Abstract
The Rous sarcoma virus Gag protein undergoes transient nuclear trafficking during virus assembly. Nuclear import is mediated by a nuclear targeting sequence within the MA domain. To gain insight into the role of nuclear transport in replication, we investigated whether addition of a “classical ” nuclear localization signal (NLS) in Gag would affect virus assembly or infectivity. A bipartite NLS derived from nucleoplasmin was inserted into a region of the MA domain of Gag that is dispensable for budding and infectivity. Gag proteins bearing the nucleoplasmin NLS insertion displayed an assembly defect. Mutant virus particles (RC.V8.NLS) were not infectious, although they were indistinguishable from wild-type virions in Gag, Gag-Pol, Env, and genomic RNA incorporation and Gag protein processing. Unexpectedly, postinfection viral DNA synthesis was also normal, as similar amounts of two-long-terminal-repeat junction molecules were detected for RC.V8.NLS and wild type, suggesting that the replication block occurred after nuclear entry of proviral DNA. Phenotypically revertant viruses arose after continued passage in culture, and sequence analysis revealed that the nucleoplasmin NLS coding sequence was deleted from the gag gene. To determine whether the nuclear targeting activity of the nucleoplasmin sequence was responsible for the infectivity defect, two critical basic amino acids in the NLS were altered. This virus (RC.V8.KR/AA) had restored infectivity, and the MA.KR/AA protein showed reduced nuclear localization, comparable to the wild-type MA protein. These data demonstrate that addition of a second NLS, which might direct MA and/or Gag into the nucleus by an alternate import pathway, is not compatible with productive virus infection.
The retroviral Gag polyprotein is sufficient to direct virus assembly, as demonstrated by its ability to form virus-like particles in the absence of all other viral gene products (reviewed in reference 39). Previously, it was thought that Gag proteins of type C retroviruses were synthesized on cytosolic ribosomes and then targeted directly to the plasma membrane, where budding occurs. However, we discovered that the Rous sarcoma virus (RSV) Gag polyprotein traffics through the nucleus (32). Nuclear import is mediated by a nuclear localization signal (NLS) within the N-terminal matrix (MA) domain. A CRM1-dependent nuclear export signal (NES) within the p10 domain of Gag mediates nuclear egress, after which Gag travels to the plasma membrane for particle release. Treatment of cells with leptomycin B (LMB), a specific inhibitor of CRM1, results in the accumulation of Gag proteins in the nucleus (32).
Nuclear import of macromolecules, including all proteins greater than 50 kDa, occurs by signal-mediated active transport across the nuclear membrane through the nuclear pore complex. The majority of proteins bearing specific NLS sequences are imported through an interaction with soluble transport factors belonging to the importin superfamily (reviewed in reference 9). The “classical” NLSs include the prototypical monopartite NLS of simian virus 40 large T antigen, which consists of a single patch of basic amino acids, and bipartite NLSs, including that of the nucleoplasmin protein, which contains two clusters of positively charged residues with an intervening spacer region of 10 amino acids (6). Nuclear import of protein cargoes containing these canonical NLSs is generally mediated by the formation of a complex between the NLS and a heterodimer of importins α and β. The importin α subunit contains an NLS-binding domain that interacts with positively charged amino acids in the NLS-containing protein (19, 23). However, certain NLS-bearing proteins are directly recognized by importin β (26, 40). Furthermore, nuclear import may be mediated by factors that are unrelated to the importin receptor family (44).
Beyond the classical NLSs, many import signals have been identified that are not enriched in basic residues and vary markedly in length and structure. Proteins bearing these atypical NLSs may be imported using the importin α/β pathway or may interact with other importin family members. For example, the NLSs of the cytomegalovirus protein UL84 and the cellular transcription factor STAT1 contain noncanonical import sequences that nonetheless utilize the importin α/β pathway (18, 20-22, 36). The human immunodeficiency virus type 1 (HIV-1) Vpr protein contains two nonclassical NLSs, and its importin-independent nuclear import is accomplished via direct interaction with the nuclear pore complex (12, 17). Thus, there is a high degree of variability in the protein signals and cellular mediators of nuclear import, resulting in a remarkably diverse array of nuclear entry pathways (4, 24, 30).
