Arbuscular mycorrhizal symbiosis compensates the transcriptional response of M. truncatula roots at low potassium level and activates specific mechanisms to tolerate long-term potassium deprivation.
Abstract
Arbuscular mycorrhizal (AM) associations enhance the phosphorous and nitrogen nutrition of host plants, but little is known about their role in potassium (K+) nutrition. Medicago truncatula plants were cocultured with the AM fungus Rhizophagus irregularis under high and low K+ regimes for 6 weeks. We determined how K+ deprivation affects plant development and mineral acquisition and how these negative effects are tempered by the AM colonization. The transcriptional response of AM roots under K+ deficiency was analyzed by whole-genome RNA sequencing. K+ deprivation decreased root biomass and external K+ uptake and modulated oxidative stress gene expression in M. truncatula roots. AM colonization induced specific transcriptional responses to K+ deprivation that seem to temper these negative effects. A gene network analysis revealed putative key regulators of these responses. This study confirmed that AM associations provide some tolerance to K+ deprivation to host plants, revealed that AM symbiosis modulates the expression of specific root genes to cope with this nutrient stress, and identified putative regulators participating in these tolerance mechanisms.
Potassium (K+) is a macronutrient required by all organisms for growth and development. In plants, K+ participates in many diverse processes, including plasma membrane polarization, growth, stomatal aperture regulation, and the acquisition of other nutrients (Wang and Wu, 2013). K+ represents 2% to 10% of the plant dry biomass, and its optimal cytoplasmic concentration for enzyme activities is around 100 to 200 mm (Leigh and Wyn Jones, 1984). Thus, a high cellular K+ concentration is vital for plants to maintain a wide range of physiological processes (Adams and Shin, 2014; Benito et al., 2014). Depending on soil type, the concentration of available K+ varies from 0.1 to 1 mm (Asher and Ozanne, 1967). Only two fractions of K+ are immediately accessible to plants: K+ in water solution and K+ in exchangeable form. These two fractions represent about 2.2% of the total amount of soil K+ (Zörb et al., 2014). This very low availability combined with the constitutive demand of plants leads to the formation of depletion zones near the root surface (Drew and Nye, 1969). Plants have developed efficient strategies to deal with limited K+ availability, including the establishment of symbiotic associations with microbes.
Arbuscular mycorrhizal (AM) fungi colonize the root cortex intercellularly and intracellularly (Brundrett, 2004; Harrison, 2005). These AM fungi belong to the fungal subphylum of Glomeromycotina and probably played a critical role in the emergence and evolution of land plants (Delaux et al., 2015; Spatafora et al., 2016). This AM symbiosis is the oldest mycorrhizal association and is found in about 65% of extant land plants, including many major crop species (Wang and Qiu, 2006). AM associations are characterized by the formation of arbuscules in the cortical cells of host plants, which are the site of nutrient exchange between the plant and the fungus (Smith and Read, 2008). The improvement of plant nutrient acquisition through AM associations becomes particularly important when hydromineral resources are scarce. In return for the nutrients provided by AM fungi, host plants provide up to 20% to 25% of their photosynthesis-derived carbohydrates to their symbionts (López et al., 2008).
The improvement of plant nutrition through AM symbiosis and the molecular basis of nutrient transfer are well characterized for phosphorus, nitrogen, and sulfur (Govindarajulu et al., 2005; Javot et al., 2007; Jin et al., 2012; Casieri et al., 2013; Courty et al., 2016; Garcia et al., 2016). However, the role of mycorrhizal symbioses in plant K+ nutrition is still understudied and misunderstood (Garcia and Zimmermann, 2014). A small number of studies reported AM fungal mediation of plant K+ nutrition. The assessment of K+ distribution in mycorrhizal plants using particle-induced x-ray emission experiments revealed a higher K+ concentration in root sections of Aster tripolium colonized by Rhizophagus irregularis than in noninoculated plants (Scheloske et al., 2004). The same trend of K+ enrichment for plants mycorrhized by various AM fungi also was observed in maize (Zea mays) root steles (Kaldorf et al., 1999), Pelargonium peltatum shoots (Perner et al., 2007), Lactuca sativa leaves (Baslam et al., 2013), and wheat (Triticum aestivum) stems (Oliveira et al., 2016). The expression of three K+ transport systems likely involved in phloem loading/unloading (ZmAKT2), xylem release (ZmSKOR), and sodium/K+ homeostasis (ZmSOS1) was differentially regulated in AM maize roots (Estrada et al., 2013). These findings suggested that AM symbioses could play a significant role in plant K+ nutrition by the reorganization of molecular responses.
If K+ amendments are not used frequently, long-term K+ deprivation can result in irreversible damage to the crop, including poor pest tolerance and significant yield losses. Soils from many parts of the world are increasingly K+ deficient, reinforcing the need for understanding how plants can cope with K+ deprivation (Moody and Bell, 2006; Römheld and Kirkby, 2010; Zörb et al., 2014). However, most studies have only investigated the short-term molecular plant responses to K+ deprivation, and not in the context of AM symbiosis. The transcriptional responses of plant roots to K+ deprivation were studied in various species, including Arabidopsis (Arabidopsis thaliana; Armengaud et al., 2004), rice (Oryza sativa; Ma et al., 2012; Shankar et al., 2013), wheat (Ruan et al., 2015), and sugarcane (Saccharum officinarum; Zeng et al., 2015). However, in all these studies, K+ was completely absent from the medium and the plants were analyzed after a relatively short period of time (1–2 weeks). Following these drastic and short-term treatments, the up-regulation of high-affinity K+ transporters often was reported. This response is a rapid and transient strategy but probably not a sustainable one if the stress is prolonged. The transcriptional response of plant roots colonized by an AM fungus during K+ deprivation is still unknown.
To address this question, we examined, to our knowledge for the first time, the effect of long-term K+ deprivation on the development, nutrition, gene expression, and AM formation using the model legume Medicago truncatula. To evaluate the impact of AM symbiosis on plant adaptation to long-term K+ deprivation, we inoculated M. truncatula plants with the AM fungus R. irregularis and watered them with high- or low-K+ solution. Plant biomass and the tissue content of various ions were determined after 6 weeks. The transcriptional profile of AM roots under K+ deprivation was investigated using RNA sequencing (RNA-seq) and revealed mycorrhiza-specific responses under K+ deprivation. Finally, a network-based prioritization analysis revealed putative regulators controlling AM symbiosis and K+ deprivation in M. truncatula.
RESULTS
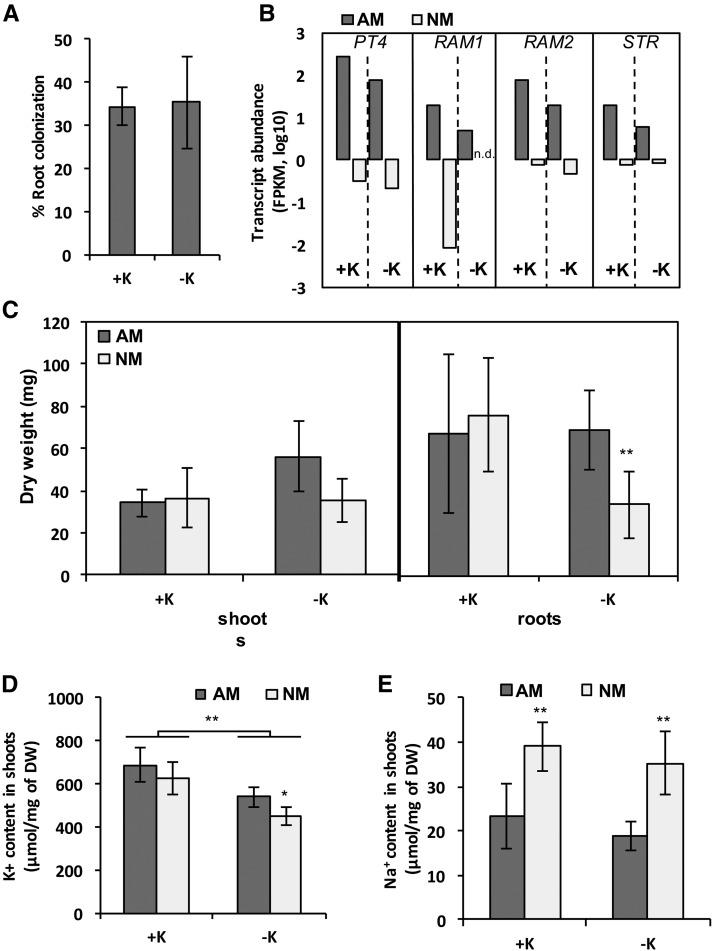
AM Symbiosis Improves the K+ Acquisition of M. truncatula
The impact of AM associations on K+ nutrition is understudied and, in particular, was never investigated in legumes. M. truncatula plants were inoculated with spores of R. irregularis (AM) or were kept nonmycorrhized (NM) for 6 weeks. All plants were watered with either high- or low-K+ solution as described previously, and various physiological parameters were measured (Fig. 1). The quantification of fungal colonization revealed that around 30% of plant roots were colonized under both K+-sufficient and K+-deficient conditions, indicating that K+ availability did not affect the establishment of AM symbiosis (Fig. 1A). The expression of four AM-specific marker genes identified by RNA-seq was determined in plants growing at high or low K+. Transcripts coding for PHOSPHATE TRANSPORTER4 (PT4), REDUCED ARBUSCULAR MYCORRHIZA1 (RAM1) and RAM2, and STUNTED ARBUSCULE (STR) were up-regulated only in AM roots at high and low K+ levels, indicating that the symbiosis is functional (Fig. 1B). Interestingly, the up-regulation of these transcripts was more pronounced in M. truncatula roots at high K+ than at low K+, suggesting a slight alteration of the AM symbiosis under K+ deprivation.
