Abstract
Using the culturomics strategy, a wide range of anaerobic bacteria was discovered including Anaeromassilibacillus senegalensis strain mt9T (= CSUR P1511 = DSM 102954), isolated from the gut microbiota of a 1-year-old Senegalese patient with kwashiorkor. This Gram-negative strain is a strictly anaerobic, spore-forming rod motile by a polar flagellum. The 3 511 289 bp long genome of this strain contains 3046 protein-coding and 49 RNA genes, including 45 tRNA and four rRNA genes, and exhibits a G+C content of 52.94%. Here we describe the features of this organism, together with the complete genome sequence and annotation.
Keywords: Anaeromassilibacillus senegalensis, culturomics, genome, human gut, taxonogenomics
Introduction
Culturomics is a new approach introduced in our laboratory in 2012 that aims to study the diversity of the microorganisms colonizing the human gut [1]. Recently culturomics was applied to explore the bacterial diversity of patients with kwashiorkor. During this study, we isolated the strain mt9T (= CSUR P1511 = DSM 102954), which belongs to a new bacterial genus within the family Clostridiaceae. This family was created by Ernst August Pribram in 1933 [2] and contains as its core the genus Clostridium and 36 other genera [3]. Clostridiaceae members are Gram positive and present low G+C% content [4]. They have been isolated from highly diverse habitats, including freshwater and marine sediments, salt lakes, various sites of the human body, manure piles, sewage sludge and different types of faeces [4].
So far, the criteria used to classify new species based on 16S rDNA sequencing and DNA-DNA hybridization (DDH), are limited due to the low cutoff between species and genus [5]. Recently we proposed a new polyphasic approach associating phenotypic characterization, matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and genotypic characteristics to better define and classify new species [6]. In addition, characterizing new species by sequencing the whole genome allows a better distinction between the closest related species, comparing the whole genome and all of its coded function [7], [8] and genomic data to become available for new bacterial species.
In this culturomics study, we isolated the strain mt9T (= CSUR P1511 = DSM 102954), which is the first representative of a new bacterial genus within the family Clostridiaceae. Here we present a summary classification and a set of features for Anaeromassilibacillus senegalensis gen. nov., sp. nov., as well as the description of its complete genomic sequence and annotation.
Materials and Methods
Sample collection
The stool specimens were collected from a 1-year-old Senegalese patient with kwashiorkor after defecation in sterile plastic containers. The stool specimen was then formed into aliquots and stored at −80°C until use. The parents of the patient provided informed consent. The study and the assent procedure were approved by the National Ethics Committee of Senegal and the local ethics committee of the IFR48 (Marseille, France) under numbers 11-017 and 09-022, respectively.
Isolation and identification of strain
The stool sample was cultivated in anaerobic conditions and preincubated in a blood culture bottle (Becton Dickinson, Le Pont-de-Claix, France) at 37°C for 15 days after a thermic shock at 80°C for 20 minutes. Then 1 mL of the enriched culture was spread on a 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l'Etoile, France) and incubated at 37°C for 72 hours. The obtained colonies were purified and identified using MALDI-TOF MS as previously described [9], [10]. This proteomic analysis was conducted using a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) with a MTP 96 MALDI-TOF target plate (Bruker). The obtained spectra were imported into MALDI BioTyper 2.0 software (Bruker) and analysed by standard pattern matching (with default parameter settings) against the 7567 references contained in our database (Bruker database incremented with our data). When the identification score was ≥1.9, the species level was determined; a score between 1.9 and 1.7 allowed identification only at the genus level; and a score <1.7 gave no identification. In this last case, the 16S rRNA gene was sequenced as previously described [11] in order to get a molecular identification. Stackebrandt and Ebers [12] determined a 98.7 and 95% similarity level threshold to define a new species and a new genus respectively without performing DDH.
Growth conditions
Growth of the strain mt9T was tested under anaerobic and microaerophilic conditions generated using GENbag anaer and GENbag microaer systems (bioMérieux) respectively, and in aerobic conditions with or without 5% CO2. Different growth temperatures including 25, 30, 37 and 45°C were tested. Optimal salt concentration was determined by growing the strain at 0, 0.5, 1 and 1.5% NaCl. Different pH conditions were tested: 6, 6.5, 7, 7.5, 8 and 8.5.
Biochemical characterization and antibiotic susceptibility test
The biochemical characteristics of the strain mt9T were studied using API ZYM, API 20 NE and API 50 CH strips (bioMérieux) according to the manufacturer's instructions. The sensitivity to antibiotics was tested using amoxicillin, amoxicillin–clavulanic acid, clindamycin, metronidazole, vancomycin, doxycycline, imipenem, rifampicin and trimethoprim/sulfamethoxazole using antibiotic discs (i2a, Montpellier, France) [13].
