Abstract
The carboxy-terminal sequence of the hepatitis B virus (HBV) core protein constitutes a nucleic acid binding domain that is rich in arginine residues and contains three serine phosphorylation sites. While dispensable for capsid assembly, this domain is involved in viral replication, as demonstrated by the effects of mutations on RNA packaging and/or reverse transcription; however, the underlying mechanisms are poorly understood. Here we tested a series of core protein mutants in which the three serine phosphorylation sites were replaced by glutamic acid, in parallel with a previously described deletion variant lacking the 19 C-terminal amino acid residues, for their ability to support viral replication in transfected hepatoma cells. Replacement of all serines and the deletion gave rise to nucleocapsids containing a smaller than wild-type DNA genome. Rather than a single-stranded DNA intermediate, as previously thought, this was a 2.0-kbp double-stranded DNA molecule derived from spliced pregenomic RNA (pgRNA). Interestingly, full-length pgRNA was associated with nucleocapsids but was found to be sensitive to nuclease digestion, while encapsidated spliced RNA and 3′ truncated RNA species were nuclease resistant. These findings suggest that HBV pgRNA encapsidation is directional and that a packaging limit is determined by the C-terminal portion of the core protein.
Hepatitis B virus (HBV) is an important human pathogen accounting for about one million deaths each year (13). The viral genome, as present in infectious virions, is a 3.2-kbp circular, partially double-stranded DNA (dsDNA) molecule that contains overlapping reading frames encoding the core protein, a reverse transcriptase (P), three surface proteins, and the X protein (20, 25).
Viral replication involves reverse transcription of a pregenomic RNA (pgRNA) intermediate inside nucleocapsids, which are formed by 180 or 240 core protein subunits (4-6, 30). Specific encapsidation of pgRNA occurs via binding of P protein to a 5′-proximal RNA stem-loop structure, termed ɛ. Subgenomic RNAs lack the 5′ ɛ and are excluded from encapsidation. Notably, however, spliced RNA species containing all sequence elements necessary for packaging and for reverse transcription have been observed in liver tissue and in transfected cells (9, 23, 26-29). Viral DNA synthesis is a highly complex process involving three translocation events of P to priming sites located at the respective 5′ and 3′ ends of the template (17). Thus, both ends must be accessible to the encapsidated P protein. The final product is a partially double-stranded, relaxed circular DNA (rcDNA). Occasionally, one of the translocation steps fails (in situ priming), resulting in a double-stranded linear molecule (17, 19, 24, 25).
The C-terminal domain of the core protein plays a crucial role in viral replication. It is rich in arginine residues and contains three serine phosphorylation sites. Though dispensable for particle assembly, it is required for pgRNA packaging (2, 3, 7, 10). Further, the phenotypes of core protein variants that are C-terminally truncated or carry mutations at the serine phosphorylation sites suggest that the C terminus is involved in HBV plus-strand DNA maturation (10, 16, 18). For instance, HBV capsids formed by the truncated variant 164 contained neither the rcDNA nor full-length dsDNA but, instead, DNAs with increased electrophoretic mobility similar to that of the single-stranded DNA (ssDNA) intermediate (18). Because these DNAs contained the termini of authentic full-length minus-strand DNA and because the packaged RNA encompassed 5′ terminal sequences of pgRNA, this was interpreted as a defect in plus-strand DNA synthesis. Similarly, substitutions of the serine residues by alanine or by glutamic acid or aspartic acid, intended to mimic nonphosphorylated and permanently phosphorylated serine, respectively, caused various defects in RNA packaging and DNA synthesis (8, 16). This concept has also been supported by studies in the duck hepatitis B virus (DHBV) model (31, 32).
However, due to the complex mixed phenotypes observed with many of these mutants, no simple scheme has emerged predicting the effects of a given mutation. Unambiguous interpretations are further complicated by the different techniques used by different investigators to analyze the same process. An instructive example is the inclusion or omission of nuclease treatments in order to digest nucleic acids that are associated with but not protected inside the viral nucleocapsid. In a recent study it was shown that DHBV core protein mutants lacking parts of the C-terminal basic domain or carrying an alanine or aspartate residue instead of one of the phosphorylatable serines formed capsids that rendered rcDNA selectively sensitive to nuclease. Hence, its absence from nuclease-treated samples was not caused by a defect in DNA synthesis but by the inability of the mutant capsid structure to effectively shield the DNA from nuclease attack (14, 15).