Figure 1.
Impact of AM symbiosis on plant fitness under K+ deprivation. A and B, The rate of fungal colonization (A) and the fragments per kilobase of transcript per million mapped reads (FPKM) of PT4, RAM1, RAM2, and STR (B) were measured on M. truncatula roots grown under high-K+ (+K) or low-K+ (−K) conditions after 6 weeks of coculture using the gridline intersection method. C, Shoot and root dry weight (DW) were measured for AM and NM plants grown under +K and −K conditions. D and E, K+ (D) and sodium (E) contents were determined by ICP-OES analysis in shoots of AM and NM plants under +K and −K conditions. Significant differences were obtained using Student’s t test between AM and NM plants (C–E) and between −K and +K conditions (C; *, P < 0.05 and **, P < 0.01). n = 6. Experiments were replicated three times independently.
Although no significant difference was observed in shoot dry biomass between AM and NM plants at high and low K+, AM plants displayed a significantly higher root dry biomass than NM plants under K+ deprivation (Fig. 1C). Sodium and K+ contents in plant roots and shoots were determined by inductively coupled plasma optical emission spectrometry (ICP-OES). The impossibility of separating the fungus from colonized tissues resulted in noncomparable ion content determination between AM and NM plant roots. As a consequence, only ion contents in the shoot are presented (Fig. 1, D and E). Both AM and NM plants watered with low-K+ solution displayed a significant lower shoot K+ content than those grown under K+-sufficient conditions (Fig. 1D). A significant reduction of K+ content was observed in NM plants in comparison with AM plants exclusively under K+ limitation (Fig. 1D).
Together, these results indicate that the mycorrhizal status helped M. truncatula deal efficiently with long-term K+ deprivation. Interestingly, the sodium content was twice higher in the shoot of NM plants than AM plants and whatever the external K+ availability, suggesting a buffering role of the fungus in preventing the accumulation of sodium cations into the plant (Fig. 1E).
AM Association Reverses Most Transcriptional Responses to K+ Deprivation and Reduces the Production of Reactive Oxygen Species in M. truncatula Roots
RNA-seq was performed on roots from AM and NM plants grown under a low- or high-K+ regime. RNAs from three to four biological replicates composed of four plants each were sequenced, and the reliability of our data set was analyzed using principal component analyses and multidimensional scaling (Supplemental Figs. S1 and S2). K-means clustering also was performed to identify transcripts following the same expression profile along each condition: AM or NM plants at high (AM+K or NM+K) or low (AM−K or NM−K) K+ levels. This analysis revealed groups of genes with condition-specific responses: for example, two clusters of genes that were mainly up-regulated (clusters 1 and 21) and two that were mainly down-regulated (clusters 8 and 20) in the AM−K condition (Supplemental Fig. S3; Supplemental Table S4).
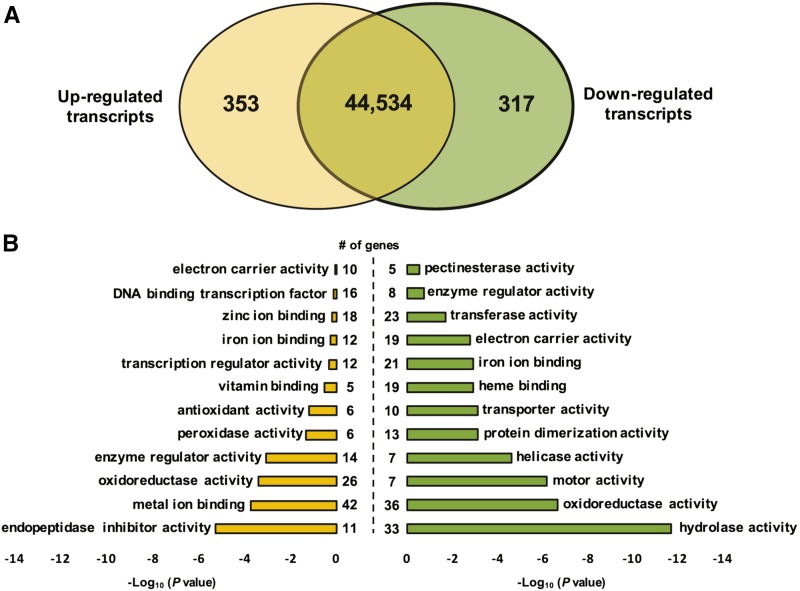
The first step was to identify transcripts specifically regulated at a low-K+ level in NM plants. Taking the NM+K condition as a control, DESeq analysis allowed the identification of 353 and 317 up- and down-regulated genes, respectively, in M. truncatula roots under K+ deprivation (Fig. 2A; Supplemental Table S1). Gene Ontology (GO) enrichment analysis identified a very significant overrepresentation of metal-binding transcripts, particularly among up-regulated transcripts (Fig. 2B). Of these proteins annotated as metal binding, one-third encoded peroxidases (six and two were up- and down-regulated, respectively) or cytochrome P450 enzymes (two and 15 were up- and down-regulated, respectively). These proteins are known to play an important role in oxidative stress responses in plants, in particular under K+ deprivation (Apel and Hirt, 2004; Hernandez et al., 2012; Wang et al., 2012). Overall, 62 transcripts encoding proteins involved in oxidative stress were differentially regulated under K+ deprivation (Supplemental Table S2), and both up- and down-regulated transcript groups were significantly enriched for oxidoreductases (Fig. 2B). Also, two-thirds of the differentially expressed hydrolases were down-regulated (Fig. 2B; Supplemental Table S3). Overall, our data revealed that many enzymes were differentially regulated in M. truncatula roots under K+ deprivation, suggesting that posttranslational regulations play an important role in the long-term adaptation to K+ deprivation.
Figure 2.
Transcriptional profiling of M. truncatula roots under K+ deprivation. A, Venn diagram showing the number of M. truncatula root genes up-regulated (yellow) and down-regulated (green) in response to K+ deprivation, based on q < 0.05 and a 1.5-fold change cutoff threshold. B, Significantly enriched GO molecular function terms for up-regulated (yellow) and down-regulated (green) genes analyzed with the AgriGO Web-based tool (http://bioinfo.cau.edu.cn/agriGO/analysis.php).
