Abstract
Retinoic acid inducible gene-I (RIG-I) receptor recognizes 5′-triphosphorylated RNA and triggers a signaling cascade that results in the induction of type-I IFN-dependent responses. Its precise regulation represents a pivotal balance between antiviral defenses and autoimmunity. To elucidate cellular cofactors that regulate RIG-I signaling, we performed two global RNAi analyses to identify both positive and negative regulatory nodes operating on the signaling pathway during virus infection. These factors were integrated with experimentally and computationally derived interactome data to build a RIG-I protein interaction network. Our analysis revealed diverse cellular processes, including the unfolded protein response, WNT signaling, and RNA metabolism, as critical cellular components governing innate responses to non-self RNA species. Importantly, we identified K-Homology Splicing Regulatory Protein (KHSRP) as a negative regulator of this pathway. We find that KHSRP associates with the regulatory domain of RIG-I to maintain the receptor in an inactive state and attenuate it’s sensing of viral RNA (vRNA). Consistent with increased RIG-I antiviral signaling in the absence of KHSRP, viral replication is reduced when KHSRP expression is knocked down both in vitro and in vivo. Taken together, these data indicate that KHSRP functions as a checkpoint regulator of the innate immune response to pathogen challenge.
Acute innate immune induction is critical for elimination of viral infection and establishing humoral immunity. Aberrant and unregulated activation of these pathways can result in excessive chemokine production and cytokine storm, chronic inflammation, and susceptibility to autoimmunity1. The cytoplasmic pattern recognition receptor (PRR) retinoic acid inducible gene I (RIG-I) detects viral infection through recognition of pathogen-associated molecular patterns (PAMPs) encoded by viral RNA (vRNA). RIG-I senses the 5’-triphosphate dsRNA produced during infection by RNA viruses such as influenza A (IAV), Sendai (SeV), and hepatitis C viruses2–7. Binding of vRNA to RIG-I triggers association with mitochondria activating signaling protein (MAVS), which leads to activation of kinases TBK1/IKKε, and the transcription factor IRF3, to induce type-I interferon (IFN) production and expression of additional antiviral responses8. Type-I IFN produced in response to RIG-I signaling then activates the secondary circuit, the JAK/STAT pathway, to induce expression of ISGs as well as RIG-I to amplify the antiviral innate response (Fig. 1a, left). It has been extensively reported that RIG-I signaling is a primary innate immune pathway induced by various RNA viruses2,5,8. A number of regulatory mechanisms have been described that ensure the precise control of RIG-I signaling responses to balance between robust activation to limit viral replication and inhibition of promiscuous activation in the absence of pathogen challenge9–15.
Figure 1. Genome-wide RNAi screens to identify regulators of the RIG-I signaling pathway.
a, Schematic representation of the RIG-I signaling pathway (left) and genome-wide siRNA screens (right) are shown. b, HEK293T ISRE-luciferase cells were transfected with indicated siRNA followed by infection with either wild type or delNS1 IAV. 24hr post infection, luciferase activities were analyzed. Results are the mean ± s.d. of three biological replicates. ***P < 0.001 (Student's t-test). Data is representation of three independent experiments. c, Primary results of genome-wide screens for positive (left panel) and negative (right panel) regulators of RIG-I signaling. ISRE-activity score values were calculated as described in RNAi Screening Analysis in Discussion. Negative controls (depicted by red circles) were assigned an activity score of 1, while positive controls (depicted by black circles) were scaled to a value 0.1. Screen hits are indicated as open circles. RSA analysis is used to generate P values; *P < 0.05, **P < 0.01, ***P < 0.001. d, Network integration of candidate RIG-I pathway regulators (permutation test p<0.001). Circles indicate protein interactions identified by GeneGo analysis. Hexagon and square shapes indicate AP-MS bait and prey interactions, respectively. Confirmed negative regulators (pink), high confidence positive regulators based on RSA cutoff (orange; p<0.01), and canonical RIG-I regulators (red and blue) are also shown. An enlarged sub network of RIG-I pathway regulators is encircled by dashed line (right). AP-MS interactions between RNAi hits and canonical RIG-I bait proteins are indicated (red edges). This sub-network is expanded using GeneGo to the first neighbor interactors of RNAi hits (indicated by blue edges; right). e, Functional enrichment of RIG-I network proteins using gene ontology resources. Nodes represent enriched functions for an annotated ontology term, and the node size indicates the number of genes that fall into that term. The pie charts embedded within the nodes represent the percentage of RIG-I positive regulators (green) and negative regulators (red) for that term. Nodes are clustered into sub-networks that encompass a representative description for the annotations.
In this study, we describe a comprehensive and systematic interrogation of cellular factors that govern RIG-I signaling through genome-wide RNAi and targeted proteomic approaches. Through computational integration of these results, we constructed a RIG-I pathway protein network, from which we identified key biological modules and nodes that govern RIG-I signaling, underscoring the involvement of discrete and parallel host cellular processes in controlling innate immune responses to viral infection. Furthermore, from these systems-level studies, we identified the RNA-binding K-Homology splicing regulatory protein (KHSRP) as a potent inhibitor of the RIG-I-dependent immune response. KHSRP associates with the regulatory domain (RD) of RIG-I, reduces vRNA association with RIG-I during viral infection, and represses RIG-I activation. We find that immunostimulatory RIG-I PAMPs displace KHSRP from RIG-I, which coincides with the triggering of RIG-I signaling. Correspondingly, depletion of KHSRP inhibits the replication of RNA viruses both in vitro and in vivo. Taken together, these findings implicate that KHSRP is a critical negative regulator of RIG-I-mediated innate sensing; preventing unsanctioned innate induction that may trigger interferonopathies or autoimmune diseases16,17.
Results
Genome-wide RNAi analysis to identify RIG-I pathway regulators
To establish a comprehensive systems-level evaluation of genes that regulate the cytoplasmic innate immune sensing of RNA viruses, we performed two genome-wide siRNA screens in HEK293T cells containing an interferon-stimulated response element (ISRE) fused to a luciferase reporter (see Methods). Infection with either deleted nonstructural 1 (NS1) protein (delNS1) IAV or wild type IAV (PR/8/34) were used to identify positive or negative regulators of RIG-I signaling, respectively (Fig. 1a). Since NS1 directly inhibits RIG-I signaling, we utilized delNS1 IAV to identify positive regulators of RIG-I-mediated innate responses18,19. A complete loss of innate induction upon depletion of RIG-I confirmed that the delNS1 IAV-induced innate response is RIG-I dependent (Fig. 1b, right panel). From this positive regulator screen, we identified many previously reported RIG-I signaling members, including MAVS, IRF3, and RIG-I itself (Fig. 1c, left panel; Supplementary Table 1), as well as additional factors involved in broader aspects of cellular functions such as viral transcription, mRNA processing and metabolism, and epigenetic regulation (see below).
