Abstract
Background
The authors have previously demonstrated highly reliable automated classification of free text computed tomography (CT) imaging reports using a hybrid system that pairs linguistic (natural language processing) and statistical (machine learning) techniques. Previously performed for identifying the outcome of orbital fracture in unprocessed radiology reports from a clinical data repository, the performance has not been replicated for more complex outcomes.
Objectives
To validate automated outcome classification performance of a hybrid natural language processing (NLP) and machine learning system for brain CT imaging reports. The hypothesis was that our system has performance characteristics for identifying pediatric traumatic brain injury (TBI).
Methods
This was a secondary analysis of a subset of 2,121 CT reports from the Pediatric Emergency Care Applied Research Network (PECARN) TBI study. For that project, radiologists dictated CT reports as free text, which were then de-identified and scanned as PDF documents. Trained data abstractors manually coded each report for TBI outcome. Text was extracted from the PDF files using optical character recognition. The dataset was randomly split evenly for training and testing. Training patient reports were used as input to the Medical Language Extraction and Encoding (MedLEE) NLP tool to create structured output containing standardized medical terms and modifiers for negation, certainty, and temporal status. A random subset stratified by site was analyzed using descriptive quantitative content analysis to confirm identification of TBI findings based upon the National Institute of Neurological Disorders and Stroke Common Data Elements project. Findings were coded for presence or absence, weighted by frequency of mentions, and past/future/indication modifiers were filtered. After combining with the manual reference standard, a decision tree classifier was created using data mining tools WEKA 3.7.5 and Salford Predictive Miner 7.0. Performance of the decision tree classifier was evaluated on the test patient reports.
Results
The prevalence of TBI in the sampled population was 159 out of 2,217 (7.2%). The automated classification for pediatric TBI is comparable to our prior results, with the notable exception of lower positive predictive value (PPV). Manual review of misclassified reports, 95.5% of which were false positives, revealed that a sizable number of false-positive errors were due to differing outcome definitions between NINDS TBI findings and PECARN clinical important TBI findings, and report ambiguity not meeting definition criteria.
Conclusions
A hybrid NLP and machine learning automated classification system continues to show promise in coding free-text electronic clinical data. For complex outcomes, it can reliably identify negative reports, but manual review of positive reports may be required. As such, it can still streamline data collection for clinical research and performance improvement.
INTRODUCTION
A well-recognized barrier to the use of electronic health records (EHR) for research is that much of the information is free-text, requiring substantial time and resources to interpret the data to allow meaningful analysis. To translate biomedical informatics tools for general use in clinical data warehouses, we propose to apply a novel computer-aided data interpretation system to generate patient-oriented outcomes data suitable for outcomes research. Our objective was to validate a previously developed computer-aided free text data collection and interpretation system1 by applying it to determine a complex clinical outcome from a large, multi-center database, which was used to derive and validate a priority pediatric clinical decision rule.
The Pediatric Emergency Care Applied Research Network (PECARN) traumatic brain injury (TBI) project was a multi-year undertaking by a national research network of 25 pediatric emergency departments (EDs).2 Using conventional methods, the investigators prospectively collected clinical data and outcomes on over 42,000 patients, including nearly 15,000 head computed tomography (CT) imaging reports. This database was manually collected and interpreted to derive and validate a clinical decision rule to guide the efficient use of CT imaging for pediatric TBI patients.
We previously demonstrated high diagnostic accuracy of a hybrid system using natural language processing (NLP) and machine learning tools for automated classification of ED CT imaging reports for the presence of orbital fractures.1 The hybrid system uses a well-established medical NLP software platform, Medical Language Extraction and Encoding (MedLEE; Columbia University, New York, NY; and Health Fidelity, Menlo Park, CA).3 In the current study, we apply this technique to detect TBI-related findings in the PECARN head injury CT reports. To determine the outcome of clinically important injuries for TBI victims, we used a modern statistical machine learning technique, decision tree classification. We compared data acquisition performance of the hybrid system to that acquired from manual coding of a subset of the high-volume multicenter free-text PECARN CT imaging database. Our long-term goal is to develop a translatable, accurate, and efficient computer-aided system that collects data suitable to perform outcomes research and quality improvement for all aspects of clinical medicine.