Morphologic and microscopic observations
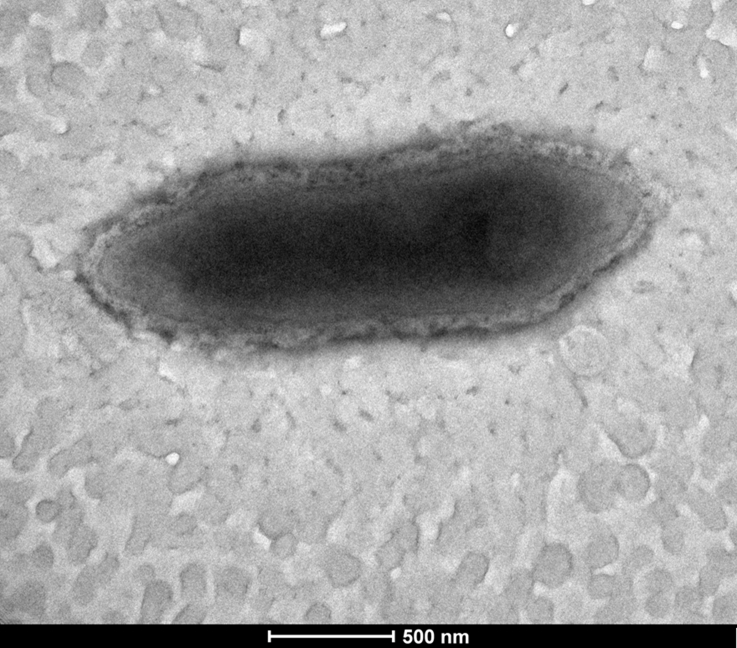
The surface colonies of the strain mt9T was observed after 72-hour incubation under anaerobic conditions at 37°C. In order to observe cell morphology, cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for at least 1 hour at 4°C. A drop of cell suspension was deposited for approximately 5 minutes on glow-discharged formvar carbon film on 400-mesh nickel grids (FCF400-Ni; Electron Microscopy Sciences (EMS), Hatfield, PA, USA). The grids were dried on blotting paper, and cells were negatively stained for 10 seconds with 1% ammonium molybdate solution in filtered water at room temperature. Electron micrographs were acquired with a Tecnai G20 Cryo (FEI Company, Limeil-Brevannes, France) transmission electron microscope operated at 200 keV. To determine the Gram stain, we used the colour Gram kit 2 (bioMérieux). Strain mt9T was observed under a DM1000 photonic microscope (Leica Microsystems, Nanterre, France) after Gram colouration as well as a fresh growing culture to assess its motility.
Fatty acid methyl ester (FAME) analysis by gas chromatography/mass spectrometry (GC/MS)
Cellular FAME analysis was performed by GC/MS. Two samples were prepared with approximately 15 mg of bacterial biomass per tube collected from several culture plates. FAME were prepared as described by Sasser [14]. GC/MS analyses were carried out as described before [15]. Briefly, FAME were separated using an Elite 5-MS column and monitored by MS (Clarus 500-SQ 8 S, PerkinElmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (National Institute of Standards and Technology (NIST), Gaithersburg, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
DNA extraction and genome sequencing
After a pretreatment by a lysozyme incubation at 37°C for 2 hours, DNA of strain mt9T was extracted on the EZ1 biorobot (Qiagen, Germantown, MD, USA) with EZ1 DNA tissues kit. The elution volume is 50 μL. Genomic DNA (gDNA) was quantified by a Qubit assay with the high sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 31.7 ng/μL.
gDNA of strain mt9T was sequenced on the MiSeq Technology (Illumina, San Diego, CA, USA) with the mate pair strategy. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). The mate pair library was prepared with 1.5 μg of gDNA using the Nextera mate pair Illumina guide. The gDNA sample was simultaneously fragmented and tagged with a mate pair junction adapter. The pattern of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1.5 to 11 kb, with an optimal size at 7.670 kb. No size selection was performed, and 600 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 804 bp on a Covaris device S2 in T6 tubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 9.39 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and sequencing run were performed in a single 39-hour run in a 2 × 251 bp read length. Total information of 6.1 Gb was obtained from a 653K/mm2 cluster density with a cluster passing quality control filters of 96.1% (12 031 000 passing filter paired reads). Within this run, the index representation for strain mt9T was determined to 9.21%. The 1 108 387 paired reads were trimmed and then assembled.
Genome annotation and comparison
Open reading frames (ORFs) were predicted using Prodigal [16] with default parameters, but the predicted ORFs were excluded if they were spanning a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank database and the Clusters of Orthologous Groups (COGs) database using BLASTP. The tRNAScanSE tool [17] was used to find tRNA genes, whereas ribosomal RNAs were found by using RNAmmer [18] and BLASTn against the GenBank database. Lipoprotein signal peptides and the number of transmembrane helices were predicted using SignalP and TMHMM respectively. ORFans were identified if their BLASTP E value was lower than 1e-03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E value of 1e-05.
We compared the genome of strain mt9T to those of other members of the order Clostridiales. Their genomes were automatically retrieved from the 16S rRNA tree using Xegen software (PhyloPattern) [19]. For each selected genome, the complete genome sequence, proteome genome sequence and ORFeome genome sequence were retrieved from the FTP of National Center for Biotechnology Information. All proteomes were analysed with proteinOrtho [20]. Then for each couple of genomes a similarity score was computed. This score is the mean value of nucleotide similarity between all couples of orthologues between the two genomes studied (average genomic identity of orthologous gene sequences; AGIOS) [21]. An annotation of the entire proteome was performed to define the distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins (using the same method as for the genome annotation). To evaluate the genomic similarity between Anaeromassilibacillus genus and closely related genera, we determined two parameters: digital DNA-DNA hybridization (dDDH), which exhibits a high correlation with DDH [22], [23], and AGIOS, which was designed to be independent from DDH. Annotation and comparison processes were performed in the Multi-Agent software system DAGOBAH [24] that included Figenix [25] libraries to provide pipeline analysis.