Conversely, to identify antagonists of RIG-I-dependent innate responses, we assayed for factors that, upon depletion, enhanced ISRE-reporter activity after wild type IAV infection (Fig. 1c, right panel; Supplementary Table 2). As expected, siRNAs targeting NS1 increased ISRE-reporter activity by 4.5-fold relative to non-targeting (nCTL) siRNA control (Fig. 1b, left panel; Supplementary Fig. 1)18. From this screen, we identified known negative regulators of RIG-I signaling, including CYLD and PIN1 (Fig. 1c, right panel), as well as a number of additional factors, which were found to be enriched in discrete orthogonal pathways: for example, the unfolded protein response (UPR), the AMPK signaling pathway, and positive regulation of WNT signaling (see below; Discussion; Supplementary Discussion)13,20.
Applying Redundant siRNA Activity (RSA; see Methods), we identified 125 putative positive (P<0.01) and 115 negative (P<0.01) regulators (Supplementary Table 1 and Table 2) of the RIG-I-dependent innate response pathway21. To better understand the biochemical relationships of these putative regulators, identified through genetic screens, in the context of RIG-I signaling, we performed the global interactome analysis of RIG-I/IRF3 pathway members using affinity-purified mass spectrometry (AP-MS; Supplementary Table 4; Methods). Canonical RIG-I signaling members (RIG-I, MAVS, TRIM25, IKKε, TBK1, IRF3), known antagonists (NLRX1, RNF5, A20, CYLD, DHX58, PIN1), and reported regulators (PCBP2, PSMA7, RNF125, STING, EYA4, DDX3X) were FLAG-tagged and subjected to AP-MS. A total of 78 proteins with high statistical confidence were identified from the AP-MS studies. Seventeen overlapped with genes identified by the RNAi screens (P<0.001) and were integrated into a RIG-I signaling network (see below)22–25.
Hits from our RNAi screens, a previously published influenza replication screen (Supplementary Table 3 Tab 1; see below), and the described AP-MS data, were used to construct an interaction network of the co-regulators that were mapped to the canonical RIG-I signaling pathway (Fig. 1d, left panel; Supplementary Table 6)26. We next extracted sub-networks that were seeded by the factors that were found in both in the RNAi and AP-MS data, and then expanded to the first interacting neighbors using curated protein interaction datasets (GeneGO MetaCore; Fig. 1d, right panel; and see Methods for details). From these networks, we identified multiple densely connected network clusters (Supplementary Fig. 2a–e). Functional enrichment analyses of 167 nodes in the network using gene ontology resources revealed more than 152 statistically enriched classifications, which could be broadly categorized into 17 related-functional groups (Supplementary Table 5) encompassing many expected terms such as regulation of viral processes (P=9.15×10−22), negative regulation of type-I IFN production (P=1.46×10−21), cellular response to cytokines (P=1.38×10−9), but also unanticipated functional groups: splicesome (P=9.95×10−19), canonical WNT signaling (P=2.87×10−3), AMPK signaling (P=1.04×10−4), and the unfolded protein response (P=1.48×10−6) (Fig. 1e; also see Discussion; Supplementary Discussion).
Taken together, integration of RNAi and proteomic analyses of the RIG-I pathway has enabled the elucidation of cross-regulatory modules that underscore the role of interdependent, but discrete, biological pathways in shaping the innate immune responses to pathogenic invasion.
Confirmation studies of the putative negative regulators on the RIG-I pathway
To avoid aberrant or prolonged activation of innate signaling that may trigger hypercytokinemia or autoimmune responses, innate immune induction must be tightly controlled, likely through a series of negative regulatory circuits27–29. To better understand the regulation of this process, we focused further studies on the identified antagonists of RIG-I signaling (see Methods; Supplementary Table 3).
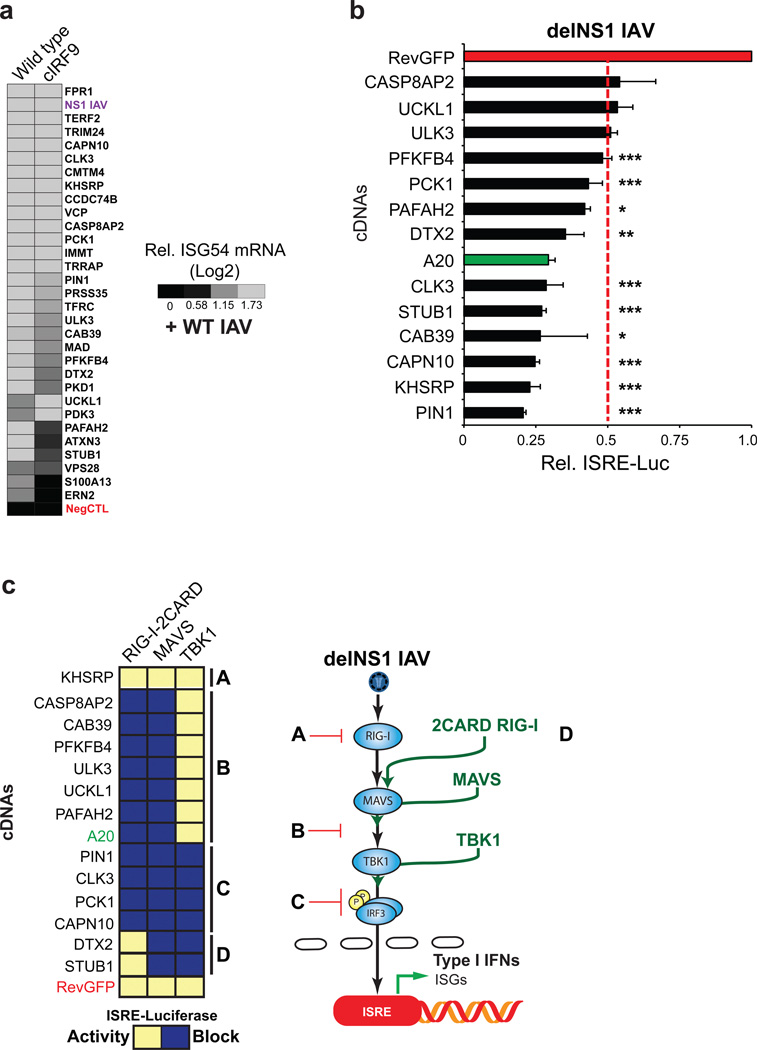
IRF3-dependent RIG-I signaling results in the induction of type-I IFNs, which in turn stimulates the JAK/STAT pathway in a secondary circuit to further enhance expression of ISGs, including ISG54. In order to distinguish negative regulators specifically affecting the primary RIG-I activation circuit from those impacting type-I IFN signaling, we generated an IRF9 knock out cell-line using CRISPR (cIRF9; Supplementary Fig. 3a)30. IRF9 is a critical component of the ISGF3 transcription complex formed upon type-I IFN signaling, and inhibition of IRF9 abolishes ISG54 induction by type-I IFN31. We tested ISG54 mRNA induction upon depletion of 30 of the confirmed factors, which were previously validated in wild type cells, in these interferon signaling-deficient cells (Fig. 2a; Supplementary Table 3 Tab 2; Methods). We found that 28 factors enhanced ISG54 expression greater than 1.5-fold in the absence of type-I IFN signaling, while 2 genes (ERN2, S100A13) fell below this cut off in cIRF9 cells (Fig. 2a; see Supplementary Table 3 Tab 2 Column F). These data suggest that while these two genes are involved in ISG54 expression exclusively through the type-I IFN signaling pathway, the remaining factors at least partially impact innate immune responses through the regulation of RIG-I signaling.
