Summary
The developmental mechanisms regulating the number of adult neural stem cells (aNSCs) are largely unknown. Here we show that the cleavage plane orientation in murine embryonic radial glia cells (RGCs) regulates the number of aNSCs in the lateral ganglionic eminence (LGE). Randomizing spindle orientation in RGCs by overexpression of Insc or a dominant-negative form of Lgn (dnLgn) reduces the frequency of self-renewing asymmetric divisions while favoring symmetric divisions generating two SNPs. Importantly, these changes during embryonic development result in reduced seeding of aNSCs. Interestingly, no effects on aNSC numbers were observed when Insc was overexpressed in postnatal RGCs or aNSCs. These data suggest a new mechanism for controlling aNSC numbers and show that the role of spindle orientation during brain development is highly time and region dependent.
Keywords: radial glial cells, short neural precursor, adult neural stem cells, SEZ, Insc, Lgn, division plane
Highlights
-
•
Randomization of the spindle orientation changes the progenitor pool composition
-
•
Overexpression of Insc or dnLgn reduces asymmetric self-renewing division of aRGCs
-
•
The change in embryonic progenitor pool leads to reduced seeding of adult NSCs
-
•
Insc influences the seeding of adult NSCs in a narrow developmental time window
Falk et al. uncover a new concept where the control of the division type in progenitor cells during embryonic development regulates the number of embryonic progenitor cells destined to become adult neural stem cells.
Introduction
The mechanisms by which tissue-specific adult somatic stem cells are formed are poorly understood. In many tissues, adult somatic stem cells are mainly quiescent during adult life and produce differentiating daughter cells only when sporadically activated (Cheung and Rando, 2013). Such quiescent adult stem cells reside in the subependymal zone (SEZ) in the lateral wall of the lateral ventricle, the largest site of ongoing adult neurogenesis in rodents (Kriegstein and Alvarez-Buylla, 2009, Ninkovic and Götz, 2013). Here, adult neural stem cells (aNSCs) give rise to a large number of neuronal progeny that migrate along the rostral migratory stream to the olfactory bulb (OB), where they differentiate into diverse interneuron types. aNSCs in the SEZ as well as the other neurogenic niche, the dentate gyrus (DG), are not only largely quiescent, but also exhibit a very limited self-renewal capacity and fast exhaustion once activated (Calzolari et al., 2015, Encinas et al., 2011, Fuentealba et al., 2015, Sierra et al., 2015). Therefore, the initial pool size of aNSCs is most crucial in determining the amount of neurogenesis over a lifetime. Importantly, the mechanisms determining the initial number of aNSCs are still mostly concealed.
aNSCs exhibit radial glia hallmarks, including apical endfeet with a primary cilium at the ventricle (Beckervordersandforth et al., 2010, Mirzadeh et al., 2008), elongated basal processes (Kokovay et al., 2010, Mirzadeh et al., 2008, Shen et al., 2008, Tavazoie et al., 2008), and the expression of many radial glial and astroglial genes (Beckervordersandforth et al., 2010, Codega et al., 2014, Llorens-Bobadilla et al., 2015) including Glast (Mori et al., 2006, Ninkovic et al., 2007) and Gfap (Doetsch et al., 1999, Giachino et al., 2014). Viral lineage tracing experiments revealed that aNSCs originate from proliferating ventricular progenitors during embryonic development (Fuentealba et al., 2015) and that the aNSC ancestors feature long basal processes at early postnatal stages (Merkle et al., 2004). Interestingly, the lineage of aNSCs in the SEZ is diverging from the lineage of the highly proliferative embryonic progenitors at midneurogenic stages (Fuentealba et al., 2015) with cells of the aNSC lineage showing only very limited proliferation thereafter (Furutachi et al., 2015). In that respect, it is intriguing to note that the lineage ancestors of aNSCs gain sonic hedgehog responsiveness around this time during embryonic development, shown by lineage tracing using Gli1CreERT2 (Ahn and Joyner, 2005) underpinning the lineage divergence.
Interestingly, the lateral ganglionic eminence (LGE), the region that generates the majority of the aNSCs in the SEZ of the lateral wall of the lateral ventricle (Young et al., 2007), shows a much higher diversity of progenitor subtypes as compared to most other CNS regions (Pilz et al., 2013). However, it is still largely unknown how these distinct subpopulations in the LGE contribute to the generation of aNSCs. Therefore, we set out to determine mechanisms regulating apical progenitor heterogeneity in the LGE and ultimately aNSC numbers. We were particularly interested in spindle positioning during M-phase as this has been implicated in the regulation of the size of stem and progenitor cell pools in a multitude of tissues by coordinating the mode of cell division (Knoblich, 2010). In many stem cell systems, oblique or horizontal cell divisions differentially distributing apico-basal components between daughter cells are involved in asymmetric stem cell self-renewing divisions (El-Hashash et al., 2011, Lechler and Fuchs, 2005, Quyn et al., 2010, Troy et al., 2012). In the nervous system, Insc- or Lgn-dependent mitotic spindle randomization has been used to favor oblique and horizontal division planes (Konno et al., 2008, Morin et al., 2007, Postiglione et al., 2011, Shitamukai et al., 2011), leading to increased delamination of daughter cells. We show here very different consequences of spindle randomization in the region that generates aNSCs with profound effects on aNSC numbers during a very limited time window.
Results
Apical Progenitor Subtypes in the LGE Differ in Their Orientation of Cell Division
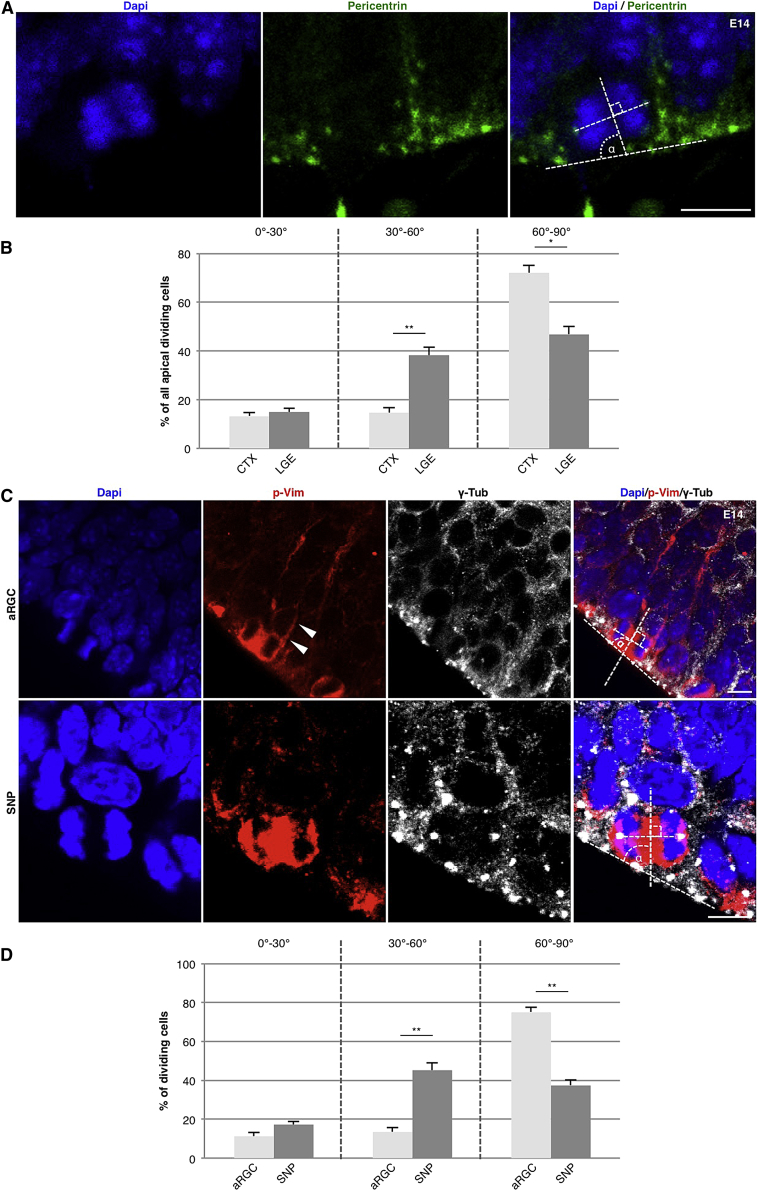
As the progenitor set up in the region generating most aNSCs is distinct from other brain regions (Pilz et al., 2013), we examined if the orientation of cell divisions is also different from other regions. We determined the cleavage angle during telophase by staining for Pericentrin (Pcnt) to visualize the centrosomes and DAPI to delineate the separated chromatids (Figure 1A; the dotted line between the chromatids shows an 81° division; hence, a vertical division 60°–90°). In contrast to other regions in the developing CNS (Kosodo et al., 2004, Noctor et al., 2008, Smart, 1973, Zamenhof, 1985), in the LGE only 47% of apical progenitors divide with a vertical orientation (60°–90°) at embryonic day (E)14 (Figures 1A and 1B). This is different from the cerebral cortex, where 72% of apical divisions are vertical (Figures 1A and 1B). Remarkably, oblique divisions (30°–60°) are more than two times more frequent in the LGE than in the cerebral cortex (Figures 1A and 1B; 15% versus 38%, respectively), while a comparable proportion divides with horizontal (0°–30°; Figures 1A and 1B) cleavage planes (cortex, 13%; LGE, 15%).
Figure 1.
Region- and Progenitor-Specific Differences in Orientation of the Cell Division
(A) Fluorescence micrographs depicting Dapi and Pericentrin stainings to localize DNA and centrosomes in late anaphase/telophase cells at the ventricular surface at E14 in the cerebral cortex. The panel to the right shows how the cleavage plane angle respective to the ventricular surface was determined, 81° in this case.
(B) Histogram showing the distribution of vertical (60°–90°), oblique (30°–60°), and horizontal (0°–30°) cleavage angles. Note that the fraction of cells dividing with an oblique angle is more than double in the LGE compared to the cerebral cortex (cortex, 62 cells from 5 embryos; LGE, 72 cells from 5 embryos).
(C) Micrographs showing immunofluorescence for p-Vim (red) and γ-tubulin (centrosomes, white) to determine the cleavage angle and the morphology of cells in M-phase simultaneously. Cells with a basal process (arrowhead) are classified as aRGCs while cells without a basal process during mitosis are SNPs. The aRGC shown divides with a cleavage angle of 78° while the SNP has a cleavage angle of 58°.