These findings prompted us to investigate the effects of mutations in the C-terminal domain of the HBV core protein on reverse transcription and on the stability of the viral nucleic acids toward in vitro nuclease treatment. Our data indicate that mutations within the C terminus of the HBV core can affect nuclease resistance of capsid-associated RNA in a size-dependent manner. Thus, full-length pgRNA was accessible to nuclease attack, precluding reverse transcription. By contrast, encapsidated spliced RNA was nuclease-resistant and was converted to short dsDNA molecules, mimicking a defect in DNA maturation.
MATERIALS AND METHODS
Plasmid constructs and transfection of hepatoma cells.
All HBV mutant constructs tested were derived from plasmid pCH-9/3091, which harbors an ayw genome. In this construct transcription of pgRNA is driven by a cytomegalovirus promoter (12, 18). A QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce nucleotide substitutions in the core gene of pCH-9/3091. All mutations were confirmed by DNA sequencing. To generate an HBV polymerase expression construct, the amino-terminal part of the core coding sequence and the packaging signal ɛ in plasmid pCH-9/3091 were deleted. For studying encapsidated RNA, the YMDD motif in the polymerase sequence was modified to YMHD by substituting aspartic acid at position 540 for histidine. This mutation blocks reverse transcription of pgRNA and its concomitant degradation (1, 11, 22).
Human hepatoma Huh-7 cells were seeded onto 10-cm dishes, cultured in Iscove's modified Dulbecco's medium containing 8% fetal calf serum, and transfected with 15 μl of Fugene 6 (Roche) and 5 μg of plasmid DNA. On day 3 posttransfection, the cells were removed from the culture dish by treatment with trypsin, resuspended in culture medium, pelleted, and resuspended in 1 ml of chilled isosmotic lysis buffer (140 mM NaCl, 1.5 mM MgCl2, 50 mM Tris-HCl [pH 8.0]) containing 0.5% Nonidet P-40. Nuclei were removed by centrifugation for 5 min at 2,000 rpm in an Eppendorf centrifuge, and the supernatant was cleared of cell debris by centrifugation for another 5 min at 14,000 rpm.
Western blot analysis.
The cytoplasmic lysates were mixed with 0.4 ml of Lämmli loading buffer containing 5% beta-mercaptoethanol and loaded on a 0.1% sodium dodecyl sulfate-12% polyacrylamide gel. Proteins were transferred onto polyvinylidene difluoride membranes (Schleicher & Schuell) by electroblotting. Proteins were detected with an HBV core-specific rabbit antiserum (3) and visualized by using an ECL Plus chemiluminescence kit (Amersham Pharmacia Biotech).
Extraction and analysis of nucleocapsid-associated viral DNA.
A 0.4-ml aliquot of cytoplasmic lysate was digested with 50 U of micrococcal nuclease (Amersham Biosciences) for 1 h at 37°C in the presence of 2 mM CaCl2. Micrococcal nuclease was inactivated by adding EDTA to a final concentration of 5 mM. Viral DNA was extracted by using a QIAamp DNA mini kit (QIAGEN) with RNase A treatment included as recommended by the manufacturer. A second 0.4-ml aliquot of cytoplasmic lysate was used for immunoprecipitation experiments. Lysates were incubated with HBV core-specific polyclonal antibodies (Dako Cytomation) coupled to protein A Sepharose at 4°C for 2 h, followed by extensive washing of the Sepharose with chilled isosmotic buffer containing 0.5% Nonidet P-40. Viral DNA was liberated from sedimented Sepharose beads by protease digestion in 0.4 ml of ATL lysis buffer (QIAamp DNA mini kit) at 56°C for 1 h. The Sepharose was removed by centrifugation, and DNA was purified by adsorption on silica columns.
DNA samples were loaded onto 1.3% agarose gels with either 3.2-, 2.0-, and 1.2-kbp linear HBV fragments or viral DNA from HBV-positive human serum included as size markers. The DNA was blotted on positively charged nylon membranes (Amersham Biosciences) and probed with a 32P-labeled full-length HBV genome or 32P-labeled oligonucleotides complementary to the 3′ end of plus-strand DNA. The sequences of the oligonucleotides were as follows: 1881, 5′-AGGTGCAATTTCCGTCCGAAG-3′; 2061, 5′-AGATGCTGTACAGACTTGGCCCCCAA-3′; 2140, 5′-CCATCTCTTTGTTTTGTTAGGG-3′; 2205, 5′-ATGTGTTCTTGTGGCAAGGAC-3′; and 2439, 5′-CGGGCAACGGGGTAAAGGTTCAGG-3′.