Figure 2. Confirmation studies of the putative negative regulators on the RIG-I pathway.
a, Confirmed RIG-I negative regulators were depleted by siRNA in wild type or CRISPR IRF9 (cIRF9) knockout HEK293T cells, followed by infection with wild type IAV at multiplicity of infection (MOI) of 2. RIG-I pathway activation was assessed by ISG54 mRNA levels using RT-qPCR. The heat map represents mean values of experimental duplicates calculated as ISG54 fold induction over the value of the non-targeting siRNA control. b, cDNAs encoding confirmed negative regulators were ectopically expressed in ISRE-luciferase HEK293T cells followed by delNS1 IAV infection and assaying for luciferase activity (see Supplementary Table 3, Tab 3). Among those factors, the expression of 13 genes resulted in a repression of reporter activity at least by 50% compared to the activity of the RevGFP negative control. Results are the mean ± s.d. of four biological replicates. Data shown here is a representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (Student's t-test) c, cDNAs in b were ectopically co-expressed in ISRE-luciferase HEK293Ts with canonical RIG-I signaling members, RIG-I 2CARD, MAVS, and TBK1. The concentration of the selected cDNAs were fixed, while canonical RIG-I signaling component cDNAs were titrated over 5 dilutions. Reporter activities were used to devise a linear regression for each candidate regulator (see Supplementary Fig. 4), and the statistical comparison (PRISM) between each antagonist cDNA slope and the slope of RevGFP control is indicated as a P value, and was used to determine whether a cDNA expression blocked (blue), or did not impact (activity-yellow), ISRE-reporter activity induced by transfection of indicated pathway signaling members (P<0.05; see Supplementary Fig. 4). A schematic is shown for pathway mapping and localization of identified groups (right). Results are representative of two independent experiments and samples were run in biological quadruplicate.
To determine whether the activity of these negative regulators were sufficient to repress RIG-I signaling, we selected 25 genes from the siRNA profiling above and ectopically expressed these factors in ISRE-reporter cells followed by delNS1 IAV infection (Fig. 2a; Supplementary Table 3 Tab 3). Among 25 tested, 13 genes were found to reduce reporter activity by at least 45% (P<0.05) relative to the value of reverse GFP (RevGFP) negative control (Fig. 2b; Supplementary Table 3 Tab 3). We hypothesize that the remaining genes that did not significantly repress RIG-I signaling may require additional cofactors for their inhibitory activities. Next, to pinpoint specific steps along the signaling pathway at which inhibition occurs by these selected factors, we employed a pathway mapping strategy based upon the inhibition profile of each identified negative regulator after induction by specific canonical RIG-I pathway members (see Supplementary Fig. 4a; see below and Methods for details). The factors were then grouped into 4 distinctive categories (Fig. 2c; Supplementary Table 3 Tab 3). For example, ectopic expression of the set denoted as Group B (CAB39, CASP8AP2, PAFAH2, ULK3, PFKFB4, UCKL1) blocked RIG-I- and MAVS-dependent, but not TBK1-induced, activation, suggesting that these genes regulate the signaling step between MAVS and TBK1 (Fig. 2c). Ectopic A20 expression has been shown to significantly block RIG-I signaling induced by SeV infection and 2CARD RIG-I-induced ISRE activation, but not with TBK1 transactivation, and served as a positive control for this group32,33. Applying analogous reasoning, we mapped the remaining candidate factors into 3 other categories, as depicted in Fig. 2c (right schematic). We did not find any candidates that exclusively regulated 2CARD RIG-I activation; however, we found 4 factors (Group C: CAPN10, CLK3, PCK1, PIN1) that act down stream of TBK1 kinase activation leading to IRF3 phosphorylation (pIRF3). PIN1 was considered as the prototypic member of this functional group, since it antagonizes IRF3 activity during RIG-I signaling13. We were unable to categorize the activities of two factors (Group D: DTX2, STUB1), since they failed to block the reporter activation by 2CARD RIG-I, but potently inhibited the activation by MAVS, TBK1, and delNS1 IAV infection (Fig. 2b–c). This may be due to differences between endogenous RIG-I activity and expression of 2CARD RIG-I construct used for pathway activation. Lastly, we identified one gene (Group A), K-Homology Splicing Regulatory Protein (KHSRP), that failed to block at any point along the pathway, (Fig. 2c, left panel; Supplementary Fig. 4a), but potently inhibited delNS1 IAV-induced signaling (Fig. 2b; Supplementary Fig. 4b), indicating that this protein may impact early steps of RIG-I activation.