(D) Histogram depicting the distribution of vertical (60°–90°), oblique (30°–60°), and horizontal (0°–30°) cleavage angles in the respective subpopulation of progenitor cells. In the LGE, the SNPs show a broader distribution of cleavage angles than aRGCs that divide largely vertically (aRGCs, 56 cells from 5 embryos; SNPs, 64 cells from 5 embryos).
Data are shown as mean ± SEM (Mann-Whitney U test; ∗p < 0.05, ∗∗p < 0.01). Scale bars, 10 μm.
Next we asked whether apical progenitor subtypes differ in their division plane. We had previously noted a large number of SNPs that lack a basal process (Gal et al., 2006, Stancik et al., 2010, Tyler and Haydar, 2013) in the LGE (Pilz et al., 2013), similar to the medial ganglionic eminence (MGE) (Petros et al., 2015). Phosphorylated Vimentin (p-Vim) is present in the cytoplasm of mitotic cells, allowing discrimination between apically anchored radial glia cells (aRGCs) (with a basal process) and SNPs (without a basal process) (Gal et al., 2006, Stancik et al., 2010, Tyler and Haydar, 2013). Interestingly, aRGCs in the LGE divide predominantly with a vertical orientation, while SNPs divide predominantly with oblique orientations (Figures 1C and 1D). To ensure that cells classified as SNPs are not falsely put in this category due to a failure of p-Vim labeling to reveal the basal process, we quantified the division angles in cells after in utero electroporation (IUE) of a membrane-tagged EGFP construct. This analysis fully confirmed that SNPs divided much more frequently in an oblique (51%) or horizontal (16%) orientation when compared to aRGCs (oblique, 12%; horizontal, 10%; Figure S1, available online).
Live Imaging of Apical Cell Divisions in the LGE
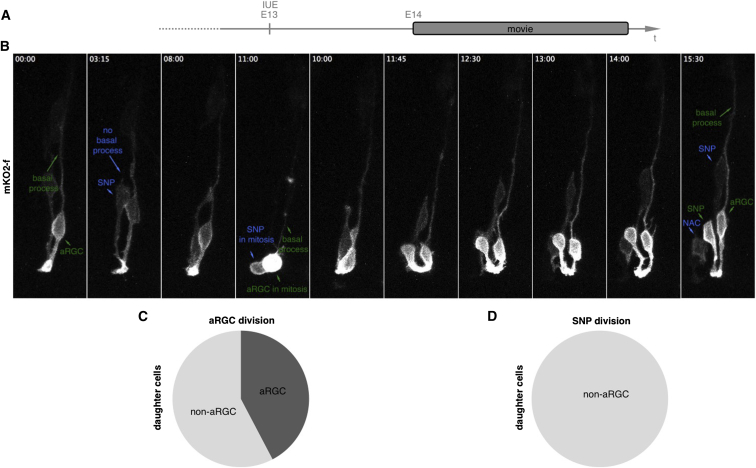
To directly determine the progeny of these different apical progenitor types in the LGE, we used live imaging after IUE of plasmids expressing membrane-tagged (farnesylated) monomeric Kusabira-Orange (mKO2-f) at E13 (Shitamukai et al., 2011) to assess their morphology (Figures 2A and 2B). Individual apically dividing cells were imaged for up to 24 hr by taking confocal stacks every 15 min in 300 μm thick slices prepared 1 day after IUE (Figures 2A and 2B). While aRGCs divided almost always in an asymmetric mode, renewing one aRGC and producing one different daughter cell type (Figures 2B and 2C; Movie S1), SNPs never gave rise to aRGCs but generated further SNPs or different non-apical cells (Figures 2B and 2D) (also see Pilz et al., 2013). Thus, aRGCs are on top of the lineage hierarchy of LGE progenitor cells.
Figure 2.
Live Imaging of Progenitor Cells Dividing at the Apical Surface in the LGE
(A) Schematic drawing depicting the experimental layout.
(B) Micrographs of a time-lapse series showing an apically dividing cell with a basal process during mitosis (green), hence an aRGC, and a cell without a long basal process (blue), therefore a SNP. The aRGC divides and produces one aRGC and one SNP while the SNP divides and gives rise to another SNP and a no longer apically anchored cell (NAC).
(C and D) Pie charts depicting the quantification of apically dividing progenitors show that the majority of aRGCs produce another aRGC (C) while no SNP was ever observed to produce an aRGC (D). SNPs mainly produce other SNPs, subapical progenitors (SAPs), or multipolar cells (together referred to as non-aRGC) (65 aRGC divisions and 17 SNP divisions were followed).
Scale bar, 10 μm.
Apical Progenitor Pool Composition Is Altered in the LGE of Inscoe Mice
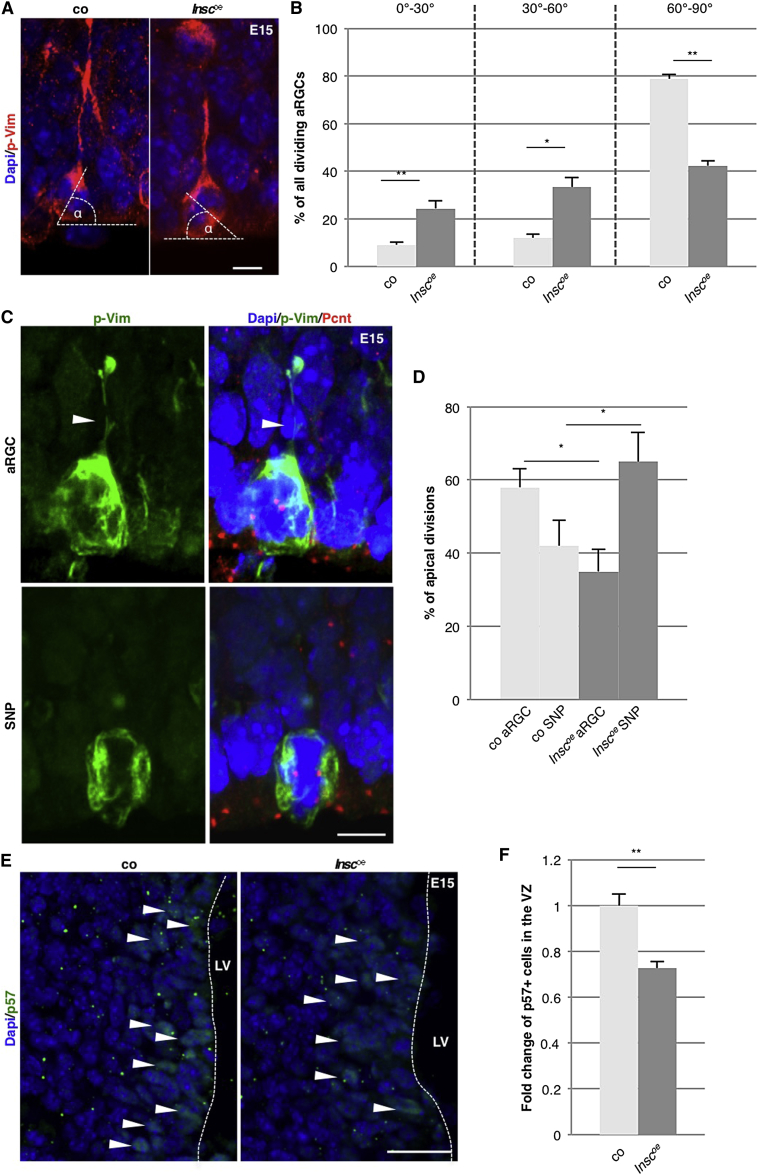
Given the progenitor subtype-specific bias for cleavage angles, we aimed to test its functional relevance by forcing alterations in the spindle orientation during M-phase. The mouse line constitutively overexpressing Insc (Inscoe) (Postiglione et al., 2011) has previously been shown to reorient spindle positioning in the developing cerebral cortex (Postiglione et al., 2011). Given that SNPs in the LGE showed almost randomized cleavage plane orientations, we assessed the effect of Insc overexpression specifically in aRGCs of the LGE utilizing p-Vim staining. As expected, more randomized cleavage planes were observed in aRGCs residing in the LGE of E15 Inscoe mice (Figures 3A and 3B). p-Vim staining performed at midneurogenesis (E15) (Figure 3C) revealed that the balance in the LGE apical progenitor pool composition was altered to more SNPs in Inscoe mice (Figure 3D). Thus, the progenitor cell type normally dividing with more oblique divisions (Figures 1C and 1D) is favored when these orientations are further increased.
Figure 3.
Randomizing Cleavage Plane Orientation and Depletion of aRGCs by Insc Overexpression in the Developing LGE
(A) Fluorescence micrographs showing dividing aRGCs with a p-Vim-positive basal process and separating chromatids. The cleavage angle was determined as indicated.
(B) Histograms depicting the distribution of cleavage plane orientation in aRGCs of the LGE in control (light gray) or Insc-overexpressing (Inscoe) mice (dark gray) at E14. Note that constitutive overexpression of Insc leads to a randomization of the division plane (control, 54 cells; Inscoe, 72 cells; each from 5 embryos). Scale bar, 10μm.
(C) Fluorescence micrographs showing apically dividing cells (aRGC with a basal process [arrowhead] and SNP without basal process) by labeling p-Vim, Pericentrin, and Dapi.
(D) Histogram depicting the quantification of different subtypes of apically dividing cells at E14 in the developing LGE of control and Inscoe animals as determined by p-Vim. Importantly, the quantification of the number of the apically dividing cells revealed that the relative proportion of aRGCs among all apically dividing cells is decreased concomitantly with a relative increase in SNPs, such that SNPs constitute the majority in Inscoe LGE while aRGCs are the majority in controls (250 cells quantified in 4 animals each genotype). Scale bar, 10 μm.
(E) Fluorescence micrograph showing p57 stainings in the LGE ventricular zone of control and Inscoe animals.
(F) Histogram depicting the quantification of p57+ cells in (E). A 27% reduction of p57+ cells was detected in the ventricular zone of Inscoe LGE (control, 226 cells; Inscoe, 187 cells). Scale bar, 40 μm.
Data are shown as mean ± SEM (Mann-Whitney U test; ∗p < 0.05, ∗∗p < 0.01).