For RSV Gag, the minimal NLS motif in MA has not been defined, but nuclear import activity depends on a complex, nonclassical sequence within the N-terminal 88 amino acids (32). It is not yet known whether MA interfaces with a member of the importin family or whether interaction with a different pathway is required for Gag nuclear transport. To determine whether the nuclear entry pathway taken by Gag plays a critical role in virus replication, it would be ideal to delete the endogenous NLS in MA; however, the nuclear import signal overlaps the plasma membrane targeting-binding domain, and deletions in this region interfere with budding. Instead, we inserted a heterologous, classical NLS derived from the nucleoplasmin protein into a dispensable region of MA. Viruses containing Gag proteins with the classical bipartite NLS, in addition to the intrinsic NLS in MA, were markedly impaired for particle assembly. Moreover, these mutant viruses had a severe infectivity defect that was overcome by the spontaneous deletion of codons required for the foreign NLS activity, suggesting selective pressure against the nucleoplasmin NES.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
Chemically transformed quail fibroblasts (QT6) were maintained as described previously (8). Proviral constructs were based on RCAN plasmids that were modified to carry the RSV Prague C gag gene derived from pATV8. Plasmids pRC.V8 (5), pMA-GFP (8), and pGag-GFP (2) have been described elsewhere. pRC.V8.NLS was constructed by annealing overlapping oligonucleotides, 5′-CTAGTAAAGCGTCCGGCAGCCACTCTGTTGGCAGGTCAAGCGAAAAAGAAA-3′ and 5′-CTAGTTTCTTTTTCGCTTGACCTGCCAACAGAGTGGCTGCCGGACGCTTTA-3′, which were ligated into the pRC.ΔMA6 plasmid (25) at the SpeI site. The nucleoplasmin NLS sequence from Xenopus laevis (31) was modified in order to insert a BstXI restriction site, resulting in the amino acid sequence KRPAATLLAGQAKKK. Plasmids pMA.NLS-GFP and pGag.NLS-GFP were created using restriction fragment exchange, as described previously (8). pMA.NLS-GFP.rev1 was constructed by amplifying proviral DNA from infected QT6 cells, digesting with Asp718, treating with Klenow, digesting with SstI, and inserting the fragment into similarly prepared pMA-GFP. Site-directed mutagenesis of the NLS sequence within the gag gene was performed using quick-change mutagenesis (Stratagene). Primers used to create MA.KR/AA-GFP were 5′-GCGGCTCGAGAACTAGTAGCGGCGCCGGCAGCCACT-3′ and 5′-AGTGGCTGCCGGCGCCGCTACTAGTTCTCGAGCCGC-3′. The KR/AA mutant has substitutions K156A and R157A, with codon numbering according to the Xenopus nucleoplasmin sequence. The SstI-BspEI restriction fragment from pMA.KR/AA-GFP was subcloned into pGag-GFP to create pGag.KR/AA-GFP. The SstII-SdaI restriction fragment from pGag.KR/AA-GFP was ligated into the digested RC.V8 plasmid, resulting in pRC.V8.KR/AA.
Confocal microscopy.
QT6 cells were transfected using the calcium phosphate method with 1 μg of DNA per 35-mm-diameter dish, as described previously (8). At 5 h posttransfection, the culture medium was changed. Six to 11 h later, cells were washed twice with Tris-buffered saline and examined using a Zeiss LSM 10 confocal microscope after excitation at 488 nm. Treatment with 20 nM LMB (Sigma) was performed for 2 h prior to imaging. For experiments in which individual cells were scored according to the subcellular distribution of green fluorescent protein (GFP)-tagged proteins, QT6 cells were imaged 18 h after transfection with a Leica TCS SP2 AOBS confocal microscope with excitation at 488 nm. At least 200 cells were scored per experiment.
Budding assays.
Duplicate plates of QT6 cells were transfected by the calcium phosphate method, as described elsewhere (8). Sixteen hours posttransfection, cells were subjected to a 30-min methionine-cysteine starvation period and metabolically radiolabeled with l-[35S]methionine-cysteine protein labeling mix (50 μCi/μl; >1,000 Ci/mmol; NEN). One set of plates was labeled for 5 min to determine intracellular Gag expression, and the other set was labeled for 2.5 h to measure virus particle release (3). Medium was removed, cells and medium samples were individually mixed with radioimmunoprecipitation assay buffer, and Gag proteins were immunoprecipitated and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described elsewhere (42). Radioactivity incorporated into the Gag and CA proteins was quantitated using a PhosphorImager (Molecular Dynamics). Budding efficiency was expressed as the ratio of extracellular Gag (CA) to intracellular Gag (Pr76).
Virus infectivity assays.
Infectivity assays were performed in one of two ways: (i) QT6 cells were transiently transfected with proviral constructs expressing wild-type or mutant sequences and passed every 3 days for 3 to 7 weeks, or (ii) equivalent numbers of virus particles obtained from transfection, as determined by reverse transcriptase (RT) activity, were added to fresh QT6 cells, which were then passed in the same manner as above. Medium was collected before each passage, and virus particles were concentrated by ultracentrifugation through a 25% sucrose cushion at 126,000 × g at 4°C for 40 min. Viral pellets were resuspended in phosphate-buffered saline and stored at −70°C. At the end of the collection period, RT assays were performed in triplicate for each sample, as described previously (5). Cellular genomic DNA was extracted from the infected or transfected cells at the end of the collection period by the addition of 50 mM Tris (pH 8.3), 100 mM EDTA, 200 mM NaCl, 10 μg of proteinase K/μl, and 0.5% sodium dodecyl sulfate for 3 h at 37°C, followed by phenol-chloroform extraction and isopropanol precipitation. The cellular DNA was used as a template for PCR amplification of the proviral gag gene with the primers 5′-CGACTAAGCAGTCCACC-3′ and 5′-AGCAGTCAATGATCACCGGA-3′, which correspond to nucleotides 329 to 345 in the 5′ untranslated region and nucleotides 1673 to 1654 in gag, respectively. The DNA products were sequenced using an ABI Prism 377 DNA sequencer (Applied Biosystems).
RPAs.
Genomic RNA from virus particles was isolated, and RNase protection assays (RPAs) were performed as previously described (8). An antisense riboprobe derived from the 3′ splice acceptor site within env (nucleotides 4998 to 5257 from pATV8) was synthesized in vitro (Riboprobe system; Promega) and used to distinguish between unspliced (263-nucleotide protected fragment) and spliced (183-nucleotide protected fragment) viral RNAs.
Postinfection products of reverse transcription.
Virus particles were purified from the culture media of transfected QT6 cells and normalized according to RT activity, as described above. Fresh QT6 cells were incubated with medium containing equivalent amounts of virus particles, and 18 h later low-molecular-weight DNA was isolated according to the Hirt protocol (11). Primers specific for two-long-terminal-repeat (2-LTR) circles and quail mitochondrial DNA were utilized for PCR analysis as described elsewhere (1, 27), except that the annealing temperature for the mitochondrial DNA primers was 59°C and all reactions proceeded for 35 cycles. PCR products were separated using 3% Nusieve GTG agarose (Cambrex), and images of gels were captured with the Eagle Eye II still video system (Stratagene).