METHODS
Study Design
This was a secondary analysis of data from a prior multi-site diagnostic imaging study on pediatric blunt head injury victims. Institutional review board approval was obtained for this secondary analysis, which was a retrospective cohort study design comparing automated classification of CT imaging reports against the reference standard of manual coding by trained data abstractors.
Study Setting and Population
The study setting and population of the original study are discussed in detail elsewhere.2 Briefly, the PECARN TBI study was a prospective cohort study of pediatric TBI patients younger than 18 years presenting to the 25 EDs in the PECARN between 2004 and 2006. Of 57,030 eligible patients, 42,412 (74.4%) were enrolled and eligible for analysis, and CT scans were obtained on 14,969 (35.3%) patients. The present study population consists of consecutive blunt head injury victims for whom head CT scanning was ordered within the Chesapeake Applied Research Network (CARN) research node subset. CT scans were obtained on 2,217 (37.1%) of the 5,987 patient cohort of the CARN node.
Head CT scans were obtained at the emergency physician’s (EP’s) discretion and interpreted by site faculty radiologists. A study pediatric radiologist, blinded to clinical presentation, made definitive interpretations of inconclusive CT scans. TBI CT findings were defined as presented in Table 1. Of the 2,217 CT scans in the CARN node, 159 (7.2%) had TBI CT findings.
Table 1.
PECARN Definition of TBI on Head CT Imaging
| Intracranial hemorrhage or contusion |
| Cerebral edema |
| Traumatic infarction |
| Diffuse axonal injury |
| Shearing injury |
| Sigmoid sinus thrombosis |
| Midline shift of intracranial contents or signs of brain herniation |
| Diastasis of the skull |
| Pneumocephalus |
| Skull fracture depressed by at least the width of the table of the skull |
Study Protocol
System Overview
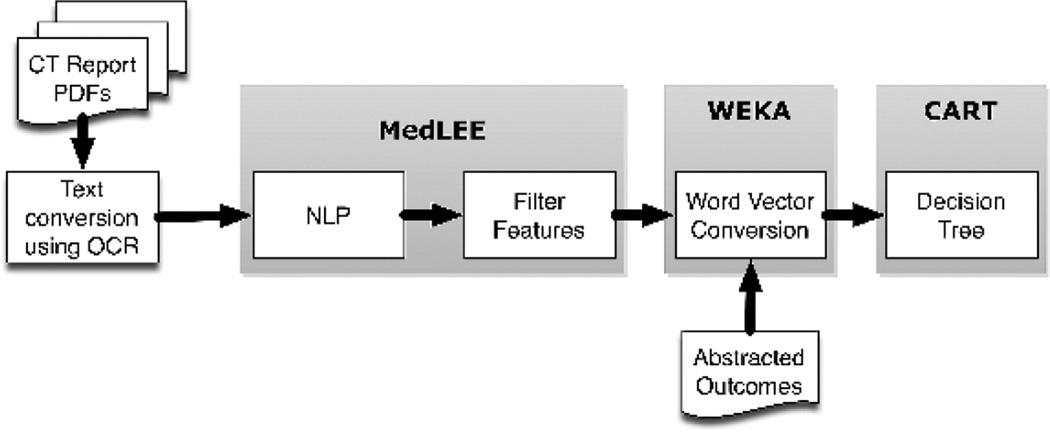
Computed tomography imaging reports were preprocessed for text conversion and then processed by NLP (Figure 1). The NLP structured output included tags to modify findings with low certainty or negation, and findings linked with patients’ histories were filtered out. The NLP-filtered findings were combined with the reference standard outcomes and then randomly divided into 50% training and 50% test sets to evaluate performance of machine learning classification.
Figure 1.
CT Reports (Pre-processing)
Computed tomography reports from the CARN node sites were provided as scanned documents in PDF format. The reports underwent optical character recognition using Adobe Acrobat Pro X to convert to text files.