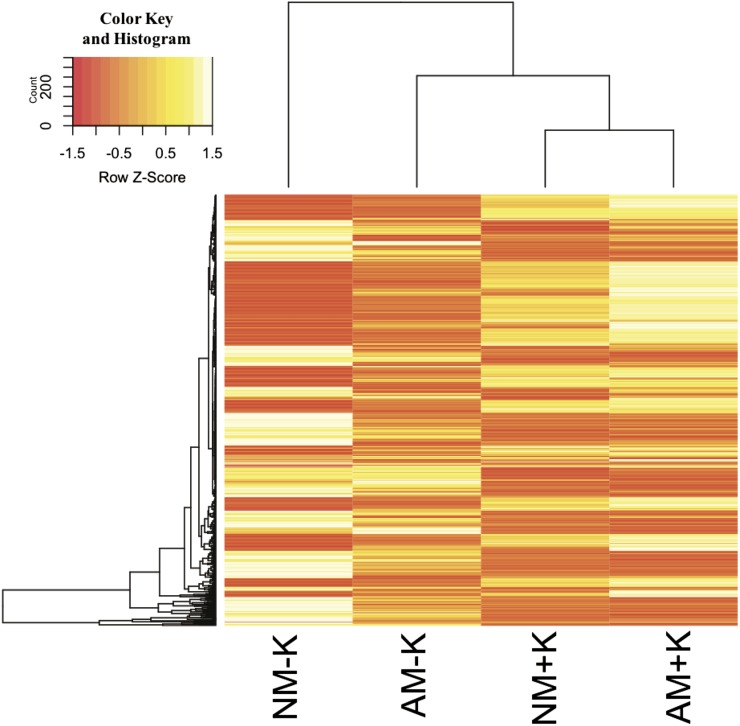
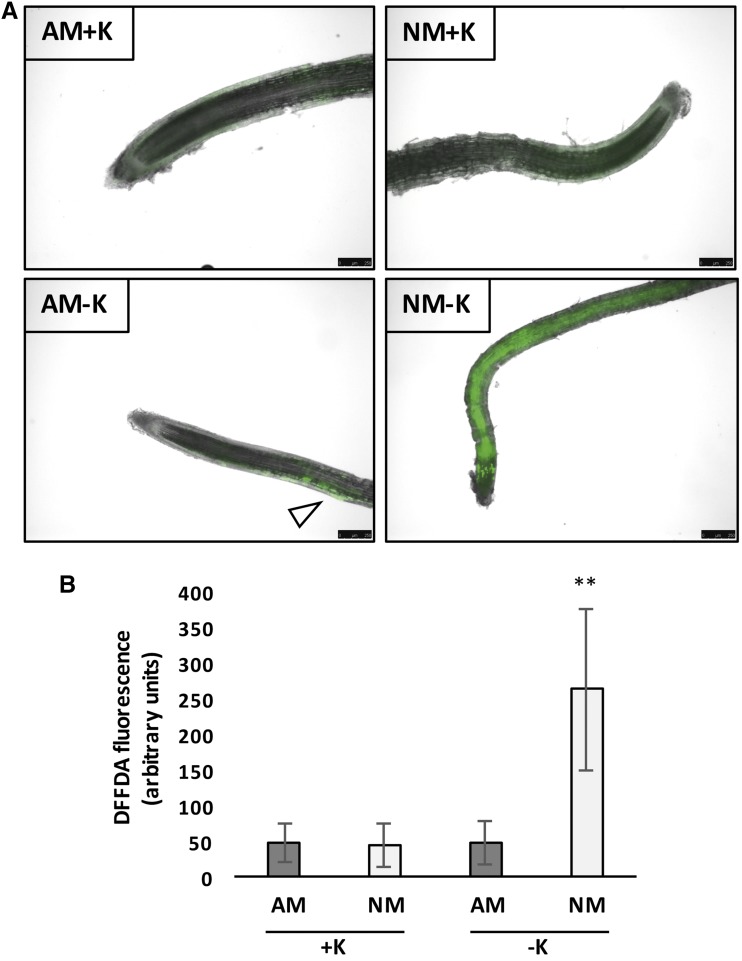
To evaluate how AM symbiosis affects the transcriptional response of M. truncatula to K+ deprivation, the expression of transcripts regulated in NM−K plants was investigated in AM−K plants. Investigating the expression of genes in AM plants at high and low K+ levels revealed that the expression of two-thirds of them (387 transcripts) was altered in AM plants under K+ deprivation (Fig. 3). As mentioned above, many of these transcripts code for proteins likely involved in oxidative stress responses. Thus, the production of reactive oxygen species (ROS) in M. truncatula roots colonized or not by R. irregularis at high or low K+ was determined using carboxy-2,7-difluorodihydrofluorescein diacetate (carboxy-H2DFFDA; Fig. 4). In NM plants, K+ deprivation significantly enhanced the production of ROS in roots. However, this effect was reduced significantly and visible in only a few cells when plants were mycorrhized (Fig. 4).
Figure 3.
Clustering and heat map of the expression values for differentially expressed genes (DEGs) in NM plants under K+ deprivation. The heat map displays DEGs identified previously in NM plants at low K+ (Fig. 2; Supplemental Table S1). An important part of the transcriptional responses of M. truncatula roots under K+ deprivation was altered during mycorrhizal association. White and red areas indicate higher and lower expression values, respectively.
Figure 4.
Impact of AM symbiosis and K+ deprivation on the production of ROS in M. truncatula. A, ROS fluorescence images are shown in AM and NM plants at high (+K) and low (−K) K+ levels. Green pseudocolor indicates ROS production. Bars = 250 μm. B, Quantified data from three representative images taken from six plants per condition. ROS were visualized by staining the roots of plants inoculated and watered with high- or low-K+ solutions for 6 weeks with 20 μm carboxy-H2DFFDA (DFFDA). **, P < 0.01.
Altogether, these observations revealed that AM symbiosis compensates the transcriptional response of M. truncatula roots and prevents the production of ROS in roots under K+-limiting conditions.
Specific Transcriptional Responses Are Triggered by AM Association under K+ Deprivation in M. truncatula Roots
Using NM+K plants as a control, DEGs regulated in AM+K, AM−K, and NM−K plants were identified to highlight specific and shared expression patterns in each condition (Supplemental Fig. S4; Supplemental Table S5). Although 230 transcripts were similarly and significantly regulated in both AM+K and AM−K plants, revealing molecular players that were involved in AM symbiosis independently of extracellular K+ availability, including various transport systems, 511 transcripts were regulated specifically in plant roots under AM−K conditions (Supplemental Fig. S5; Supplemental Table S5). A GO enrichment analysis revealed an overrepresentation of genes involved in redox homeostasis, plant cell wall formation, and responses to oxidative stress (Fig. 5A). Interestingly, five transcripts encoding for transport systems were up-regulated in AM plants under K+ deprivation: an ortholog of the plasma membrane K+/H+ exchanger AtCHX20 from Arabidopsis (Medtr7g099800.1), a putative major facilitator superfamily (MFS) transporter (Medtr2g081930.1), a mitochondrial phosphate transporter (Medtr6g033280.1), and vacuolar malate (Medtr4g133230.1) and ion (Medtr4g094332.1) transporters (Fig. 5B; Supplemental Fig. S6). Among the 30 most DEGs in AM−K plants (Table I), a clade A type 2C protein phosphatase (Medtr5g009370.1) was expressed and could be involved in the posttranslational regulation of K+ transport systems. These results suggest that AM symbiosis activates specific mechanisms to tolerate long-term K+ deprivation, including transport mechanisms to likely facilitate the acquisition of K+.
Figure 5.
GO enrichment and heat map of the expression values for DEGs in transport activity in AM plants under K+ deprivation. A, Significantly enriched GO terms in AM−K plants specifically in comparison with NM+K plants. Dashed arrows indicate that all the GO levels are not represented. B, The heat map displays DEGs in transport activity specifically up-regulated in AM−K plants. White and red areas indicate higher and lower expression values, respectively.