KHSRP negatively regulates RIG-I signaling through interaction with the RD domain of RIG-I involved in PAMP sensing
KHSRP depletion by siRNA enhanced ISG54 expression upon delNS1 IAV and Sendai Virus (SeV) infections by 5–6 fold, similar to knockdown of IRF3 antagonist PIN1 (Fig. 3a). Moreover, KHSRP depletion was also sufficient to partially reverse NS1 antagonism during wild type IAV infection, as demonstrated by 14-fold ISG54 mRNA induction compared to non-targeting control (Supplementary Fig. 5a). Next, we evaluated the role of KHSRP in primary cells that are more relevant to IAV pathogenesis. KHSRP and PIN1 were selectively depleted in cultured undifferentiated normal human bronchial-tracheal epithelial cells (NHBE), and then infected with delNS1 IAV or SeV. KHSRP-depleted primary cells displayed enhanced expression of ISG54 (2–3 fold) compared to non-targeting control cells (P<0.01) after viral challenge (Fig. 3b). Similar to ISG54 mRNA, KHSRP depletion in both 293T and NHBE resulted in enhanced IFNβ mRNA expression post viral infection (Supplementary Fig. 5b; and 6b).
Figure 3. KHSRP negatively regulates RIG-I signaling.
a, HEK293T were transfected with the indicated siRNAs and subsequently infected with delNS1 IAV at MOI of 0.7, MOI of 1.4, or 40 hemagglutination units (HAU/mL) SeV, as indicated. ISG54 expression, analyzed by RT-qPCR (left), and knockdown efficiency of KHSRP, verified by western blot analysis (right), are shown. b, Primary normal human bronchial-tracheal epithelial cells (NHBE) were subjected to analogous analysis as in a. c, Wild type or CRISPR-mediated IRF9 knock out (cIRF9) HEK293T cells depleted of KHSRP were assessed for ISG54 induction relative to non-targeting control (nCTL; left) or IRF3 phosphorylation status (western; right) upon delNS1 infection. Error bars represent means ± s.d., biological triplicates. Data on ISG54 induction is a representative of three independent experiments. ***P < 0.001 (Student's t-test). Western analysis on the phospho-IRF3 status has been subjected to two independent experiments. d, HEK293T cells transfected with a cDNA encoding KHSRP or RevGFP control were infected with delNS1 IAV, and ISG54 mRNA levels were determined. The values indicated are relative to the value of RevGFP control (left). A dose range of KHSRP or negative control (LacZ) cDNAs (1.5ug, 0.9ug, 0.3ug) were transfected, followed by infection with delNS1 IAV at MOI 0.7 for 12 hours. Western blot analysis on the lysates was conducted with the indicated antibodies (right). e, wild type or CRISPR-mediated KHSRP knock out (cKHSRP) cells were either transfected with mock or Flag-tagged RIG-I cDNA, and subsequently infected with 40 HAU/mL SeV for 12 hours. ISG54 induction (left) and western blot analysis (right) are shown. Error bars represent means ± s.d., biological triplicates. Data is a representative of two independent experiments. ***P < 0.001 (Student's t-test).
KHSRP has been previously reported to regulate the stability of various mRNA species encoded by immune regulatory genes, including type-I IFNs34,35. To establish that the enhanced ISG54 expression in the absence of KHSRP does not require the type-I IFN signaling circuit, we utilized both cIRF9 knock out cells and a type-I IFN neutralization approach, which relies on treatment with an IFNAR2 decoy to uncouple RIG-I signaling from IFN feedback (Supplementary Fig. 3b). Both the loss of IRF9 and inhibition of IFN signaling utilizing the IFNAR2 decoy treatment did not alter the enhanced ISG54 expression in the absence of KHSRP (Supplementary Fig. 3b), indicating that KHSRP negatively regulates the innate response through direct regulation of RIG-I signaling.
Furthermore, we found that ectopic expression of KHSRP inhibited ISG54 induction (Fig. 3d left) and ISRE-reporter activation (Fig. 2b) during delNS1 IAV infection by 60% and 75%, respectively. Importantly, ectopic KHSRP expression also inhibited IRF3 phosphorylation (S386) and STAT1 phosphorylation (pSTAT), key steps in RIG-I/IFN signaling, in a dose-dependent manner relative to control (LacZ; Fig. 3d right). Conversely, knock out of KHSRP using CRISPR (cKHSRP) enhanced ISG54 expression upon IAV challenge by 7-fold (Supplementary Fig. 6a; and 6b, left panel), or SeV infection by 40-fold, compared to wild type cells in a RIG-I-dependent manner (Fig. 3e, left). Since cKHSRP cells showed elevated RIG-I levels upon SeV infection compared to wild type cells (Fig. 3e, middle panel), it was difficult to assess whether enhanced ISG54 expression in cKHSRP is due to enhanced RIG-I signaling, or increased RIG-I protein levels. We therefore examined a system wherein RIG-I levels were equalized by ectopic RIG-I expression. We find that cKHSRP cells, even with ectopic expression of RIG-I, still showed enhancement in ISG54 induction (Fig. 3e, left).