Notably, in the cerebral cortex (Konno et al., 2008, Postiglione et al., 2011) and the chicken spinal cord (Das and Storey, 2012, Morin et al., 2007), an increase in delaminating progenitors was observed after Insc overexpression. To determine if this was also the case in the LGE, we quantified the overall number of mitoses at apical and non-apical positions using the mitotic markers p-Vim (Figure S2) and phosphorylated histone H3 (pH3; data not shown). In profound contrast to the cerebral cortex and spinal cord, no change in the fraction of apical versus non-apical mitosis was detectable in the LGE of Inscoe animals (Figure S2B). Moreover, no effect on apical adhesion and polarity as assessed by N-cadherin and β-catenin stainings (Figures S3A–S3C) could be observed in the LGE of Inscoe mice at E15. Furthermore, cell density was unchanged (Figure 3D), suggesting that in the LGE, adhesion is not affected by Insc overexpression. Importantly, co-IUE of ZO1-GFP, together with a membrane-tagged mKO2, shows that SNPs are anchored at the apical side during interphase (Figure S3E), and N-cadherin staining in GFP-labeled mitotic SNPs (Figure S3F) showed that anchoring is maintained also during M-phase. Moreover, co-IUE of the ciliary maker Arl13b-RFP demonstrates that SNPs maintain a functional apical endfoot with the cilium being localized at the apical membrane (Figure S3G). Together, these data demonstrate that apical anchoring is not altered in Inscoe animals and that SNPs remain integrated at the apical surface.
Reduced Numbers of p57+ Cells in the LGE of Inscoe Mice
Given the profound changes in the composition of apical progenitors, we next examined their proliferation behavior by quantifying Ki67+ cells. Equal numbers of cells were Ki67+ in the LGE of control and Inscoe animals at E15 (Figures S2C and S2D). Likewise, BrdU labeling showed no difference between genotypes (Figures S2C and S2D). To further specifically probe for changes in a rare subpopulation of slowly dividing cells, we examined the number of cells labeled by high levels of p57, a factor that has recently been implicated in the generation of aNSCs (Furutachi et al., 2015). Interestingly, we observed a significant decrease in the number of p57+ cells in the LGE ventricular zone of Inscoe embryos (Figures 3E and 3F), suggesting that the slow-dividing ancestors of aNSCs are affected by Inscoe.
Acute Insc Overexpression by IUE Alters Fate of Apical Progenitors toward More Tα1-GFP+ SNPs
Given the intriguing phenotype observed in the LGE of Inscoe mice, we were concerned that earlier effects of the overexpression may cause these changes in LGE progenitor composition. As a first step to determine earlier defects, we examined the LGE in Inscoe mice at an earlier stage, E12 (Figures S4A–S4C). At this stage, however, the progenitor pool composition (Figures S4A–S4C) was not affected in the Inscoe LGE compared to controls. Indeed, as observed previously in the developing cortex (Postiglione et al., 2011), at this early stage the division plane in the LGE apical progenitor pool was similar in Inscoe to control mice (Figures S4D and S4E).
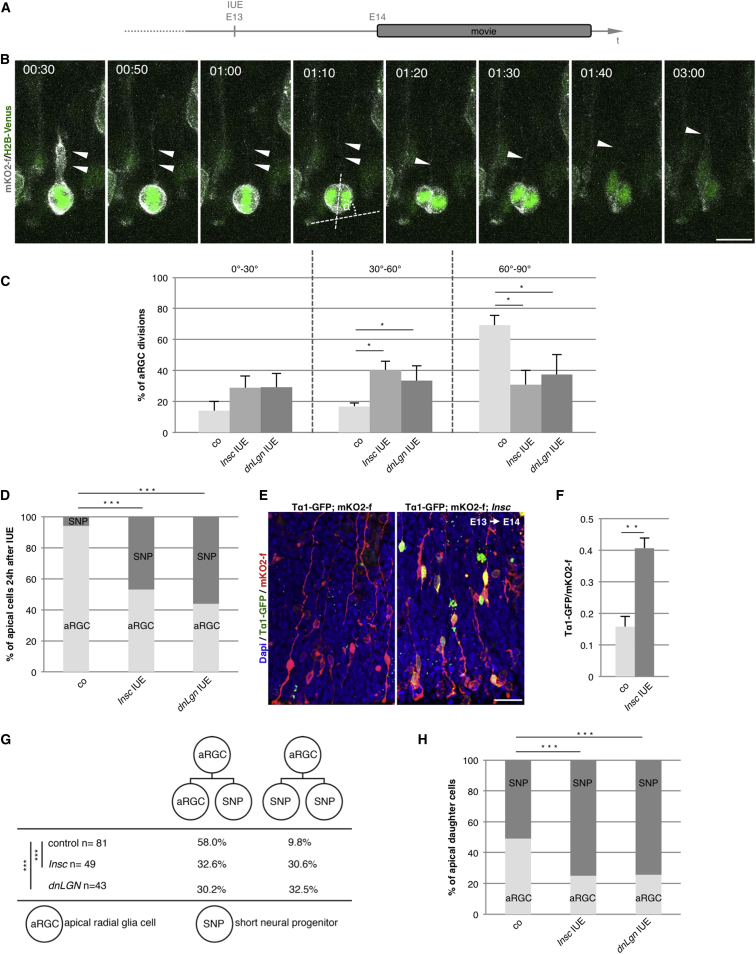
We next used IUE to acutely overexpress Insc (Konno et al., 2008, Shitamukai et al., 2011) in a wild-type environment. By simultaneous labeling with mKO2-f and H2B-Venus, we could follow the cell morphology and the chromatin by live imaging (Figures 4A and 4B). One day after IUE of Insc (at E14), randomization of the division plane was observed (Figures 4C) to a similar extent as in the LGE of the transgenic Inscoe mice (Figures 3A and 3B). In full agreement with the phenotype observed in the Inscoe mice, aRGCs were decreased and SNPs were increased already 1 day after electroporating Insc (Figure 4D). However, so far we could classify SNPs only based on morphology (apical division and lack of basal process). IUE now allowed determining if there is a fate change toward this progenitor subtype by using the Tα1-GFP reporter, as previously described in SNPs in the cerebral cortex (Gal et al., 2006). Indeed, a significant increase in Tα1-GFP+ cells among the mKO2-f+ cells was detected upon Insc IUE (Figures 4E and 4F), demonstrating that cells change not only morphology but also fate after Insc IUE.
Figure 4.
Live Imaging Reveals Preferential Generation of SNPs after Randomization of the Cleavage Plane
(A) Experimental paradigm for live imaging of apically anchored progenitor cells.
(B) Still images of cells labeled by IUE at E13 with mKO2-f (membrane; white) and H2B-venus (chromatin; green) and imaged every 10 min for 24 hr in slices of the LGE prepared at E14. Note the basal and apical process (arrowhead) in the starting cell. This aRGC is dividing with an angle of 81° and produces another aRGC with a basal process, as seen at 01:50, and a SNP.
(C) Quantification of cleavage planes of cells observed by live imaging as shown in (B) after co-electroporations of control, Insc, or the dominant-negative form of Lgn (dnLgn). Note that both Insc and dnLgn randomized the division plane (Mann-Whitney U test, ∗p < 0.05).
(D) Histogram depicting the proportion of aRGCs and SNPs after overexpression of either Insc or dnLgn for 24 hr, i.e., at the beginning of live imaging, shows an increase in SNPs at the expense of aRGCs (chi-square test, ∗∗∗p < 0.001).
(E and F) Co-electroporation of CAG-mKO2-f with Tα1-GFP reveals an increase of progenitor cells expressing the Tα1 promoter after Insc overexpression (F) as compared to control (E) (Mann-Whitney test, ∗∗p < 0.01).
(G) Lineage analysis of the daughter cells derived from an aRGC shows a decrease in asymmetric divisions producing one aRGC and one SNP with concomitant increase in symmetric divisions producing two SNPs from one aRGC (chi-square test, ∗∗∗p < 0.001) (control, 81 cells; Insc, 49 cells; dnLgn, 43 cells; at least 5 animals in each condition).
(H) Histograms illustrating the fate of aRGC daughter cells produced as a fraction of all cells produced. A marked increase in SNP generation upon Insc or dnLgn manipulation was observed by live imaging with a concurrent decrease of aRGCs (chi-square test, ∗∗∗p < 0.01).
Data in (C) and (F) are shown as mean ± SEM. Scale bar, 10 (B) and 40 μm (G).
The increase in numbers of SNPs may also be caused by increased proliferation of SNPs. We therefore performed live imaging of the electroporated cells as described above (Figure 2) to determine the cell division frequency and progeny of cells after Insc IUE (Figure 4G). No change in the number of divisions within a 20 hr observation window was detectable between controls and Insc IUE (aRGCs; control, 63% of n = 81; Insc IUE, 59% of n = 49; SNPs; control, 75% of n = 8; Insc IUE, 73% of n = 35). Intriguingly, 1 day after Insc IUE aRGCs were already significantly biased toward the generation of SNPs (Figure 4H). This was due to an increase in symmetric SNP generation (Movie S2) (control, 10% of n = 81; Insc IUE, 31% of n = 49), while in controls the vast majority of aRGC divisions were dividing asymmetrically, generating two different daughter cells (88% of n = 81): typically one aRGC and one SNP (control, 58% of n = 81; Insc IUE, 33% of n = 49) (Figure 4G) (Movie S1; more examples in Movie S4). As our live imaging observations can also rule out any effects of selective proliferation (as described above) or selective cell death (data not shown; Figures S5A–S5E; Movie S3), we conclude that Insc overexpression directly alters the aRGC progeny by affecting the mode of cell division. In pronounced contrast to other stem cell systems (Ben-Yair et al., 2011, Konno et al., 2008), more oblique and horizontal divisions decrease the self-renewing asymmetric division mode and rather favor the symmetric generation of two SNPs from an aRGC.