Table I. List of the top 30 transcripts specifically up-regulated in mycorrhizal plants under K+ deprivation (AM−K condition).
| Gene Identifier | Locus Name in Mt4.0v1 | Mean Read No. | Log2 Ratio, AM−K Versus NM−K |
|---|---|---|---|
| Medtr5g0113400.1 | Subtilase family protein | 11.207 | 8.4835 |
| Medtr1g0907370.1 | OPC-8:0 CoA ligase1 | 4.84641 | 6.855 |
| Medtr1g0907070.1 | SKU5 similar3 | 5.20123 | 6.31504 |
| Medtr2g0635600.1 | HSP20-like chaperones superfamily protein | 1.07509 | 2.86477 |
| Medtr1g0872000.1 | Adenine nucleotide α-hydrolase-like superfamily protein | 1.64562 | 2.64534 |
| Medtr4g0112300.1 | Late embryogenesis abundant protein (LEA) family protein | 5.10893 | 2.4046 |
| Medtr3g4669800.1 | AGAMOUS-like80 | 1.85953 | 2.27527 |
| Medtr8g0429000.1 | Root hair specific12 | 0.816833 | 2.14694 |
| Medtr4g0291900.1 | Peroxidase superfamily protein | 3.51445 | 2.10615 |
| Medtr3g0888450.1 | Thiamine diphosphate-binding fold superfamily protein | 0.939511 | 2.02649 |
| Medtr7g1082500.1 | Thioredoxin superfamily protein | 51.2384 | 1.97414 |
| Medtr6g0883200.1 | Xyloglucan endotransglucosylase/hydrolase26 | 0.766958 | 1.87461 |
| Medtr5g0258000.1 | Unknown protein | 3.00168 | 1.75596 |
| Medtr7g0920900.1 | Unknown protein | 1.44917 | 1.73762 |
| Medtr1g0834400.1 | Dormancy/auxin-associated family protein | 22.4113 | 1.72544 |
| Medtr4g1024500.1 | Expansin A7 | 0.911692 | 1.69244 |
| Medtr1g0237000.1 | Cytochrome P450, family 83, subfamily B, polypeptide 1 | 1.00486 | 1.68266 |
| Medtr3g0850200.1 | Protein of unknown function (DUF3339) | 5.66561 | 1.65014 |
| Medtr7g1012700.1 | Unknown protein | 4.36525 | 1.64898 |
| Medtr6g0846400.1 | Cold-regulated47 | 4.58727 | 1.6426 |
| Medtr1g0987600.1 | Unknown protein | 2.99537 | 1.6257 |
| Medtr4g0990100.1 | Plant invertase/pectin methylesterase inhibitor superfamily protein | 2.63717 | 1.61548 |
| Medtr7g1099200.1 | Galactinol synthase1 | 0.0000541 | 1.60318 |
| Medtr3g1029700.1 | Unknown protein | 1.68067 | 1.56649 |
| Medtr1g1108700.1 | B-box-type zinc finger protein with CCT domain | 0.713362 | 1.55694 |
| Medtr4g0551700.1 | HXXXD-type acyltransferase family protein | 10.5731 | 1.55133 |
| Medtr4g0639400.1 | Protein kinase1B | 1.34305 | 1.54197 |
| Medtr2g4377700.1 | Peroxidase superfamily protein | 3.42251 | 1.53209 |
| Medtr5g0093700.1 | Highly ABA-induced PP2C gene2 | 1.25488 | 1.5162 |
| Medtr2g0702000.1 | Glutathione S-transferase TAU25 | 1.10787 | 1.4356 |
Identification of Putative Regulators for the Long-Term Adaptation of M. truncatula to K+ Deprivation in AM and NM Roots
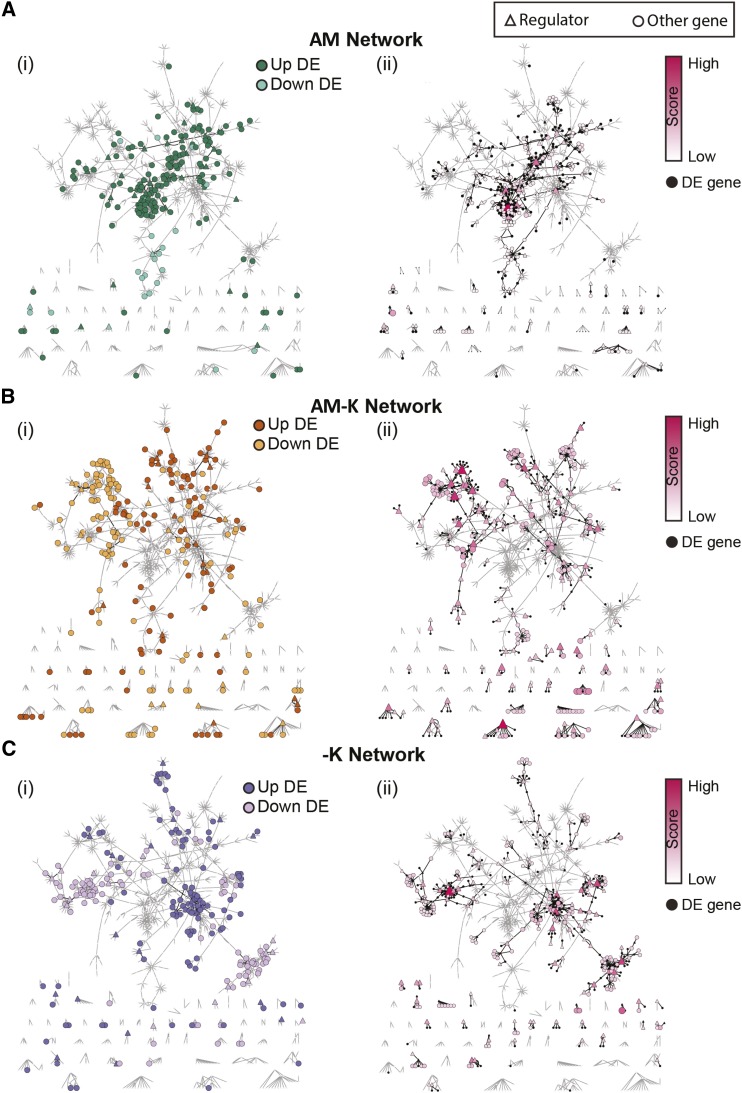
We sought to identify regulatory networks associated with the AM and K+ DEG sets described previously (Supplemental Fig. S4) by comparing them with our global M. truncatula regulatory network (Marx et al., 2016). This network was inferred using the MERLIN algorithm (Roy et al., 2013). We defined five condition-specific gene sets, each corresponding to a bar in Supplemental Figure S4 and Supplemental Table S5. To assess the connectivity patterns, we overlaid the gene sets on the regulatory network (Supplemental Methods S1). Figure 6 shows regulatory network connections involving the DEGs and their prioritized network neighbors, summarized into three major categories: genes differentially expressed in AM roots regardless of K+ level (Fig. 6Ai), genes specifically differentially expressed in the AM−K condition (Fig. 6Bi), and genes differentially expressed under K+ deprivation regardless of AM status (Fig. 6Ci). Genes within each gene set were statistically enriched for direct interactions compared with randomly chosen genes, controlling for the degree (Z ≥ 7.09; Supplemental Table S6). Within each condition, genes with the same direction of expression change (up- or down-regulated) represented distinct network components (Fig. 6, i images) and had significantly shorter network distances than pairs of genes with opposite expression patterns (Mann-Whitney rank-sum test P < 0.001; Supplemental Fig. S7). This result indicates that genes that were induced represent distinct components of a pathway compared with genes that were repressed. Distinct regulatory systems are likely involved in controlling the up- and down-regulated components of each DEG set.
Figure 6.
Network analysis of DEGs. Shown are inferred regulatory interactions spanning DEGs and genes prioritized by the diffusion kernel. In each image, the same network is shown (i.e. same edges and nodes), but they are colored differently to highlight DEG sets or top prioritized genes. A, Network for AM plants, regardless of K+ level (bars I and II in Supplemental Fig. S4). i, Up- and down-regulated DEGs as dark and light green nodes. ii, Genes prioritized by the diffusion kernel are colored from white to red; size and red intensity are proportional to the prioritization score. B, Network for the AM−K condition (bar III in Supplemental Fig. S4). i, Up- and down-regulated DEGs as dark and light orange. ii, Prioritized genes. C, Network for K+ deprivation (−K), regardless of mycorrhizal status (bars IV and V in Supplemental Fig. S4). i, Up- and down-regulated DEGs as dark and light purple. ii, Prioritized genes. Regulators (transcription factors and signaling proteins) are indicated with triangles, whereas all other genes are represented as circles. Small white nodes are genes (DEG or prioritized) in other conditions. Black edges are between genes relevant to the condition of interest, and gray edges are between genes from other conditions. For details, see Supplemental Methods S1, Supplemental Figure S8, and Supplemental Tables S1 and S2.
Two complementary network-based approaches were used to identify important regulatory genes associated with responses to AM and low-K+ conditions: diffusion kernel-based prioritization and statistical enrichment of targets of regulators in DEG sets. While the diffusion kernel approach can prioritize any gene in the network, the gene set enrichment analysis focuses on regulatory proteins such as transcription factors and signaling proteins. The diffusion kernel-based approach identified several candidate genes from AM roots putatively involved in the response to K+ deprivation (Fig. 6Bii; Table II). For example, among the top 25 predicted genes for the AM−K DEG set were nine kinases, including a calcineurin B-like (CBL)-interacting kinase (Medtr2g105010.1) and an ethylene response factor (Medtr4g078710.1; Table II). Full prioritization results for all gene sets are available in Supplemental Table S7.