Results
Strain identification and phylogenetic analyses
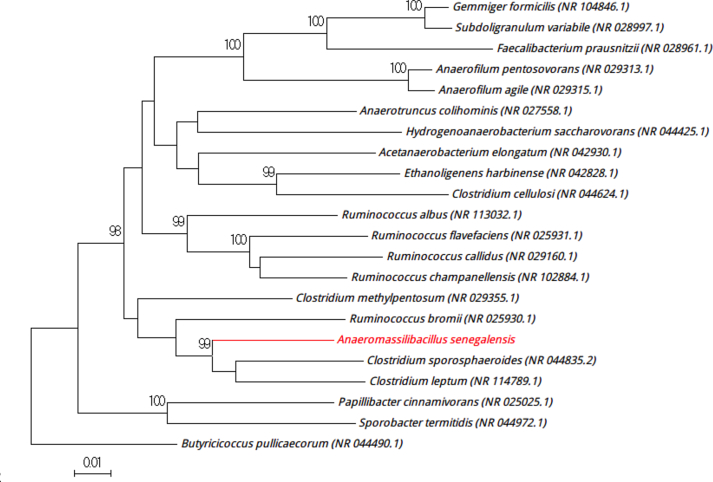
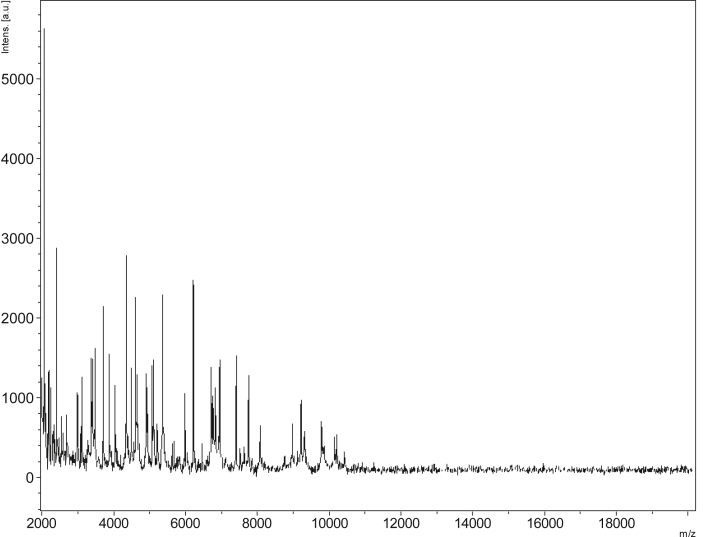
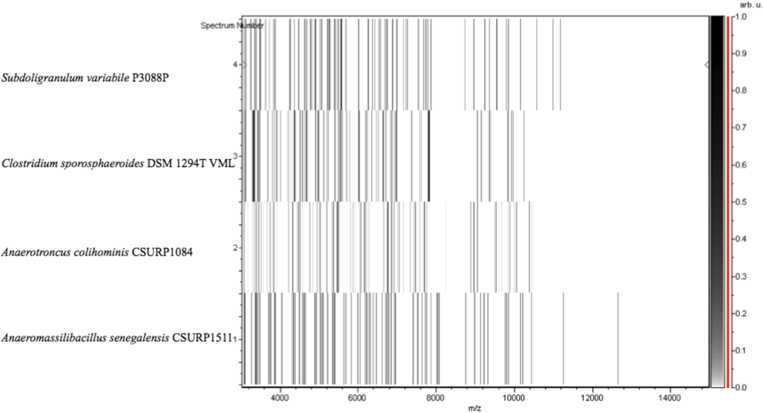
Strain mt9T (Table 1) was isolated in January 2015 by anaerobic cultivation at 37°C on 5% sheep's blood–enriched Columbia agar after a thermic shock and 15-day preincubation on blood culture bottle. MALDI-TOF MS results showed that the score obtained was under 1.7 for the strain mt9T, thus suggesting that our isolate was not a member of a known species listed in our database. 16S rRNA gene sequencing and comparison demonstrate that the strain mt9T (LN866991) exhibited 93.4% nucleotide sequence similarity with Clostridium leptum (NR114789) (Fig. 1). This value was lower than the 95% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers [12] to delineate a new genus without carrying out DDH. We incremented our database with the spectrum from strain mt9T (Fig. 2). Spectrum differences with other species are shown in Fig. 3.
Table 1.
Classification and general features of Anaeromassilibacillus senegalensis strain mt9T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum | Firmicutes |
| Class | Clostridia |
| Order | Clostridiales |
| Family | Clostridiaceae |
| Genus | Anaeromassilibacillus |
| Species | Anaeromassilibacillus senegalensis |
| Type strain | mt9T |
| Gram stain | Negative |
| Cell shape | Rod |
| Motility | Motile |
| Sporulation | sporulating |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
Fig. 1.
Phylogenetic tree highlighting position of Anaeromassilibacillus senegalensis strain mt9T relative to other type strains. 16S rRNA sequences accession numbers are indicated in parentheses. Sequences were aligned by Muscle 3.8.31 with default parameters, and phylogenetic inferences were obtained using neighbour-joining method with 500 bootstrap replicates within MEGA6 software. Only bootstraps >95% are shown. Scale bar represents 0.01% nucleotide sequence divergence.
Fig. 2.
Reference mass spectrum from Anaeromassilibacillus senegalensis strain mt9T.
Fig. 3.
Gel view comparing Anaeromassilibacillus senegalensis strain mt9T to closely related genera. Gel view displays raw spectra of loaded spectrum files arranged in pseudo-gel-like look. X-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. The color bar and the right y-axis indicate the relation between the color, the peak displayed and the peak intensity in arbitrary units. Displayed species are indicated on the left.
