Abstract
Mutations in the UGT1A1 gene have been implicated in Gilbert syndrome, which shows mild hyperbilirubinemia, and a more aggressive childhood subtype, Crigler–Najjar syndrome. To date, more than 100 variants have been found in the UGT1A1 gene. Among them, UGT1A1*28 and UGT1A1*6 have been reported to be associated with severe toxicities in patients treated with irinotecan-based chemotherapy by increasing the dose of SN-38 (7-ethyl-10-hydroxycamptothecin), an active form of irinotecan. Many association studies and meta-analyses have demonstrated the contribution of UGT1A1*28 and UGT1A1*6 polymorphisms to the toxicities caused by irinotecan-based therapy. The aim of this review was to evaluate the impact of these variants upon the toxicities and the efficacy of irinotecan-based chemotherapy.
Keywords: UGT1A1, irinotecan, chemotherapy, toxicity, response, survival
Introduction
Irinotecan hydrochloride, inhibiting topoisomerase I, is one of the key anticancer drugs in chemotherapy for several cancers such as colorectal cancer, lung cancer, gastric cancer, and gynecologic cancers.1–4 The patients treated with irinotecan occasionally experience severe neutropenia and delayed diarrhea; however, the occurrence of these adverse reactions has been unpredictable and largely unexplained.5 An active metabolite of irinotecan, SN-38 (7-ethyl-10-hydroxycamptothecin), is glucuronidated by uridine diphosphate glucuronosyltransferase 1As (UGT1As), such as UGT1A1, and is inactivated by forming the SN-38 glucuronide (SN-38G). Among these UGT1A enzymes, UGT1A1 protein has the highest ability to glucuronidate SN-38.6 Various studies have demonstrated a relationship between UGT1A1 genotypes affecting SN-38 pharmacokinetics and the experienced toxicity.7 The transport pathway of irinotecan is shown in Figure 1. In addition to UGT1A1 polymorphism, polymorphisms of carboxylesterase (CES) and ATP-binding cassette (ABC) genes have been reported to affect the metabolism of irinotecan.8,9 In this review, the impact of UGT1A1 genotypes on irinotecan treatment will be discussed.
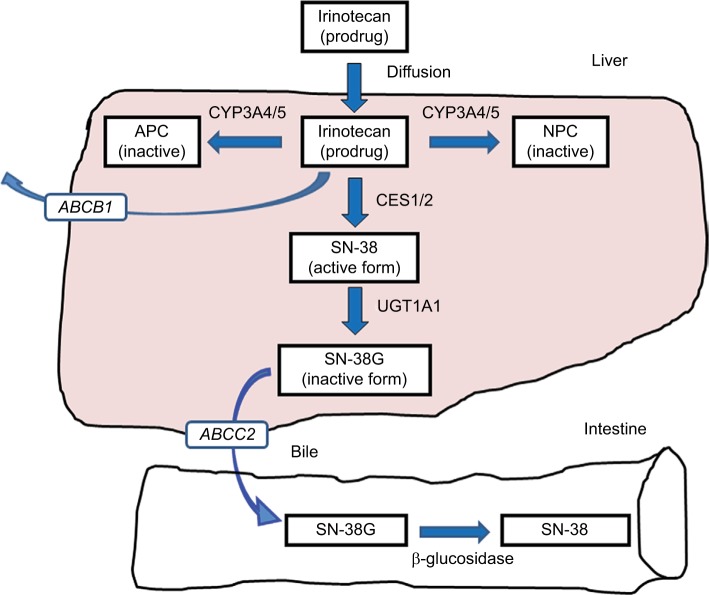
Figure 1.
Transport pathway of irinotecan.
Notes: A prodrug, irinotecan, is moved into hepatic cells by passive effusion. Activation to an active form, SN-38, is mediated by CES1/2. Deactivation to an inactive form, SN-38G (glucuronidated SN-38), is mainly mediated by UGT1A1. A part of SN-38G is hydrolyzed to SN-38 by bacterial β-glucosidase. Moreover, irinotecan is degraded to APC and NPC metabolites by CYP3A4/5. Irinotecan and metabolites are transported by P-glycoprotein, a protein of the cell membrane that pumps foreign substances out of cells, which is encoded by ABCB1 gene. Biliary excretion of SN-38G is mainly mediated by the MRP2/ABCC2.
Abbreviations: APC, 7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyloxycamptothecin; CES1/2, carboxylesterases 1 and 2; CYP3A4/5, cytochrome P450 isoforms 3A4 and 3A5; MRP2/ABCC2, multidrug resistance associated protein-2; NPC, 7-ethyl-10-(4-amino-1-piperidino) carbonyloxycamptothecin; SN-38, 7-ethyl-10-hydroxycamptothecin; UGT1A1, uridine diphosphate glucuronosyltransferase 1A1.