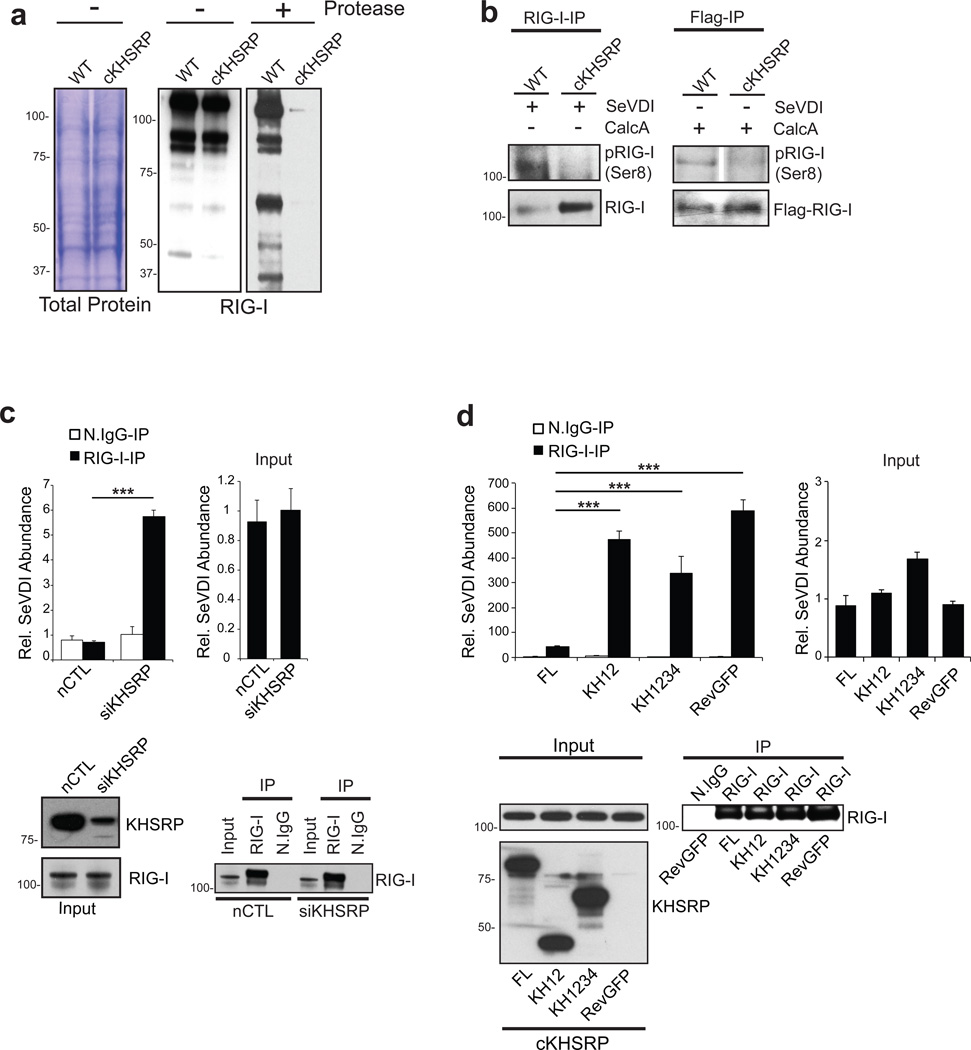
To further explore the structural and functional basis of KHSRP antagonism on RIG-I signaling, we examined the biochemical interaction between RIG-I and KHSRP. IFN-pretreated HEK293T cells, which have induced endogenous RIG-I protein levels, were utilized to immunoprecipitate RIG-I, and subsequently evaluate levels of co-purifying endogenous KHSRP. Under basal conditions, we observed a stable association between RIG-I and KHSRP. However, the two proteins were found to disassociate upon SeV infection (Fig. 4a, top right), or transfection of SeV defective interfering RNA (SeV DI), a potent viral PAMP (Fig. 4a, bottom right and left, respectively)36. A similar interaction dynamic was observed between ectopically expressing HA-RIG-I and V5-KHSRP (Fig. 4b, left panel). We next conducted a systematic truncation analysis to further delineate the interface of interaction for both proteins. We observed that the RIG-I helicase domain (H) alone failed to pull-down KHSRP, while both RD alone and HRD domains efficiently immunoprecipitated the KHSRP protein (Fig. 4b). The interaction was also reconfirmed via reciprocal immunoprecipitation of V5-KHSRP and blotting for HA-RIG-I (Supplementary Fig. 7). Conversely, co-immunoprecipitation analysis of truncated KHSRP proteins revealed that interaction with RIG-I is mediated by the C-terminal domain (CTD) of KHSRP (Fig. 4c). Importantly, reconstitution of KHSRP knock out cells (cKHSRP) with the full-length protein, but not the truncations lacking the CTD (i.e KH12 and KH1234), was able to reinstate KHSRP-mediated negative regulation on RIG-I signaling after challenge with either IAV or SeV infection (Fig. 4d). These data indicate that the physical interaction between the KHSRP CTD and RIG-I RD domains mediates repression of the signaling pathway.
Figure 4. KHSRP associates with regulatory domain of RIG-I.
a, Non-targeting control, or KHSRP siRNAs, were transfected into HEK293Ts and then stimulated by transfection of in vitro-transcribed SeVDI vRNA for 6 hours. ISG54 mRNA induction was accessed by RT-qPCR (left). HEK293Ts, pre-stimulated with 1000U/mL IFNβ for 16 hours to induce RIG-I protein levels, were challenged with SeV (40 HAU/mL) for 20 hours (top right panel), or transfected with SeVDI vRNA for 6 hours (bottom right panel), and subjected to immunoprecipitation of endogenous RIG-I. Immunoprecipitants were probed for KHSRP. Inputs represent 1% (top right) and 10% (bottom right) of binding reaction, respectively. b, V5-tagged KHSRP and HA-tagged RIG-I full-length (FL) or truncation mutants [helicase (H), regulatory domain (RD) and helicase plus RD (HRD)] were co-expressed in HEK293Ts for 36 hours followed by immunoprecipitation of HA-RIG-I and probed for V5-KHSRP association. Where indicated, HA-RIG-I (FL) and V5-KHSRP co-expressing cells were also subjected to SeVDI vRNA transfection for 5 hours prior to immunoprecipitation. Schematic of RIG-I domains are shown at the bottom. *ns denotes a non-specific band. c, HA-RIG-I (FL) and FLAG-KHSRP full-length (FL) or domain mutants (KH12, KH34, KH34C, KH1234 and CTD) were co-expressed and subjected to co-immunoprecipitation analysis, as indicated. Schematic of KHSRP domain mutants are shown at the bottom. Arrows on the blot depicts FLAG-KHSRP-CTD. d, cKHSRP cells, reconstituted with either KHSRP FL, truncation mutants (KH12 and KH1234), or RevGFP control, were infected with 0.7 MOI of delNS1 IAV (left) or 40 HAU SeV (right) for 24 hours. ISG54 mRNA expression relative to RevGFP reconstitution was determined. Error bars represent means ± s.d., biological triplicates. Data is a representative of three independent experiments. **P<0.01, ***P < 0.001 (Student's t-test). The reconstituted cells were also analyzed for KHSRP (FL and truncation mutants) expression (bottom).
KHSRP maintains a closed inactive conformation of RIG-I
In an attempt to further understand how KHSRP regulates RIG-I activity, we evaluated the impact of KHSRP depletion on specific steps of RIG-I activation. Under basal conditions, RIG-I exists in a closed conformation, coincident with phosphorylation at the Ser8 position43. Upon sensing of the vRNA PAMP after infection, RIG-I undergoes a conformational change that results in the exposure of the N-terminal CARD domain, a state that exhibits a higher sensitivity to protease digestion. In addition, dephosphorylation of the Ser8 residue and subsequent K63-linked K172 polyubiquitination by TRIM25, results in RIG-I recruitment to MAVS, and successive pathway activation4,10,37. We initially evaluated the impact of KHSRP on the conformation of RIG-I. Both wild type and cKHSRP cell lysates expressing RIG-I were subjected to limited trypsin digestion, and sensitivity of RIG-I to protease digestion was assessed. Although comparable RIG-I levels were detected prior to treatment with trypsin, RIG-I was observed to possess a significantly higher sensitivity to trypsin treatment in KHSRP-deficient cells (cKHSRP). These data indicate that, in the absence of KHSRP, RIG-I adopts a conformational state that is consistent with an activated receptor state (Fig. 5a). Consistent with this observation, RIG-I, both in the presence and absence of stimulus, harbors lower phosphorylation levels in the absence of KHSRP (Fig. 5b). These data suggests that KHSRP binding to RIG-I results in maintenance of an inactive closed receptor conformation that promotes antagonism of the signaling pathway.