Medical Language Extraction and Encoding Overview
MedLEE was chosen as the NLP module because it is one of the most widely used NLP software packages in the medical research community,4 and has previously successfully interpreted findings from free-text radiology procedure reports, including pediatric populations and head CT imaging for stroke and facial trauma.1,5,6 It is available under both commercial and academic licenses. MedLEE parses text using a grammar to recognize syntactic and semantic patterns, generating structured text with contextual modifiers that are organized in tables and assigned to Unified Medical Language System (UMLS) codes, specifically Concept Unique Identifiers (CUIs).7
Lexicon Modification
To adapt MedLEE for new clinical applications, its lexicon, abbreviations, and section names can be extended dynamically to reflect the terms and organization seen in the documents to be interpreted. This is necessary because of the need for disambiguation, where terms have different meanings in different contexts (e.g. “ventricle” in an echocardiogram report is anatomically different from “ventricle” in a CT head report). Unlike most prior methods for lexicon verification, we used descriptive quantitative content analysis to review MedLEE interpretation of TBI CT findings. Descriptive quantitative content analysis exerts prospective rigor to the process of lexicon coverage, affording a measure of objectivity, reliability, and reproducibility.8 Using a subset of 200 PECARN CT reports randomly sampled using Stata 10.1 and stratified by study site, two investigators (KY, WBC) identified content associated with presence or absence of each TBI CT finding definition, based upon the National Institute of Neurological Disorders and Stroke Common Data Elements project.9 Content analysis was performed using NVivo 9 (QSR International, Victoria, Australia).
Feature Selection Filtering
MedLEE output includes problems, findings, and procedures with associated modifiers that report specific body locations, certainty, and temporal status (Figure 2). We used the certainty and temporal status modifiers to include only likely acute findings, filtering out findings associated with historical or chronic temporal status modifiers. Findings associated with negated and low-probability certainty modifiers were included with a preceding “no_” modifier.
Figure 2.
Sample MedLEE and filtered feature outputs
Postprocessing Using Waikato Environment for Knowledge Analysis
Waikato Environment for Knowledge Analysis (WEKA; Waikato University, Hamilton, New Zealand) is an open-source collection of machine learning algorithms for data mining tasks written in Java.10 We solely used WEKA 3.7.5 for postprocessing the filtered feature sets from NLP output compiled with the reference standard outcomes of acute orbital fracture. The output was in attribute relation file format (arff), where each line represents one report with its associated outcome, which underwent conversion into word vector representations.11 The word vector representations combined unigram words and UMLS CUI phrases, with count data to weight frequency of findings within a report.
Decision Tree Classification
We used decision trees for classification because of their explicit rule-based output, which can be easily evaluated for content validity. We used the Classification and Regression Trees (CART) module of Salford Predictive Miner 6.6 (Salford Systems, San Diego, CA) to generate decision trees using the word vector attributes as predictors, without explicit constraints, minimum performance cutoffs, or maximum number of nodes. The goal was to generate a parsimonious tree that was robust to varying penalties applied to false positive or false negative cases (misclassification costs). We opted to use training and testing sets to evaluate performance instead of cross-validation, because cross-validation allows the system to train on all the data, which would not be possible in the real world. We wanted to continue our prior pragmatic approach of seeing how decision tree classifiers would perform if they were only allowed to train using a subset of data with known outcomes, and then applied to the remaining testing subset heretofore unknown to the system.1
Measurements
Automated Classification Performance
We used a 2 by 2 table to report performance of the automated classification system, using the manually abstracted PECARN TBI findings on CT as the reference standard. We report sensitivity, specificity, positive predictive value, and negative predictive value, with 95% confidence intervals.
Misclassification Analysis
Any CT report that was misclassified either in the training or test set was reviewed by two study investigators independently. Misclassifications were categorized inductively for the nature of the error. Expected categories included text conversion errors, dictation errors, report ambiguity, MedLEE NLP errors, and decision tree classification errors.
Data Analysis
A precision-based sample size for the lexicon modification was calculated to achieve a desired precision of 0.1 for a character level inter-rater agreement of Cohen’s kappa. With an alpha of 0.05, power of 0.90, and standard deviation (SD) ± 0.32 (based upon a content analysis test run of 84 PECARN CT reports), we used Stata 10.1 to estimate that 107 patient reports were needed to achieve a desired precision of 0.1 for the kappa coefficient.