UGT1A1 polymorphisms and disease susceptibility
Mutations in the UGT1A1 gene have been implicated in Gilbert’s syndrome, which shows mild hyperbilirubinemia, and a more aggressive childhood subtype, Crigler–Najjar syndrome.10,11 A common cause of decreased UGT1A1 activity is the insertion of a TA in the TATA box at the promoter region of the UGT1A1 gene, which was named as UGT1A1*28.10 Individuals with homozygous UGT1A1*28 had higher levels of serum bilirubin compared with those with heterozygous UGT1A1*28 or the wild-type allele.10
Gilbert’s syndrome, also known as constitutional hepatic dysfunction or familial nonhemolytic jaundice, is an inherited disorder of the liver resulting in an overabundance of bilirubin. Most of the patients with Gilbert’s syndrome are asymptomatic; however, they sometimes present with episodes of mild intermittent jaundice due to predominantly unconjugated hyperbilirubinemia. Crigler–Najjar syndrome is a rare, but more severe, disorder of bilirubin metabolism and is divided into two distinct forms (types I and II) based upon the severity of the disease. Gilbert’s syndrome is part of a continuous spectrum of altered glucuronidation that extends to the fatal Crigler–Najjar disease.
Gilbert’s syndrome is primarily linked to UGT1A1*28 variants, but other variants in the promoter and coding regions are also involved in the predisposition of the disease.12 To date, more than 100 variants have been identified in the UGT1A1 gene.13 Among these polymorphisms, the clinically important variants are listed in Table 1.14–19
Table 1.
UGT1A1 allelic variants and their biologic impact
| Denomination | Variants14 | Allele frequency (ethnicity)15,16 | Expression level | Enzymatic activity | Clinical consequence |
|---|---|---|---|---|---|
| UGT1A1*1 | (TA)6TA | Common allele | 100% | 100% | None |
| TATA box polymorphisms | |||||
| UGT1A1*28 | c.–39_–40 ins TA: (TA)7TA | 29–45% (Caucasians); 42–51% (Africans); 16% (Asians) | Reduced | Reduced | Gilbert’s syndrome, Crigler–Najjar syndrome17 |
| Polymorphisms in the promoter region | |||||
| UGT1A1*60 | c.–3279 T>G | 23–39% (Caucasian); 15% (African Americans); 17% (Asians) | Reduced | Unchanged | Gilbert’s syndrome, Crigler–Najjar syndrome18 |
| Polymorphisms in exon 1 | |||||
| UGT1A1*6 | c.211 G>A p.Gly71Arg | 15–20% (Asians) | Unchanged | Reduced | Gilbert’s syndrome, Crigler–Najjar syndrome19 |
| UGT1A1*27 | c.686 C>A p.Pro229Gln | 5–28% (Asians) | Unchanged | Reduced | Gilbert’s syndrome, Crigler–Najjar syndrome19 |
Recently, a large population-based cohort study, the Rotterdam Study,20 investigated the association between UGT1A1 genotype and incidence of coronary heart disease (CHD). However, in this study, neither bilirubin nor UGT1A1*28 genotype was associated with development of CHD. Another large trial evaluating 1,780 unrelated individuals aged more than 24 years suggested that homozygous UGT1A1*28 alleles and higher serum level of bilirubin were related with lower risk of cardiovascular disease (CVD).21 Serum bilirubin has a protective effect on CVD and CVD-related disease. It seems that individuals with Gilbert syndrome and UGT1A1*28 allele and having moderate elevation of serum bilirubin could have a lower risk of CHD and CVD.
UGT1A1*28 allele and efficacy of irinotecan-based therapy
Emerging data on the role of genetic variants in the UGT1A1 gene confirm that the UGT1A1*28 allele is associated with severe toxicities in irinotecan-based chemotherapy.22 Additionally, it seems that patients with the allele were also associated with better outcome, despite severe toxicities.22 A study by Toffoli et al,22 conducted in 238 patients with metastatic colorectal cancers, showed that *28/*28 cases had a better response rate and progression-free survival compared with *1/*1 cases. However, most of the other studies evaluating survival according to UGT1A1 genotypes failed to show the significance of UGT1A1 variants in terms of survival. A meta-analysis by Dias et al,23,24 evaluating 10 studies using irinotecan-based chemotherapy, revealed that there was no significant efficacy in terms of response rate, progression-free survival, and overall survival. Additionally, another meta-analysis by Liu et al25 also confirmed that the UGT1A1 genotype could not be a predictor for response rate and survival. These results might reflect a lower dose intensity of irinotecan in patients with *28/*28 or *1/*28 alleles, due to severe toxicities. Representative studies evaluated in these meta-analyses are listed in Table 2.22,26–36
Table 2.