Table II. Top prioritized genes for AM plants in K+ deprivation.
Shown are the top 25 prioritized genes for the AM−K DEG set. Prioritization (Rank and Score) columns give the rank and score from the diffusion kernel analysis. Target Enrichment indicates whether a prioritized gene in this list also has target enrichment in a DEG set; nonregulators are designated NA in this column.
| Gene Identifier | Locus Name in Mt4.0v1 | Rank (AM−K) | Score (AM−K) | Target Enrichment |
|---|---|---|---|---|
| Medtr3g115620.1 | Wuschel-related homeobox protein | 1 | 0.634459 | AM−K, down |
| Medtr7g005400.1 | Somatic embryogenesis receptor kinase-like protein | 2 | 0.578335 | AM−K, down |
| Medtr2g087090.1 | Ser/Thr-kinase Nek4 | 3 | 0.497066 | AM−K, down |
| Medtr2g089440.1 | S-locus lectin kinase family protein | 4 | 0.454545 | |
| Medtr4g078710.1 | Ethylene response factor | 5 | 0.396948 | |
| Medtr4g058015.1 | Electron transporter, putative | 6 | 0.389796 | AM−K, down |
| Medtr1g080210.1 | Light-dependent short-hypocotyl protein | 7 | 0.389508 | AM−K, down |
| Medtr1g097580.1 | LRR receptor-like kinase | 8 | 0.369039 | |
| Medtr5g014640.1 | bHLH DNA-binding family protein | 9 | 0.352642 | |
| Medtr4g133938.1 | Nuclear transcription factor Y protein | 10 | 0.328559 | AM−K, up |
| Medtr2g105010.1 | CBL-interacting kinase | 11 | 0.326467 | −K, up |
| Medtr1g107460.1 | LRR receptor-like kinase family protein | 12 | 0.321412 | |
| Medtr3g115500.1 | Receptor Ser/Thr kinase | 12 | 0.321412 | |
| Medtr5g012490.1 | Eukaryotic aspartyl protease family protein | 12 | 0.321412 | |
| Medtr4g128990.1 | Receptor-like kinase | 15 | 0.304361 | AM−K, down |
| Medtr7g084000.1 | Light-regulated protein, putative | 16 | 0.298936 | AM−K, up |
| Medtr3g097150.1 | Aspartic proteinase nepenthesin | 17 | 0.29324 | AM−K, down |
| Medtr5g014520.1 | bHLH DNA-binding family protein | 18 | 0.291187 | |
| Medtr3g069590.1 | RHO guanyl-nucleotide exchange factor | 19 | 0.28769 | AM−K, down |
| Medtr4g117040.1 | Cys-rich RLK (receptor-like kinase) protein | 20 | 0.283186 | |
| Medtr2g087390.1 | FAF-like protein | 21 | 0.279036 | NA |
| Medtr2g087430.1 | DUF3049 family protein | 21 | 0.279036 | NA |
| Medtr4g068780.1 | CASP-like protein | 21 | 0.279036 | NA |
| Medtr8g027040.1 | Cytochrome P450 family 78 protein | 21 | 0.279036 | NA |
| Medtr8g008820.1 | Receptor-like kinase plant | 25 | 0.273366 |
Statistical enrichment of regulatory targets in the DEG sets identified 56 putative regulators (Supplemental Table S7; Supplemental Fig. S8; Supplemental Methods S1) that included both new and known proteins such as MYB, BHLH, and AP2/ERF transcription factors as well as chromatin-modifying enzymes. For example, targets from a CBL-interacting kinase (Medtr2g105010.1; nine targets), a Cys desulfurase (Medtr8g093560.1; seven targets), and a GATA transcription factor (Medtr3g109760.1; 24 targets) were enriched for up-regulated genes in response to K+ deprivation in both AM−K and NM−K conditions, suggesting their involvement in the tolerance to low-K+ stress. Similarly, targets from a dehydration-responsive element-binding protein (Medtr1g019110.1; six targets), a C2H2-type zinc finger protein (Medtr3g102980.1; three targets), and a leucine-rich repeat (LRR) receptor-like kinase (Medtr8g014970.1; 10 targets) were specifically enriched for up-regulated genes in the AM−K condition. Finally, 29 and 17 targets from a cyclin-dependent kinase (Medtr8g092290.1) and a Tyr kinase (Medtr7g116650.1), respectively, were enriched for up-regulated genes in AM roots regardless of extracellular K+ level, suggesting a major role of these proteins in the regulation of AM symbiosis.
Interestingly, the regulators identified by both methods were consistent with each other. The 56 regulators identified by the enrichment analysis received diffusion kernel percentile ranks of 95 or greater compared with all ranked transcription factors and signaling proteins in the regulatory network. Each method also was able to identify some unique prioritized genes that the other would miss or rank lower. Overall, prioritization and regulator enrichment approaches revealed promising candidates for further functional analyses of the regulatory network controlling responses to changes in AM fungi and K+ availability in M. truncatula.
DISCUSSION
K+ Deprivation Affects Growth, K+ Acquisition, and Gene Expression in M. truncatula Roots
K+ is an essential macronutrient for plants. Its limitation contributes to slowing down many vital processes. Plants elicit rapid responses to strong K+ deprivation by inducing plasma membrane hyperpolarization and the up-regulation of high-affinity transporters (Schachtman and Shin, 2007). Prolonged K+ deficiency has been reported previously to limit root development (Armengaud et al., 2004; Shin and Schachtman, 2004; Kellermeier et al., 2013), which was observed in our experiments (Fig. 1). Transcriptional analysis of plants facing short-term and extreme K+ deprivation revealed an increase in the expression of K+ transporters and channels from the HAK, HKT, and AKT families (Armengaud et al., 2004; Ma et al., 2012; Shankar et al., 2013; Ruan et al., 2015; Zeng et al., 2015). In our study, we analyzed the impact of long-term K+ deprivation in the model legume M. truncatula. In our case, no K+ transport system was differentially regulated in response to long-term K+ deprivation. Because the goal of our study was also to evaluate how a prolonged low-K+ treatment impacts AM symbiosis, and reciprocally how the AM status alters the plant responses to K+ deprivation, the stress condition was 0.05 mm external K+ over 6 weeks. This treatment was obviously very different from that in previous studies in which short and extreme K+ stresses were applied. Therefore, our results suggest that the up-regulation of HAK, HKT, and AKT transport systems observed in other plant species is probably a rapid and transient response to an intense stress.
It is well established that K+ deficiency contributes to the accumulation of ROS in plant roots, leading to a general oxidative stress (Shin and Schachtman, 2004; Kim et al., 2010; Hernandez et al., 2012). Long-term K+ deprivation in M. truncatula resulted in the differential expression of many transcripts related to oxidative stress, including peroxidases, cytochrome P450 enzymes, and reductases. Most of the peroxidases were up-regulated under K+ deficiency in M. truncatula roots. Interestingly, Kim et al. (2010) hypothesized that the up-regulation of peroxidases could be the origin of ROS production observed in Arabidopsis roots. The accumulation of ROS production in M. truncatula roots detected under long-term K+ deprivation supports this view and validates the in silico prediction we made with our RNA-seq approach. Overall, many enzymes were differentially regulated in stressed plants, suggesting that posttranscriptional mechanisms may be involved mainly in the tolerance of K+ deprivation (Schachtman and Shin, 2007).