Preferential Generation of SNPs from aRGCs upon dnLgn Overexpression by IUE
In order to employ an independent approach for randomizing spindle orientation, we electroporated a dominant-negative form of Lgn, dnLgn. Lgn provides attachment for the astral microtubules and hence ensures alignment of the cell division orientation with the apico-basal polarity (Konno et al., 2008, Morin et al., 2007, Shitamukai et al., 2011). It has been previously shown in murine and chicken developing CNS that dnLgn expression results in randomization of the cleavage plane orientation (Konno et al., 2008, Morin et al., 2007, Shitamukai et al., 2011). In the cerebral cortex, this resulted in delamination of many progenitors from the apical surface (Konno et al., 2008, Shitamukai et al., 2011). Here we would predict a rather different outcome, namely, an increase of SNPs upon dnLgn IUE in the LGE. Indeed, we observed a randomization of cleavage planes at the apical surface (Figure 4C), an increase in SNPs (Figure 4D), and an increased number of SNPs generated from aRGCs (Figures 4G and 4H) after IUE of dnLgn and live imaging. As observed after Insc IUE, with dnLgn IUE we also did not detect a change in the number of mitoses in aRGCs (control, 63% of n = 81; dnLgn IUE, 62% of n = 43) or SNPs (control, 75.0% of n = 8; dnLgn IUE, 69% of n = 21) within 20 hr. Similar to Insc manipulations, the symmetric SNP-generating divisions were increased (control, 10% of n = 81; dnLgn IUE, 33% of n = 43) and the asymmetric divisions producing one aRGC and one SNP were decreased (control, 58% of n = 81; dnLgn IUE, 30% of n = 43). Thus, expression of dnLgn phenocopies the effects seen upon both acute and persistent Insc overexpression, further supporting the concept that the orientation of cell division regulates the progenitor subtypes at the apical surface in the LGE.
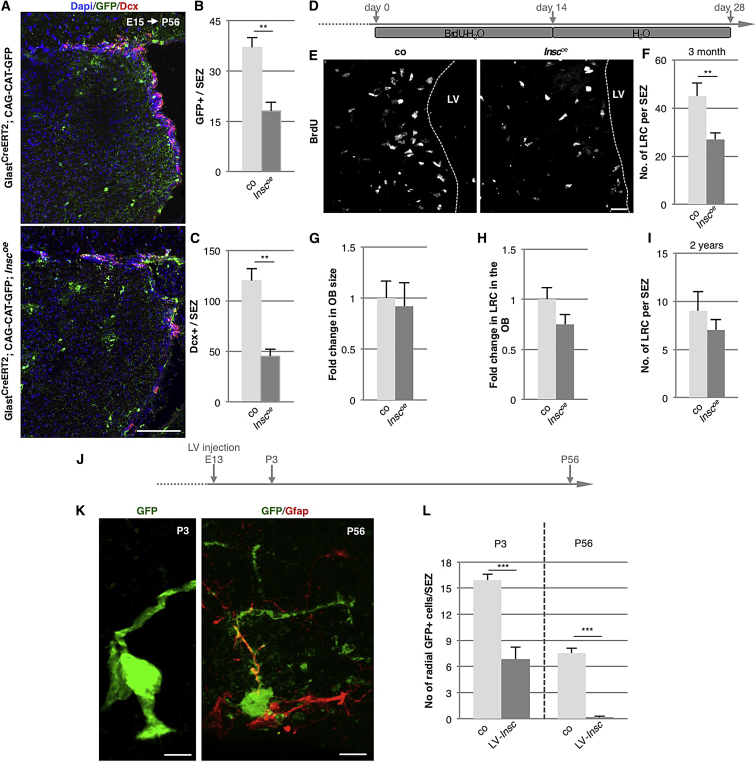
Reduced Numbers of Adult SEZ NSCs and a Decrease in Adult Neurogenesis in Inscoe Mice
Given the profound alterations in progenitor composition during development and the reduction on p57+ cells in Inscoe LGE, we next aimed to determine the consequences for adult neurogenesis. Toward this aim, we returned to the Inscoe mouse line and used genetic lineage tracing with the GlastCreERT2 (Mori et al., 2006) and CAG-CAT-GFP reporter lines (Nakamura et al., 2006). After tamoxifen induction at E15, we followed the progeny (GFP+) of cells expressing the radial glia marker Glast at E15 into the SEZ of 2-month-old mice. Remarkably, GFP+ cells were dramatically reduced in the SEZ of Inscoe mice to less than half of controls (Figures 5A and 5B). This was accompanied by a significant and severe reduction in doublecortin (Dcx)+ neuronal progenitor cells to 38% of the controls (Figures 5A and 5C), suggesting that there is an overall decrease in adult SEZ neurogenesis in Inscoe mice. Note that a similar decrease in Dcx+ neuronal progenitors was also observed in Inscoe mice not crossed to GlastCreERT2 mice and hence not treated with tamoxifen to induce recombination (Figure S7). Next, we employed a label-retaining paradigm of 2 weeks labeling the dividing cells with BrdU, followed by a 2 week chase period without BrdU (Figure 5D). This protocol allows the fast-dividing cells to dilute the BrdU label and the labeled post-mitotic neuronal progeny to leave the SEZ and migrate to the OB. Primarily slow-dividing aNSCs keep the label and stay within the SEZ. Indeed, the number of label-retaining NSCs in the SEZ of Inscoe mice is strongly diminished (Figures 5E and 5F).
Figure 5.
Reduced Numbers of Adult NSCs and Neurogenesis in the SEZ of Inscoe Mice
(A) Fluorescence micrographs depicting GFP+ cells in the adult SEZ labeled by administration of tamoxifen to GlastCreERT2; CAG-CAT-GFP animals at E15 and Dcx+ neuroblasts.
(B and C) Histograms showing quantifications of recombined cells (GFP+) and Dcx+ cells in the SEZ of control and Inscoe mice. Note the reduced number of GFP+ cells in the SEZ in Inscoe mice (B), indicating reduced progeny of embryonic RGCs (control, n = 134 cells; Inscoe, n = 97 cells; 4 animals in each condition) and the reduced number of Dcx+ neuroblasts (C) in the SEZ of Inscoe mice (control, n = 439 cells; Inscoe, n = 328 cells; 5 animals in each condition; Mann-Whitney test, ∗∗p < 0.01). Quantifications were calculated as number of cells per SEZ section.
(D) Experimental paradigm to uncover label-retaining cells (LRCs) in the SEZ.
(E) Fluorescence micrographs of LRCs in the SEZ of control and Inscoe mice.
(F) Histograms depicting the number of LRCs in 3-month-old animals per SEZ section normalized to the control, which is significantly reduced in Inscoe animals (control, n = 734 cells; Inscoe, n = 697 cells; 5 animals in each condition).
(G and H) Histograms depicting size measurements (G) or quantifications of the number of LRCs found in the OB (H) that were not affected in Inscoe.
(I) Interestingly, the number of LRCs was no longer significantly changed in the SEZ of 2-year-old Inscoe mice when compared to controls (control, n = 440 cells; Inscoe, n = 457 cells; 5 animals in each condition). Quantifications were calculated as number of cells per SEZ section (Mann-Whitney test, ∗∗p < 0.01).
(J) Experimental paradigm for the lentiviral delivery of Insc.
(K) Confocal pictures of cells with a radial morphology in the SEZ at P3 and P56.
(L) Histograms showing the quantification of the number infected cells (GFP+) with a radial morphology at P3 and P56 per SEZ section. Note the strong reduction of cells with a radial morphology in embryos infected with LV-Insc (P3; control, n = 191 cells; LV-Insc, n = 82 cells; P56; control, n = 90; LV-Insc, n = 2; 6 hemispheres each condition).
Data are shown as mean ± SEM (Mann-Whitney test, ∗∗∗p < 0.001).
Conversely, neither the number of BrdU+ cells in the OB (Figure 5H) nor its size (Figure 5G) was significantly changed despite the reduced neurogenic output of the SEZ. As many of the newly arriving neurons are subject to cell death (Mouret et al., 2009, Yamaguchi and Mori, 2005), we stained for cleaved and hence activated (a) caspase 3 to determine possible differences in cell death between the genotypes. Indeed, the number of activated caspase 3 positive cells is reduced to 54% of the control in the OB of Inscoe mice (Figure S5F). Thus, improved survival counterbalances the reduced neuronal progenitor output, rendering further support to cell death exerting homeostatic control over the numbers of new neurons (Mouret et al., 2008, Petreanu and Alvarez-Buylla, 2002).
To investigate how sustained overexpression of Insc in these transgenic mice would affect aNSCs over the course of adult life, we applied the label-retaining protocol in 2-year-old animals. As described previously (Maslov et al., 2004), the number of LRCs is reduced in the aged SEZ (Figure 5I) when compared to young adult (3 months old) animals (Figure 5F). Notably, however, over the course of time the number of LRCs in the aged SEZ has become rather similar between control and constitutive Inscoe animals (Figure 5I).
To test whether the change in aNSC number is due to indirect effects of the constitutive overexpression of Insc or due to direct effects on the progenitor lineage, we turned to acute overexpression using lentiviral delivery of Insc. We injected lentivirus coding for GFP or Insc-IRES-GFP at E13 and quantified the number of GFP+ cells with a radial morphology (apical endfoot at the ventricular surface and a basal process) typical for NSCs in the SEZ at postnatal (postnatal day 3 [P3]) and adult stages (P56) (Figures 5J and 5K). Virtually all GFP+ cells with a radial NSC-like morphology in the SEZ were also positive for Gfap at P3 and P56. Strikingly, Insc overexpression led to a marked reduction of cells with these NSC criteria in the SEZ (Figure 5L), corroborating that Insc overexpression is regulating the number of aNSCs by cell-intrinsic mechanisms rather than indirect extrinsic effects.