Severe toxicities according to UGT1A1*28 genotyping
| Study | No of patients | Regimens | Irinotecan dose (mg/m2) | Neutropenia G3–4, %
|
Diarrhea G3–4, %
|
Efficacy | ||
|---|---|---|---|---|---|---|---|---|
| Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
|||||
| *28/*28 vs *1/*1 | *1/*28 vs *1/*1 | *28/*28 vs *1/*1 | *1/*28 vs *1/*1 | |||||
| Rouits et al26 | 73 | FOLFIRI, mFOLFIRI | 85/1w, 180/2w | 23.33 (3.08–177.04) | 6.22 (1.58–24.47) | 2.70 (0.39–18.92) | 1.69 (0.44–6.43) | – |
| Marcuello et al27 | 95 | IRI-Tomudex, etc | 80/w, 180/2w, 350/3w | – | – | 11.00 (2.27–53.37) | 2.36 (0.85–6.57) | RR, no difference (NS) |
| Toffoli et al22 | 250 | FOLFIRI, mFOLFIRI | 180/2w | 2.08 (0.60–7.28) | 1.99 (0.91–4.38) | 0.86 (0.10–7.49) | 2.52 (0.93–6.81) | RR 40% (*1/*1), 42% (*1/*28), 67% (*28/*28) (p=0.03) |
| Côté et al28 | 89 | FOLFIRI | 180/2w | 6.40 (1.20–34.20) | 1.88 (0.58–6.11) – | – | – | DFS 52% (*1/*1), 42% (*1/*28), 87% (*28/*28) (p=0.06, NS) |
| Kweekel et al29 | 218 | IRI, IRI-Cape | 250 or – 350/3w | – | – | IRI 11.14 (0.89–140.12) IRI-Cape 2.08 (0.53–8.14) |
IRI 1.63 – (0.51–5.21) | |
| Braun et al30 | 326 | IRI, IRI-5Fu | 300–350/3w, 180/2w | IRI 1.73 (0.16–18.40) IRI–5Fu 1.07 (0.22–5.25) |
IRI 1.28 (0.52–3.13) IRI–5Fu 2.82 (0.69–11.58) |
IRI 1.20 (0.12–11.91) IRI–5Fu 2.75(0.49–15.38) |
IRI-Cape 1.06 – (0.46–2.46) | – |
| Ferraldeschi et al31 | 92 | FOLFIRI, IRI-Cape, etc | 180/2w, 250 or 350/3w | 2.53 (0.40–16.15) | 2.14 (0.66–7.03) | 14.00 (1.09–179.00) | IRI 0.48 (0.08–2.74) IRI–5Fu 1.24 (0.32–4.77) | – |
| McLeod et al32 | 212 | IFL, IROX | 100–125/1w, 200/3w | IFL 3.04 (0.44–20.91) IROX 11.28 (2.51–50.70) | IFL 1.71 (0.40–7.26) IROX 1.66 (0.47–5.88) | IFL 1.94 (0.47–8.01) IROX 1.88 (0.50–7.05) | 3.32 (0.33–33.25) | RR of IROX, lower in *28/*28 (p=0.02) |
| Martinez-Balibrea et al33 | 149 | FOLFIRI, FUIRI | 80/1w, 180/2w | 4.00 (1.12–14.32) | 1.67 (0.67–4.20) | 4.96 (1.49–16.55) | IFL 0.97 (0.37–2.52) IROX 1.08 (0.45–2.63) 1.38 (0.63–3.04) | RR, PFS, OS, no difference (NS) |
| Glimelius et al34 | 136 | FLIRI, Lv-5Fu-IRI | 180/2w | 6.88 (1.70–27.75) | 1.75 (0.55–5.56) | 1.42 (0.15–13.79) | 1.85 (0.47–7.25) | RR, no difference (NS) |
| Shulman et al35 | 214 | TEGAFIRI, XELIRI, etc | 80/w, 180/biweekly | 5.43 (1.50–19.67) | 1.53 (0.48–4.85) | 0.66 (0.22–1.95) | 0.59 (0.30–1.18) | – |
| Lamas et al36 | 100 | IRI-Cape, FOLFIRI | 100 or 125/w, 180/biweekly | 0.80 (0.09–7.25) | 2.49 (0.85–7.29) | 0.28 (0.01–5.22) | 0.57 (0.14–2.29) | – |
Abbreviations: CI, confidence interval; RR, response rate; DFS, disease-free survival, PFS, progression-free survival; OS, overall survival; NS, not significant; FOLFIRI, 5-fluorouracil, leucovorin, irinotecan; mFOLFIRI, modified FOLFIRI; IRI-Tomudex, irinotecan plus tomudex, IRI, irinotecan; IRI-Cape, irinotecan plus capecitabine; IRI-5Fu, irinotecan plus 5-fluorouracil; IFL, irinotecan plus 5-fluorouracil; IROX, irinotecan plus oxaliplatin; FUIRI, irinotecan plus high-dose 5-fluorouracil; FLIRI, irinotecan plus bolus 5-fluorouracil/leucovorin; Lv-5Fu-IRI, irinotecan, plus bolus/infused 5-fluorouracil/leucovorin; TEGAFIRI, uracil, ftorafur, leucovorin, and irinotecan; XELIRI, capecitabine plus irinotecan.
UGT1A1*28 allele and the toxicities of irinotecan-based therapy
Many studies have evaluated toxicities in patients treated with irinotecan-based therapy according to UGT1A1*28 genotypes.22,26–36 Table 2 summarizes representative studies evaluating the incidence of neutropenia and diarrhea. In terms of neutropenia, approximately half of these studies suggested a significant contribution of *28/*28 alleles to severe toxicities. A study by Kweekel et al29 analyzing high-dose irinotecan regimens (250 or 350 mg/m2) revealed that patients with *28 allele had a significantly higher rate of febrile neutropenia compared with *1/*1 cases.