Phenotypic characterization
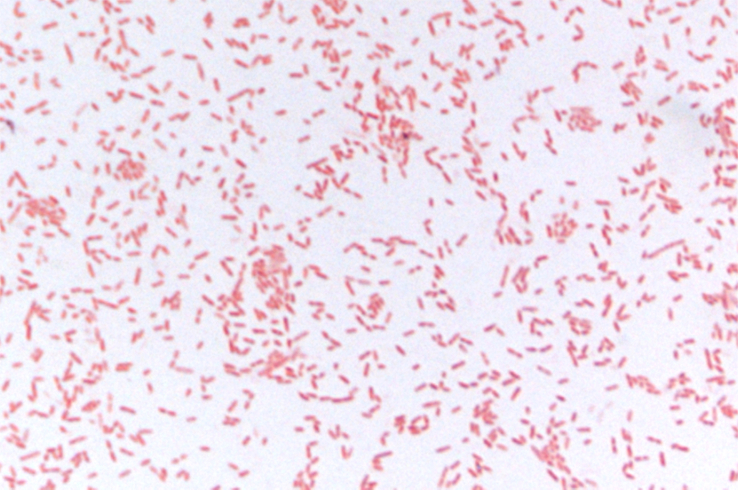
Colonies of the strain mt9T were 1 to 2 mm in diameter on Columbia agar, and opaque white in colour. Cells were Gram negative, rod shaped (Fig. 4), motile and spore-forming, 2.40 μm in length and 0.9 μm in diameter (Fig. 5). Strain mt9T grows in a range from 25 to 37°C, with an optimum at 37°C. NaCl is not required for growth, but strain mt9T was only able to grow between 0 and 0.5% NaCl. The optimum pH for growth was 7 (pH range, 6–8.5).
Fig. 4.
Gram staining of Anaeromassilibacillus senegalensis strain mt9T.
Fig. 5.
Electron micrographs of Anaeromassilibacillus senegalensis strain mt9T using Tecnai G20 transmission electron microscope (FEI Company) at operating voltage of 200 keV. Scale bar = 500 nm.
Strain mt9T does not exhibit catalase or oxidase activities. Using the API ZYM gallery, positive reactions were observed for alkaline phosphatase, esterase, esterase lipase, valine arylamidase, acid phosphatase, naphtol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase and α-mannosidase. The API 50CH gallery showed positive reactions for d-ribose, d-adonitol, dulcitol, d-sorbitol, amygdalin, arbutin, esculin, d-cellobiose, d-maltose, inulin, d-raffinose, amidone, glycogen, xylitol, gentiobiose, d-tagatose and potassium 5-ketogluconate. Using the API 20 NE, enzymatic activities were observed for gelatin hydrolysis and β-galactosidase. Urease reaction, nitrate reduction and indole production were negative.
Antibiotic susceptibility test demonstrated that strain mt9T is susceptible to vancomycin, doxycycline, ceftriaxone, imipenem, amoxicillin–clavulanic acid, penicillin and rifampicin, and is resistant to trimethoprim/sulfamethoxazole, ciprofloxacin, oxacillin, cefoxitin, colistin and fosfomycin. The phenotypic and biochemical features of the strain mt9T were compared to the features of closely related species (Table 2). The major fatty acids were hexadecanoic acid (32%), tetradecanoic acid (27%) and 9-octadecenoic acid (26%). Minor amounts of unsaturated, branched and other saturated fatty acids were also detected (Table 3).
Table 2.
Differential characteristics of Anaeromassilibacillus senegalensis DSM 102954, Clostridium thermobutyricum DSM 4928, Clostridium cellulolyticum H10, Clostridium leptum DSM 753, Clostridium sporosphaeroides DSM 1294, and Ethanoligenens harbinense YUAN-3 [26], [27], [28]
| Property | A. senegalensis | C. sporosphaeroides | C. cellulolyticum | C. leptum | E. harbinense | C. thermobutyricum |
|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.9 | 0.5–0.6 | 1.5 | 0.6–0.8 | 0.4–0.8 | 1.6–2.1 |
| Oxygen requirement | Strictly anaerobic | Strictly anaerobic | Strictly anaerobic | Strictly anaerobic | Strictly anaerobic | Strictly anaerobic |
| Gram stain | Negative | Positive | Positive | Positive | Positive | Negative |
| Motility | Motile | Nonmotile | Motile | Nonmotile | Motile | Nonmotile |
| Endospore formation | + | + | + | + | − | + |
| Indole | − | − | − | NA | + | − |
| Production of: | ||||||
| Alkaline phosphatase | + | NA | NA | NA | − | + |
| Catalase | − | NA | − | NA | − | − |
| Oxidase | − | NA | NA | NA | − | − |
| Nitrate reductase | − | − | − | NA | − | − |
| Urease | − | − | − | NA | + | NA |
| β-galactosidase | + | NA | NA | NA | − | − |
| N-acetyl-glucosamine | + | NA | NA | NA | + | − |
| Acid from: | ||||||
| l-Arabinose | − | − | NA | NA | − | + |
| Ribose | + | − | NA | w | + | + |
| Mannose | − | − | NA | − | + | − |
| Mannitol | − | − | NA | − | − | NA |
| Sucrose | − | NA | NA | w | + | − |
| d-Glucose | − | NA | NA | NA | + | + |
| d-Fructose | − | NA | NA | NA | + | + |
| d-Maltose | + | NA | NA | NA | + | + |
| d-Lactose | − | NA | NA | NA | + | + |
| G+C content (%) | 52.94 | 53.5 | 41 | 51 | 47.8 | 37 |
| Habitat | Human gut | Human gut | Compost | Human gut | Environment | Environment |
+, positive result; −, negative result; w, weakly positive result; NA, data not available.
Table 3.