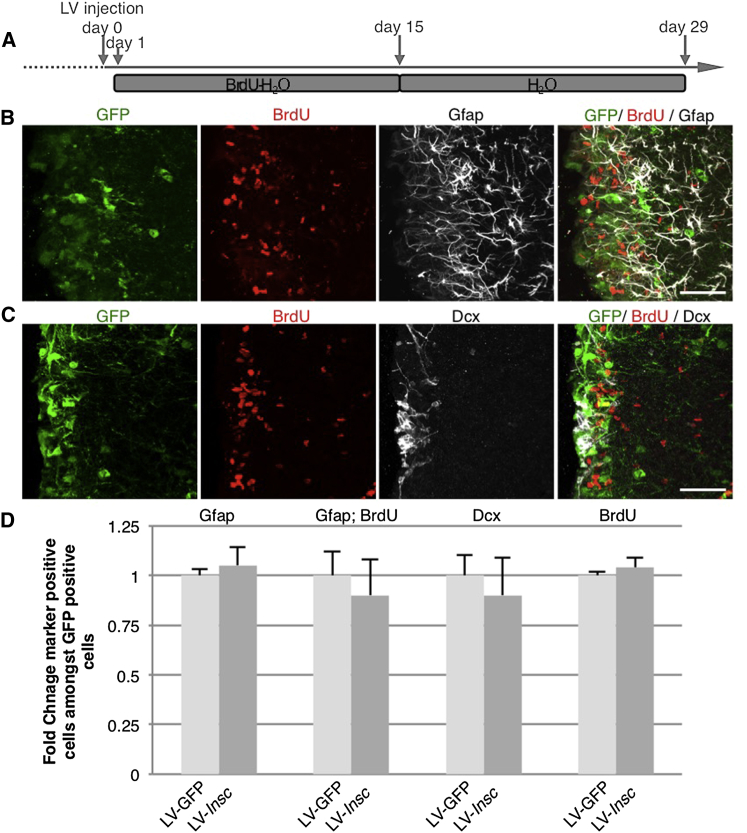
No Effect of Acute Insc Overexpression in the Adult SEZ
To directly determine to what extent Insc overexpression would also affect aNSCs in the adult, we injected the lentivirus described above into the SEZ followed by a label-retaining paradigm (Figure 6A). If the role of Insc were the same as at embryonic stages, we would expect a fast depletion of aNSCs. However, the proportion of GFP+ cells positive for the aNSC marker Gfap (Figures 6B and 6D) was similar after control or Insc transduction 29 days after infection. Moreover, neither the proportion of LRCs, nor the number of LRCs/Gfap double-positive cells, nor the proportion of Dcx+ neuroblasts, was affected in any detectable manner by acute overexpression of Insc in the adult SEZ (Figures 6B–6D), suggesting that aNSCs are not influenced by Insc overexpression.
Figure 6.
No Effect on aNSCs and Progeny after Insc Manipulation at Adult Stages
(A) Experimental paradigm used for acute manipulation of Insc levels in aNSCs. Lentiviruses coding for Insc or corresponding GFP controls were injected in 3-month-old wild-type animals, followed by a label-retaining paradigm.
(B and C) Fluorescence micrographs depicting GFP+ (infected) cells co-stained for the NSC marker Gfap, BrdU (LRC) (B), and the neuroblast marker Dcx (C).
(D) Histograms showing that the acute manipulation of Insc in adult NSCs does not have any impact on these populations (control, n = 187 cell; Insc, n = 121 cells; 6 hemispheres from 3 animals in each condition).
Data are shown as mean ± SEM (Mann-Whitney test, p > 0.05). Scale bar, 40 μm (B and C).
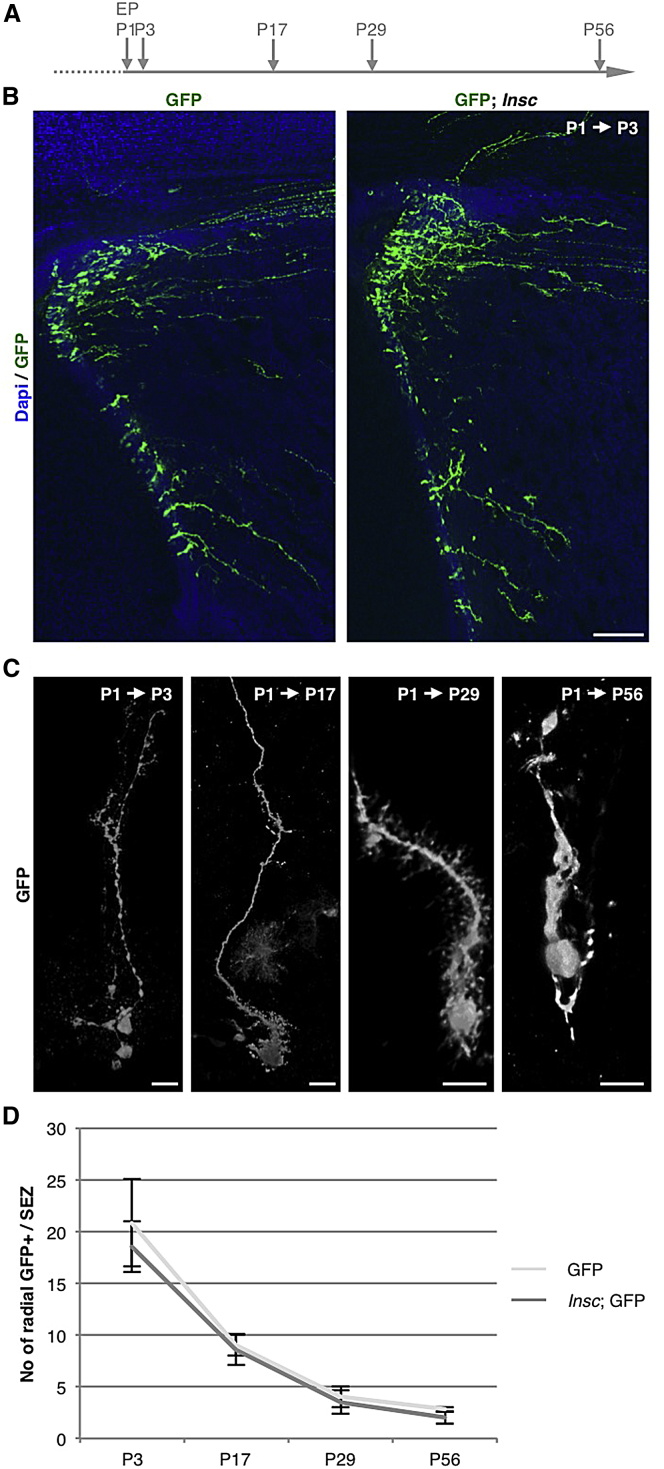
No Effect of Acute Insc Overexpression at Early Postnatal Stages
Given the lack of effects upon acute Insc overexpression in the adult SEZ as opposed to the pronounced effect at embryonic stages, we aimed to determine when sensitivity to Insc overexpression would end. Toward this end, we electroporated pups at P1 with plasmids coding for GFP or GFP and Insc (Figures 7A–7C) and examined the cells at different time points from postnatal to adult stages. At 2 days after electroporation, many GFP+ cells exhibited a radial morphology in both control and Insc electroporated conditions (Figures 7B and 7C). We also quantified the number of cells with the NSC criteria described above (apical anchoring and radial morphology with a basal process; virtually all of these cells were Gfap+) at P17, P29, and P56 in the SEZ and found no change in the number of cells with a radial morphology (Figure 7D). Thus, already at early postnatal stages Insc overexpression has no effect on the progenitor pool.
Figure 7.
No Effect on Progenitor Cell Pool after Insc Manipulation at Early Postnatal Stages
(A) Experimental paradigm used for postnatal electroporation (EP).
(B) Fluorescence micrographs showing an overview of control electroporations (CAG-GFP) and co-electroporations with CAG-Insc at P3.
(C) High-power picture of 3D renderings of apically anchored radial cells at P3, P17, P29, and P56.
(D) Graph depicting the number GFP+ cells per SEZ section maintaining a radial morphology at P3, P17, P29, and P56. Note that there is no statistical difference detectable (P3; control, n = 617 cells; Insc EP [electroporation], n = 588 cells; 6 animals each condition; P17; control, n = 348 cells; Insc EP, n = 291 cells; 8 animals each condition; P29; control, n = 147 cells; Insc EP, n = 138 cells; 8 animals each condition; P56 and P29; control, n = 82 cells; Insc EP, n = 63 cells; 6 animals each condition).
Data are shown as mean ± SEM (Mann-Whitney test, p > 0.05). Scale bar, 80 μm (B) and 15 μm (C).
Taken together, these findings demonstrate that Insc overexpression in progenitor cells of the embryonic LGE affects the embryonic progenitor cell pool in a narrow time window during later embryonic stages (no changes at E12, but effects at E14/15), but no longer at postnatal or adult stages. These changes ultimately result in a reduced seeding of aNSCs and thereby influence the amount of adult SEZ neurogenesis.
Discussion
Cell-Type Differences in Spindle Positioning and Insc Function
Here we show that cleavage plane orientation is a key factor for regulating the number of aNSCs. Surprisingly, randomization of the division plane does not increase asymmetric cell division as in other systems (Ben-Yair et al., 2011, Konno et al., 2008), but has rather the opposite effect in the developing LGE. Randomizing the cleavage plane during mitosis by perturbing either Insc levels or Lgn function increases the frequency of SNP production in the LGE. This demonstrates that the maintenance of the aRGC fate, the cells with a more stereotypic positioning of centrosomes during mitosis, is dependent on this stereotypic vertical cell division in the mother cell. Classically, epithelial cells would self-renew in a planar (here vertical) orientation of cell division with equal distribution of basal and apical determinants to the daughter cells (Alexandre et al., 2010, Das and Storey, 2012, El-Hashash et al., 2011, Goulas et al., 2012, Lechler and Fuchs, 2005). Insc promotes non-planar and hence asymmetric divisions, as observed, e.g., in the developing epidermis (Williams et al., 2014). However, in the mammary epithelium Insc levels have recently been shown to be regulated by Snail, and high levels of Insc promote stem cell numbers by promoting symmetric stem cell self-renewing divisions (Ballard et al., 2015).
Conversely, here we find that overexpression of Insc also promotes symmetric cell divisions in the LGE, but favors the generation of non-stem cell SNPs. Importantly, we observe the same phenotype when interfering with Lgn function. In the developing CNS, Lgn has been shown to be concentrated at the lateral contacts of neuroepithelial cells and aRGCs (Konno et al., 2008, Peyre et al., 2011) and to promote planar/vertical cell divisions. Accordingly, when we express dnLgn the orientation of cell divisions is also randomized, as upon Insc overexpression. Live imaging of these cells in slices of the LGE showed that this promotes in both cases a switch of aRGCs from asymmetric self-renewing divisions to symmetric divisions generating two SNPs.
It is important to note, however, that we can only predict the daughter cell type from the orientation of the cell division at the population level. In other words, if a cell divides with oblique orientation it is more likely to generate SNPs, but not every single cell that divides with oblique angle generates SNPs. This is an important difference to the rather invariable predictive behavior at the single-cell level of, e.g., Drosophila neuroblasts (Cabernard and Doe, 2009).