RESULTS
The recent discovery of a nuclear trafficking step in the life cycle of RSV has raised intriguing questions concerning possible roles for nuclear transport in retroviral replication. A report from our laboratory described a mutant of RSV Gag that does not undergo nuclear trafficking and is noninfectious, suggesting that elimination of nuclear trafficking is detrimental to virus replication (32). However, it was not known whether the opposite effect—enhancement of the nuclear trafficking of Gag—would have any consequence for virus assembly or infectivity. We considered that if nuclear trafficking were indeed a critical step in virus replication, it would be likely to be tightly controlled; thus, altering the nuclear entry properties of Gag might not be tolerated. On the other hand, enhancing the nuclear import of the mature MA protein during the early steps of infection might promote import of the preintegration complex (PIC), potentially increasing infectivity. To address these questions, we examined the consequences of inserting an ectopic NLS derived from the nucleoplasmin protein into the MA/Gag protein.
Subcellular distribution of MA-GFP fusion proteins.
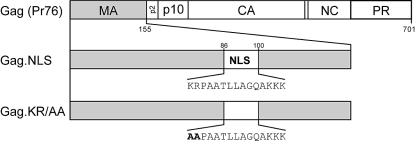
The 15-amino-acid bipartite NLS sequence was inserted in place of a dispensable region of the MA sequence, between codons 86 and 100 (25) (Fig. 1). The endogenous NLS in MA could not be deleted, since the sequence needed for nuclear localization overlaps the Gag membrane-binding domain (41) and we have not yet been able to separate these distinct endogenous targeting signals by a genetic approach (33; L. Z. Scheifele, E. P. Ryan, and L. J. Parent, unpublished data). A variant of the nucleoplasmin sequence that was predicted to be defective in nuclear import activity (KR/AA) (Fig. 1) was also inserted into the same region of MA. By using this control, we could determine whether any effects of the heterologous sequences on protein trafficking or virus replication were due to the presence of the additional nuclear targeting activity of the inserted NLS.
FIG. 1.
Schematic diagram of wild-type and mutant MA proteins. The RSV Gag polyprotein is depicted at the top, with the MA, p2, p10, CA, NC, and PR domains indicated. Gag.NLS contains a bipartite NLS derived from nucleoplasmin inserted between positions 86 and 100 of the MA domain. Gag.NLS.KR/AA contains substitutions of K to A and R to A as shown.
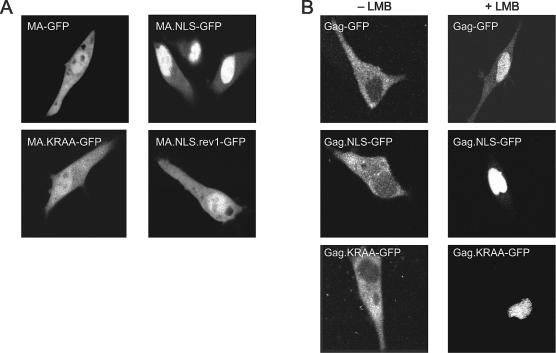
To determine whether the nucleoplasmin NLS functions in the context of the MA protein, we examined the subcellular localization of wild-type and mutant MA-GFP fusion proteins in living cells by using confocal microscopy. We expected that the NLS-containing MA protein would have an enhanced nuclear pool if the inserted nucleoplasmin NLS were active. As shown in Fig. 2A, this was the case. The MA-GFP fusion protein was distributed throughout the cell with accumulation in the nucleus, as reported previously (8), while the MA.NLS-GFP protein appeared almost entirely within the nuclear compartment (Fig. 2A). Thus, the inserted bipartite NLS enhanced the nuclear localization of RSV MA. In contrast, the subcellular distribution of MA.KR/AA-GFP was similar to that of the wild-type MA protein, indicating that substitution of alanines for positively charged residues in the upstream basic cluster of the nucleoplasmin NLS reduced nuclear targeting activity.
FIG. 2.
Subcellular localization of wild-type and mutant MA-GFP and Gag-GFP fusion proteins. (A) Live QT6 cells expressing wild-type or mutant MA-GFP fusion proteins were examined by fluorescence confocal microscopy 18 to 24 h after transfection. (B) QT6 cells transfected with plasmid DNAs expressing wild-type or mutant Gag-GFP fusion proteins were either untreated (−LMB) or treated (+LMB) with 20 nM LMB for 2 h and examined by confocal microscopy.
While the confocal microscopy images shown in Fig. 2A are representative of the population of cells expressing each of the fusion proteins, there was some heterogeneity in the distribution of fluorescence. To quantitate these differences, at least 200 individual cells were scored according to the subcellular localization of the GFP fusion proteins (Table 1). This analysis revealed that MA-GFP was located equally throughout the nucleus and cytoplasm in ∼52% of cells (Fig. 2A, top left panel), while it appeared to be more concentrated within the nucleus in ∼48% of cells. For MA.NLS-GFP, a higher percentage of cells showed nearly complete nuclear localization (∼86%) (Fig. 2A, top right panel), indicating more efficient targeting to the nuclear compartment. To determine whether the enhanced nuclear localization of MA.NLS-GFP was dependent on the integrity of the nucleoplasmin NLS, distribution of the MA.KR/AA-GFP protein was examined. For this protein, there was a reduction in the percentage of cells with nuclear localization (∼37%), indicating that disruption of the upstream basic cluster interfered with nuclear import (Fig. 2A, bottom left panel). Western blot analysis of whole-cell lysates revealed that each of the MA-GFP fusion proteins was stably expressed and there was little or no detectable free GFP protein (data not shown).