Cellular fatty acid composition (%)
| Fatty acid | Name | Mean relative %a |
|---|---|---|
| 16:0 | Hexadecanoic acid | 32.3 ± 1.4 |
| 14:0 | Tetradecanoic acid | 26.5 ± 0.9 |
| 18:1n9 | 9-Octadecenoic acid | 25.8 ± 2.2 |
| 18:1n7 | 11-Octadecenoic acid | 4.0 ± 0.6 |
| 18:2n6 | 9,12-Octadecadienoic acid | 3.0 ± 0.4 |
| 18:0 | Octadecanoic acid | 2.5 ± 0.3 |
| 15:0 | Pentadecanoic acid | 1.5 ± 0.4 |
| 16:1n7 | 9-Hexadecenoic acid | 1.2 ± 0.0 |
| 14:1 | Tetradecanoic acid | TR |
| 12:0 | Dodecanoic acid | TR |
| 15:0 iso | 13-methyl-Tetradecanoic acid | TR |
| 13:0 | Tridecanoic acid | TR |
| 17:0 anteiso | 14-methyl-Hexadecanoic acid | TR |
| 15:0 anteiso | 12-methyl-tetradecanoic acid | TR |
TR, trace amounts <1%.
Mean peak area percentage.
Genome properties
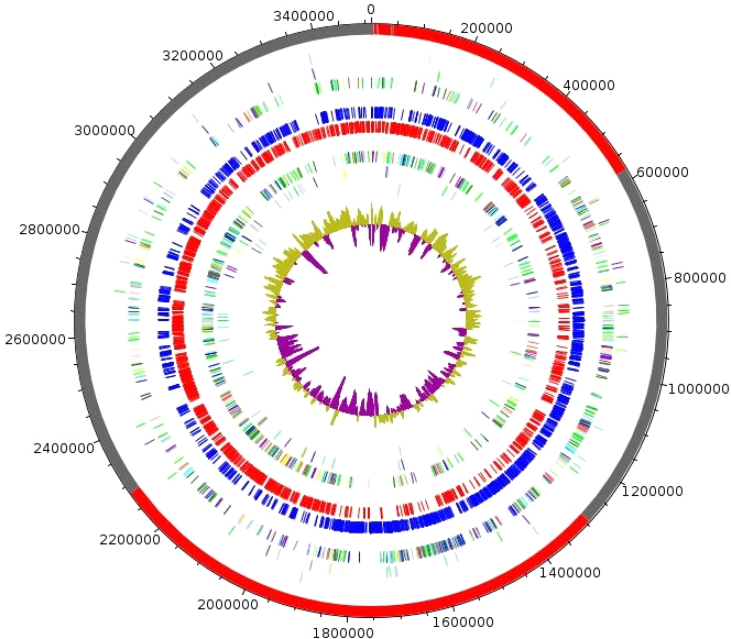
The 3 511 289 bp long genome contains 52.94% of G+C content (Fig. 6 and Table 4) and is composed of 12 scaffolds (composed of 15 contigs). Of the 3095 predicted genes, 3046 were protein-coding genes and 49 were RNAs (two genes are 5S rRNA, one gene is 16S rRNA, one gene is 23S rRNA and 45 genes are tRNA genes). A total of 1773 genes (58.21%) were assigned as putative function (by COGs or by NR BLAST). Four hundred genes were identified as ORFans (13.13%). The remaining genes were annotated as hypothetical proteins (749 genes, 24.59%). Table 5 summarizes the distribution of genes into COGs functional categories. The genome sequence is deposited in European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) under accession number CWIP00000000.
Fig. 6.
Graphical circular map of genome. From outside to centre: contigs (red/grey), COGs category of genes on forward strand (three circles), genes on forward strand (blue circle), genes on reverse strand (red circle), COGs category on reverse strand (three circles), G+C content. COGs, Clusters of Orthologous Groups database.
Table 4.
Nucleotide content and gene count levels of genome
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of totala | |
| Size (bp) | 3 511 289 | 100 |
| G+C content (%) | 1 858 158 | 52.93 |
| Coding region (bp) | 3 051 966 | 86.92 |
| Total genes | 3095 | 100 |
| RNA genes | 49 | 1.58 |
| Protein-coding genes | 3046 | 100 |
| Genes with function prediction | 1773 | 58.20 |
| Genes assigned to COGs | 1526 | 50.09 |
| Genes with peptide signals | 413 | 13.55 |
| Genes with transmembrane helices | 578 | 18.97 |
| ORFans genes | 400 | 13.13 |
| Genes associated with PKS or NRPS | 5 | 0.16 |
| No. of antibiotic resistance genes | 0 | 0 |
COGs, Clusters of Orthologous Groups database.; NRPS, nonribosomal peptide synthase; PKS, polyketide synthase.
Total is based on either size of genome in base pairs or total number of protein-coding genes in annotated genome.