CNS Region-Specific Differences in Spindle Positioning and Insc Function
Interestingly, the effects of spindle reorientation are not the same in the cerebral cortex and the LGE. In the Inscoe cerebral cortex, basal mitoses and Tbr2+ basal progenitors were increased (Postiglione et al., 2011), while the fraction of non-apical mitoses was not significantly changed in the LGE. Consistent with the effects of Inscoe in the cerebral cortex, perturbing Lgn function in the cortex also caused an increase in oblique and horizontal divisions with the basal daughter cells delaminating and more progenitors dividing at non-apical positions (Das and Storey, 2012, Konno et al., 2008, Morin et al., 2007). This is very different in the LGE, with aRGCs generating more SNPs that remain apically anchored, but are not NSCs. SNPs directly generate two post-mitotic neurons in the cerebral cortex (Stancik et al., 2010), while their progeny continue to divide in the LGE (Pilz et al., 2013). The progeny of LGE SNPs are intermediate and amplifying progenitors and eventually young neurons that ultimately migrate into the striatum, as shown by short-term fate mapping using the Tα1-GFP reporter (Figure S6). Conversely, some of the aRGC progeny are self-renewing (normally, without additional manipulations) as some aRGCs are found at the apical surface using the BLBP-driven reporter several days later (Figure S6). Upon Insc overexpression, many more cells were Tα1-GFP+, demonstrating clear changes in cell fate, despite the progeny initially still being anchored at the apical surface.
SNPs differ from aRGCs by lacking a basal process (Gal et al., 2006, Pilz et al., 2013). Importantly, the basal process of aRGCs not only serves as a scaffold for migrating neurons but is also implicated in signaling events (Fietz et al., 2010, Shitamukai et al., 2011, Tsunekawa et al., 2012) and has been shown to be asymmetrically inherited between daughter cells (Miyata et al., 2001). Often the daughter cell retaining the basal process maintains the self-renewal capacity (Konno et al., 2008, LaMonica et al., 2013, Shitamukai et al., 2011), suggesting that there are cues at the basal side or within the basal process controlling aRGC self-renewal. Basally localized integrins (Fietz et al., 2010, Stenzel et al., 2014) and CyclinD2 (Tsunekawa et al., 2012) promote progenitor cell proliferation and hence self-renewal (Fietz et al., 2010, Stenzel et al., 2014). Thus, the lack of a basal process is in line with the lack of long-term self-renewal of SNPs. It is noteworthy that we did not observe a regrowth of a basal process from SNPs once the basal process was lost in live-imaging experiments, demonstrating the irreversibility of this process. Taken together, these observations suggest that the maintenance of the basal process (Shitamukai et al., 2011) is involved in regulating the number of aNSCs in the future SEZ.
Notably, aRGCs are the only source for further aRGCs, as SNPs never produced aRGCs and therefore cannot replenish the aRGC pool. The reduction of aRGCs acting as stem cells at the top of the lineage at E15 accordingly results in a reduced number of aRGCs at E18 (Figures S4F–S4H) and at postnatal stages (Figures 5J–5L). The relative increase in SNPs and their progeny compensates the reduced aRGC numbers by amplifying the lineage and, accordingly, the output. Indeed, the size of the striatum is not affected in Inscoe mice. In light of this, the long-lasting effects on aNSCs due to transient changes in the embryonic progenitor pool are even more striking, further corroborating the specific contribution of each progenitor cell population to the developing tissue, most importantly the selective contribution of aRGC maintenance to the seeding of aNSCs.
Time-Specific Role of Spindle Positioning and Insc Function
To our surprise, the above-described effects of Insc overexpression were restricted to a relatively narrow time window. While both in the developing cortex and epidermis previous work showed that Insc does not regulate the orientation of cell divisions at early stages (Postiglione et al., 2011, Williams et al., 2014), as is also the case in the E12 LGE (Figures S4A–S4E), we show here that this window also closes again at the end of embryogenesis. While in the LGE Insc overexpression rapidly and profoundly affects progenitor composition at E14/15, favoring SNPs at the expense of aRGCs, this is no longer the case when Insc is overexpressed by the same means (electroporation of the same plasmid) at early postnatal stages. Likewise, overexpression of Insc in the adult SEZ by lentiviral vectors did not affect aNSC numbers or proliferation, while overexpression with the same means during development led to a reduction of aNSCs. This is important, as changing the mode of division in a system with limited self-renewal capacity (Calzolari et al., 2015, Fuentealba et al., 2015) would rather reduce the output of neurons, with little effect on aNSC numbers, which deplete anyway after about three to four cell divisions. However, acute overexpression of Insc in aNSCs also shows an equal number of neuroblasts, suggesting that adult SEZ cells are refractory to Insc overexpression and that the reduced neuroblast numbers in adult Inscoe mice rather result from the reduced number of aNSCs due to the defects at the time when aNSCs are set aside. Thus, this study demonstrates that the seeding of aNSCs in the SEZ is modulated by spindle positioning during a critical window in embryonic development, but no longer at postnatal or adult stages. Given the limited self-renewal of aNSCs in both the SEZ and DG (Calzolari et al., 2015, Encinas et al., 2011, Fuentealba et al., 2015, Sierra et al., 2015), determining the number of aNSCs is particularly critical as it directly influences the amount of adult neurogenesis. Indeed, the reduced numbers of aNSCs results in reduced numbers of neuroblasts, the progeny of aNSCs. However, the final number of neurons in the OB is unchanged due to increased survival of adult-born neurons that counterbalances the reduced neuroblast output (Figures 5G and 5H). This finding further supports the concept that cell death exerts homeostatic control over the numbers of new neurons (Mouret et al., 2008, Petreanu and Alvarez-Buylla, 2002). However, having reduced aNSC numbers may be deleterious due to faster depletion in aging and reduced restorative capacity after brain injury. After an injury such as stroke, the SEZ contributes to the healing process by increased formation of migrating astrocytes or neuroblasts and recruitment to the injury site (Arvidsson et al., 2002, Benner et al., 2013). This response occurs over many months (Chapman et al., 2015, Thored et al., 2009) and hence would most likely be severely compromised by reduced NSC numbers.
Remarkably, at least some of the aNSCs are derived from slowly dividing progenitor cells during embryonic development (Fuentealba et al., 2015, Furutachi et al., 2015). These data not only fit to our previous and present observations based on live imaging that aRGCs in the LGE are slowest to divide (Pilz et al., 2013), but also explain why aRGCs do not get depleted even faster when generating more SNPs. Moreover, these findings are consistent with the temporal restriction of the phenotype in Inscoe mice. Very slow-dividing, label-retaining cells appear between E13 and E15 in the LGE (Furutachi et al., 2015), around the time when we see the peak of alterations in progenitor numbers and subtype specification in the Inscoe LGE. Moreover, p57 was suggested to be a key factor in driving some LGE progenitors into quiescence and hence the emergence of aNSCs (Furutachi et al., 2015). These data are consistent with the reduction of p57+ cells in the LGE ventricular zone of the LGE accompanying the reduction of the future aNSCs in Inscoe mice.
Surprisingly, our findings suggest that at the time when future aNSCs are set aside, vertical cell divisions of aRGCs are required for their formation. This could either happen by a symmetric division in which one aRGC gives rise to two future aNSCs, or by an asymmetric division in which one aRGC produces only one future aNSC and one daughter cell with a different fate. While we do not yet know the exact mode of cell division for the aRGCs generating quiescent future aNSCs, we demonstrate here that overexpression of Insc and dnLgn promotes aRGCs to generate more SNPs in a symmetric division mode, thereby depleting aRGCs. Thus, the spindle orientation machinery component Insc exerts highly region- and time-specific effects on the seeding of aNSCs and does so by a surprising effect on promoting symmetric generation of non-NSCs that remain, however, apically anchored. Taken together, our analysis hence unravels a novel effect of Insc specific to the region of the CNS that generates most of the aNSCs.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BrdU | Abcam | Cat#: ab6326; RRID: AB_305426 |
| β-catenin | BD Biosciences | Cat#: 610153; RRID: AB_397554 |
| Cleaved caspase 3 | Cell Signaling Technology | Cat#: 9661; RRID: AB_2341188 |
| Dcx | Millipore | Cat#: AB2253; RRID: AB_1586992 |
| γ-Tubulin | Sigma-Aldrich | Cat#: T6557; RRID: AB_477584 |
| Gfap | Sigma-Aldrich | Cat#: G3893; RRID: AB_477010 |
| GFP | Aves Labs | Cat#: GFP_1020; RRID: AB_10000240 |
| Ki67 | Abcam | Cat# ab15580; RRID: AB_443209 |
| mKO2 | MBL International | Cat#: M168-3; RRID: AB_10597268 |
| N-Cadherin | BD Biosciences | Cat#: 610921; RRID: AB_398236 |
| Pericentrin | Covance Research Products | Cat#: PRB-432C; RRID: AB_2313709 |
| Phospho-Histone H3 | Millipore | Cat# 06-570; RRID: AB_310177 |
| phospho-Vimentin | MBL International | Cat#: D076-3; RRID: AB_592963 |
| p57Kip2 | Sigma-Aldrich | Cat#: P0357; RRID: AB_260850 |
| Sox2 | R & D Systems | Cat#: MAB2018; RRID: AB_358009 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BrdU | Sigma-Aldrich | Cat#: B5002 |
| Tamoxifen | Sigma-Aldrich | Cat#: T5648 |
| Corn oil | Sigma-Aldrich | Cat#: C8267 |
| cell matrix type I-A | Nitta Gelatin/Wako | Cat#: 631-00651 |
| Nylon filter | Millipore | Cat#: PICM0RG50 |
| Experimental Models: Organisms/Strains | ||
| Inscoe | (Postiglione et al., 2011) | MGI:5304352 |
| CAG-CAT-GFP | (Nakamura et al., 2006) | RRID: IMSR_JAX:024636 |
| GlastCreERT2 | (Mori et al., 2006) | MGI:3830051 |
| Swiss | Janvier | RjOrl:SWISS |
| C57Bl6/J | Charles River | RRID: IMSR_JAX:000664 |
| Recombinant DNA | ||
| pCAG-LoxP-mKO2-f | (Shitamukai et al., 2011) | N/A |
| pCAG-LoxP-EGFP | (Shitamukai et al., 2011) | N/A |
| pCAG-Insc | (Konno et al., 2008) | N/A |
| pCAG-dnLgn | (Shitamukai et al., 2011) | N/A |
| pCAG-ZO1-EGFP | (Konno et al., 2008) | N/A |
| pCAG-Cre | (Shitamukai et al., 2011) | N/A |
| pCAG-H2B-Venus | kind gift of Ingo Burtscher and Heiko Lickert, Institute of Diabetes and Regeneration Research, Helmholtz Zentrum München, 85764 Neuherberg, Germany | ingo.burtscher@helmholtz-muenchen.de |
| pCAG-Arl13b-RFP | kind gift of Ingo Burtscher and Heiko Lickert; Institute of Diabetes and Regeneration Research, Helmholtz Zentrum München, 85764 Neuherberg, Germany | ingo.burtscher@helmholtz-muenchen.de |
| pTα1-EGFP | (Gal et al., 2006) | N/A |
| pBlbp-EGFP | (Gal et al., 2006) | N/A |
| pCAGGGS-EGFP | (Boutin et al., 2008) | N/A |
| Software and Algorithms | ||
| Fiji | https://fiji.sc | RRID: SCR_002285 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Magdalena Götz (magdalena.goetz@helmholtz-muenchen.de).