TABLE 1.
Subcellular localization of GFP fusion proteins in transfected QT6 cellsa
| GFP fusion protein | Localization
|
||
|---|---|---|---|
| Nuclear | Cytoplasmic | Diffuse | |
| MA | 48 ± 7 | 0 | 52 ± 8 |
| MA.NLS | 86 ± 1 | 0 | 14 ± 1 |
| MA.KR/AA | 37 ± 2 | 0 | 63 ± 7 |
| Untreated | |||
| Gag | 0 | 99 ± 2 | 1 ± 0 |
| Gag.NLS | 0 | 99 ± 1 | 1 ± 0 |
| Gag.KR/AA | 0 | 99 ± 1 | 1 ± 0 |
| LMB treated | |||
| Gag | 97 ± 2 | 0 | 3 ± 2 |
| Gag.NLS | 98 ± 1 | 0 | 2 ± 1 |
| Gag.KR/AA | 98 ± 1 | 0 | 2 ± 1 |
At least 200 cells were scored for protein localization in each of three independent experiments. The number of cells in each category is shown as a percentage of the total number of cells observed ± the standard deviation.
Nuclear sequestration of Gag-GFP proteins.
Following nuclear import, the RSV Gag polyprotein is transported back into the cytoplasm via the CRM1 nuclear export pathway. Treatment of cells expressing the wild-type Gag-GFP fusion protein with LMB, a specific inhibitor of CRM1, leads to the accumulation of Gag proteins within the nucleus (32). To determine whether insertion of the nucleoplasmin NLS affected nuclear trafficking of Gag, the subcellular distribution of Gag-GFP was observed in the absence or presence of LMB. In approximately 99% of untreated cells, Gag-GFP was localized throughout the cytoplasm with punctate accumulation at the plasma membrane (Fig. 2B and Table 1). Because insertion of the nucleoplasmin NLS into the MA protein enhanced the nuclear localization of MA-GFP, we expected that the nuclear concentration of Gag.NLS-GFP might also be increased, even without LMB treatment. Surprisingly, Gag.NLS-GFP was dispersed throughout the cytoplasm with plasma membrane foci and appeared indistinguishable from wild-type Gag-GFP. Therefore, the bipartite NLS did not appear to result in the accumulation of Gag in the nucleus, at least under steady-state conditions. Furthermore, addition of the heterologous NLS in Gag did not interfere with the plasma membrane targeting activity of the MA domain. As anticipated, the distribution of Gag.KR/AA-GFP was similar to that of wild-type Gag-GFP. Thus, the p10 NES is effective in mediating export, even in the presence of both the native nuclear import signal in MA and the additional inserted nucleoplasmin NLS.
Although the nucleoplasmin NLS facilitated enhanced nuclear import of MA, it was possible that it did not function in the context of the full-length Gag protein. To determine whether the NLS-modified Gag protein underwent nuclear transport, cells expressing each Gag-GFP fusion protein were treated with LMB for 2 h prior to examination. For Gag.NLS-GFP, Gag.KR/AA-GFP, and Gag-GFP, the fusion proteins were sequestered in the nucleus, indicating that at least one of the import signals (the endogenous NLS in MA and/or the ectopic nucleoplasmin NLS) was functional, and nuclear export continued to be mediated via the CRM1 pathway (Fig. 2B and Table 1). Taken together, these results indicate that the presence of the nucleoplasmin NLS sequence does not affect the export of Gag via the NES activity of p10.
Budding efficiency of wild-type and nucleoplasmin NLS-containing viruses.
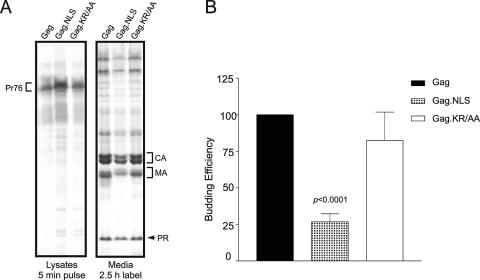
Although the Gag.NLS protein did not appear to accumulate in the nucleus under steady-state conditions, a more dynamic assay might detect differences in intracellular trafficking that are attributable to the nucleoplasmin sequence. For example, if Gag.NLS were targeted more efficiently to the nucleus due to the nucleoplasmin NLS insertion, then the mutant Gag protein might be less available in the cytoplasm to direct particle assembly. To address this possibility, genes encoding the Gag.NLS and Gag.KR/AA proteins were transferred into a proviral vector, and budding efficiency was compared to that of the wild-type virus, RC.V8. As shown in Fig. 3A, in each case the Gag polyprotein precursor (Pr76gag) was expressed in cell lysates, and Gag cleavage products (CA, MA, and PR) were detected in the culture medium. Proteolytic processing of Gag appeared to be normal for the mutants, although the MA.NLS and MA.KR/AA MA proteins migrated more slowly than wild-type MA due to their increased positive charge.
FIG. 3.
Budding efficiency of NLS mutant viruses in avian cells. (A) QT6 cells were transiently transfected with the indicated wild-type or mutant plasmids, metabolically labeled for either 5 min (left panel) or 2.5 h (right panel), and lysed, and viral proteins were immunoprecipitated from lysates and growth media by using polyclonal RSV antiserum. (B) The amount of Gag polyprotein (Pr76) isolated from cell lysates after the 5-min pulse and the amount of CA released into the medium after the 2.5-h labeling period were quantified by phosphorimager analysis. Budding efficiency was calculated by comparing the amount of CA released into the medium to the amount of Pr76 produced in cells. Budding efficiency for the wild type was assigned a value of 100, and mutant budding values were compared to the wild-type value. Each bar represents the average of eight independent experiments. The budding efficiency of RC.V8.NLS was statistically different from that of wild type (RC.V8) (P < 0.0001).