Table 5.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % of total | Description |
|---|---|---|---|
| J | 142 | 4.66 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 197 | 6.47 | Transcription |
| L | 102 | 3.35 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 19 | 0.62 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 54 | 1.77 | Defense mechanisms |
| T | 59 | 1.94 | Signal transduction mechanisms |
| M | 80 | 2.63 | Cell wall/membrane biogenesis |
| N | 5 | 0.16 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 22 | 0.72 | Intracellular trafficking and secretion |
| O | 49 | 1.61 | Posttanslational modification, protein turnover, chaperones |
| C | 81 | 2.66 | Energy production and conversion |
| G | 112 | 3.68 | Carbohydrate transport and metabolism |
| E | 146 | 4.79 | Amino acid transport and metabolism |
| F | 49 | 1.61 | Nucleotide transport and metabolism |
| H | 69 | 2.27 | Coenzyme transport and metabolism |
| I | 38 | 1.25 | Lipid transport and metabolism |
| P | 77 | 2.53 | Inorganic ion transport and metabolism |
| Q | 20 | 0.66 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 216 | 7.09 | General function prediction only |
| S | 124 | 4.07 | Function unknown |
| — | 1520 | 49.90 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Genome comparison
The genomic characteristics of strain mt9T were compared to closely related species (Table 6). The draft genome sequence of strain mt9T (3.51 MB) is smaller than those of Oscillibacter valericigenes, Clostridium cellulolyticum, Flavonifractor plautii, Acetivibrio cellulolyticus, Clostridium clariflavum and Clostridium papyrosolvens (4.47, 4.07, 3.81, 6.15, 4.90 and 4.40 MB respectively), but larger than those of Ethanoligenens harbinense, Faecalibacterium prausnitzii and Clostridium leptum (3.01, 3.32 and 2.82 MB respectively). The G+C content of strain mt9T (52.94%) is smaller than those of O. valericigenes, E. harbinense, F. prausnitzii and F. plautii (53.19, 55.56, 56.82 and 61.07% respectively), but larger than those of C. cellulolyticum, C. leptum, A. cellulolyticus, C. clariflavum and C. papyrosolvens (37.40, 50.25, 35.55, 35.72 and 37.13% respectively). The gene content of strain mt9T (3046) is smaller than those of O. valericigenes, C. cellulolyticum, F. plautii, A. cellulolyticus, C. clariflavum and C. papyrosolvens (4723, 3390, 4278, 5082, 3892 and 3867 respectively), but larger than those of E. harbinense, F. prausnitzii and C. leptum (2701, 2756 and 2482 respectively).
Table 6.
Genome comparison of closely related species to Anaeromassilibacillus senegalensis strain mt9T
| Organism | INSDC | Size (Mb) | G+C (%) | Protein-coding genes |
|---|---|---|---|---|
| Anaeromassilibacillus senegalensis strain mt9T | CWIP00000000 | 3.51 | 52.93 | 3046 |
| Oscillibacter valericigenes strain Sjm18–20 | AP012044 | 4.47 | 53.19 | 4723 |
| Clostridium cellulolyticum strain H10 | CP001348 | 4.07 | 37.40 | 3390 |
| Flavonifractor plautii strain ATCC_29863 | ADLO00000000.1 | 3.81 | 61.07 | 4278 |
| Clostridium clariflavum strain DSM 19732 | CP003065.1 | 4.90 | 35.72 | 3892 |
| Clostridium papyrosolvens strain DSM 2782 | ACXX00000000.2 | 4.40 | 37.13 | 3867 |
| Acetivibrio cellulolyticus strain CD2 | AEDB00000000.2 | 6.15 | 35.55 | 5082 |
| Ethanoligenens harbinense strain YUAN-3 | CP002400.1 | 3.01 | 55.56 | 2701 |
| Faecalibacterium prausnitzii strain ATCC_27768 | ACOP00000000.2 | 3.32 | 56.82 | 2756 |
| Clostridium leptum strain DSM 753 | ABCB00000000.2 | 2.81 | 50.25 | 2482 |
INSDC, International Nucleotide Sequence Database Collaboration.
Strain mt9T shared respectively 866, 787, 841, 789, 766, 818, 681, 780 and 772 orthologous genes with F. plautii, A. cellulolyticus, C. leptum, C. papyrosolvens, E. harbinense, O. valericigenes, F. prausnitzii, C. cellulolyticum and C. clariflavum. AGIOS values ranged from 52.27 to 87.31% among compared species except strain mt9T. The comparison of the strain mt9T to other species resulted in AGIOS values ranging from 57.07% with A. cellulolyticus to 67.67% with C. leptum (Table 7). dDDH was estimated between strain mt9T's genome and the other compared genomes, and results ranged from 19.6% with F. plautii to 30.4% with C. clariflavum (Table 8). These values are in the range of the dDDH values of compared species except our strain (18.8–32.2%) and under the 70% threshold used to delineate a new species [22], thus confirming the status of the strain mt9T as a previously unknown member of the Clostridiaceae family.
Table 7.
Numbers of orthologous proteins shared between genomes (upper right)a
| Flavonifractor plautii | Acetivibrio cellulolyticus | Clostridium leptum | Anaeromassilibacillus senegalensis | Clostridium papyrosolvens | Ethanoligenens harbinense | Oscillibacter valericigenes | Faecalibacterium prausnitzii | Clostridium cellulolyticum | Clostridium clariflavum | |
|---|---|---|---|---|---|---|---|---|---|---|
| F. plautii | 4278 | 795 | 760 | 866 | 817 | 734 | 906 | 701 | 800 | 773 |
| A. cellulolyticus | 52.27 | 5082 | 745 | 787 | 1348 | 824 | 822 | 601 | 1328 | 1345 |
| C. leptum | 62.04 | 58.15 | 2482 | 841 | 760 | 729 | 729 | 630 | 757 | 735 |
| A. senegalensis | 62.70 | 57.07 | 67.67 | 3046 | 789 | 766 | 818 | 681 | 780 | 772 |
| C. papyrosolvens | 53.17 | 66.65 | 58.44 | 57.65 | 3867 | 826 | 818 | 600 | 1534 | 1269 |
| E. harbinense | 63.64 | 55.08 | 63.42 | 64.45 | 55.74 | 2701 | 744 | 605 | 809 | 792 |
| O. valericigenes | 67.12 | 55.30 | 62.08 | 62.83 | 55.64 | 62.71 | 4723 | 665 | 817 | 786 |
| F. prausnitzii | 65.36 | 52.95 | 61.97 | 62.78 | 53.97 | 63.62 | 63.58 | 2756 | 603 | 584 |
| C. cellulolyticum | 53.41 | 66.63 | 58.47 | 57.76 | 87.31 | 55.69 | 55.95 | 53.76 | 3390 | 1247 |
| C. clariflavum | 53.18 | 77.09 | 58.65 | 57.72 | 66.68 | 55.75 | 55.76 | 53.51 | 66.81 | 3892 |
Average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left) and numbers of proteins per genome (bold).