Experimental Model and Subject Details
C57BL/6J, GlastCreERT2 (Mori et al., 2006), CAG-CAT-GFP (Nakamura et al., 2006) and Inscoe (Postiglione et al., 2011) mice were bred in the animal facility of the Helmholtz Center Munich. The day of the vaginal plug was considered embryonic day (E) 0. Animal experiments were performed according to the institutional and legal guidelines and were approved by the government of Upper Bavaria. Swiss mice were bred in the animal facility of the Centre National de la Recherche Scientifique in Marseille. Animal experiments were carried out in accordance to European Communities Council Directive and approved by French ethical committees.
Method Details
BrdU and Tamoxifen administration
For detection of slow dividing cells in the adult SEZ, 5-bromodeoxyuridine (BrdU; Sigma, B5002) was given in drinking water (1 mg/ml) for 2 weeks, followed by 2 weeks with BrdU-free drinking water. BrdU labeling during development was performed by i.p. injection of 10 μl/g body weight of the pregnant mother of a 3.07 g/ml BrdU solution. Tamoxifen (Sigma, T5648) was dissolved at 20 mg/ml in corn oil (Sigma, C8267). 100 μg Tamoxifen was administrated per g of body weight of the pregnant mother. Note that a transient reduction in the number of proliferative cells was sporadically observed immediately after Tamoxifen administration during development.
Immunostainings
Postnatal animals were transcardially perfused with PBS followed by 4% Paraformaldehyde (PFA; (Roth, 0335.3)) in PBS. Brains were dissected and fixed overnight in 4% PFA at 4°C. E12 – E18 brains were dissected and fixed in 4% PFA for 4-14 hr at 4°C. The tissue was cryoprotected in 30% sucrose (Roth, 9097.2) in PBS, embedded in Tissue-Tek OCT (Sakura, 4583) and cut to 50 μm sections. For immunostainings the sections were blocked with blocking solution (TBS containing 10% Goat-Serum (Life Technologies, 16210072) and 0.2% Triton X-100 (Roth, 3051.2)) for 1 hr. Primary antibodies were applied overnight at 4°C in blocking solution. Primary antibodies used: BrdU (Abcam, rat, ab6326, 1:250), β-catenin (BD Biosciences, mouse IgG1, 610153, 1:1000), cleaved caspase 3 (Cell Signaling Technology, rabbit, 9661, 1:250), Dcx (Millipore, guineapig, AB2253, 1:1000), γ-Tubulin (Sigma, mouse IgG1, T6557, 1:250), Gfap (Sigma, mouse IgG1, G3893, 1:500), GFP (Aves, chicken IgY, GFP-1020, 1:1000), Ki67 (Abcam, rabbit, ab15580, 1:250) mKO2 (MBL International, mouse IgG1, M168-3, 1:500), N-Cadherin (BD Bioscience, mouse IgG1, 610921, 1:500) Pericentrin (Covance, rabbit, PRB-432C, 1:250), pH3 (Millipore, 06-570, 1:500) p-Vim (MBL, mouse IgG2b, D076-3, 1:500), p57 (Sigma, rabbit, P0357, 1:250), Sox2 (R&D, mouse IgG2a, MAB2018, 1:250). For BrdU staining sections were pre-treated with 2 N HCl at 37°C for 30 min followed by incubation in borate buffer (Na2B4O7, 0.1 M, pH 8.5) 2x15 min before starting the staining protocol. Alexa-conjugated secondary antibodies (Invitrogen) were used for visualization at a dilution of 1:500. GFP stainings for the short-term lineage tracing experiments were done with the TSA Plus System from Perkin Elmer according to the manufacturer’s instructions. Sections were counterstained with DAPI (Sigma, D9542, 1:1000) and mounted in Aqua-Polymount (Polyscience, 18606-20) and analyzed using an FV1000 Olympus laser scanning confocal.
In utero manipulations
Timed pregnant females were anaesthetized by i.p. injection of Fentanyl (0.05 mg/kg), Midazolam (5 mg/kg) and Medetomidine (0.5 mg/kg). Plasmids were mixed with FastGreen FCF 1% w/v, injected into the lateral ventricle of the developing brain and electroporated with 5 unipolar pulses at 36V for 100ms separated for 400ms by an Electro-Square-Porator ECM830 (BTX Harvard Apparatus). Anesthesia was terminated by s.c. injection of Buprenorphine (0.1 mg/kg), Atipamezole (2.5 mg/kg), Flumazenil (0.5 mg/kg). Plasmids injected were coding for pCAG-LoxP-mKO2-f, pCAG-LoxP-EGFP (Shitamukai et al., 2011), pCAG-H2B-venus and pCAG-Arl13b-RFP (kind gift of Ingo Burtscher and Heiko Lickert), pCAG-Insc (Konno et al., 2008), pCAG-dnLgn (Konno et al., 2008, Shitamukai et al., 2011), pCAG-ZO1-EGFP (Konno et al., 2008) at 1 μg/μl and pCAG-Cre at 5 ng/μl for sparse labeling (Shitamukai et al., 2011).
Postnatal electroporation
P1 pups from swiss mice were electroporated as described previously (Boutin et al., 2008). 2 μL of DNA solution containing either pCAGGS-eGFP (Boutin et al., 2008) or pCAGGS-eGFP and pCAG-Insc (Konno et al., 2008) was injected at final concentration 5 μg/μl in PBS into the lateral ventricle. Electroporation was performed using the CUY21 edit device and the 10-mm tweezers electrodes (CUY650P10, Nepagene) coated with conductive gel (Signagel, Parker Laboratories).
Time-lapse imaging
Slices of electroporated embryonic LGE were prepared and imaged as described previously (Pilz et al., 2013, Shitamukai et al., 2011). Briefly, slices were cut at 300 μm thickness on a vibratome (Leica VT 1200S) in ice-cold DMEM/F12 GlutaMax (Invitrogen) oxygenated with 95% O2, embedded in cell matrix type I-A (Nitta Gelatin) on a nylon filter (Millicell) and then cultured in DMEM/F12 GlutaMax (Invitrogen), 5% Fetal Calf Serum (GIBCO), 5% Horse Serum (GIBCO), N2 supplement (Invitrogen), B27 supplement (Invitrogen) and PenStrep (Invitrogen). The slice were kept in an atmosphere with 40% O2, 5% CO2 at 37°C.
Viral constructs, Virus production and Stereotactic injection
Lentiviral vectors expressing either IRES-GFP (LV-GFP) or Insc-IRES-GFP (LV-Insc) under the CAG promoter are based on a previously described vector system (Pfeifer et al., 2001) and were produced as described previously (Pfeifer et al., 2002). For stereotaxic injections, 10 week old mice were anesthetized as described above and injected with 1 μL of viral suspension at the coordinates 0.6 (anteroposterior), 1.2 (mediolateral), and 2–1.7 (dorsoventral) relative to bregma. For embryonic injections mice were anesthetized as described above and injected with 1 μL of viral suspension mixed with FastGreen FCF 1% w/v as described for in utero electroporations.
Quantification and Statistical Analysis
For the quantification of p-Vim z stacks of 10 optical sections 2 μm each from an Olympus FV1000 laser scanning confocal were analyzed.
To determine the cleavage angle of apical progenitors confocal stacks of Pericentrin or γ-Tubulin stained cells were taken. The cleavage plane was estimated to be perpendicular to the line between the two centrosomes in late anaphase or telophase. The cleavage angle in turn is determined to be the angle between cleavage plane and the local apical surface. We obtained very similar results by this method to the 3D analysis previously described (Jüschke et al., 2014, Postiglione et al., 2011) (data not shown). In time-lapse movies and in Figure 3A, the cleavage plane was determined using the separating sister-chromatids in late anaphase or telophase.
Quantifications are represented as mean ± SEM and statistical significance was tested using nonparametric Mann-Whitney U test or Chi-square test as indicated. ∗p < 0.05 was considered significant, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author Contributions
S.F. and M.G. designed the project and experiments. G.-A.P. made the original discovery of altered cleavage angles in the LGE. S.F., S.B., and J.N. performed the experiments and analyzed the data; M.P.P. and J.A.K. provided the Inscoe mouse line. S.F. and M.G. wrote the manuscript. S.B., H.C., J.N., G.-A.P., M.P.P., and J.A.K. corrected the manuscript.
Acknowledgments
We are particularly grateful to Fumio Matsuzaki and Tarik Haydar for plasmids used for IUE, and to Victor Borrell and Marisa Karow for excellent comments on the manuscript and experimental suggestions. We would also like to thank Timucin Öztürk, Andrea Steiner-Mezzadri, and Sabine Ulbricht for excellent technical assistance and the members of the Götz lab for discussions. This work was funded by the SFB870, the ERC ChroNeuroRepair 340793, and the SPP1757 to M.G. and by an SNF post-doctoral fellowship (PA00P3_139709) to S.F. H.C. received funding from Agence National pour la Recherche (ANR, AtMir). Work in the J.A.K. laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund (Z_153_B09), and an advanced grant from the European Research Council (ERC).
Published: February 22, 2017
Footnotes
Supplemental Information includes seven figures and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.neuron.2017.02.009.
Supplemental Information
Related to Figures 2 and 4.
Related to Figure 4.
Related to Figure 4.
Related to Figure 4. The aRGC labeled with green is asymmetrically dividing producing one aRGC and one SNP. The aRGC labeled in blue is producing one SNP and one cell that is no longer apically anchored (NAC = non-apical cell).