Budding efficiency was calculated by comparing the amount of CA released into the medium during a 2.5-h labeling period with the amount of Gag polyprotein (Pr76) produced within the cells during a 5-min pulse-labeling period. Budding efficiency for wild-type virus was assigned a relative value of 100. Insertion of the nucleoplasmin NLS (RC.V8.NLS) resulted in a significant reduction in particle assembly to approximately 30% of wild type (P < 0.0001). This result is consistent with the possibility that the virus assembly pathway is altered due to the nuclear targeting properties of the nucleoplasmin NLS. Moreover, disruption of basic residues in the bipartite NLS restored budding to near wild-type levels (RC.V8.KR/AA).
Infectivity in avian cells.
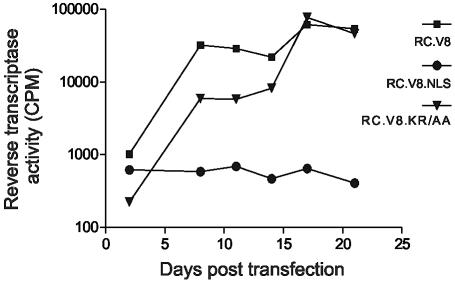
Even though RC.V8.NLS displayed a defect in virus assembly, it was possible that alteration of the dynamics of nuclear transport was inconsequential to virus replication. However, insertion of a heterologous NLS into RSV MA might be expected to enhance infectivity of the virus, as was observed for spleen necrosis virus (SNV). For SNV, insertion of a heterologous NLS into MA resulted in the efficient infection of nondividing cells (28). To determine whether the classic bipartite NLS affected RSV replication, QT6 cells were transfected with wild-type or RC.V8.NLS proviral constructs. Equivalent numbers of virus particles were collected and added to fresh cells. Establishment of infection was determined by assaying RT activity in media samples collected over a 21-day period. As shown in Fig. 4, RC.V8.NLS was unable to establish persistent infection in actively dividing QT6 cells.
FIG. 4.
Infectivity of the RC.V8.NLS viral mutant. QT6 cells were transfected with either the wild-type, RC.V8.NLS, or RC.V8.KR/AA proviral constructs and passaged in culture every 3 days for 21 days. Medium was collected prior to each passage, pelleted through a 25% sucrose cushion, and frozen. At the end of the 3-week period, RT assays on all of the stored samples were performed in triplicate and average values were determined for each.
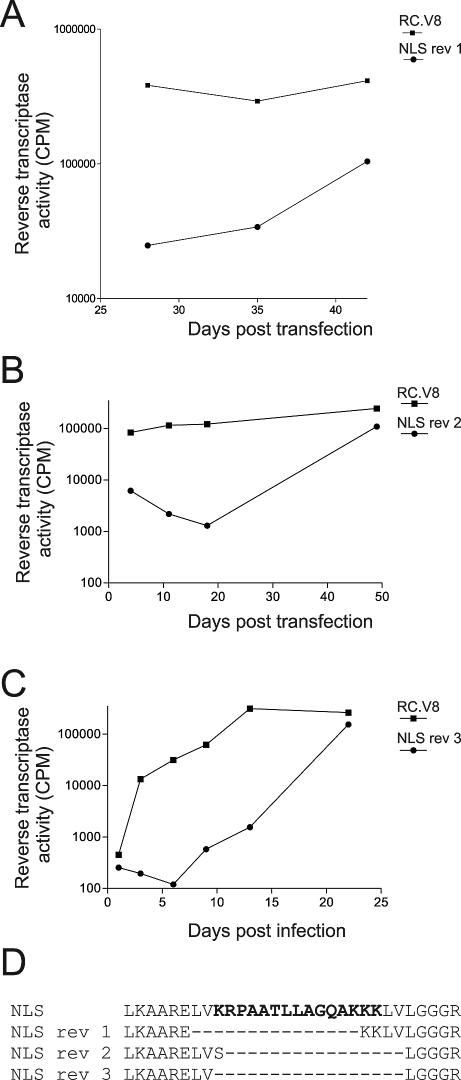
Unexpectedly but reproducibly, infectious viruses arose during continuous passage of RC.V8.NLS in QT6 cells. Three independent infectious viruses were isolated, and in each case analysis of viral sequences revealed that the nucleoplasmin-derived NLS had been eliminated (Fig. 5A, B, and C). Two of the reversion events resulted in the deletion of the entire NLS sequence with maintenance of flanking residues, while one revertant maintained the last two lysine residues of the NLS (Fig. 5D). When the MA sequence of revertant 1 was fused to enhanced GFP and protein localization was examined, the MA.NLS.rev1-GFP protein showed the same localization as wild-type MA-GFP (Fig. 2A).
FIG. 5.
Isolation and sequencing of viral revertants. (A and B) QT6 cells were transiently transfected with proviral constructs expressing the wild-type or mutant sequences and passaged every 3 days for periods ranging from 21 to 49 days, and media were analyzed as described in the legend for Fig. 4. (C) Following transfection of QT6 cells, equivalent amounts of virus particles, as determined by RT activity, were added to fresh QT6 cells, which were then passaged and analyzed as described for panels A and B. (D) Genomic DNA was extracted from the transfected cells (from panels A and B) or infected cells (from panel C) and subjected to PCR amplification of the gag gene, and the PCR products were analyzed by dideoxy sequencing. The predicted amino acid sequence of the NLS mutant and three sequences of phenotypically revertant viruses isolated from independent experiments are shown. The bold letters represent the nucleoplasmin NLS sequence, and the dashed lines indicate amino acids that were deleted from the revertant viruses.