An aliquot of purified viral DNA was amplified with Taq DNA polymerase by using forward primer 18 (5′-AAAGAATTTGGAGCTACTGTGGAG-3′) and reverse primer 2855 (5′-CCGGCAGATGAGAAGGCACAGAC-3′) by 23 cycles of 30 s at 94°C, 30 s at 55°C, and 3 min at 72°C. The primer designations correspond to their positions in the genome relative to the core start codon. The amplification products were separated on an agarose gel and visualized by ethidium bromide staining. The 1.6-kbp PCR product was excised and purified by adsorption on silica columns. Manual sequencing was performed with 32P-labeled forward primer 399 (5′-ACCACCAAATGCCCCTATCC-3′) by using a ThermoSequenase kit (USB).
Extraction and detection of viral RNA.
Cytoplasmic lysates from transfected cells were prepared and processed by immunoprecipitation as described above. To disrupt viral nucleocapsids, 0.7 ml of RLT lysis buffer (RNeasy Kit; QIAGEN) was added to 0.2 ml of Sepharose suspension followed by vigorous mixing. Sepharose was removed by centrifugation, and RNA was extracted from the supernatant by using silica columns. A second 0.2-ml aliquot of cytoplasmic lysate or Sepharose suspension was treated with 5 U of micrococcal nuclease for 30 min at 37°C in the presence of 2 mM CaCl2. Thereafter, EDTA was added to a final concentration of 5 mM and the mixture was placed on ice before adding 0.7 ml of chilled RLT lysis buffer. RNA was purified by adsorption on silica columns, digested with RNase-free DNase (Roche) and purified for a second time.
RNA samples were resolved on MOPS (morpholinepropanesulfonic acid)-buffered 1% agarose gels containing 1% formaldehyde, followed by capillary transfer onto positively charged nylon membranes and hybridization with either a 32P-labeled full-length HBV probe or 32P-labeled PCR fragments encompassing genome position 556 to 871 and genome position 2537 to 2855, respectively.
A second aliquot of purified RNA was converted to cDNA by oligo(dT)18 priming or random hexamer priming by using SuperScript II reverse transcriptase (Invitrogen). The cDNA was amplified by 24 PCR cycles with forward primer 18 (5′-AAAGAATTTGGAGCTACTGTGGAG-3′) or 1546 (5′-CTTCTCTCAATTTTCTAGGGG-3′) in combination with reverse primer 1335 (5′AATACAGGCCTCTCACTCTGG-3′) or 2855 (5′CCGGCAGATGAGAAGGCACAGAC-3′).
RESULTS
Viral replicative intermediates in mutant nucleocapsids.
Based on earlier studies that had suggested that constitutive serine phosphorylation (mimicked by glutamic acid) or C-terminal deletion of HBV core at position 164 is associated with an apparent defect in viral DNA synthesis (16, 18), we analyzed three mutants bearing glutamic acid substitutions as well as the previously described C-terminally truncated core mutant (Fig. 1). The core deletion mutant is silent with respect to the overlapping polymerase open reading frame (ORF) (18), while the glutamic acid substitutions caused concomitant changes in the polymerase ORF (Fig. 1). In addition we included two mutants mimicking nonphosphorylated serine without changing the polymerase ORF (Fig. 1, ASS and SSP).
FIG. 1.
Mutant HBV core proteins. Serine phosphorylation sites at position 155, 162, and 170 are in bold letters. The wild-type (WT) sequence of the genotype ayw core protein is displayed in single-letter code. Nomenclature of the mutants refers to substitutions for the respective serine residues as shown. The asterisk marks the artificial stop codon in mutant 164. Mutants ASS, SSP, and 164 are silent with respect to the overlapping polymerase ORF. The glutamic acid mutations in core cause the following mutations in P: G19P, R27S, and NR(34/35)KS.
The plasmid constructs were transfected into Huh-7 human hepatoma cells and analyzed for core protein expression and viral DNA synthesis. As shown in Fig. 2A, core protein was readily detectable by Western blotting in all cases. Newly synthesized viral DNA was prepared from nuclease-treated cytoplasmic lysates. Figure 2B illustrates that the viral polymerase tolerated the amino acid exchanges associated with the S155E and S162E substitutions, and the respective mutant core proteins supported viral replication. In contrast, the viral polymerase was inactivated by the amino acid exchanges associated with the S170E mutation in the core gene. Viral replication, however, was successfully rescued by cotransfection of a wild-type polymerase expression plasmid.