Table 8.
Pairwise comparison of Anaeromassilibacillus senegalensis with nine other speciesa
| Anaeromassilibacillus senegalensis | Oscillibacter valericigenes | Clostridium cellulolyticum | Flavonifractor plautii | Clostridium clariflavum | Clostridium papyrosolvens | Acetivibrio cellulolyticus | Ethanoligenens harbinense | Faecalibacterium prausnitzii | Clostridium leptum | |
|---|---|---|---|---|---|---|---|---|---|---|
| A. senegalensis | 100% ± 00 | 23.1% ± 2.5 | 29.0% ± 2.5 | 19.6% ± 2.4 | 30.4% ± 2.5 | 30.0% ± 2.5 | 28.5% ± 2.5 | 23.2% ± 2.5 | 25.6% ± 2.5 | 21.0% ± 2.4 |
| O. valericigenes | 100% ± 00 | 25.7% ± 2.5 | 20.5% ± 2.4 | 26.8% ± 2.5 | 27.1% ± 2.5 | 26.6% ± 2.5 | 27.5% ± 2.5 | 19.6% ± 2.4 | 25.5% ± 2.5 | |
| C. cellulolyticum | 100% ± 00 | 25.8% ± 2.5 | 32.2% ± 2.5 | 28.1% ± 2.5 | 29.1% ± 2.5 | 25.7% ± 2.4 | 24.7% ± 2.5 | 27.6% ± 2.5 | ||
| F. plautii | 100% ± 00 | 27.5% ± 2.4 | 25.7% ± 2.5 | 29.0% ± 2.5 | 18.8% ± 2.4 | 20.4% ± 2.4 | 21.0% ± 2.4 | |||
| C. clariflavum | 100% ± 00 | 23.6% ± 2.5 | 21.0% ± 2.4 | 25.9% ± 2.5 | 28.4% ± 2.5 | 29.5% ± 2.5 | ||||
| C. papyrosolvens | 100% ± 00 | 23.6% ± 2.5 | 26.4% ± 2.5 | 25.8% ± 2.5 | 26.1% ± 2.5 | |||||
| A. cellulolyticus | 100% ± 00 | 27.1% ± 2.5 | 25.2% ± 2.5 | 26.2% ± 2.5 | ||||||
| E. harbinense | 100% ± 00 | 20.1% ± 2.4 | 22.2% ± 2.5 | |||||||
| F. prausnitzii | 100% ± 00 | 28.7% ± 2.5 | ||||||||
| C. leptum | 100% ± 00 |
DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HSP, high-scoring pair.
Comparison made using GGDC, formula 2 (DDH estimates based on identities/HSP length). Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with 16S rRNA and phylogenomic analyses as well as GGDC results.
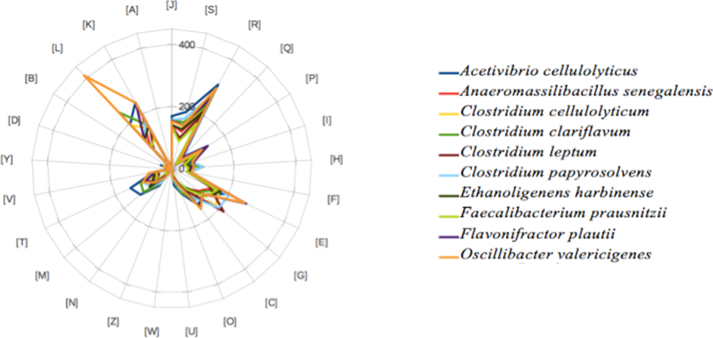
The distribution of genes into COGs categories was similar, but not identical, in all compared genomes (Fig. 7). For instance, O. valericigenes presented more genes dedicated to the DNA replication, recombination and repair.
Fig. 7.
Distribution of functional classes of predicted genes according to clusters of orthologous groups of proteins of Anaeromassilibacillus senegalensis strain mt9T and other strains.
Conclusion
The new polyphasic approach based on phenotypic, phylogenetic and genomic analyses (taxonogenomics) represented here support the affiliation of the strain mt9T to a new bacterial genus. Consequently, we formally propose the creation of the genus Anaeromassilibacillus gen. nov., and Anaeromassilibacillus senegalensis gen. nov., sp. nov., is the first representative of this new genus for which the strain mt9T is the type strain. This strain was isolated from the stool specimen of a 1-year-old patient with kwashiorkor living in Senegal.
Description of Anaeromassilibacillus gen. nov.
Anaeromassilibacillus gen. nov. (a.na.e′ro.mas.si.li.ba.cil′lus, N.L. masc. n., anaero, from anaêr, the Greek term for absence of oxygen, massili for Massilia, the Latin name of Marseille, and bacillus, ‘little rod’). Cells are Gram-negative, motile, strictly anaerobic, spore-forming, rod-shaped bacilli. Strain mt9T do not exhibited catalase or oxidase activities. Habitat is human gut. The type species is Anaeromassilibacillus senegalensis gen. nov., sp. nov.