Although it was likely that reversion occurred because of selective pressure against the nuclear targeting activity of the nucleoplasmin NLS, it was also possible that insertion of any foreign peptide sequence in this region might disrupt virus infectivity. Therefore, we examined the ability of the RC.V8KR/AA mutant, which contains a nonfunctional nucleoplasmin sequence, to propagate in culture. Importantly, RC.V8.KR/AA productively infected QT6 cells, and sequence analysis of the integrated DNA revealed that the virus had maintained the nucleoplasmin-derived sequence (Fig. 4 and data not shown).
Analysis of noninfectious virus particles.
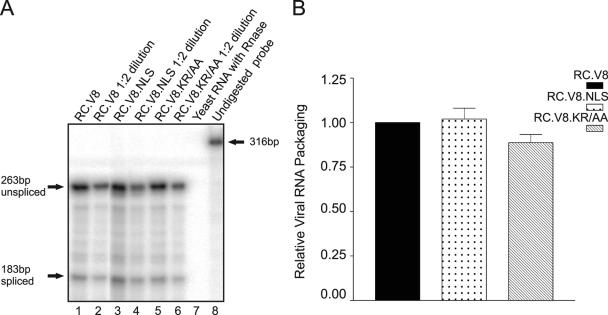
Biochemical analysis of the RC.V8.NLS viruses was undertaken in an attempt to identify a defect that could explain the loss of infectivity. To assess genomic RNA encapsidation, virus particles were isolated from the medium of transfected cells and normalized according to RT activity, and RNA extracted from equivalent numbers of particles was analyzed in an RPA with an env-specific probe (8). Viral RNA packaging for the wild type was assigned a relative value of 1.0, and for each mutant RNA packaging efficiency was expressed in comparison to that of the wild type (Fig. 6). Insertion of neither the intact nor mutated nucleoplasmin NLS sequence into Gag altered viral RNA incorporation (RC.V8.NLS and RC.V8.KR/AA, respectively). In addition, incorporation of spliced and unspliced viral RNA species was similar in the mutant and wild-type particles, indicating that splicing of the primary viral transcript was not affected (Fig. 6 and data not shown) (14). The RC.V8.NLS and RC.V8.KR/AA virus particles were also normal for Env content by immunoblot analysis with an anti-gp37 antibody (data not shown). The CA-to-RT ratio was preserved in the mutants, suggesting that Gag-Pol incorporation was unaffected (data not shown).
FIG. 6.
Quantitative viral RNA packaging. Virus-associated RNA was extracted from virus particles, which were collected from the media of transiently transfected QT6 cells. (A) RPAs were performed with a probe which spans the 3′ env splice site to distinguish between spliced (183-bp) and unspliced (263-bp) viral RNA. As a control, 5 μg of yeast RNA was added to the reaction mixture to indicate that an adequate concentration of RNase A/T1 was used (lane 7). Undigested probe is shown as a 316-bp band (lane 8). A twofold dilution of each viral RNA was included to demonstrate that the RPA was in the linear range (lanes 2, 4, and 6). (B) Viral RNA packaging efficiency for the wild type (RC.V8) was assigned a value of 1.0, and the ratios for each mutant are shown relative to the wild-type ratio. Each bar represents the average of at least three independent experiments.
Postinfection products of reverse transcription.
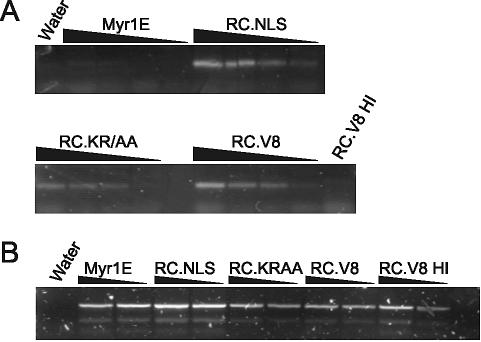
Because the biochemical analysis of RC.V8.NLS particles failed to find a cause for the infectivity defect, we wondered whether there was a defect early in infection. To evaluate the ability of the mutant virus to complete reverse transcription postentry, a PCR-based assay was utilized. QT6 cells were incubated with equivalent amounts of virus particles produced by transient transfection of proviral constructs. Eighteen hours later, cells were lysed and low-molecular-weight DNA was isolated (11) and analyzed with PCR primers specific for 2-LTR circle junctions (27). These primers do not amplify proviral plasmid DNA, and so contamination from transfected DNA was not a concern. Unexpectedly, 2-LTR circle junction DNA was readily detected for the noninfectious RC.V8.NLS mutant, as well as for the replication-competent RC.V8.KR/AA and RC.V8 viruses (Fig. 7A). The PCR products were sequenced from each reaction mixture, confirming that the 215-bp band represented a precise U5-U3 junction (data not shown). Additionally, primers specific for the gag gene were used to amplify the same DNA, and the products were sequenced to verify the presence of the original mutations for RC.V8.NLS and RC.V8.KR/AA and the wild-type sequence for RC.V8. Mitochondrial DNA was amplified from each of the same samples and, as shown in Fig. 7B, the efficiency of low-molecular-weight DNA isolation was similar from each cell lysate.
FIG. 7.