FIG. 2.
(A) Mutant core protein levels in transfected cells. Cells were transfected as indicated at the top of each lane and analyzed by Western blotting by using an HBV core-specific polyclonal rabbit antibody. Note that all mutants produce similar levels of core protein. (B) Replication competence of mutant capsids. Cells were transfected as indicated at the top of each lane. Mutants SSE and EEE were cotransfected with a polymerase expression plasmid (Pol+). Viral DNA was prepared from micrococcal nuclease-treated cell lysates and visualized by Southern blotting. Numbers on the right side indicate the positions of the 3.2-, 2.0-, and 1.2-kbp size marker fragments (M).
All mutants, except for mutant ESS, produced less viral DNA than the wild-type control. In addition, viral DNA maturation in mutants EES, SSP, and SSE seemed to be impaired to some extent because, in particular, larger DNA species were underrepresented. Strikingly, mutants EEE and 164 produced mostly DNA migrating at the position of the 2.0-kbp marker fragment, while larger DNA species were virtually absent (Fig. 2).
Next we asked whether the apparent absence of high-molecular-weight DNA in mutants EEE and 164 was due to nuclease treatment during the extraction procedure. To this end, viral DNA was prepared from immunoprecipitated capsids without prior nuclease treatment and compared side by side to DNA extracted from nuclease-treated lysates.
As shown in Fig. 3, the high-molecular-weight DNA species were undetectable, irrespective of the purification procedure applied. Instead, low-molecular-weight DNA migrating close to but not exactly at the position of ssDNA was detectable. Essentially the same results were obtained in other experiments where the viral DNA was directly extracted from cytoplasmic lysate without previous immunoprecipitation or nuclease treatment (data not shown). Furthermore, the use or omission of micrococcal nuclease treatment had no effect on viral DNA species associated with mutants ASS, ESS, EES, SSE, and SSP, and the patterns of viral DNAs isolated from cell nuclei or culture supernatants were similar to those from cytoplasmic lysates (data not shown).
FIG. 3.
Viral DNA associated with mutant capsids EEE and 164 is not nuclease sensitive. Viral DNA was prepared from immunoprecipitated nucleocapsids (IP) without prior nuclease treatment or from micrococcal nuclease-treated cell lysates (MN). Transfected plasmid (Pl) was not completely removed during immunoprecipitation in the case of mutant EEE and remained visible as a high-molecular-weight signal. Native (nat) and denatured (den) DNA extracted from HBV-positive human serum was loaded as a size marker to localize the positions of rcDNA and ssDNA.
Molecular structure of capsid-associated viral DNA.
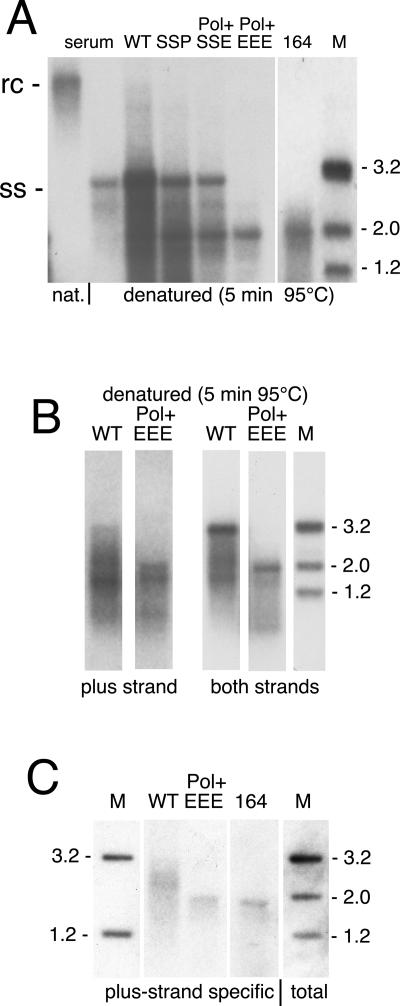
Four mutants were chosen for further analysis of the fast-migrating DNA species, two of which produce predominantly the 2-kbp species (EEE and 164) and the other two produce both the 2- and 3.2-kbp species (SSP and SSE). To determine whether viral DNA associated with mutant capsids was single- or double-stranded, the migration pattern of purified DNA was analyzed after heat denaturation. As shown in Fig. 4A, wild-type, mutant SSP, and mutant SSE yielded full-length ssDNA. In addition, lower-molecular-weight DNA species were detectable in all samples tested. Surprisingly, the dominant ssDNA species associated with mutants EEE and 164 comigrated with the denatured 2.0-kbp marker fragment and thus migrated significantly faster than denatured serum DNA, suggesting it was a 2.0-kbp dsDNA molecule. This was confirmed by hybridization with a plus-strand-specific probe (Fig. 4B and C) and by restriction enzyme digestion of nondenatured DNA with NcoI and NsiI, which yielded fragments of the expected sizes (data not shown).