References
- Ahn S., Joyner A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Alexandre P., Reugels A.M., Barker D., Blanc E., Clarke J.D. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat. Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- Arvidsson A., Collin T., Kirik D., Kokaia Z., Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat. Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- Ballard M.S., Zhu A., Iwai N., Stensrud M., Mapps A., Postiglione M.P., Knoblich J.A., Hinck L. Mammary stem cell self-renewal is regulated by Slit2/Robo1 signaling through SNAI1 and mINSC. Cell Rep. 2015;13:290–301. doi: 10.1016/j.celrep.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Tripathi P., Ninkovic J., Bayam E., Lepier A., Stempfhuber B., Kirchhoff F., Hirrlinger J., Haslinger A., Lie D.C. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Ben-Yair R., Kahane N., Kalcheim C. LGN-dependent orientation of cell divisions in the dermomyotome controls lineage segregation into muscle and dermis. Development. 2011;138:4155–4166. doi: 10.1242/dev.065169. [DOI] [PubMed] [Google Scholar]
- Benner E.J., Luciano D., Jo R., Abdi K., Paez-Gonzalez P., Sheng H., Warner D.S., Liu C., Eroglu C., Kuo C.T. Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature. 2013;497:369–373. doi: 10.1038/nature12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C., Diestel S., Desoeuvre A., Tiveron M.C., Cremer H. Efficient in vivo electroporation of the postnatal rodent forebrain. PLoS ONE. 2008;3:e1883. doi: 10.1371/journal.pone.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C., Doe C.Q. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev. Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Calzolari F., Michel J., Baumgart E.V., Theis F., Götz M., Ninkovic J. Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat. Neurosci. 2015;18:490–492. doi: 10.1038/nn.3963. [DOI] [PubMed] [Google Scholar]
- Chapman K.Z., Ge R., Monni E., Tatarishvili J., Ahlenius H., Arvidsson A., Ekdahl C.T., Lindvall O., Kokaia Z. Inflammation without neuronal death triggers striatal neurogenesis comparable to stroke. Neurobiol. Dis. 2015;83:1–15. doi: 10.1016/j.nbd.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P., Silva-Vargas V., Paul A., Maldonado-Soto A.R., Deleo A.M., Pastrana E., Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.M., Storey K.G. Mitotic spindle orientation can direct cell fate and bias Notch activity in chick neural tube. EMBO Rep. 2012;13:448–454. doi: 10.1038/embor.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Caillé I., Lim D.A., García-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- El-Hashash A.H., Turcatel G., Al Alam D., Buckley S., Tokumitsu H., Bellusci S., Warburton D. Eya1 controls cell polarity, spindle orientation, cell fate and Notch signaling in distal embryonic lung epithelium. Development. 2011;138:1395–1407. doi: 10.1242/dev.058479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Encinas J.M., Michurina T.V., Peunova N., Park J.H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz S.A., Kelava I., Vogt J., Wilsch-Bräuninger M., Stenzel D., Fish J.L., Corbeil D., Riehn A., Distler W., Nitsch R., Huttner W.B. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Miya H., Watanabe T., Kawai H., Yamasaki N., Harada Y., Imayoshi I., Nelson M., Nakayama K.I., Hirabayashi Y., Gotoh Y. Slowly dividing neural progenitors are an embryonic origin of adult neural stem cells. Nat. Neurosci. 2015;18:657–665. doi: 10.1038/nn.3989. [DOI] [PubMed] [Google Scholar]
- Gal J.S., Morozov Y.M., Ayoub A.E., Chatterjee M., Rakic P., Haydar T.F. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J. Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C., Basak O., Lugert S., Knuckles P., Obernier K., Fiorelli R., Frank S., Raineteau O., Alvarez-Buylla A., Taylor V. Molecular diversity subdivides the adult forebrain neural stem cell population. Stem Cells. 2014;32:70–84. doi: 10.1002/stem.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas S., Conder R., Knoblich J.A. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüschke C., Xie Y., Postiglione M.P., Knoblich J.A. Analysis and modeling of mitotic spindle orientations in three dimensions. Proc. Natl. Acad. Sci. USA. 2014;111:1014–1019. doi: 10.1073/pnas.1314984111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E., Goderie S., Wang Y., Lotz S., Lin G., Sun Y., Roysam B., Shen Q., Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T., Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kosodo Y., Röper K., Haubensak W., Marzesco A.M., Corbeil D., Huttner W.B. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica B.E., Lui J.H., Hansen D.V., Kriegstein A.R. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat. Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Maslov A.Y., Barone T.A., Plunkett R.J., Pruitt S.C. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J. Neurosci. 2004;24:1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F.T., Tramontin A.D., García-Verdugo J.M., Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. USA. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z., Merkle F.T., Soriano-Navarro M., Garcia-Verdugo J.M., Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Kawaguchi A., Okano H., Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Mori T., Tanaka K., Buffo A., Wurst W., Kühn R., Götz M. Inducible gene deletion in astroglia and radial glia—a valuable tool for functional and lineage analysis. Glia. 2006;54:21–34. doi: 10.1002/glia.20350. [DOI] [PubMed] [Google Scholar]
- Morin X., Jaouen F., Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- Mouret A., Gheusi G., Gabellec M.M., de Chaumont F., Olivo-Marin J.C., Lledo P.M. Learning and survival of newly generated neurons: when time matters. J. Neurosci. 2008;28:11511–11516. doi: 10.1523/JNEUROSCI.2954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret A., Lepousez G., Gras J., Gabellec M.M., Lledo P.M. Turnover of newborn olfactory bulb neurons optimizes olfaction. J. Neurosci. 2009;29:12302–12314. doi: 10.1523/JNEUROSCI.3383-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Colbert M.C., Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Götz M. Fate specification in the adult brain—lessons for eliciting neurogenesis from glial cells. BioEssays. 2013;35:242–252. doi: 10.1002/bies.201200108. [DOI] [PubMed] [Google Scholar]
- Ninkovic J., Mori T., Götz M. Distinct modes of neuron addition in adult mouse neurogenesis. J. Neurosci. 2007;27:10906–10911. doi: 10.1523/JNEUROSCI.2572-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S.C., Martínez-Cerdeño V., Kriegstein A.R. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L., Alvarez-Buylla A. Maturation and death of adult-born olfactory bulb granule neurons: role of olfaction. J. Neurosci. 2002;22:6106–6113. doi: 10.1523/JNEUROSCI.22-14-06106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros T.J., Bultje R.S., Ross M.E., Fishell G., Anderson S.A. Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 2015;13:1090–1095. doi: 10.1016/j.celrep.2015.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyre E., Jaouen F., Saadaoui M., Haren L., Merdes A., Durbec P., Morin X. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 2011;193:141–154. doi: 10.1083/jcb.201101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Brandon E.P., Kootstra N., Gage F.H., Verma I.M. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc. Natl. Acad. Sci. USA. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A., Ikawa M., Dayn Y., Verma I.M. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl. Acad. Sci. USA. 2002;99:2140–2145. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz G.A., Shitamukai A., Reillo I., Pacary E., Schwausch J., Stahl R., Ninkovic J., Snippert H.J., Clevers H., Godinho L. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat. Commun. 2013;4:2125. doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione M.P., Jüschke C., Xie Y., Haas G.A., Charalambous C., Knoblich J.A. Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–284. doi: 10.1016/j.neuron.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quyn A.J., Appleton P.L., Carey F.A., Steele R.J., Barker N., Clevers H., Ridgway R.A., Sansom O.J., Näthke I.S. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Shen Q., Wang Y., Kokovay E., Lin G., Chuang S.M., Goderie S.K., Roysam B., Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 2008;3:289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitamukai A., Konno D., Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Martín-Suárez S., Valcárcel-Martín R., Pascual-Brazo J., Aelvoet S.A., Abiega O., Deudero J.J., Brewster A.L., Bernales I., Anderson A.E. Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell. 2015;16:488–503. doi: 10.1016/j.stem.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart I.H. Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: a pilot study based on recording the number, location and plane of cleavage of mitotic figures. J. Anat. 1973;116:67–91. [PMC free article] [PubMed] [Google Scholar]
- Stancik E.K., Navarro-Quiroga I., Sellke R., Haydar T.F. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J. Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel D., Wilsch-Bräuninger M., Wong F.K., Heuer H., Huttner W.B. Integrin αvβ3 and thyroid hormones promote expansion of progenitors in embryonic neocortex. Development. 2014;141:795–806. doi: 10.1242/dev.101907. [DOI] [PubMed] [Google Scholar]
- Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., Garcia-Verdugo J.M., Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thored P., Heldmann U., Gomes-Leal W., Gisler R., Darsalia V., Taneera J., Nygren J.M., Jacobsen S.E., Ekdahl C.T., Kokaia Z., Lindvall O. Long-term accumulation of microglia with proneurogenic phenotype concomitant with persistent neurogenesis in adult subventricular zone after stroke. Glia. 2009;57:835–849. doi: 10.1002/glia.20810. [DOI] [PubMed] [Google Scholar]
- Troy A., Cadwallader A.B., Fedorov Y., Tyner K., Tanaka K.K., Olwin B.B. Coordination of satellite cell activation and self-renewal by Par-complex-dependent asymmetric activation of p38α/β MAPK. Cell Stem Cell. 2012;11:541–553. doi: 10.1016/j.stem.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa Y., Britto J.M., Takahashi M., Polleux F., Tan S.S., Osumi N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012;31:1879–1892. doi: 10.1038/emboj.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler W.A., Haydar T.F. Multiplex genetic fate mapping reveals a novel route of neocortical neurogenesis, which is altered in the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2013;33:5106–5119. doi: 10.1523/JNEUROSCI.5380-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Ratliff L.A., Postiglione M.P., Knoblich J.A., Fuchs E. Par3-mInsc and Gαi3 cooperate to promote oriented epidermal cell divisions through LGN. Nat. Cell Biol. 2014;16:758–769. doi: 10.1038/ncb3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Mori K. Critical period for sensory experience-dependent survival of newly generated granule cells in the adult mouse olfactory bulb. Proc. Natl. Acad. Sci. USA. 2005;102:9697–9702. doi: 10.1073/pnas.0406082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.M., Fogarty M., Kessaris N., Richardson W.D. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 2007;27:8286–8296. doi: 10.1523/JNEUROSCI.0476-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamenhof S. Quantitative studies of mitoses in cerebral hemispheres of fetal rats. Brain Res. 1985;352:306–309. doi: 10.1016/0165-3806(85)90120-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figures 2 and 4.
Related to Figure 4.
Related to Figure 4.
Related to Figure 4. The aRGC labeled with green is asymmetrically dividing producing one aRGC and one SNP. The aRGC labeled in blue is producing one SNP and one cell that is no longer apically anchored (NAC = non-apical cell).