Figure 5. KHSRP maintains RIG-I in an inactive state and attenuate it’s sensing of vRNA.
a, Whole cell lysates prepared from wild type or cKHSRP cells were subjected to a limited trypsin digestion. A commassie stain of mock treated and anti-RIG-I blot of both untreated and trypsin digested lysates are shown. b, Wild type or cKHSRP cells transfected with SeVDI vRNA for 5 hrs were subjected to immunoprecipitation against endogenous RIG-I, followed by blotting for Serine 8 phosphorylated RIG-I (left). These cell lines were also transfected with FLAG-RIG-I in the absence of SeVDI RNA and subjected to immunoprecipitation against FLAG (right). Where indicated, cells were treated with 100 nM calculinA for 45 mins before harvesting. c, cIRF9 cells transfected with KHSRP or non-targeting control (nCTL) siRNAs were infected by SeV (40 HAU/ml) for 18 hours and subjected to CLIP assay (see Methods for details). SeVDI vRNAs in both input and anti-RIG-I or normal IgG IPs were quantified by RT-qPCR. In the IPs, the relative abundance of SeVDI RNA was calculated from the value of associating SeVDI vRNAs, relative to the value of normal IgG in siKHSRP lysates. To calculate levels of vRNA in the input, SeVDI vRNA abundance was calculated relative to the input value of siKHSRP. KHSRP and RIG-I expression in the input, and the level of RIG-I in the IPs, are shown by western blot analysis (bottom). d, cKHSRP cells reconstituted with full length (FL), truncated mutants KH12 or KH1234 KHSRP, or RevGFP were treated with 1000U/mL IFNβ for 15 hours and subsequently transfected with 1ug SeVDI RNA for 5 hours. Samples were prepared and relative abundance was calculated as in c, but are represented relative to the SeVDI RNA value of RevGFP for IPs and input. For c–d, error bars represent means ± s.d. four technical replicates. Three independent experiments were conducted and a representative is shown here. ***P < 0.001 (Student's t-test). Reconstitution of FL KHSRP, KHSRP truncated mutants, and the RIG-I expression in the inputs and IPs were also analyzed by western (bottom).
KHSRP inhibits RIG-I association with vRNA and inhibit viral replication
We next investigated whether KHSRP can modulate RIG-I sensing of vRNA PAMPs. Both wild type and KHSRP-depleted cells, either infected with SeV (Fig. 5c) or transfected with SeV DI (Fig. 5d), were subjected to in vivo UV cross-linking immunoprecipitation (CLIP) against endogenous RIG-I. RIG-I binding of immunogenic SeV defective-interfering (DI) vRNA was assessed using RT-qPCR analysis of the immunoprecipitants (see Methods)7. Both wild type and KHSRP-depleted cells contained equivalent levels of RIG-I protein in the input and immunoprecipitated samples (Fig. 5c, bottom right panel). The levels of SeV DI RNA were also comparable in the inputs (Fig. 5c top right panel). However, in the absence of KHSRP, a significantly greater level of vRNA was found to associate with RIG-I, in comparison to wild type cells (Fig. 5c, top left panel). Conversely, reconstitution of cKHSRP knockout cells with full-length KHSRP, but not KHSRP mutants (KH12 or KH1234) or RevGFP, inhibited RIG-I association with vRNA, suggesting that KHSRP can also regulate the binding of the vRNA PAMP to RIG-I (Fig. 5d), possibly through competition for binding to the RIG-I RD.
Lastly, we asked whether KHSRP modulation of RIG-I signaling impacted IAV replication. HEK293T cells were transfected with siRNAs against KHSRP and IAV nucleoprotein (NP), a viral protein necessary for its transcription and replication. Subsequently, the cells were infected with IAV and viral replication levels were assessed by measuring viral titers in the supernatant. Consistent with an inhibitory role on RIG-I signaling, KHSRP depletion resulted in reduced IAV replication by 2-logs, levels comparable to NP knockdown, at 36 and 48 hours post infection (Fig. 6a). In addition, a similar inhibition on IAV and VSV replication were observed in KHSRP-depleted NHBE and HEK293T cells, respectively (Supplementary Fig. 8). The impact of KHSRP depletion on IAV replication was further investigated in vivo. Towards this end, wild type C57BL/6 mice were administered peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs) targeting KHSRP expression in the lung for 2 consecutive days, as we have previously described38. The inhibitory effect of PPMOs on KHSRP expression was validated by western blot analysis (Fig. 6b, right). On the following day, mice were challenged with PR8 IAV intranasally, and 6 days post-infection, viral titers were measured in the lungs (Fig. 6b, left). Mice showed 3-fold attenuation in IAV titers upon KHSRP knockdown. Taken together, these data indicate that KHSRP-mediated regulation of innate responses impacts RNA virus replication in both cell culture and in vivo models.
Figure 6. KHSRP inhibits replication of RNA viruses.
a, KHSRP, NP IAV, and nCTL siRNAs were transfected into HEK293Ts. After 36 hours, the cells were infected with wild type H1N1 PR/8/34 IAV (MOI of 0.1) for one hour. At the indicated time points, viral titer in the supernatants was measured using plaque assays. Titer values for each siRNA time point were normalized to the 0 hpi titer. Error bars represent means ± s.d. biological triplicate. A representative of six independent experiments is presented here (left). Wild type or cKHSRP HEK293Ts were infected with MHV-68 luciferase reporter virus (MOI 0.5) for the indicated time points and luciferase activity was measured to assess MHV replication (right). Error bars represent means ± s.d., biological triplicates. Data shown here is a representative of at least two independent experiments. ns indicates not significant, *P < 0.05, (Student's t-test). b, Wild type 5–6 week old C57BL/6 male mice were given 100ug KHSRP, non-targeting PPMOs, or PBS (5 mice per treatment) for two consecutive days. Subsequently they were infected with 500 pfu H1N1 PR/8/34 IAV intranasally. At day 6, lungs were harvested and homogenized for viral plaque assays to measure titers. Error bars represent means ± s.d., of five mice. Data shown here is a representative of two independent experiments. ***P < 0.001 (Student's t-test). KHSRP knockdown was also confirmed by western in the lung homogenates (right). c, Model for RIG-I regulation by KHSRP. In a basal state, KHSRP is bound to the RIG-I through the latter’s RD domain, and maintains an inactive closed conformation of RIG-I accompanied by high level of Serine 8 phosphorylation, which results in a dimished interaction with ligand. Upon introduction of activating levels of PAMPs, KHSRP dissociates with RIG-I, resulting in a conformational change of the receptor, induction of dephosphorylation, and activation of the signaling cascade.