Sample size for testing MedLEE performance was determined by the precision of the confidence interval (CI) around the sensitivity, knowing that the CARN research node head CT data set has a positive TBI CT report prevalence of 7.2%. Assuming sensitivity and specificity of the system is similar to the prior study1 (93% and 97% respectively), to determine the sensitivity to within 5%, we needed to enroll 1,348 total patients. Sample size requirements were calculated using PASS 2004 (NCSS, Utah).
RESULTS
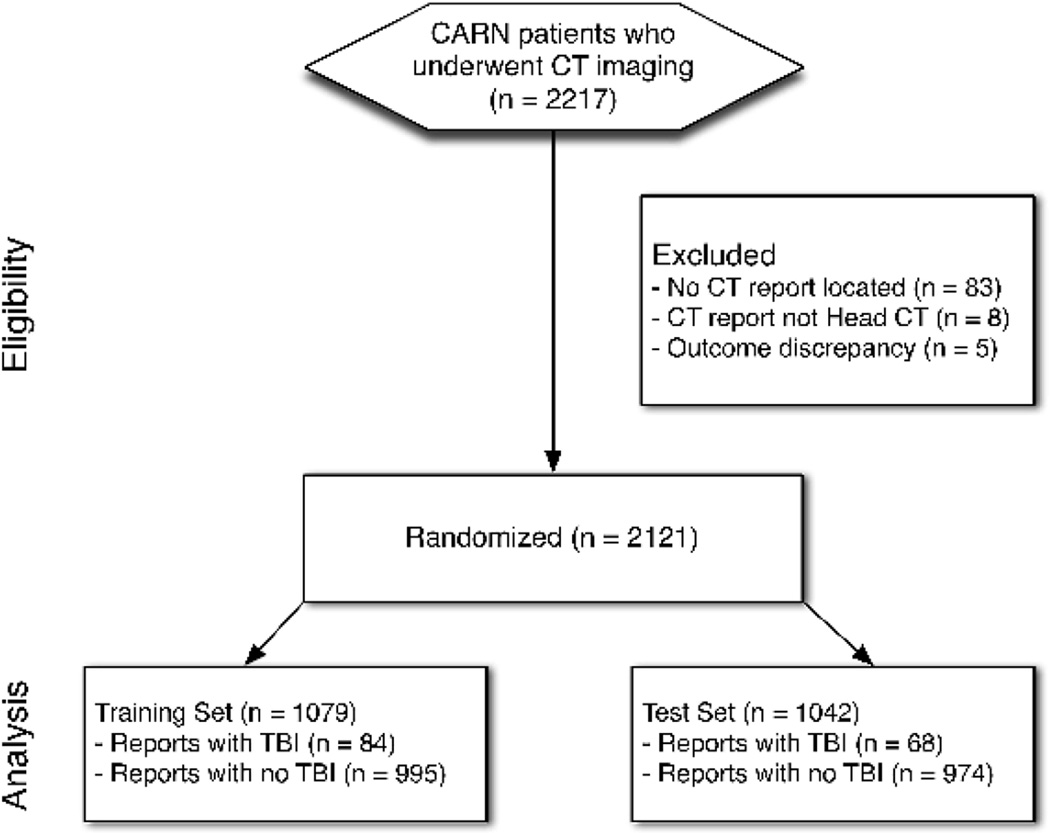
Of the 2,217 CTs performed through the CARN node, CT report PDF files were located for 2,134 (96.3%) (Figure 3). Of those, eight were not head CT reports. We excluded an additional five cases in which the reference standard was miscoded (four positives miscoded as negative and one negatives miscoded as positive). The remaining 2,121 CARN node head CT report PDF files were successfully converted to text files, which underwent processing using the MedLEE NLP platform. Content analysis performance was analyzed using stringent character level agreement for thematic coding, as well as simple agreement as to whether the report was positive or negative for TBI findings. Within 107 reports across 53 positive and negative TBI themes, the kappa for character level agreement was 0.79 (95% CI = 0.78 to 0.80), and the kappa for simple agreement was 0.88 (95% CI = 0.71 to 1.00).
Figure 3.