FIG. 4.
Effect of heat denaturation on electrophoretic mobility of viral DNA. Cells were transfected as indicated at the top of each lane. DNA samples were prepared from micrococcal nuclease-treated cell lysates and denatured by heating for 5 min at 95°C before loading the agarose gel (A and B) or loaded without prior denaturation (C). Native (nat) and denatured serum-derived HBV DNA was applied for reference in panel A. Southern blots were hybridized with a full-length HBV probe, hybridizing to both minus- and plus-strand HBV DNA (A; B, right; and C, total), or with 32P-labeled oligonucleotides complementary to the 3′ end of plus strand DNA (B and C). Numbers on the right side indicate the positions of heat-denatured or native 3.2-, 2.0-, and 1.2-kb size marker fragments (M).
For a more detailed analysis we performed PCR amplification experiments with primers spanning most of the HBV genome (position 18 in the core reading frame through position 2855 in the X protein reading frame). Cloned DNA, which served as a positive control, yielded the expected 2.8-kbp fragment (Fig. 5A). Viral DNA from wild-type transfected cells generated the 2.8-kbp fragment as well. However, additional smaller fragments were visible, which were absent in the plasmid control. Strikingly, the 2.8-kbp fragment was undetectable in mutants EEE and 164; instead, the most predominant amplification product was a 1.6-kbp fragment.
FIG. 5.
Detection of HBV-DNA derived from spliced pgRNA. (A) Viral DNA was amplified by PCR by using the primer pair 18/2855 which spans most of the genome including the intron sequence as depicted in the graphic map in panel C. (B) Sequence analysis of the 1.6-kbp amplification product derived from mutant EEE reveals deletion of a 1.2-kbp intron sequence. Note that the core mutations are upstream of position 548 and do not affect the splice site itself.
Sequence analysis of the 1.6-kbp fragment revealed a 1,223-nucleotide deletion from position 548 in the core reading frame to position 1772 in the surface reading frame (Fig. 5B). The junction sequence was identical to previously published sequences of spliced pgRNA molecules (23). Therefore, the predominant viral DNA species produced by mutants 164 and EEE is a 2.0-kbp dsDNA derived from packaged spliced pgRNA. Additional low-molecular-weight DNA species were detected in all cases, most likely representing minor splice variants as described before (9, 23).
Shortened genomes were also found in wild-type capsids and the other mutants. In the case of mutants SSP and SSE, these DNA species were clearly detectable by Southern blotting, since full-length genomes were underrepresented (Fig. 2 and 4). Shortened genomes associated with wild-type capsids were barely visible in the Southern blots, because of a strong background of replicative intermediates derived from full-length pgRNA. Nevertheless, PCR analysis clearly demonstrated that genomes derived from spliced RNA were present in wild-type capsids as well (Fig. 5A).
Characterization of capsid-associated RNA species.
Finally, we tested the mutant core particles for their ability to package viral RNA. Since encapsidated RNA is degraded during minus-strand DNA synthesis, we introduced a mutation in the reverse transcriptase domain of the viral polymerase, which prevents DNA synthesis but does not affect the RNA packaging function of the viral polymerase (22). To remove nonencapsidated mRNA, the capsids were either purified by immunoprecipitation or treated with micrococcal nuclease prior to RNA extraction and Northern blotting.