Discussion
The innate immune response plays a pivotal role in the defense against infection. However, uncontrolled chronic activation of innate immune signaling can lead to cellular damage and contribute to the pathogenesis of a range of inflammatory diseases39,40. Conversely, a vigorous and rapid response to microbial infection is required for pathogen clearance and initiation of humoral immunity. Thus, intricate networks of regulatory mechanisms are in place to prevent the promiscuous activation of cellular innate immune signaling pathways, yet to also ensure robust activation upon pathogen encounter. In this study, utilizing a series of unbiased global approaches, we systematically profiled the genetic and biochemical landscape of the RIG-I-dependent innate response to influenza A virus infection. Integrative computational analysis was used to establish an extended signaling hierarchy that contained 125 positive and 187 negative regulators of the RIG-I-dependent response.
Based on network analysis of the global datasets, we identified a number of previously unappreciated biological processes and pathways that govern RIG-I-dependent innate responses, including energy metabolism (AMPK pathway; P=1.04×10−4), mRNA splicing (SF3A and SF3B; P=9.95×10−19), and canonical WNT signaling (β-catenin activity; Supplementary Discussion). Interestingly, this analysis found a significant enrichment of genes that function to regulate the unfolded protein response (UPR) during ER stress (ATXN3, STUB1, and VCP; P=1.48×10−6; Fig. 1d–e). Viral infection and replication can burden normal ER functions and trigger stress responses41. This ER disturbance induces the unfolded protein response (UPR) to mitigate ER stress load42. Misfolded proteins are sensed by the ER membrane protein IRE1α to activate UPR, and, interestingly, IRE1α has been reported to mediated the cleavage of cytoplasmic mRNAs to drive induction of RIG-I activation and type-I IFN responses43–46. The co-chaperone and E3 ligase STUB1 and the de-ubiquitinating protein ATXN3 function as negative regulators of the UPR47–49. Our study identified both STUB1 and ATXN3 also function as negative regulators of both RIG-I signaling and IFN signaling further supporting a critical functional coupling of RIG-I/IFN signaling and the UPR.
RIG-I sensing of non-self RNA represents a pivotal step in triggering an innate immune cascade. In this study, we found that KHSRP specifically dampens the ability of RIG-I to sense viral PAMPs. Our data indicates that KHSRP forms a stable complex with RIG-I in the absence of viral PAMP. The formation of this complex is mediated by the regulatory domain (RD) of RIG-I, which is also involved in the recognition of RNA PAMPs (Fig. 4)50. Our data suggest a model wherein KHSRP interaction with the RIG-I RD maintains RIG-I in an inactive conformation, which may hinder PAMP association with RIG-I (see Fig. 6c). Consistent with a model in which KHSRP and PAMP RNAs compete for access to the RD of RIG-I, binding of RIG-I to Sendai virus RNA increases in the absence of KHSRP (Fig. 5c and 5d), while introduction of Sendai virus RNA reverses the biochemical interaction of RIG-I and KHSRP (Fig. 4a and 4b). We hypothesize that KHSRP antagonism of RIG-I enhances the threshold for pathway activation, and augments the ability of the receptor to distinguish immunogenic and non-immunogenic RNA species. However, additional studies are required to provide a more detailed understanding of the PAMP-dependent regulation of KHSRP association with RIG-I, and the consequences of this interaction upon RIG-I binding to RNA ligands, and subsequent impact on receptor activation.
Taken together, this study reveals the global landscape of proteins, regulatory modules, and pathways that influence RIG-I-mediated innate immune signaling. Network-based analyses of these systems-level data enabled the identification of intricate cross-talk between these signaling modifiers and canonical regulators of the RIG-I signaling apparatus. These circuits likely function in concert to ensure a robust and timely response to pathogen invasion, while limiting activation by sub-threshold stimuli or attenuating prolonged responses to appropriate ligands. We anticipate that these data will be a critical resource for the community to better understand the molecular regulation that enables the precise induction of innate immune processes and promotes short-term viral control, as well as long-term protection against pathogen challenge. Importantly, deregulation of these circuits may also underlie auto-immunity and inflammatory disease. Therefore, development of therapeutics that target these regulators may facilitate the development of next-generation immune-mediated antivirals, vaccine adjuvants, and therapies for autoimmune disease.
Methods
Cell culture
Wild type HEK293T was purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 1% penicillin-streptomycin-glutamine, 1× HEPES, and 1× NEAA. Authentication of these cells was performed on the GenePrint 10 System from Promega for analysis of short tandem repeat (STR) on genomic DNA. The STR profiles were analyzed against databases of known STR profiles (ATCC, RIKEN, JCRB, and DSMZ) and returned the best match. This was conducted by the SBP Genomics Core. HEK293T ISRE-luciferase cells were gifted from the Garcia-Sastre lab and have not been authenticated51. Normal Human Bronchial/Tracheal Epithelial (NHBE) cells were purchased from ATCC and sourced from one donor and cultured in commercially available airway epithelial cell basal medium supplemented with cell growth kit (ATCC). All tissues used for isolation of these cells are obtained under informed consent and conform to HIPAA standards to protect the privacy of the donor’s personal health information as conferred by ATCC (PCS-300-010). These primary cells have not been authenticated. A recombinant human IFNβ (Avonex) or a recombinant human IFN-α/β R2 Fc chimera (R&D Systems) was used where indicated. Doxycycline inductions of KHSRP flag-tagged constructs were conducted at 5 ug/mL for at least 24 hours. Calculin A (Invitrogen; PHZ1044) treatment was conducted as described37. All cells were tested and confirmed to be free of mycoplasma contamination.