Several studies evaluating 5-fluorouracil, leucovorin, irinotecan (FOLFIRI) regimen also reported significantly higher incidence of severe neutropenia in cases with *28/*28 alleles.26,28,32–34 Some reports suggested significant association between diarrhea and *28/*28 alleles27,31,33 Several meta-analyses have examined the impact of the *28 allele on the toxicities of irinotecan-based therapy. A study by Hoskins et al,37 evaluating 821 cases, revealed that severe hematological toxicities were more frequently observed in *28/*28 patients, when the irinotecan doses were high (>250 mg/m2) or intermediate (150–250 mg/m2). However, the risk was not elevated in patients treated with low doses of irinotecan (<150 mg/m2).37 Another study by Hu et al38 reported that the *28/*28 genotype was associated with an increased risk of neutropenia not only at medium (response rate [RR] =2.0, 95% confidence interval [CI] =1.6–2.5, p<0.01) or high doses (RR =7.2, 95% CI =3.1–16.8, p<0.01) of irinotecan but also at low doses (RR =2.4, 95% CI =1.3–4.4, p<0.01) from the results of meta-analyses evaluating 1,998 patients. Additionally, a study by Liu et al39 confirmed that patients with *28/*28 genotype had higher incidence of neutropenia compared with *1/*1 or *1/*28 genotype cases, in addition to suggesting that patients with *1/*28 genotype had significantly higher rate of severe neutropenia compared with *1/*1 genotype cases (odds ratio [OR] =1.84, 95% CI =1.24–2.72, p<0.01).
UGT1A1*6 allele and efficacy or toxicities of irinotecan-based therapy
The most frequent and important variant in the Asian population is UGT1A1*6, which is rarely found among Caucasians. Representative studies evaluating UGT1A1*6 and clinical outcomes in patients treated with irinotecan-based therapy are listed in Table 3.40–50
Table 3.
Severe toxicities according to UGT1A1*6 genotyping
| Study | No of patients | Regimens | Irinotecan dose | Neutropenia G3–4 (%)
|
Diarrhea G3–4 (%)
|
Efficacy | ||
|---|---|---|---|---|---|---|---|---|
| Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
Odds ratio (95% CI)
|
|||||
| *6/*6 vs *1/*1 | *1/*6 vs *1/*1 | *6/*6 vs *1/*1 | *1/*6 vs *1/*1 | |||||
| Jada et al40 | 45 | IRI | 375/3w | – | – | 7.75 (0.40–149.70) | 1.11 (0.11–11.49) | |
| Sai et al41 | 49 | IRI | 60/1w, 100, 125, 150/2w | 28.00 (0.92–851.54) | 8.00 (0.82–78.47) | – | – | |
| Takano et al42 | 30 | IRI-cisplatin | 60/1w | – | 11.20 (1.73–72.30) | – | 16.00 (1.45–176.45) | – |
| Seo et al43 | 39 | FOLFIRI | 150/2w | – | 0.97 (0.24–3.90) | – | 0.56 (0.10–3.08) | RR, OS, no difference (NS) |
| Onoue et al44 | 133 | IRI-platinum, FOLFIRI | <60, >100 | 7.78 (1.36–44.50) | 4.27 (1.71–10.62) | – | – | – |
| Satoh et al45 | 73 | IRI | 150/2w | 9.33 (1.96–44.49) | 0.85 (0.16–4.51) | 18.18 (0.68–483.94) | – | – |
| Okuyama et al46 | 39 | FOLFIRI | 150/2w | 4.04 (0.15–108.57) | 1.93 (0.44–8.42) | – | – | – |
| Wang et al47 | 130 | FOLFIRI, IFL | 180/2w 125/1w | 0.59 (0.06–5.54) | 0.97 (0.44–2.13) | 4.47 (0.16–30.12) | 1.99 (0.77–5.13) | PFS, OS, no difference (NS) |
| Gao et al48 | 276 | FOLFIRI, XELIRI | 180/2w | 4.07 (1.50–11.04) | 2.02 (1.06–3.86) | 1.98 (0.40–9.77) | 0.90 (0.27–2.95) | RR, no difference (NS) |
| Gao et al49 | 133 | IRI-cisplatin, FOLFIRI, etc | 180/2w | 6.40 (1.30–31.60) | 3.34 (1.43–7.79) | 2.25 (0.23–21.86) | 1.13 (0.27–4.75) | – |
| Ichikawa et al50 | 1,376 | FOLFIRI, IRIS, etc | 150/2w 125/2w | 3.34 (2.19–5.10) | 1.64 (1.21–2.17) | 1.91 (NS) | 0.84 (NS) | – |
Abbreviations: CI, confidence interval; RR, response rate; PFS, progression-free survival; OS, overall survival; NS, not significant; IRI, irinotecan; FOLFIRI, 5-fluorouracil, leucovorin, irinotecan; IFL, irinotecan plus 5-fluorouracil; XELIRI, capecitabine plus irinotecan; IRIS, irinotecan plus S-1.