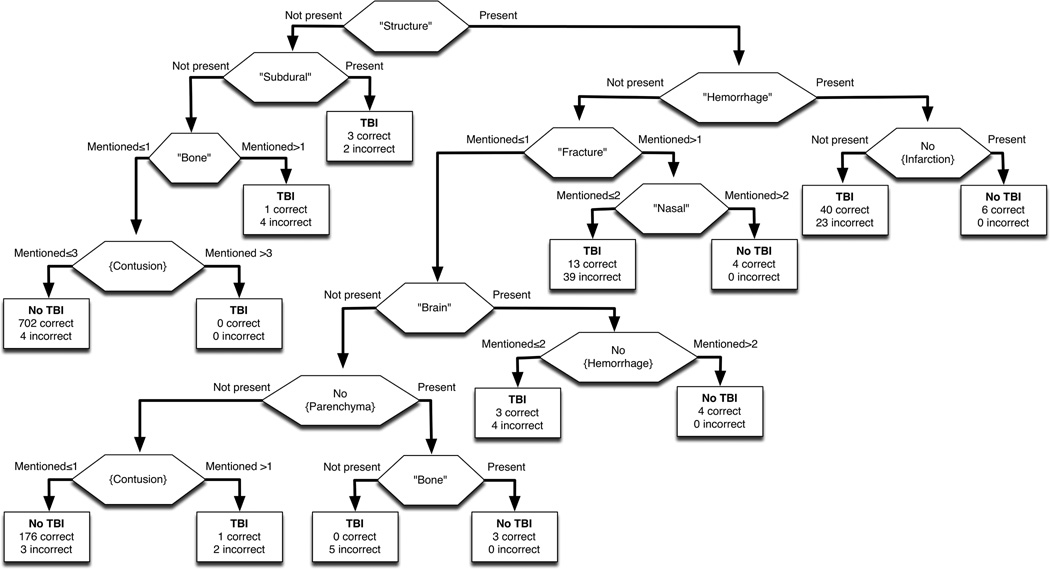
The performance of automated classification of PECARN head CT reports is provided in Table 2, with comparison to the prior performance of the system for detection of orbital fractures.1 The parsimonious CART tree was robust to varying misclassification costs (Figure 4). Misclassification analysis identified a total of 154 reports (7.3% of CT reports), with 147 (95.5%) being false positives. The categorization of misclassification is provided in Table 3.
Table 2.
Automated classification performance
| Test Characteristic | Pediatric TBI (95% CI) | Orbital Fracture1 (95% CI) |
|---|---|---|
| Sensitivity (recall) | 0.897 (0.801–0.953) | 0.933 (0.897–0.959) |
| Specificity | 0.919 (0.912–0.923) | 0.969 (0.964–0.973) |
| PPV (precision) | 0.436 (0.389–0.463) | 0.816 (0.785–0.839) |
| NPV | 0.992 (0.985–0.996) | 0.990 (0.985–0.994) |
NPV = negative predictive value; PPV = positive predictive value
Figure 4.
Table 3.
Misclassification categorization (from both test and training sets)
| Misclassification Reason | Number (%) |
|---|---|
| False negatives (from 1,829 coded negative) | 7 (0.4) |
| Decision tree misclassification | 7 (100) |
| False positives (from 292 coded positive) | 147 (50.3) |
| Abnormal but not PECARN TBI | 53 (36.1) |
| Report ambiguity | 12 (8.2) |
| Report dictation error | 6 (4.1) |
| Text conversion error | 3 (2.0) |
| MedLEE misread | 27 (18.4) |
| Decision tree misclassification | 46 (31.3) |
MedLEE = Medical Language Extraction and Encoding; PECARN = Pediatric Emergency Care Applied Research Network; TBI = traumatic brain injury
DISCUSSION
Our study adhered to the methodological standards highlighted in a prior systematic review on the use of NLP for automated classification of radiology.12 Specifically, in keeping with the prior study that established this hybrid approach of NLP and machine learning,1 this external validation study used a trained reference standard for the outcome of interest, and reports rigorous statistics for system evaluation. Improvements on the prior methodological approach included use of a robust qualitative approach to training the system for new tasks using content analysis, and provided a table of double-coded misclassification analysis results.
