, Suh MC (2006) Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol Genet Genomics 276: 351–368 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Kim JK, Shin JS, Suh MC (2007) The SebHLH transcription factor mediates trans-activation of the SeFAD2 gene promoter through binding to E- and G-box elements. Plant Mol Biol 64: 453–466 [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupke T, Hernández-Acosta P, Culiáñez-Macià FA (2003) 4′-Phosphopantetheine and coenzyme A biosynthesis in plants. J Biol Chem 278: 38229–38237 [DOI] [PubMed] [Google Scholar]
- Li Q, Zheng Q, Shen W, Cram D, Fowler DB, Wei Y, Zou J (2015) Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27: 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C. (2000) Adaptive evolution of seed oils in plants: accounting for the biogeographic distribution of saturated and unsaturated fatty acids in seed oils. Am Nat 156: 442–458 [DOI] [PubMed] [Google Scholar]
- Lou Y, Schwender J, Shanklin J (2014) FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J Biol Chem 289: 17996–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozinsky S, Yang H, Forseille L, Cook GR, Ramirez-Erosa I, Smith MA (2014) Characterization of an oleate 12-desaturase from Physaria fendleri and identification of 5'UTR introns in divergent FAD2 family genes. Plant Physiol Biochem 75: 114–122 [DOI] [PubMed] [Google Scholar]
- Martínez-Rivas JM, Sánchez-García A, Sicardo MD, García-Díaz MT, Mancha M (2003) Oxygen-independent temperature regulation of the microsomal oleate desaturase (FAD2) activity in developing sunflower (Helianthus annuus) seeds. Physiol Plant 117: 179–185 [Google Scholar]
- Martz F, Kiviniemi S, Palva TE, Sutinen ML (2006) Contribution of omega-3 fatty acid desaturase and 3-ketoacyl-ACP synthase II (KASII) genes in the modulation of glycerolipid fatty acid composition during cold acclimation in birch leaves. J Exp Bot 57: 897–909 [DOI] [PubMed] [Google Scholar]
- May P, Liao W, Wu Y, Shuai B, McCombie WR, Zhang MQ, Liu QA (2013) The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat Commun 4: 2145. [DOI] [PubMed] [Google Scholar]
- Mendes A, Kelly AA, van Erp H, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013) bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Miquel M, James D Jr, Dooner H, Browse J (1993) Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc Natl Acad Sci USA 90: 6208–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K (2000) Trienoic fatty acids and plant tolerance of high temperature. Science 287: 476–479 [DOI] [PubMed] [Google Scholar]
- Murata N, Ishizaki-Nishizawa O, Higashl S, Hayashl H, Tasaka Y, Nishida I (1992) Genetically engineered alteration in the chilling sensitivity of plants. Nature 356: 710–713 [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill CM, Morgan C, Hattori C, Brennan M, Rosas U, Tschoep H, Deng PX, Baker D, Wells R, Bancroft I (2012) Towards the genetic architecture of seed lipid biosynthesis and accumulation in Arabidopsis thaliana. Heredity (Edinb) 108: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Quin JB, Bourassa L, Zhang D, Shockey JM, Gidda SK, Fosnot S, Chapman KD, Mullen RT, Dyer JM (2010) Temperature-sensitive post-translational regulation of plant omega-3 fatty-acid desaturases is mediated by the endoplasmic reticulum-associated degradation pathway. J Biol Chem 285: 21781–21796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearcy RW. (1978) Effect of growth temperature on the fatty acid composition of the leaf lipids in Atriplex lentiformis (Torr.) Wats. Plant Physiol 61: 484–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Sánchez-García A, Gotor C, Romero LC, Martínez-Rivas JM, Mancha M (2007) Temperature-dependent endogenous oxygen concentration regulates microsomal oleate desaturase in developing sunflower seeds. J Exp Bot 58: 3171–3181 [DOI] [PubMed] [Google Scholar]
- Román Á, Andreu V, Hernández ML, Lagunas B, Picorel R, Martínez-Rivas JM, Alfonso M (2012) Contribution of the different omega-3 fatty acid desaturase genes to the cold response in soybean. J Exp Bot 63: 4973–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-García A, Mancha M, Heinz E, Martínez-Rivas JM (2004) Differential temperature regulation of three sunflower microsomal oleate desaturase (FAD2) isoforms overexpressed in Saccharomyces cerevisiae. Eur J Lipid Sci Technol 106: 583–590 [Google Scholar]
- Sanyal A, Linder CR (2012) Quantitative trait loci involved in regulating seed oil composition in Arabidopsis thaliana and their evolutionary implications. Theor Appl Genet 124: 723–738 [DOI] [PubMed] [Google Scholar]
- Sanyal A, Linder CR (2013) Plasticity and constraints on fatty acid composition in the phospholipids and triacylglycerols of Arabidopsis accessions grown at different temperatures. BMC Plant Biol 13: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SD, Zou J, Weselake RJ (2016) Abiotic factors influence plant storage lipid accumulation and composition. Plant Sci 243: 1–9 [DOI] [PubMed] [Google Scholar]
- Slack CR, Roughan PG (1978) Rapid temperature-induced changes in the fatty acid composition of certain lipids in developing linseed and soya-bean cotyledons. Biochem J 170: 437–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack CR, Roughan PG, Balasingham N (1978) Labelling of glycerolipids in the cotyledons of developing oilseeds by [1-14C]acetate and [2-3H]glycerol. Biochem J 170: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang GQ, Novitzky WP, Griffin HC, Huber SC, Dewey RE (2005) Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J 44: 433–446 [DOI] [PubMed] [Google Scholar]
- van Erp H, Kelly AA, Menard G, Eastmond PJ (2014) Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol 165: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CT, Xu YN (2010) The 5′ untranslated region of the FAD3 mRNA is required for its translational enhancement at low temperature in Arabidopsis roots. Plant Sci 179: 234–240 [Google Scholar]
- Wang T, Xing J, Liu X, Liu Z, Yao Y, Hu Z, Peng H, Xin M, Zhou DX, Zhang Y, et al. (2016) Histone acetyltransferase general control non-repressed protein 5 (GCN5) affects the fatty acid composition of Arabidopsis thaliana seeds by acetylating fatty acid desaturase3 (FAD3). Plant J 88: 794–808 [DOI] [PubMed] [Google Scholar]
- Willemot C, Hope HJ, Williams RJ, Michaud R (1977) Changes in fatty acid composition of winter wheat during frost hardening. Cryobiology 14: 87–93 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cao Z, Xu F, Huang Y, Chen M, Guo W, Zhou W, Zhu J, Meng J, Zou J, et al. (2012) Analysis of gene expression profiles of two near-isogenic lines differing at a QTL region affecting oil content at high temperatures during seed maturation in oilseed rape (Brassica napus L.). Theor Appl Genet 124: 515–531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.