Viral DNA synthesis postinfection. QT6 cells were infected with the indicated virus, low-molecular-weight DNA was isolated 18 h later, and PCR analysis was performed using primers specific for 2-LTR circle junctions (A) and quail mitochondrial DNA (B). Decreasing amounts of DNA in serial twofold dilutions were used as the template for each PCR, as indicated by the black triangles. Wild-type virus (RC.V8) was heat inactivated at 65°C (RC.V8 HI) for 20 min prior to infection, and Hirt supernatant DNA was used undiluted as a negative control for the 2-LTR circle PCR primers and as a positive control for the quail mitochondrial DNA PCR primers.
DISCUSSION
In these experiments, we examined the effects of insertion of an additional NLS in MA/Gag on RSV replication. The nucleoplasmin NLS interfered with both particle assembly and virus replication (RC.V8.NLS). It is likely that the effects of the NLS insertion were attributable to the nuclear targeting activity of the sequence, because substitution of critical basic residues restored budding and infectivity (RC.V8.KR/AA). These data suggest that altering the nuclear trafficking properties of MA and/or Gag interferes with processes that are crucial to the virus life cycle.
Nuclear transport is a highly regulated and tightly controlled process. The integrity of the nucleus must be maintained and its contents protected, and so the nuclear envelope is selective about allowing cytosolic macromolecules access into the nuclear compartment. To gain entry, proteins typically contain an NLS that is recognized by the family of import receptors (importins) that mediate transport through the nuclear pore. In addition to the classical NLSs that enter via importin α/β interactions, many noncanonical signals have been identified that interface with other importin molecules. Thus, there are diverse pathways utilized for nuclear entry, and distinct cellular factors are involved. Whether the specific route into the nucleus has any bearing on the subsequent subnuclear localization or activities of the imported protein is not well understood but remains an intriguing question (29, 37).
Many viruses have developed strategies for entering the nucleus at particular stages of their replication cycle. In the case of RSV, the Gag polyprotein undergoes active nuclear import during the assembly phase of infection, and nuclear shuttling of Gag is an intrinsic part of the production of progeny virions (32). In this report, we found that addition of a classical NLS to MA/Gag interferes with both budding and infectivity, and there is selection against the ectopic import signal. One explanation for the decrease in budding efficiency is that the nucleoplasmin NLS might be stronger than the native NLS in Gag. Although the Gag.NLS-GFP protein did not accumulate in the nucleus under steady-state conditions (e.g., in the confocal microscopy experiments), the significant reduction in budding suggests that the Gag.NLS protein might have increased dwell time in the nucleus relative to wild-type Gag, leaving fewer molecules available in the cytoplasm to carry out virus assembly. Alternatively, perhaps the assembly defect was due to an alteration in the nuclear import pathway used by Gag.NLS; entry through the classical importin α/β pathway might be dominant over the usual import pathway used by Gag, leading to a block in virus particle formation. If the import pathway affects later intranuclear events, then perhaps the mutant lacks some type of posttranslational modification (e.g., ubiquitination or sumoylation) or it is prevented from interacting with a cellular cofactor that is required for subsequent membrane targeting or particle release. It is unlikely that plasma membrane targeting per se was disrupted in Gag.NLS, since the NLS insertion was distant from the membrane-binding (M) domain, and the protein did not accumulate in intracytoplasmic compartments, as reported for other M domain mutants (2, 32).
In addition, we must consider that the negative impact on virus replication might not be the result of altered Gag trafficking during virus assembly. Instead, the bipartite NLS introduced into the MA domain might exert its influence during the early steps of virus infection. Following receptor binding and uncoating, the preintegration complex (PIC) must gain access to the nucleus. Recent reports that RSV infects nondividing cells, albeit with low efficiency (10, 13), in conjunction with the identification of NLSs within integrase (IN) (15, 16), RT (43), and MA (32), suggest possible roles for these viral proteins in active import of the RSV PIC. Adding an ectopic NLS and, therefore, changing the nuclear entry pathway of MA, might ultimately affect nuclear targeting of the PIC. If MA plays other postentry roles, modification of its targeting signal might interfere with other events, including regulation of reverse transcription, integration, or viral transcription.
In support of this idea, our initial investigation of the infectivity defect of the nucleoplasmin NLS-containing virus particles yielded intriguing results. The RC.V8.NLS virus particles appeared to be normal in their Gag protein processing, Gag-Pol incorporation, Env packaging, and viral RNA content, yet they are replication incompetent. While there may be a gross structural problem, this seems unlikely, since changing just two basic residues in the NLS restored normal nuclear trafficking of Gag as well as infectivity, and this region of MA can be deleted without affecting infectivity. Most unexpected was the finding that RC.V8.NLS virus particles were capable of synthesizing full-length proviral DNA, and 2-LTR circle junction molecules were readily detected. Because the mutant virus RC.V8.NLS completes reverse transcription yet is unable to establish a productive infection, circular forms of proviral DNA accumulate as dead-end products that are not integrated. This result suggests that the mutant RC.V8.NLS PIC undergoes nuclear entry (although this assumption was challenged by the recent report that 2-LTR circles were isolated from the cytoplasm early after infection with murine leukemia virus [38]). This finding also supports the idea that for RSV, the MA protein might contribute to nuclear entry of the PIC; if so, then the enhanced or altered nuclear entry properties of the RC.V8.NLS MA protein appear to have adverse consequences on integration of the provirus.