AM Symbiosis Modulates the Plant Responses to K+ Deprivation
The role of endomycorrhizal and ectomycorrhizal associations in plant K+ nutrition has been debated for decades. Although some studies described an improvement of K+ acquisition in plants colonized by mycorrhizal fungi, others presented apparently contradictory results (for review, see Garcia and Zimmermann, 2014). The transport of nutrients from the external medium to the host plant through mycorrhizal structures has been investigated using radioactive isotopes (He et al., 2009; Smith et al., 2011). Because K+ isotopes are not stable enough to track K+ movement from the soil to the mycorrhizal plants, some studies have used rubidium as an analog tracer (Rygiewicz and Bledsoe, 1984; Hawkes and Casper, 2002). Further experiments will be needed in M. truncatula using rubidium to evaluate the actual transport of K+ from the soil to the mycorrhizal plants. However, in a previous publication on ectomycorrhizae, we demonstrated an improvement of maritime pine (Pinus pinaster) K+ nutrition when colonized by an ectomycorrhizal fungus only under K+ deprivation (Garcia et al., 2014). In this study, the shoot K+ content of M. truncatula also was significantly higher in AM plants compared with NM plants but only at low K+ levels and after 6 weeks of coculture. This effect should be investigated in other plant species. We can assume that under K+-sufficient conditions, host plants can acquire external K+ by themselves. However, when K+ ions in solution have been limiting for a relatively long period of time, AM fungi may help their hosts to acquire K+ from the soil. Our analysis revealed that M. truncatula plants colonized by R. irregularis displayed increases of biomass and K+ ion content as well as specific transcriptional responses to the low-K+ regime. These results confirmed that AM symbiosis was able to provide specific adaptation mechanisms to the host plant to tolerate long-term K+ deprivation.
ROS have been detected in legume roots colonized by both AM fungi and nitrogen-fixing bacteria, particularly in colonized cells (Salzer et al., 1999; Fester and Hause, 2005; Puppo et al., 2013). Moreover, some NADPH oxidase-encoding genes were recently found to be up-regulated in colonized cortical cells of M. truncatula roots, suggesting their role in arbuscule development (Belmondo et al., 2016a, 2016b). Our results revealed that the two-thirds of M. truncatula root transcriptional responses to low K+ were absent in mycorrhizal plants, including many enzymes putatively involved in ROS production. The ROS accumulation observed in NM roots was correlated with the external K+ availability but not with the AM symbiosis, suggesting specific ROS production in response to K+ deprivation. This accumulation was reduced significantly in AM plants at low K+, indicating that AM symbiosis helped M. truncatula to cope with long-term K+ deprivation.
Also, NM plants accumulated much more sodium in shoots than AM plants. Sodium can be toxic to the cells at a high level. By preventing its accumulation, AM symbiosis may protect M. truncatula from salt stress by buffering the uptake of sodium. Although the role of AM associations on salt tolerance was suggested in other plants (e.g. basil [Ocimum basilicum; Zuccarini and Okurowska, 2008], olive [Olea europaea; Porras-Soriano et al., 2009], and maize [Estrada et al., 2013]), further experiments will be needed to unravel the molecular mechanisms of this adaptation in M. truncatula.
The uptake of nutrients from inner symbiotic structures requires the specific expression and regulation of plasma membrane transport systems in colonized cortical cells (Casieri et al., 2013; Garcia et al., 2016). Some plant transporters were described to transport phosphorus or nitrogen from AM fungi to plant cells in arbuscules (Harrison et al., 2002; Breuillin-Sessoms et al., 2015). One study in the legume Lotus japonicus reported a 44-fold up-regulation of a K+ transporter in AM roots compared with NM roots (Guether et al., 2009). However, in transcriptome studies of mycorrhizal roots from M. truncatula, no K+ transporter was ever found up-regulated in AM roots under the standard K+ regime (Gomez et al., 2009; Gaude et al., 2012; this study). In our study, we found that some genes encoding putative transporters were up-regulated in mycorrhizal plants under K+ deprivation, particularly a putative K+/H+ exchanger (CHX; Medtr7g099800.1). A close homolog of this CHX protein in Arabidopsis was found expressed in guard cells (Padmanaban et al., 2007). Other members of the CHX family were regulated in starved K+ roots, such as AtCHX17 (Cellier et al., 2004). As a consequence, further analyses will be required in M. truncatula to determine if and how this CHX can transport K+ during AM symbiosis under long-term K+ deprivation.
Network Analysis Identified Putative Regulators Controlling the Tolerance of Mycorrhizal Roots to K+ Deprivation
The MERLIN algorithm (Roy et al., 2013) was used to infer an M. truncatula regulatory network from publicly available gene expression data representing a wide array of conditions and tissues. Such a network analysis allows us to identify regulatory connections between DEGs but also to predict which specific regulators (signaling proteins and transcription factors) may control the expression of genes of interest. In our study, this approach identified differentially regulated genes and prioritized putative regulators that could be involved in controlling the tolerance of AM plants to K+ deprivation. Among the top 30 up-regulated genes in AM−K roots, a clade A type 2C protein phosphatase (Medtr5g009370.1) was identified. This gene encodes an ortholog of AtPP2CA in Arabidopsis involved in the regulation of the weak-rectifying K+ channel AKT2 (Chérel et al., 2002) as well as of the uptake and efflux Shaker K+ channels AKT1 and GORK, respectively (Lee et al., 2007; Lan et al., 2011; Lefoulon et al., 2016). Both prioritization and target enrichment analyses suggested the importance of a CBL-interacting kinase (Medtr2g105010.1) in AM−K plant roots. CBL-interacting kinases are major regulators of nutrient transporters, abscisic acid responses, and K+ homeostasis (D’Angelo et al., 2006; Ho et al., 2009; Hashimoto et al., 2012; Liu et al., 2013; Ragel et al., 2015). This observation suggests a role of Medtr2g105010.1 in the posttranslational regulation of transport systems in M. truncatula roots colonized by AM fungi to promote the symbiotic acquisition of K+. The targets of regulators involved in plant adaptation to oxidative stress also were predicted in AM−K roots and included a Cys desulfurase (Medtr8g093560.1) and a dehydration-responsive element-binding protein (Medtr1g019110). Dehydration-responsive element-binding proteins are involved in salt and hydric stresses, which are consequences of K+ deprivation in plants, and the overexpression of one of them in Arabidopsis (AtDREBP2C) improved its tolerance to global oxidative stress (Hwang et al., 2012).
Altogether, our study revealed that AM symbiosis allows host plants to cope with long-term K+ deprivation. This tolerance mechanism probably involves the regulation of a gene network to protect host plants against oxidative stress but also mechanisms to facilitate K+ uptake from the soil through symbiotic structures. Further studies will be needed to validate these predictions and to fully decipher the role of AM associations in plant K+ nutrition.
MATERIALS AND METHODS
Plant Growth Conditions, Fungal Inoculation, and Long-Term K+ Deficiency Treatment
Medicago truncatula ‘Jemalong A17’ seeds were acid scarified and surface sterilized, plated on 1% (w/v) agar supplemented with 1 µg mL−1 GA3, vernalized at 4°C for 4 d, and allowed to germinate overnight at room temperature. The germinated seedlings were placed for 2 weeks on modified Fahräeus medium as described previously (Catoira et al., 2000). The plants were transferred to pots filled with Turface and inoculated with 400 spores of Rhizophagus irregularis. Plants were watered regularly with standard (+K; 3.75 mm K+) Long Ashton solution [3.75 mm KNO3, 2 mm Ca(NO3)2·4H2O, 7.5 μm NaH2PO4·H2O, 1 mm MgSO4·7H2O, 0.05 mm NaCl, 5 μm MnSO4, 0.5 μm CuSO4, 1 μm ZnSO4, 16.5 μm H3BO3, and 0.1 μm Na2MoO4] or low-K+ (−K; 0.05 mm K+) solution [0.05 mm KNO3, 3.85 mm Ca(NO3)2·4H2O, 7.5 μm NaH2PO4·H2O, 1 mm MgSO4·7H2O, 0.05 mm NaCl, 5 μm MnSO4, 0.5 μm CuSO4, 1 μm ZnSO4, 16.5 μm H3BO3, and 0.1 μm Na2MoO4]. The plants were harvested after 6 weeks of treatment.
Mycorrhizal Quantification, Dry Weight Determination, and Ion Content in Plants
Shoots and roots of 6-week-old M. truncatula plants growing in K+-sufficient or K+-deficient conditions were harvested separately. Mycorrhizal colonization rates were assessed using the gridline intersection method on roots stained previously with Sheaffer ink (McGonigle et al., 1990). Fresh weight was determined, and roots and shoots were dried at 60°C for 1 week for dry weight determination. The dried samples were sent to the Soil & Forage Analysis Laboratory (University of Wisconsin-Madison) to determine the K+, sodium, phosphorus, and calcium contents in plant tissues by ICP-OES.