Most of these studies were mainly focused on the toxicities of the regimens,40,42,44–46,49,50 and quite a few studies reported the clinical outcomes such as response rate and survival.39,47,48 Some studies reported that there were no significant associations between *6 alleles and the efficacy, including response rate and survival.40,44,45 Among the studies listed, almost all the studies reported significant relationship between UGT1A1*6/*6 and severe neutropenia, compared with *1*1 cases. Additionally, half of the studies suggested significantly higher incidence of severe neutropenia in patients with UGT1A1*1/*6.42,44,48–50 A study evaluating a combination therapy with irinotecan and cisplatin reported an increased risk of severe diarrhea in patients with *1/*6 alleles.39
A meta-analysis evaluating mainly Asian studies reported that patients with *6/*6 alleles had increased incidences of severe neutropenia with both high/medium (OR =3.95, 95% CI =2.05–7.64, p<0.01) and low doses (OR =9.64, 95% CI =2.05–45.28, p<0.01) of irinotecan. This trend was also observed in patients with *1/*6 alleles compared with *1/*1 cases: OR =4.42 for low dose, and OR =1.55 for high/intermediate dose of irinotecan.51
Genotype-based dose modification studies
Accumulated evidence suggests that optimal doses of irinotecan according to UGT1A1 genotype are needed. Several dose-finding studies have been published; however, most of the studies were dose modifications of the FOLFIRI regimens (Table 4).52–56 Three studies evaluating irinotecan doses in FOLFIRI showed that the maximal tolerated dose (MTD) in patients with *1/*1, *1/*28, and *1/*6 alleles was higher than the standard doses of the FOLFIRI regimen.52–54 The MTD in the *1/*1 patients was also higher than that of patients with *1/*28 and *1/*6 alleles, and the MTD in patients with *28/*28, *6/*6, and *28/*6 alleles was lower than the current standard doses of the FOLFIRI regimen.53,54 In the Asian population, incorporation of UGT1A1*6 in addition to UGT1A1*28 would be needed for the safety of irinotecan-based chemotherapy. All these results suggested that patients with heterozygous UGT1A1 variants, in addition to those with homozygous UGT1A1 variants, had lower MTD of irinotecan compared with those with wild-type alleles.
Table 4.
Genotype-based dose-finding studies
| Study | Chemotherapy | Genotype | Starting dose, mg/m2 | Results, mg/m2 |
|---|---|---|---|---|
| Toffoli et al52 | FOLFIRI | *1/*1 | 215 | MTD =370 |
| *1/*28 | 215 | MTD =310 | ||
| Marcuello et al53 | FOLFIRI | *1/*1 | 180 | MTD =390 |
| *1/*28 | 110 | MTD =340 | ||
| *28/*28 | 90 | MTD =130 | ||
| Kim et al54 | FOLFIRI | *1/*1 | 240 | MTD ≧330 |
| *1/*28, *1/*6 | 240 | MTD =300 | ||
| *28/*28, *6/*6, *6/*28 | 240 | MTD ≧150 | ||
| Hazama et al55 | Irinotecan (every 2w) + doxifluridine | *1/*1 | 70 | MTD >150; RD =150 |
| *1/*28 | 70 | MTD =100; RD =70 | ||
| Lu et al56 | FOLFIRI + bevacizumab | *1/*1 | 180 | MTD =260 |
| *1/*28 | 180 | MTD =240 | ||
| *28/*28 | 120 | MTD =210 |
Abbreviations: MTD, maximal tolerated dose; RD, recommended dose; FOLFIRI, 5-fluorouracil, leucovorin, irinotecan.
Current recommendation for UGT1A1 genotyping in daily practice
The US Food and Drug Administration recommends on the irinotecan drug label that patients with the *28/*28 genotype should receive a lower starting dose of irinotecan.57 Additionally the recommendation also noted that “the precise dose reduction in this patient population is not known, and subsequent dose modifications should be considered based on individual patient tolerance to treatment”.57
According to European Society for Medical Oncology (ESMO) guidelines, testing for UGT1A1 polymorphisms should be considered only if severe toxicity potentially related to treatment with irinotecan occurs. The ESMO guideline noted that testing for UGT1A1 is particularly important when irinotecan is used at high doses (300–350 mg/m2) but of less importance when it is administered at lower doses (125–180 mg/m2).58
According to the Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines, it is especially desirable to test for a UGT1A1 genetic polymorphism before administering irinotecan to patients with a high serum bilirubin level, elderly patients, patients whose general condition is poor (eg, performance status 2 [PS2]), and patients in whom severe toxicity (especially neutropenia) developed after the previous administration of irinotecan.59 The guidelines also noted that “irinotecan toxicity cannot be predicted with certainty on the basis of the presence of a UGT1A1 genetic polymorphism alone”, and that “it is essential to monitor patients’ general condition during treatment and to manage adverse drug reactions carefully, irrespective of whether a genetic polymorphism is detected”.