Modification of the pathway of nuclear import for several other retroviral proteins has been shown to affect virus replication. The avian retrovirus SNV does not normally infect nondividing cells (7). However, insertion of the HIV-1 MA NLS into the SNV MA sequence confers enhanced viral replication in stationary cells. Similarly, NLSs derived from simian virus 40 T antigen or nucleoplasmin inserted into the murine leukemia virus IN protein disrupted virus infectivity, while NLSs from HIV-1 MA, RSV IN, or the M9 sequence of hnRNP were compatible with virus replication (34, 35). These results provide strong support for the idea that the route and regulation of nuclear entry for viral proteins is a critical determinant of virus infectivity.
These observations in other retroviral systems together with our findings in RSV lend credence to the hypothesis that it is the altered nuclear import activity of the MA protein that is detrimental to virus replication. Considering that the import factors that associate with classical NLSs, like nucleoplasmin, might be different from those utilized by the nonclassical NLS in MA, it might be utilization of the nucleoplasmin nuclear import pathway itself that interferes with the virus life cycle, presumably affecting integration. Targeting MA (and/or Gag) using the nucleoplasmin-derived NLS, in addition to the intrinsic nuclear import signal in MA, might change the route of nuclear entry and the cellular import factors engaged in transport, thereby altering the intranuclear destination of the viral protein (29, 37), so that subsequent nuclear events are unfavorable for virus replication.
Acknowledgments
This work was supported by a grant from the National Institutes of Health to L.J.P. (R01 CA76534). L.J.P. is an MDRFA scholar at the Penn State College of Medicine.
We thank Sue Delos and Judy White for kindly sharing the anti-gp37 antibody. We appreciate the contributions of Anita Hopper and Lisa Scheifele for scientific interactions and critical reviews of the manuscript and Rebecca Craven for insightful discussions. This research utilized the core facilities at the Penn State College of Medicine.
REFERENCES
- 1.Cairns, T. M., and R. C. Craven. 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J. Virol. 75:242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callahan, E. M., and J. W. Wills. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 13:310-319. [DOI] [PubMed] [Google Scholar]
- 5.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 7.Fritsch, E., and H. M. Temin. 1977. Formation and structure of infectious DNA of spleen necrosis virus. J. Virol. 21:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 10.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins, Y., M. McEntee, K. Weis, and W. C. Greene. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz, R. A., R. W. Terry, and A. M. Skalka. 1986. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 59:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukolj, G., R. A. Katz, and A. M. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 17.Le Rouzic, E., A. Mousnier, C. Rustum, F. Stutz, E. Hallberg, C. Dargemont, and S. Benichou. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 277:45091-45098. [DOI] [PubMed] [Google Scholar]
- 18.Lischka, P., G. Sorg, M. Kann, M. Winkler, and T. Stamminger. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77:3734-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 20.McBride, K. M., G. Banninger, C. McDonald, and N. C. Reich. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melen, K., R. Fagerlund, J. Franke, M. Kohler, L. Kinnunen, and I. Julkunen. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193-28200. [DOI] [PubMed] [Google Scholar]
- 22.Melen, K., L. Kinnunen, and I. Julkunen. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276:16447-16455. [DOI] [PubMed] [Google Scholar]
- 23.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 24.Nakielny, S., M. C. Siomi, H. Siomi, W. M. Michael, V. Pollard, and G. Dreyfuss. 1996. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp. Cell Res. 229:261-266. [DOI] [PubMed] [Google Scholar]
- 25.Nelle, T. D., and J. W. Wills. 1996. A large region within the Rous sarcoma virus matrix protein dispensable for budding and infectivity. J. Virol. 70:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmeri, D., and M. H. Malim. 1999. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74:164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parveen, Z., A. Krupetsky, M. Engelstadter, K. Cichutek, R. J. Pomerantz, and R. Dornburg. 2000. Spleen necrosis virus-derived C-type retroviral vectors for gene transfer to quiescent cells. Nat. Biotechnol. 18:623-629. [DOI] [PubMed] [Google Scholar]
- 29.Pemberton, L. F., J. S. Rosenblum, and G. Blobel. 1999. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol. 145:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 31.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 32.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 99:3944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheifele, L. Z., J. D. Rhoads, and L. J. Parent. 2003. Specificity of plasma membrane targeting by the Rous sarcoma virus gag protein. J. Virol. 77:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seamon, J. A., M. Adams, S. Sengupta, and M. J. Roth. 2000. Differential effects of C-terminal molecular tagged integrase on replication competent Moloney murine leukemia virus. Virology 274:412-419. [DOI] [PubMed] [Google Scholar]
- 35.Seamon, J. A., K. S. Jones, C. Miller, and M. J. Roth. 2002. Inserting a nuclear targeting signal into a replication-competent Moloney murine leukemia virus affects viral export and is not sufficient for cell cycle-independent infection. J. Virol. 76:8475-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senger, B., G. Simos, F. R. Bischoff, A. Podtelejnikov, M. Mann, and E. Hurt. 1998. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serhan, F., M. Penaud, C. Petit, T. Leste-Lasserre, S. Trajcevski, D. Klatzmann, G. Duisit, P. Sonigo, and P. Moullier. 2004. Early detection of a two-long-terminal-repeat junction molecule in the cytoplasm of recombinant murine leukemia virus-infected cells. J. Virol. 78:6190-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 40.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 19:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verderame, M. F., T. D. Nelle, and J. W. Wills. 1996. The membrane-binding domain of the Rous sarcoma virus Gag protein. J.Virol. 70:2664-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner, S., P. Hindmarsh, M. Napirei, K. Vogel-Bachmayr, and B. M. Wohrl. 2002. Subcellular localization and integration activities of Rous sarcoma virus reverse transcriptase. J. Virol. 76:6205-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, D. H., A. H. Corbett, H. M. Kent, M. Stewart, and P. A. Silver. 1997. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol. Cell. Biol. 17:3755-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]