The amount and pattern of total cytoplasmic viral RNA species were similar for wild-type and mutants EES, EEE, and 164 (Fig. 6A). Capsid-associated viral RNA was less abundant for EES, EEE, and 164 compared to wild-type, but again the pattern of RNA species was similar in all samples (Fig. 6B, left panel). Nuclease treatment did not significantly alter the pattern of RNA species in wild-type and EES capsids (Fig. 6B, left panel). Strikingly, however, high-molecular-weight capsid-associated RNA was selectively nuclease-sensitive for EEE and 164 (Fig. 6B, right panel).
FIG. 6.
Analysis of total and capsid-associated RNA. Cells were transfected as indicated at the top of each lane. All plasmids code for a reverse transcriptase-deficient polymerase. (A) Total cytoplasmic RNA extracted from cytoplasmic lysates without micrococcal nuclease treatment. Note that similar amounts of pgRNA were produced in all cases. (B) Capsid-associated RNA isolated from immunoprecipitated capsids (IP) in the absence or presence of nuclease treatment (MN) followed by Northern blotting. Free mRNA was removed completely with both protocols as evidenced by the absence of a GAPDH (glyceraldehyde-3-phosphate dehydrogenase)-specific hybridization signal. The left panel shows a 10% aliquot of total RNA isolated from cytoplasmic lysate of wild-type transfected cells. Threefold more RNA from nuclease-treated 164 capsids was loaded relative to the amount loaded in the other lanes to highlight the absence of full-size pgRNA. sgRNA, cytoplasmic subgenomic RNA.
The RNA species present in nuclease-treated capsids were characterized in more detail by reverse transcription PCR. As shown in Fig. 7A, a large PCR fragment derived from full-length pgRNA was amplified in the case of wild-type and mutant EES but not in the case of mutants EEE and 164. In contrast, small fragments derived from spliced RNA were present in all samples. The PCR amplification experiments also revealed the existence of RNA molecules covering the 5′ part of pgRNA as well as the intron region (Fig. 7B). Additional hybridization assays with subgenomic probes confirmed the presence of these various short RNA molecules in wild-type as well as mutant 164 capsids. As shown in Fig. 8, pgRNA was readily detectable in the cytoplasm of transfected cells and coprecipitated with HBV core protein in both cases. Again, full-length pgRNA produced by mutant 164 but not by wild type was sensitive to exogenous nuclease. By using a subgenomic probe detecting the 5′ end of pgRNA and intron sequences but not subgenomic RNA, short RNA molecules were clearly detectable in immunoprecipitated and in nuclease-treated capsids (Fig. 8 A and B, left panels). In contrast, by using a subgenomic probe detecting the 3′ end of pgRNA, spliced RNA, and cytoplasmic subgenomic RNA but not intron sequences, short RNA species were hardly detectable in nuclease-treated capsids. In conclusion, the nuclease-resistant small RNA molecules represent spliced RNA as well as significant amounts of 3′ truncated pgRNA.
FIG. 7.
Molecular structure of encapsidated RNA. Viral RNA was extracted from nuclease-treated capsids and amplified by reverse transcription PCR as follows: oligo(dT) priming followed by PCR amplification with the primer pair 18 and 2855 (A) and random hexamer priming followed by PCR-amplification with the primer pair 18 and 1335 or 1556 and 2855 (B). Graphic map depicts the expected PCR products and the major RNA splice sites (C). Note that primers 1335 and 1546 bind within the intron sequence.
FIG. 8.
Characterization of encapsidated viral RNA by hybridization with subgenomic probes. Cells were transfected with plasmids coding for wild-type core or core mutant 164 and a reverse transcriptase-deficient polymerase. RNA was prepared from cytoplasmic lysates (CP) or immunoprecipitated capsids (IP). An aliquot of immunoprecipitated capsids was treated with micrococcal nuclease prior to extraction of RNA (+MN). For panel B, RNA was visualized by Northern blotting by using probes specific to the 5′ end or 3′ end of pgRNA as schematically depicted in panel C. The right-hand blot in panel B is a long exposure of the Northern blot shown in the middle of the panel. sgRNA, cytoplasmic subgenomic RNA.