In the USA, single agent irinotecan (350 mg/m2, triweekly, monotherapy) is usually used as one of the “irinotecan-based therapies”, so the doses of irinotecan are usually higher than in Europe (180 mg/m2, biweekly, combination) or Japan (150 mg/m2, biweekly, combination). Although the recommendations for UGT1A1 genotyping are different according to the doses of irinotecan which are clinically often used in daily practice, clinical usefulness should be always considered in all patients who receive irinotecan-based therapy.
Conclusion
Emerging data confirmed an increased risk of severe toxicities, such as neutropenia, in patients with UGT1A1*28 and/or UGT1A1*6 genotype when the patients received irinotecan-based chemotherapy. Homozygous variants and double heterozygous variants showed a higher risk of severe toxicities compared with single heterozygous variants. However, genotype-based studies suggest that MTD is clearly lower in patients with heterozygous UGT1A1 variants compared with those with wild-type alleles. Further clinical studies that include heterozygous UGT1A1 variants, in addition to homozygous variants, are needed to evaluate the clinical utility of UGT1A1 genotyping in patients treated with irinotecan-based therapy. On the other hand, although severe toxicities were clearly evident when the dose of irinotecan was high or intermediate, the incidence of these toxicities was significantly higher even when the dose of irinotecan was lower. Furthermore, clinical significance in terms of tumor response or survival was not found according to UGT1A1 genotypes. Further investigations, such as genotype-based therapy, are needed for increasing the efficacy and decreasing the toxicities for patients receiving irinotecan-based therapy.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Stintzing S, Modest DP, Rossius L, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol. 2016;17(10):1426–1434. doi: 10.1016/S1470-2045(16)30269-8. [DOI] [PubMed] [Google Scholar]
- 2.Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med. 2002;346(2):85–91. doi: 10.1056/NEJMoa003034. [DOI] [PubMed] [Google Scholar]
- 3.Boku N, Yamamoto S, Fukuda H, et al. Fluorouracil versus combination of irinotecan plus cisplatin versus S-1 in metastatic gastric cancer: a randomised phase 3 study. Lancet Oncol. 2009;10(11):1063–1069. doi: 10.1016/S1470-2045(09)70259-1. [DOI] [PubMed] [Google Scholar]
- 4.Sugiyama T, Okamoto A, Enomoto T, et al. Randomized phase III trial of irinotecan plus cisplatin compared with paclitaxel plus carboplatin as first-line chemotherapy for ovarian clear cell carcinoma: JGOG3017/GCIG trial. J Clin Oncol. 2016;34(24):2881–2887. doi: 10.1200/JCO.2016.66.9010. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T, Nishida T, Kumagai S, et al. Combination therapy with irinotecan and cisplatin as neoadjuvant chemotherapy in locally advanced cervical cancer. Br J Cancer. 1999;81(1):95–98. doi: 10.1038/sj.bjc.6690656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer L, King CD, Whitington PF, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101(4):847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Innocenti F, Kroetz DL, Schuetz E, et al. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J Clin Oncol. 2009;27(16):2604–2614. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merali Z, Ross S, Paré G. The pharmacogenetics of carboxylesterases: CES1 and CES2 genetic variants and their clinical effect. Drug Metabol Drug Interact. 2014;29(3):143–151. doi: 10.1515/dmdi-2014-0009. [DOI] [PubMed] [Google Scholar]
- 9.De Mattia E, Toffoli G, Polesel J, et al. Pharmacogenetics of ABC and SLC transporters in metastatic colorectal cancer patients receiving first-line FOLFIRI treatment. Pharmacogenet Genomics. 2013;23(10):549–557. doi: 10.1097/FPC.0b013e328364b6cf. [DOI] [PubMed] [Google Scholar]
- 10.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med. 1995;333(18):1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 11.Bosma PJ, Chowdhury NR, Goldhoorn BG, et al. Sequence of exons and the flanking regions of human bilirubin-UDP-glucuronosyltransferase gene complex and identification of a genetic mutation in a patient with Crigler-Najjar syndrome, type I. Hepatology. 1992;15(5):941–947. doi: 10.1002/hep.1840150531. [DOI] [PubMed] [Google Scholar]
- 12.Kadakol A, Ghosh SS, Sappal BS, Sharma G, Chowdhury JR, Chowdhury NR. Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler-Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutat. 2000;16(4):297–306. doi: 10.1002/1098-1004(200010)16:4<297::AID-HUMU2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 13.Pharmacogenomics Knowledge. Implementation (PharmGKB) [web-page on the Internet]. Haplotypes for UGT1A1 (UGT Alleles Nomenclature Page) [Accessed January 12, 2017]. Available from: https://www.pharmgkb.org/haplotypeSet/ PA166115840.