RNA Isolation and Sequencing
Frozen root tissues were ground in liquid nitrogen, and total RNA was extracted from the resulting powder using the PureLink RNA Mini Kit (Thermo Fisher) and treated with TURBO DNase (Thermo Fisher). RNA quality was assessed using 2100 BioAnalyzer technology (Agilent Technologies). Six micrograms of total RNA was used to construct poly(A) selection libraries using the Illumina TruSeq RNA Sample Preparation kit; 100-nucleotide single-end reads were obtained on an Illumina HiSeq2000 platform using TruSeq version 3 (University of Wisconsin Biotechnology Center DNA Sequencing Facility).
RNA-seq Analysis, Annotations, Cutoff, and Analysis
RNA-seq data were processed to obtain DEGs using the TopHat2-SE, Cufflinks2, Cuffmerge2, and Cuffdiff2 applications from the iPlant Collaborative platform (http://www.iplantcollaborative.org). The DESeq Bioconductor package version 3.3 for R (Anders and Huber, 2010) was used for the analysis of DEGs. Cuffdiff2 sequencing data were analyzed using the cummeRbund package to generate principal component and multidimensional scaling analyses on biological and technical replicates. K-means clustering also was performed using the cummeRbund package for R with K = 25. For further analyses, DEGs were called by DESeq using a 1.5-fold change cutoff threshold and false discovery rate-adjusted q < 0.05. DEGs were annotated with the Mt4.0v1 version of the M. truncatula ‘Jemalong A17’ genome (http://jcvi.org/medicago/index.php). GO enrichment analysis was conducted with the AgriGO Web-based tool (http://bioinfo.cau.edu.cn/agriGO/analysis.php) using the Mt4.0v1 version of the M. truncatula genome. The Venn diagram was obtained using the Bioinformatics & Evolutionary Genomics Web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Finally, clustering and heat maps were generated on R with scripts based on the heatmap.2 function as available in the gplots Bioconductor package. The data discussed in this article have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through the Gene Expression Omnibus Series accession number GSE94266 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94266).
ROS Detection and Measurement
To detect ROS, the root samples of AM and NM 6-week-old plants grown under K+-sufficient or K+-deficient conditions were incubated with 20 μm carboxy-H2DFFDA (Thermo Fisher) for 20 min in high- or low-K+ solution, respectively. The roots were washed twice with high- or low-K+ solution before microscopy visualization. All fluorescence images were captured using a Leica DMi8 microscope and a Leica DFC365 FX camera. ROS fluorescence was quantified and converted into pseudocolor images using the ImageJ software program. Background noise was subtracted from the fluorescence intensity value for quantification.
Regulatory Network Analysis and Visualization
The base network visualized in all parts of Figure 6 was selected from a global inferred M. truncatula regulatory network (Marx et al., 2016) by extracting any interaction between a DEG and top n computationally prioritized genes for each K+ and AM condition, where n is the size of the experimentally identified DEG set. For visual simplicity, connected components involving only one edge were omitted. Network images were created using Cytoscape (Shannon et al., 2003). For full details of the regulatory network analyses, including prioritization and regulator gene set enrichment, see Supplemental Methods S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers GSE94266.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Principal component analysis of the changes in transcript abundance in AM and NM M. truncatula roots under high- and low-K+ regimes.
Supplemental Figure S2. Multidimensional scaling plot showing the two-dimensional distribution of the samples.
Supplemental Figure S3. K-means cluster analysis of DEGs of M. truncatula roots under high- and low-K+ regimes.
Supplemental Figure S4. Impact of AM symbiosis on the transcriptional profiling of M. truncatula roots under K+ deprivation.
Supplemental Figure S5. Representative clustering and heat map of expression values for DEG in transport activity specifically regulated by AM symbiosis independently of K+ availability.
Supplemental Figure S6. Phylogenetic analysis of the cation/H+ exchanger Medtr7g099800.1.
Supplemental Figure S7. Network connectivity statistics suggest differential regulation for induced and repressed gene sets.
Supplemental Figure S8. Predicted regulators of mycorrhizal and low-K+ gene sets represent relevant pathways and propose candidates for future study.
Supplemental Table S1. List of up- and down-regulated transcripts in NM M. truncatula roots under K+ deprivation.
Supplemental Table S2. List of differentially expressed oxidative stress-related transcripts in NM M. truncatula roots under K+ deprivation.
Supplemental Table S3. List of differentially expressed hydrolase transcripts in NM M. truncatula roots under K+ deprivation.
Supplemental Table S4. List of transcripts categorized in the 25 clusters of the K-means cluster analysis (Supplemental Fig. S4).
Supplemental Table S5. List of transcripts differentially regulated in M. truncatula roots in comparison with the NM−K condition.
Supplemental Table S6. Edge density analysis results for input gene sets.
Supplemental Table S7. Genes ranked by diffusion kernel and enrichment-based prioritization analyses.
Supplemental Methods S1. Regulatory network-based interpretation and prioritization of DEGs.
Supplementary Material
Acknowledgments
We thank Dr. Sabine Zimmermann for constructive comments on the article and the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing RNA-seq facilities and services.
Glossary
- AM
arbuscular mycorrhizal
- RNA-seq
RNA sequencing
- NM
nonmycorrhized
- ICP-OES
inductively coupled plasma optical emission spectrometry
- ROS
reactive oxygen species
- GO
Gene Ontology
- DEG
differentially expressed gene
Footnotes
This work was supported by the National Science Foundation (grant no. NSF-IOS 1331098 to J.-M.A. and CAREER award grant no. NSF-DBI 1350677), by a Sloan Foundation research fellowship, by University of Wisconsin-Madison startup funds to S.R., and by the Environmental Protection Agency (grant no. 83573701).
Articles can be viewed without a subscription.
References
- Adams E, Shin R (2014) Transport, signaling, and homeostasis of potassium and sodium in plants. J Integr Plant Biol 56: 231–249 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher C, Ozanne P (1967) Growth and potassium content of plants in solution cultures maintained at constant potassium concentrations. Soil Sci 103: 155–161 [Google Scholar]
- Baslam M, Garmendia I, Goicoechea N (2013) The arbuscular mycorrhizal symbiosis can overcome reductions in yield and nutritional quality in greenhouse-lettuces cultivated at inappropriate growing seasons. Sci Hortic (Amsterdam) 164: 145–154 [Google Scholar]
- Belmondo S, Calcagno C, Genre A, Puppo A, Pauly N, Lanfranco L (2016a) The Medicago truncatula MtRbohE gene is activated in arbusculated cells and is involved in root cortex colonization. Planta 243: 251–262 [DOI] [PubMed] [Google Scholar]
- Belmondo S, Calcagno C, Genre A, Puppo A, Pauly N, Lanfranco L (2016b) NADPH oxidases in the arbuscular mycorrhizal symbiosis. Plant Signal Behav 11: e1165379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B, Haro R, Amtmann A, Cuin TA, Dreyer I (2014) The twins K+ and Na+ in plants. J Plant Physiol 171: 723–731 [DOI] [PubMed] [Google Scholar]
- Breuillin-Sessoms F, Floss DS, Gomez SK, Pumplin N, Ding Y, Levesque-Tremblay V, Noar RD, Daniels DA, Bravo A, Eaglesham JB, et al. (2015) Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27: 1352–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett M. (2004) Diversity and classification of mycorrhizal associations. Biol Rev Camb Philos Soc 79: 473–495 [DOI] [PubMed] [Google Scholar]
- Casieri L, Ait Lahmidi N, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, Courty PE, Garcia K, Charbonnier M, Delteil A, et al. (2013) Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza 23: 597–625 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellier F, Conéjéro G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F (2004) Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J 39: 834–846 [DOI] [PubMed] [Google Scholar]
- Chérel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courty PE, Doidy J, Garcia K, Wipf D, Zimmermann SD (2016) The transportome of mycorrhizal systems. In Martin F, ed, Molecular Mycorrhizal Symbiosis. John Wiley & Sons, Hoboken, NJ, pp 239–256 [Google Scholar]
- D’Angelo C, Weinl S, Batistic O, Pandey GK, Cheong YH, Schültke S, Albrecht V, Ehlert B, Schulz B, Harter K, et al. (2006) Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J 48: 857–872 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, Sekimoto H, Nishiyama T, Melkonian M, Pokorny L, et al. (2015) Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA 112: 13390–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Nye PH (1969) The supply of nutrient ions by diffusion to plant roots in soil. Plant Soil 31: 407–424 [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Aroca R, Maathuis FJM, Barea JM, Ruiz-Lozano JM (2013) Arbuscular mycorrhizal fungi native from a Mediterranean saline area enhance maize tolerance to salinity through improved ion homeostasis. Plant Cell Environ 36: 1771–1782 [DOI] [PubMed] [Google Scholar]