DISCUSSION
Collectively, our data indicate that the physical lack of core protein sequences downstream of position 164, as well as substitution in the full-length core protein of all three phosphorylatable serines with acidic residues, results in an inability of the mutant capsids to properly encapsidate full-length pgRNA. While pgRNA is still associated with such capsids, it is sensitive to exogenous nuclease and inaccessible to the reverse transcriptase activity inside core particles. Consequently, the 3.2-kbp viral genome cannot be formed. Strikingly, capsid-associated internally shortened RNAs are nuclease resistant and can serve as a template for reverse transcription. Hence, these mutant capsids provide a highly selective environment for the reverse transcription of spliced variants of the pgRNA. Such spliced RNAs and their reverse-transcribed DNA counterparts were also present in wild-type cores and in all other core variants tested in this study, although they were partially covered up by the excess of full-size pgRNA-derived DNAs. The precise mechanism accounting for the selective protection of short RNAs in mutant capsids remains to be further elucidated. Conceivably, host nucleic acids might compete with pgRNA for packaging in mutant capsids. Further, the morphology of mutant capsids might play a role. Thus, it seems possible that mutant capsids might preferentially be formed by 180 or by 240 core proteins.
These data shed new light on several previously published reports describing the effects of mutations in the C-terminal domain of HBV core protein. C-terminal deletions were investigated in studies by Nassal (18) and by Beames and Lanford (2). In the Nassal study the increased mobility of viral DNA formed by mutant 164 was ascribed to the presence of an intact minus-strand DNA with some incomplete plus-strand DNA derived from in situ priming. Notably, our conclusion that this molecule is, instead, a short, spliced pgRNA-derived dsDNA is fully consistent with all experimental results in that study; furthermore, it explains the reported failure of exogenously added avian myeloblastosis virus reverse transcriptase to generate the expected 3.2-kbp double-stranded HBV genome when added to DNA templates extracted from 164 capsids. Our data are also compatible with a study by Beames and Lanford, who, using a core protein variant truncated at amino acid position 163, reported that the corresponding capsids contained RNA that was significantly smaller than pgRNA. Notably, immunoprecipitated capsids were treated with nuclease before RNA extraction and Northern blotting; therefore, mutant 163 indeed seems to behave similarly to mutant 164 in protecting preferentially smaller RNAs.
The effects of site-specific mutations on packaging and replication were investigated by several groups, including Lan et al. (16) as well as Gazina et al. (8). The study by Lan et al. included an HBV core protein mutant with all three serine residues replaced by glutamic acid, as described here. Mutant capsids reportedly packaged pgRNA but failed to synthesize viral DNA. We suggest that the respective assay formats might explain the apparent discrepancy with the results presented here. Thus, primer extension analysis was utilized to detect pgRNA. However, this method does not discriminate between full-length pgRNA and spliced or 3′ trimmed pgRNA. The reported absence of rcDNA is compatible with our results, and DNA molecules derived from spliced RNA may have gone unnoticed because of the overall low levels of replication in the reported assays. In any case, our data clearly show that the EEE HBV core mutant supports reverse transcription. Gazina et al. (8) have described mutations in the C-terminal domain of the HBV core protein displaying site-specific as well as synergistic effects on the extent of RNA packaging. While the size and potential nuclease sensitivity of capsid-associated RNA were not assessed, strongly reduced RNA packaging was observed in a mutant bearing aspartic acid substitutions of all three serine residues. This result is highly compatible with our findings regarding mutant EEE, which were attributable in part to the nuclease sensitivity of full-length pgRNA.
Interestingly, recent findings in the DHBV system have revealed that the 3′ region of DHBV pgRNA is underrepresented in wild-type DHBV nucleocapsids and that this phenotype is partially due to micrococcal nuclease digestion during the extraction procedure (21). Our data suggest that capsid-associated HBV pgRNA was sensitive to exogenous nucleases in mutant capsids but not in wild-type capsids. Further, an internal deletion of HBV pgRNA strongly reduced its susceptibility to micrococcal nuclease digestion. Finally, we found unspliced 3′ truncated pgRNA molecules in both mutant and wild-type capsids. This would suggest a model in which pgRNA packaging proceeds from the 5′ end to its 3′ end with the 19 most C-terminal amino acids of HBV core, with its serine residues being instrumental for packaging substrates of greater than 2 kb in length. The fact that no evidence for directional, size-dependent pgRNA packaging was observed for DHBV may relate to the substantially different size of the respective internal pgRNA deletions (150 nucleotides in the DHBV variant [21] versus 1.2 kb in spliced HBV genomes).