Our results are consistent with the conclusion of the prior systematic review, which noted that studies looking at more complex outcomes tend to underperform.12 Since that 2010 publication, more studies have been performed using hybrid techniques to evaluate complex outcomes. To our knowledge, there are no prior published studies on classification of TBI findings on head CT reports, although an abstract was presented at a conference in 2013.14 A popular example of a complex outcome has been pulmonary embolism, where location and acuity are important aspects for defining the disease and guiding clinical management.13,15
In a study by Yu et al., an enriched preprocessed sample of the Findings and Impression sections of 10,330 CT pulmonary artery (CTPA) reports from a single center were analyzed using the Narrative Information Linear Extraction-based NLP platform and logistic classifier machine learning approach.15 Although the performance metrics of the Yu et al. study were impressive (precisions between 0.79 to 0.96, recalls between 0.91 to 0.96), it should be noted that this was a derivation study without an independent testing sample, and the authors used bootstrapped performance estimates. To address the complex characterization of pulmonary embolus, the Yu et al. study made multiple binary evaluations across each aspect of the pulmonary embolus (central location, non-subsegmental, non-acute). The study authors noted that the description of acuity was often not explicit in a given sentence in the CT report (“hidden”), and so the automated classification system suffered. In our study, misclassification analysis revealed a general problem with report ambiguity (Table 3). In addition, certain aspects of the injury findings of the PECARN TBI criteria (such as degree of displacement of a skull fracture) were often not explicitly reported, and therefore hard to detect by our automated classification approach, leading to an “Abnormal but not PECARN TBI” misclassification. It should be noted that this CT report issue does not detract from the original PECARN TBI study wherein each patient had a chart review performed and the clinical decision instrument was created to predict clinically-important outcomes, not solely abnormal CT findings.2
A study be Pham et al. used a Naïve Bayes Classifier as a baseline, and either support vector machines or maximum entropy algorithms for the machine learning step to detect pulmonary embolism.13 They studied 573 preprocessed, manually annotated and sectioned CTPA reports in French from a single institution. The outcome determination and annotation tasks were performed by a single person without any validity checks, and oversampling was used despite an 83% training set. With that in mind, the performance on an independent 17% testing sample was impressive (precision of 1.00 and recall of 0.95). In contrast to both the Pham et al. and Yu et al. studies, our study did not manually preprocess CT reports beyond review of text conversion from the original PDF files. Our study used multicenter, consecutive CT reports without enrichment, without manual annotation, and without manually removing sections. While this was meant to reflect a more pragmatic use of automated classification, it did contribute to misclassifications by MedLEE as the History or Indication sections were sometimes misread as findings (Table 3).
Unlike prior studies that use “black box” machine learning classifiers like the Pham et al. study, use of decision trees allows evaluation of specific node classifiers. Furthermore, logistic model classifiers like those employed in the Yu et al. study can only be evaluated for relative contribution to classification, whereas in our study, each node can be examined for face validity and the classification can be replicated by hand. Nodes in our decision tree (Figure 3) included mentions of intracranial hemorrhage without infarction, contusions without mention of subdural or bones, hemorrhage in the context of describing brain structures, and fractures not involving decision tree creation to compare to high sensitivity or high specificity decision trees, and the final decision tree used in our study was robust to misclassification cost.
For the stated purpose of automated classification applied in a pragmatic way, the results demonstrate utility in efficiently classifying negative CTs as those with no TBI findings. Given that most naturally-occurring CT report databases have far more normal reports than abnormal ones (low prevalence populations like in our study), we would prefer to have ambiguity of positives requiring manual review than the other way around. When considering the use of such automated classification beyond research purposes, flagging potentially abnormal CT reports for review by the ordering physician would be such an application. This would be a useful application, since no patient in the PECARN TBI study with a negative head CT had clinically important TBI.16
LIMITATIONS
It is worth highlighting that the misclassification table reveals that a sizable number of false positive CT reports did have NINDS criteria for TBI but not PECARN TBI criteria. This is an important limitation for our study design, as we used the NINDS criteria rather than the PECARN TBI criteria to perform the lexicon modifications of MedLEE for the automated classification system. We chose this route because the content analysis step required a robust set of category definitions that had terminology depth, and converting the CT report to re-usable structured text is one of the purported advantages of NLP.12 The mismatch in criteria was an expected risk, and performance clearly would have been better if the outcome was also defined using the NINDS criteria for TBI.