- Fester T, Hause G (2005) Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza 15: 373–379 [DOI] [PubMed] [Google Scholar]
- Garcia K, Delteil A, Conéjéro G, Becquer A, Plassard C, Sentenac H, Zimmermann S (2014) Potassium nutrition of ectomycorrhizal Pinus pinaster: overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of both K+ and phosphorus in the host plant. New Phytol 201: 951–960 [DOI] [PubMed] [Google Scholar]
- Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty PE (2016) Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci 21: 937–950 [DOI] [PubMed] [Google Scholar]
- Garcia K, Zimmermann SD (2014) The role of mycorrhizal associations in plant potassium nutrition. Front Plant Sci 5: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F (2012) Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J 69: 510–528 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar-Hill Y (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823 [DOI] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Harrison MJ. (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Eckert C, Anschütz U, Scholz M, Held K, Waadt R, Reyer A, Hippler M, Becker D, Kudla J (2012) Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J Biol Chem 287: 7956–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes CV, Casper BB (2002) Lateral root function and root overlap among mycorrhizal and nonmycorrhizal herbs in a Florida shrubland, measured using rubidium as a nutrient analog. Am J Bot 89: 1289–1294 [DOI] [PubMed] [Google Scholar]
- He X, Xu M, Qiu GY, Zhou J (2009) Use of 15N stable isotope to quantify nitrogen transfer between mycorrhizal plants. J Plant Ecol 2: 107–118 [Google Scholar]
- Hernandez M, Fernandez-Garcia N, Garcia-Garma J, Rubio-Asensio JS, Rubio F, Olmos E (2012) Potassium starvation induces oxidative stress in Solanum lycopersicum L. roots. J Plant Physiol 169: 1366–1374 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hwang JE, Lim CJ, Chen H, Je J, Song C, Lim CO (2012) Overexpression of Arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. Mol Cells 33: 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Liu J, Liu J, Huang X (2012) Forms of nitrogen uptake, translocation, and transfer via arbuscular mycorrhizal fungi: a review. Sci China Life Sci 55: 474–482 [DOI] [PubMed] [Google Scholar]
- Kaldorf M, Kuhn AJ, Schröder WH, Hildebrandt U, Bothe H (1999) Selective element deposits in maize colonized by a heavy metal tolerance conferring arbuscular mycorrhizal fungus. J Plant Physiol 154: 718–728 [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Ciani S, Schachtman DP (2010) A peroxidase contributes to ROS production during Arabidopsis root response to potassium deficiency. Mol Plant 3: 420–427 [DOI] [PubMed] [Google Scholar]
- Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S (2011) Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol Plant 4: 527–536 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefoulon C, Boeglin M, Moreau B, Véry AA, Szponarski W, Dauzat M, Michard E, Gaillard I, Chérel I (2016) The Arabidopsis AtPP2CA protein phosphatase inhibits the GORK K+ efflux channel and exerts a dominant suppressive effect on phosphomimetic-activating mutations. J Biol Chem 291: 6521–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentration for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH (2013) A protein kinase, calcineurin B-like protein-interacting protein kinase9, interacts with calcium sensor calcineurin B-like protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol 161: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López MF, Dietz S, Grunze N, Bloschies J, Weiss M, Nehls U (2008) The sugar porter gene family of Laccaria bicolor: function in ectomycorrhizal symbiosis and soil-growing hyphae. New Phytol 180: 365–378 [DOI] [PubMed] [Google Scholar]
- Ma TL, Wu WH, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx H, Minogue CE, Jayaraman D, Richards AL, Kwiecien NW, Siahpirani AF, Rajasekar S, Maeda J, Garcia K, Del Valle-Echevarria AR, et al. (2016) A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat Biotechnol 34: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Moody PW, Bell MJ (2006) Availability of soil potassium and diagnostic soil tests. Soil Res 44: 265–275 [Google Scholar]
- Oliveira RS, Rocha I, Ma Y, Vosátka M, Freitas H (2016) Seed coating with arbuscular mycorrhizal fungi as an ecotechnological approach for sustainable agricultural production of common wheat (Triticum aestivum L.). J Toxicol Environ Health A 79: 329–337 [DOI] [PubMed] [Google Scholar]
- Padmanaban S, Chanroj S, Kwak JM, Li X, Ward JM, Sze H (2007) Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol 144: 82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner H, Schwarz D, Bruns C, Mäder P, George E (2007) Effect of arbuscular mycorrhizal colonization and two levels of compost supply on nutrient uptake and flowering of pelargonium plants. Mycorrhiza 17: 469–474 [DOI] [PubMed] [Google Scholar]
- Porras-Soriano A, Soriano-Martín ML, Porras-Piedra A, Azcón R (2009) Arbuscular mycorrhizal fungi increased growth, nutrient uptake and tolerance to salinity in olive trees under nursery conditions. J Plant Physiol 166: 1350–1359 [DOI] [PubMed] [Google Scholar]
- Puppo A, Pauly N, Boscari A, Mandon K, Brouquisse R (2013) Hydrogen peroxide and nitric oxide: key regulators of the legume-rhizobium and mycorrhizal symbioses. Antioxid Redox Signal 18: 2202–2219 [DOI] [PubMed] [Google Scholar]
- Ragel P, Ródenas R, García-Martín E, Andrés Z, Villalta I, Nieves-Cordones M, Rivero RM, Martínez V, Pardo JM, Quintero FJ, et al. (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol 169: 2863–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Kirkby EA (2010) Research on potassium in agriculture: needs and prospects. Plant Soil 335: 155–180 [Google Scholar]
- Roy S, Lagree S, Hou Z, Thomson JA, Stewart R, Gasch AP (2013) Integrated module and gene-specific regulatory inference implicates upstream signaling networks. PLOS Comput Biol 9: e1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L, Zhang J, Xin X, Zhang C, Ma D, Chen L, Zhao B (2015) Comparative analysis of potassium deficiency-responsive transcriptomes in low potassium susceptible and tolerant wheat (Triticum aestivum L.). Sci Rep 5: 10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygiewicz PT, Bledsoe CS (1984) Mycorrhizal effects on potassium fluxes by northwest coniferous seedlings. Plant Physiol 76: 918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer P, Corbière H, Boller T (1999) Hydrogen peroxide accumulation in Medicago truncatula roots colonized by the arbuscular mycorrhiza-forming fungus Glomus intraradices. Planta 208: 319–325 [Google Scholar]
- Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Scheloske S, Maetz M, Schneider T, Hildebrandt U, Bothe H, Povh B (2004) Element distribution in mycorrhizal and nonmycorrhizal roots of the halophyte Aster tripolium determined by proton induced x-ray emission. Protoplasma 223: 183–189 [DOI] [PubMed] [Google Scholar]
- Shankar A, Singh A, Kanwar P, Srivastava AK, Pandey A, Suprasanna P, Kapoor S, Pandey GK (2013) Gene expression analysis of rice seedling under potassium deprivation reveals major changes in metabolism and signaling components. PLoS ONE 8: e70321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Jakobsen I, Grønlund M, Smith FA (2011) Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol 156: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Read D (2008) Mycorrhizal Symbiosis, Ed 3 Academic Press, New York, NY [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, et al. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108: 1028–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Qiu YL (2006) Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363 [DOI] [PubMed] [Google Scholar]
- Wang C, Chen H, Hao Q, Sha A, Shan Z, Chen L, Zhou R, Zhi H, Zhou X (2012) Transcript profile of the response of two soybean genotypes to potassium deficiency. PLoS ONE 7: e39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64: 451–476 [DOI] [PubMed] [Google Scholar]
- Zeng Q, Ling Q, Fan L, Li Y, Hu F, Chen J, Huang Z, Deng H, Li Q, Qi Y (2015) Transcriptome profiling of sugarcane roots in response to low potassium stress. PLoS ONE 10: e0126306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörb C, Senbayram M, Peiter E (2014) Potassium in agriculture: status and perspectives. J Plant Physiol 171: 656–669 [DOI] [PubMed] [Google Scholar]
- Zuccarini P, Okurowska P (2008) Effects of mycorrhizal colonization and fertilization on growth and photosynthesis of sweet basil under salt stress. J Plant Nutr 31: 497–513 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.