- 14.OMIM [webpage on the Internet] UDP-Glycosyltransferase 1 Family, Polypeptide A1. UGT1A1; [Accessed January 12, 2017]. Available from: http://omim.org/entry/191740. [Google Scholar]
- 15.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A. 1998;95(14):8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33(3):458–465. doi: 10.1124/dmd.104.001800. [DOI] [PubMed] [Google Scholar]
- 17.Maruo Y, Nakahara S, Yanagi T, et al. Genotype of UGT1A1 and phenotype correlation between Crigler-Najjar syndrome type II and Gilbert syndrome. J Gastroenterol Hepatol. 2016;31(2):403–408. doi: 10.1111/jgh.13071. [DOI] [PubMed] [Google Scholar]
- 18.Lee JS, Wang J, Martin M, et al. Genetic variation in UGT1A1 typical of Gilbert syndrome is associated with unconjugated hyperbilirubinemia in patients receiving tocilizumab. Pharmacogenet Genomics. 2011;21(7):365–374. doi: 10.1097/FPC.0b013e32834592fe. [DOI] [PubMed] [Google Scholar]
- 19.Udomuksorn W, Elliot DJ, Lewis BC, Mackenzie PI, Yoovathaworn K, Miners JO. Influence of mutations associated with Gilbert and Crigler-Najjar type II syndromes on the glucuronidation kinetics of bilirubin and other UDP-glucuronosyltransferase 1A substrates. Pharmacogenet Genomics. 2007;17(12):1017–1029. doi: 10.1097/FPC.0b013e328256b1b6. [DOI] [PubMed] [Google Scholar]
- 20.Bosma PJ, van der Meer IM, Bakker CT, et al. UGT1A1*28 allele and coronary heart disease: the Rotterdam Study. Clin Chem. 2003;49(7):1180–1181. doi: 10.1373/49.7.1180. [DOI] [PubMed] [Google Scholar]
- 21.Lin JP, O’Donnell CJ, Schwaiger JP, et al. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation. 2006;114(14):1476–1481. doi: 10.1161/CIRCULATIONAHA.106.633206. [DOI] [PubMed] [Google Scholar]
- 22.Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24(19):3061–3068. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- 23.Dias MM, McKinnon RA, Sorich MJ. Impact of the UGT1A1*28 allele on response to irinotecan: a systematic review and meta-analysis. Pharmacogenomics. 2012;13(8):889–899. doi: 10.2217/pgs.12.68. [DOI] [PubMed] [Google Scholar]
- 24.Dias MM, Pignon JP, Karapetis CS, et al. The effect of the UGT1A1*28 allele on survival after irinotecan-based chemotherapy: a collaborative meta-analysis. Pharmacogenomics J. 2014;14(5):424–431. doi: 10.1038/tpj.2014.16. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association between UGT1A1*28 polymorphisms and clinical outcomes of irinotecan-based chemotherapies in colorectal cancer: a meta-analysis in Caucasians. PLoS One. 2013;8(3):e58489. doi: 10.1371/journal.pone.0058489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouits E, Boisdron-Celle M, Dumont A, Guérin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clin Cancer Res. 2004;10(15):5151–5159. doi: 10.1158/1078-0432.CCR-03-0548. [DOI] [PubMed] [Google Scholar]
- 27.Marcuello E, Altés A, Menoyo A, Del Rio E, Gómez-Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br J Cancer. 2004;91(4):678–682. doi: 10.1038/sj.bjc.6602042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Côté JF, Kirzin S, Kramar A, et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin Cancer Res. 2007;13(11):3269–3275. doi: 10.1158/1078-0432.CCR-06-2290. [DOI] [PubMed] [Google Scholar]
- 29.Kweekel DM, Gelderblom H, Van der Straaten T, et al. UGT1A1*28 genotype and irinotecan dosage in patients with metastatic colorectal cancer: a Dutch Colorectal Cancer Group study. Br J Cancer. 2008;99(2):275–282. doi: 10.1038/sj.bjc.6604461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun MS, Richman SD, Thompson L, et al. Association of molecular markers with toxicity outcomes in a randomized trial of chemotherapy for advanced colorectal cancer: the FOCUS trial. J Clin Oncol. 2009;27(33):5519–5528. doi: 10.1200/JCO.2008.21.6283. [DOI] [PubMed] [Google Scholar]
- 31.Ferraldeschi R, Minchell LJ, Roberts SA, et al. UGT1A1*28 genotype predicts gastrointestinal toxicity in patients treated with intermediate-dose irinotecan. Pharmacogenomics. 2009;10(5):733–739. doi: 10.2217/pgs.09.20. [DOI] [PubMed] [Google Scholar]
- 32.McLeod HL, Sargent DJ, Marsh S, et al. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28(20):3227–3233. doi: 10.1200/JCO.2009.21.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Balibrea E, Abad A, Martínez-Cardús A, et al. UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. Br J Cancer. 2010;103(4):581–589. doi: 10.1038/sj.bjc.6605776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glimelius B, Garmo H, Berglund A, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11(1):61–71. doi: 10.1038/tpj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shulman K, Cohen I, Barnett-Griness O, et al. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117(14):3156–3162. doi: 10.1002/cncr.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamas MJ, Duran G, Balboa E, et al. The value of genetic polymorphisms to predict toxicity in metastatic colorectal patients with irinotecan-based regimens. Cancer Chemother Pharmacol. 2012;69(6):1591–1599. doi: 10.1007/s00280-012-1866-2. [DOI] [PubMed] [Google Scholar]