A persistently vexing problem for automated classification is ambiguity of source documents. Most NLP approaches have qualifiers that measure certainty, but often the machine learning classifiers need to make a binary determination. A number of CT head reports lacked detail to meet PECARN criteria, such as the common example mentioned in Discussion of a “mildly depressed skull fracture” which could not be classified as meeting PECARN criteria for depth being more than the width of the skull table. But given the degree of false positives for this task, perhaps a second-step machine learning classifier with a “manual review” category could be employed.
As general purpose automated classification systems need some degree of modification for the specific task at hand (TBI, appendicitis, tuberculosis), and custom-built automated classification systems are developed within a specific context, generalizability is always a concern. The original study that used the hybrid approach we validated here was identifying orbital fractures in CT reports sourced from a single hospital. For this study, we sourced reports from multiple hospitals across the Eastern seaboard, which would increase reporting variation and hopefully create a more generalizable product. That said, we would recommend sampling CT reports at every site to evaluate performance using the methodological approach outlined in this study.
CONCLUSIONS
A hybrid natural language processing and machine learning automated classification system continues to show promise in coding free-text electronic clinical data. For complex outcomes, this approach of training an automated classification system can reliably identify negative reports, but manual review of positive reports may be required. As such, it can still streamline data collection for clinical research and performance improvement.
Acknowledgments
Financial Support: This publication is supported through the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program, grants UL1TR000075 and KL2TR000076. The CTSA program is led by the NIH's National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
MedLEE was developed with support from the National Library of Medicine (R01LM010016 and R01LM008635)
The authors would like to gratefully acknowledge the assistance of Dr. Carol Friedman, and Lyudmila Ena in the execution and completion of this project.
References
- 1.Yadav K, Sarioglu E, Smith M, Choi H-A. Automated outcome classification of emergency department computed tomography imaging reports. Acad Emerg Med. 2013;20:848–854. doi: 10.1111/acem.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuppermann N, Holmes JF, Dayan PS, et al. Identification of children at very low risk of clinically-important brain injuries after head trauma: a prospective cohort study. Lancet. 2009;374:1160–1170. doi: 10.1016/S0140-6736(09)61558-0. [DOI] [PubMed] [Google Scholar]
- 3.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–144. [PubMed] [Google Scholar]
- 5.Elkins JS, Friedman C, Boden-Albala B, Sacco RL, Hripcsak G. Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review. Comput Biomed Res. 2000;33:1–10. doi: 10.1006/cbmr.1999.1535. [DOI] [PubMed] [Google Scholar]
- 6.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. J Biomed Inform. 2005;38:314–321. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krippendorff KH. Content Analysis: An Introduction to Its Methodology. 2nd. Thousand Oaks, CA: Sage Publications, Inc; 2003. [Google Scholar]
- 9.Grinnon ST, Miller K, Marler JR, et al. National Institute of Neurological Disorders and Stroke Common Data Element Project - approach and methods. Clinical Trials. 2012;9:322–329. doi: 10.1177/1740774512438980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall M, Frank E, Holmes G, Pfahringer B, Reutmann P, Witten IH. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10–18. [Google Scholar]
- 11.Salton G. Automatic text processing. Boston, MA: Addison-Wesley; 1989. [Google Scholar]
- 12.Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. J Am Med Inform Assoc. 2010;17:646–651. doi: 10.1136/jamia.2009.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pham A-D, Névéol A, Lavergne T, et al. Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC Bioinformatics. 2014;15:266–266. doi: 10.1186/1471-2105-15-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson J, Alkasab T, Yeh D, Schaefer P. Utility assessment of repeat head CT in the setting of mild traumatic brain injury using a natural language processing tool. Radiological Society of North America 2013 Scientific Assembly and Annual Meeting; December 1 – December 6, 2013; Chicago IL. [Google Scholar]
- 15.Yu S, Kumamaru KK, George E, et al. Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform. 2014;52:386–393. doi: 10.1016/j.jbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes JF, Borgialli DA, Nadel FM, et al. Do children with blunt head trauma and normal cranial computed tomography scan results require hospitalization for neurologic observation? Ann Emerg Med. 2011;58:315–322. doi: 10.1016/j.annemergmed.2011.03.060. [DOI] [PubMed] [Google Scholar]