Alternatively, the discordant results may reflect genuine differences in core protein function related to the considerable sequence divergence between the mammalian and avian hepadnaviral core proteins. In support of the latter view, Gazina et al. found no significant impact of site-specific mutations within the DHBV core C terminus on pgRNA packaging (8). Further, previous work on mutations of the DHBV core C terminus has revealed a destabilizing effect at a later stage of nucleocapsid maturation, with selective nuclease sensitivity of capsid-associated rcDNA but not ssDNA (15, 16). It should be noted, however, that mutational studies introduce permanent modifications that cannot reflect dynamic changes in the phosphorylation state during nucleocapsid assembly and disassembly. Accordingly, an enhanced nuclease sensitivity of HBV pgRNA in mutant capsids will make it more difficult than with DHBV to detect potential effects on rcDNA synthesis. Nevertheless, it seems possible that reversible conformational changes within the C terminus of hepadnaviral core proteins associated with pgRNA encapsidation might also be operative in releasing the viral genome during infection.
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (We 1365/5-1) and the Bundesministerium für Bildung und Forschung (01K19951) to F. von Weizsäcker.
The technical assistance of Bettina Muchow, Bayer AG, and Christine Rösler, University of Freiburg, is gratefully acknowledged.
REFERENCES
- 1.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology 194:597-607. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 64:3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttcher, B., S. A. Wynne, and R. A. Crowther. 1997. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386:88-91. [DOI] [PubMed] [Google Scholar]
- 5.Conway, J. F., N. Cheng, A. Zlotnick, P. T. Wingfield, S. J. Stahl, and A. C. Steven. 1997. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature 386:91-94. [DOI] [PubMed] [Google Scholar]
- 6.Crowther, R. A., N. A. Kiselev, B. Böttcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 7.Gallina, A., F. Bonelli, L. Zentilin, G. Rindi, M. Muttini, and G. Milanesi. 1989. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol. 63:4645-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Günther, S., G. Sommer, A. Iwanska, and H. Will. 1997. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238:363-371. [DOI] [PubMed] [Google Scholar]
- 10.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 66:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 12.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, J. H., and D. S. Chen. 2002. Global control of hepatitis B virus infection. Lancet Infect. Dis. 2:395-403. [DOI] [PubMed] [Google Scholar]
- 14.Köck, J., S. Wieland, H. E. Blum, and F. von Weizsäcker. 1998. Duck hepatitis B virus nucleocapsids formed by N-terminally extended or C-terminally truncated core proteins disintegrate during viral DNA maturation. J. Virol. 72:9116-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köck, J., M. Kann, G. Pütz, H. E. Blum, and F. von Weizsäcker. 2003. Central role of a serine phosphorylation site within duck hepatitis B virus core protein for capsid trafficking and genome release. J. Biol. Chem. 278:28123-28129. [DOI] [PubMed] [Google Scholar]
- 16.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 17.Liu, N., R. Tian, and D. D. Loeb. 2003. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proc. Natl. Acad. Sci. USA 100:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassal, M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassal, M., and H. Schaller. 1993. Hepatitis B virus replication. Trends Microbiol. 1:221-228. [DOI] [PubMed] [Google Scholar]
- 20.Nassal, M. 1999. Hepatitis B virus replication: novel roles for virus-host interactions. Intervirology 42:100-116. [DOI] [PubMed] [Google Scholar]
- 21.Ostrow, K. M., and D. D. Loeb. 2004. Underrepresentation of the 3′ region of the capsid pregenomic RNA of duck hepatitis B virus. J. Virol. 78:2179-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosmorduc, O., M. A. Petit, S. Pol, F. Capel, F. Bortolotti, P. Berthelot, C. Brechot, and D. Kremsdorf. 1995. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology 22:10-19. [PubMed] [Google Scholar]
- 24.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer, G., F. van Bommel, and H. Will. 2000. Genotype-specific synthesis and secretion of spliced hepatitis B virus genomes in hepatoma cells. Virology 271:371-381. [DOI] [PubMed] [Google Scholar]
- 27.Su, T. S., C. J. Lai, J. L. Huang, L. H. Lin, Y. K. Yauk, C. M. Chang, S. J. Lo, and S. H. Han. 1989. Hepatitis B virus transcript produced by RNA splicing. J. Virol. 63:4011-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terre, S., M. A. Petit, and C. Brechot. 1991. Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J. Virol. 65:5539-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, H. L., P. J. Chen, S. J. Tu, M. H. Lin, M. Y. Lai, and D. S. Chen. 1991. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J. Virol. 65:1680-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 31.Yu, M., and J. Summers. 1991. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J. Virol. 65:2511-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, M., and J. Summers. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J. Virol. 68:4341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]