- 37.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 38.Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16(15):3832–3842. doi: 10.1158/1078-0432.CCR-10-1122. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. Pharmacogenomics J. 2014;14(2):120–129. doi: 10.1038/tpj.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jada SR, Lim R, Wong CI, et al. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98(9):1461–1467. doi: 10.1111/j.1349-7006.2007.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sai K, Saito Y, Sakamoto H, et al. Importance of UDP-glucuronosyltransferase 1A1*6 for irinotecan toxicities in Japanese cancer patients. Cancer Lett. 2008;261(2):165–171. doi: 10.1016/j.canlet.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Takano M, Kato M, Yoshikawa T, et al. Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology. 2009;76(5):315–321. doi: 10.1159/000209335. [DOI] [PubMed] [Google Scholar]
- 43.Seo BG, Kwon HC, Oh SY, et al. Comprehensive analysis of excision repair complementation group 1, glutathione S-transferase, thymidylate synthase and uridine diphosphate glucuronosyl transferase 1A1 polymorphisms predictive for treatment outcome in patients with advanced gastric cancer treated with FOLFOX or FOLFIRI. Oncol Rep. 2009;22(1):127–136. [PubMed] [Google Scholar]
- 44.Onoue M, Terada T, Kobayashi M, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol. 2009;14(2):136–142. doi: 10.1007/s10147-008-0821-z. [DOI] [PubMed] [Google Scholar]
- 45.Satoh T, Ura T, Yamada Y, et al. Genotype-directed, dose-finding study of irinotecan in cancer patients with UGT1A1*28 and/or UGT1A1*6 polymorphisms. Cancer Sci. 2011;102(10):1868–1873. doi: 10.1111/j.1349-7006.2011.02030.x. [DOI] [PubMed] [Google Scholar]
- 46.Okuyama Y, Hazama S, Nozawa H, et al. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol. 2011;41(4):477–482. doi: 10.1093/jjco/hyr001. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Shen L, Xu N, et al. UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol. 2012;18(45):6635–6644. doi: 10.3748/wjg.v18.i45.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Zhou J, Li Y, Lu M, Jia R, Shen L. UGT1A1 6/28 polymorphisms could predict irinotecan-induced severe neutropenia not diarrhea in Chinese colorectal cancer patients. Med Oncol. 2013;30(3):604. doi: 10.1007/s12032-013-0604-x. [DOI] [PubMed] [Google Scholar]
- 49.Gao J, Zhou J, Li Y, et al. Associations between UGT1A1*6/*28 polymorphisms and irinotecan-induced severe toxicity in Chinese gastric or esophageal cancer patients. Med Oncol. 2013;30(3):630. doi: 10.1007/s12032-013-0630-8. [DOI] [PubMed] [Google Scholar]
- 50.Ichikawa W, Uehara K, Minamimura K, et al. An internally and externally validated nomogram for predicting the risk of irinotecan-induced severe neutropenia in advanced colorectal cancer patients. Br J Cancer. 2015;112(10):1709–1716. doi: 10.1038/bjc.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng L, Li M, Hu J, et al. UGT1A1*6 polymorphisms are correlated with irinotecan-induced toxicity: a system review and meta-analysis in Asians. Cancer Chemother Pharmacol. 2014;73(3):551–560. doi: 10.1007/s00280-014-2382-3. [DOI] [PubMed] [Google Scholar]
- 52.Toffoli G, Cecchin E, Gasparini G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28(5):866–871. doi: 10.1200/JCO.2009.23.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marcuello E, Páez D, Paré L, et al. A genotype-directed phase I-IV dose-finding study of irinotecan in combination with fluorouracil/leucovorin as first-line treatment in advanced colorectal cancer. Br J Cancer. 2011;105(1):53–57. doi: 10.1038/bjc.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim KP, Hong YS, Lee JL, et al. A phase I study of UGT1A1 *28/*6 genotype-directed dosing of irinotecan (CPT-11) in Korean patients with metastatic colorectal cancer receiving FOLFIRI. Oncology. 2015;88(3):164–172. doi: 10.1159/000368674. [DOI] [PubMed] [Google Scholar]
- 55.Hazama S, Nagashima A, Kondo H, et al. Phase I study of irinotecan and doxifluridine for metastatic colorectal cancer focusing on the UGT1A1*28 polymorphism. Cancer Sci. 2010;101(3):722–727. doi: 10.1111/j.1349-7006.2009.01428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu CY, Huang CW, Hu HM, et al. Prognostic advantage of irinotecan dose escalation according to uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) genotyping in patients with metastatic colorectal cancer treated with bevacizumab combined with 5-fluorouracil/leucovorin with irinotecan in a first-line setting. Transl Res. 2014;164(2):169–176. doi: 10.1016/j.trsl.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Camptosar [prescribing information] [webpage on the Internet] [Accessed January 12, 2017]. Available from: http://labeling.pfizer.com/ShowLabeling.aspx?id=533.
- 58.Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. A personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516. doi: 10.1093/annonc/mds236. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, Itabashi M, Shimada Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20(2):207–239. doi: 10.1007/s10147-015-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]