Description of Anaeromassilibacillus senegalensis sp. nov.
Anaeromassilibacillus senegalensis (se.ne.gal.en′sis, L. masc. adj., senegalensis, ‘originating from Senegal,’ the country from which the stool sample was collected).
Cells are Gram-negative, motile, spore-forming, rod-shaped bacilli with a length of 2.4 μm and a diameter of 0.9 μm. Colonies were 1 to 2 mm in diameter on Columbia agar, and opaque white in colour. A. senegalensis strain mt9T is strictly anaerobic, grows at 37°C (optimum) and pH 7 (optimum) and NaCl is not required. A. senegalensis strain mt9T does not exhibit catalase or oxidase activities. Positive reactions were observed for alkaline phosphatase, esterase, esterase lipase, valine arylamidase, acid phosphatase, naphtol-AS-BI-phosphohydrolase, galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, d-ribose, d-adonitol, dulcitol, d-sorbitol, amygdalin, arbutin, esculin, d-cellobiose, d-maltose, inulin, d-raffinose, amidone, glycogen, xylitol, gentiobiose, d-tagatose and potassium 5-ketogluconate. Enzymatic activities were observed for gelatin hydrolysis and β-galactosidase. Urease reaction, nitrate reduction and indole production were negative. Antibiotic susceptibility test showed that A. senegalensis strain mt9T is susceptible to vancomycin, doxycycline, ceftriaxone, imipenem, amoxicillin–clavulanic acid, penicillin, and rifampicin, and resistant to trimethoprim/sulfamethoxazole, ciprofloxacin, oxacillin, cefoxitin, colistin and fosfomycin. The major fatty acid is hexadecanoic acid (32%).
The 3 511 289 bp long genome exhibits a G+C content of 52.94%. The 16S rRNA and genome sequences are deposited in EMBL-EBI under accession numbers LN866991 and CWIP00000000, respectively. The type strain mt9T (= CSUR P1511 = DSM 102954) was isolated from the gut microbiota of a 1-year-old Senegalese patient with kwashiorkor. Habitat is human gut.
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Pribram E. F. Deuticke; 1933. Klassifikation der schizomyceten (bakterien): Versuch einer wissenschaftlichen Klassifikation der bakterien auf botanischer grundlage. [Google Scholar]
- 3.Cato E.P., George W.L., Finegold S.M. Genus Clostridium Prazmowski 1880, 23 AL. In: Sneath P.H.A., Mair N.S., Sharpe M.E., Holt J.G., editors. Vol. 2. Williams & Wilkins; Baltimore, MD: 1986. pp. 1141–1200. (Bergey's manual of systematic bacteriology). [Google Scholar]
- 4.Vos P., Garrity G., Jones D., Krieg N.R., Ludwig W., Rainey F.A. Vol. 3. Springer Science & Business Media; Amsterdam: 2011. Bergey's manual of systematic bacteriology. The Firmicutes. [Google Scholar]
- 5.Rosselló-Mora R. DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. In: Stackebrandt E., editor. Molecular identification, systematics, and population structure of prokaryotes. Springer-Verlag; Berlin: 2006. pp. 23–50. [Google Scholar]
- 6.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Lagier J.C., El Karkouri K., Mishra A.K., Robert C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci. 2013;7:399–412. doi: 10.4056/sigs.3396830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hugon P., Mishra A.K., Lagier J.C., Nguyen T.T., Couderc C., Raoult D. Non-contiguous finished genome sequence and description of Brevibacillus massiliensis sp. nov. Stand Genomic Sci. 2013;8:1–14. doi: 10.4056/sigs.3466975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 10.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 13.Leclercq R., Cantòn R., Brown D.F., Giske C.G., Heisig P., MacGowan A.P. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19:141–160. doi: 10.1111/j.1469-0691.2011.03703.x. [DOI] [PubMed] [Google Scholar]
- 14.Sasser M. Microbial ID; Newark, NY: 2006. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) [Google Scholar]
- 15.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouret P., Thompson J.D., Pontarotti P. PhyloPattern: regular expressions to identify complex patterns in phylogenetic trees. BMC Bioinformatics. 2009;10:298. doi: 10.1186/1471-2105-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64(Pt 2):384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 22.Auch A.F., von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouret P., Paganini J., Dainat J., Louati D., Darbo E., Pontarotti P. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. In: Pontarotti P., editor. Evolutionary biology—concepts, biodiversity, macroevolution and genome evolution. Springer-Verlag; Berlin: 2011. pp. 71–87. [Google Scholar]
- 25.Gouret P., Vitiello V., Balandraud N., Gilles A., Pontarotti P., Danchin E.G. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiegel J., Kuk S.-U., Kohring G.W. Clostridium thermobutyricum sp. nov., a moderate thermophile isolated from a cellulolytic culture, that produces butyrate as the major product. Int J Syst Evol Microbiol. 1989;39:199–204. [Google Scholar]
- 27.Lagier J.C., Bibi F., Ramasamy D., Azhar E.I., Robert C., Yasir M. Non contiguous-finished genome sequence and description of Clostridium jeddahense sp. nov. Stand Genomic Sci. 2014;9:1003–1019. doi: 10.4056/sigs.5571026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing D., Ren N., Li Q., Lin M., Wang A., Zhao L. Ethanoligenens harbinense gen. nov., sp. nov., isolated from molasses wastewater. Int J Syst Evol Microbiol. 2006;56(Pt 4):755–760. doi: 10.1099/ijs.0.63926-0. [DOI] [PubMed] [Google Scholar]