RNAi, CRISPR, and mutagenesis
Lipofectamine RNAiMAX (Thermo-Fisher) was used to transfect siRNAs. The following siRNAs were used: KHSRP_5 (Qiagen; SI00300587), KHSRP_3 (Qiagen; SI00054691), PIN_6 (Qiagen; SI02662667), DDX58_6 (Qiagen; SI03019646), and influenza A NP (5'-GGAUCUUAVUUCUUCGGAG-3')52. The following non-targeting (nCTL) siRNAs were used as negative controls: 1776 (5'-AAGCGTTCGTGCATAGCTAAG-3’), 1777 (5’-AAGCGTTCGTCCTATGATCGA-3’) and firefly luciferase (GL3; 5'-AACTTACGCTGAGTACTTCGA-3’). The influenza A NS1 was custom designed and available upon request.
CRISPR IRF9 (cIRF9) and KHSRP (cKHSRP) HEK293T cell lines were generated according to the published methods53. Briefly, gene specific guide RNAs (gRNA) were designed using the publically available online design algorithm tool (http://crispr.genome-engineering.org/) and the following sgRNAs were used: ATACAGCTAAGACCATGTTC (IRF9), and CTTTCGCCGACGCCGTGCAG (KHSRP). gRNAs were cloned into pX330 plasmid (Addgene) and transfected into HEK293T cells. The pX330 (pX330-U6-Chimeric_BB-CBh-hSpCas9) was a gift from Feng Zhang (Addgene plasmid # 42230)30. Single cell clones of the CRISPR cells were expanded and confirmed for both protein loss and genomic mutations.
V5-tagged or FLAG-tagged, wild type KHSRP and truncation mutants were generated by sub-cloning KHSRP cDNA into pLenti-V5-TOPO (Life Technologies) or pEasiLV (a gift from Michael Malim), respectively54. The following mutants were sub-cloned:
FL (a.a. 1–710), KH12 (a.a. 1–305), KH34 (a.a. 323–503), KH34C (a.a. 323–710), CTD (a.a. 503–710), and KH1234 (a.a. 1–503). Specifically, V5-tagged wild type KHSRP and truncation mutants were generated using full length KHSRP (FL) forward primer: 5’-CACCATGTCGGACTACAGCACGGGAG-3’ and specific reverse primers that removed the STOP codon and truncated the protein from the C-terminal end:
FL: 5’-ATCTCCTCTTGAACAGATG-3’, KH1234: (KH1234) 5’-CTGGTTGAAGGGCCCAGGATTG-3’, and KH12: 5’-TCCGCCAATCCGAGATCCG-3’.
In addition, KHSRP domain mutants and GFP were also cloned into the dox-inducible N-terminal 3×FLAG-tagged construct pEasiLV using In-Fusion HD (Clontech) cloning according to manufacturer protocol. The PCR forward and reverse primers for In-Fusion cloning are:
eGFP: 5’-GGGGCGGCCGCTCGAgtgagcaagggcgaggag-3’ and 5’-GCTGCAGATGCATCTCGATCActtgtacagctcgtccatgc-3’
FL: 5’-GGGGCGGCCGCTCGAtcggactacagcacgggag-3’ and 5’-GCTGCAGATGCATCTCGATCAatctcctcttgaacagatgaaaag-3’
KH12: 5’-GGGGCGGCCGCTCGAtcggactacagcacgggag-3’ and 5’-GCTGCAGATGCATCTCGATCAtccgccaatccgagatccgta-3’
KH34: 5’-GGGGCGGCCGCTCGAtacggatctcggattggc-3’ and 5’-GCTGCAGATGCATCTCGATCActggttgaagggcccagg-3’
KH34C: 5’-GGGGCGGCCGCTCGAtacggatctcggattggc-3’ and 5’-GCTGCAGATGCATCTCGATCAatctcctcttgaacagatgaaaag-3’
CTD: 5’-GGGGCGGCCGCTCGAcctgggcccttcaaccag-3’ and 5’-GCTGCAGATGCATCTCGATCAatctcctcttgaacagatgaaaag-3’
KH1234: 5’-GGGGCGGCCGCTCGAtcggactacagcacgggag-3’ and 5’-GCTGCAGATGCATCTCGATCActggttgaagggcccagg-3’.
The N-terminal HA-tagged RIG-I domain mutants (FL, H, RD, HRD) were a gift from the Garcia-Sastre lab36. The reverse GFP (RevGFP) was constructed by subcloning GFP cDNA into CMV-Sport6 (Invitrogen) in the reverse orientation served as a negative control cDNA in inhibition assays.
Virus and infections
Cantell Sendai Virus (SeV), H1N1 PR/8/34 (PR8), and H1N1 NS1-deleted PR8 (delNS1 PR8) influenza A (IAV) viruses were a gift from the Garcia-Sastre lab and amplified using 10 day embryonated chicken eggs. MHV-68 viruses were a gift from the laboratories of Ren Sun and Ting-Ting Wu (University of California, Los-Angeles). The PR8 IAV infections were conducted at multiplicity of infection (MOI) of 2 unless otherwise specified. The delNS1 PR8 IAV infections were conducted at MOI of 0.7. IAV infections were supplemented with 3.5 µg/mL TPCK (Sigma) in DMEM. SeV infections were conducted at 40 hemagglutinating units (HAU)/ml.
Genome-wide siRNA screening
Genome-wide libraries (Qiagen, IDT, Invitrogen, GNF Druggable) comprising 98,737 synthetic siRNAs targeting 19,628 unique human genes were pre-arrayed in 384-well plates (0.5 pmol/well) such that each well contained a pool of two siRNAs in a well, with 4–6 total siRNAs targeting one gene in duplicates. The library matrix was introduced into HEK293T-ISRE luciferase reporter cells through reverse transfection method with Lipofectamine RNAiMAX (Invitrogen) in duplicate 384-well plates. After 48 hours, the cells were infected with H1N1 PR8 virus (RIG-I negative regulators) or delNS1 PR8 (RIG-I positive regulators) virus at a MOI of 1.0 and 0.7, respectively. BriteLite Plus (PerkinElmer) was added after 24 hours and relative luminescence for each well was analyzed on the Viewlux plate reader (Perkin-Elmer). In parallel screening plates, cytotoxicity was evaluated by measuring ATP levels relative to controls with CellTiterGlo (Promega).
RNAi Screening Analysis
To help mitigate differences of ISRE-luciferase activity between plates, all well values were first normalized to the plate median signal. Using a scaling normalization, wells were then set to the positive control siRNA (RIG-I siRNA) at an arbitrary value of 0.1, and the negative control siRNAs (non-targeting siRNAs) at 1.0 for the RIG-I positive regulator screen. Wells with lower values correspond to pathway components required for RIG-I signaling during delNS1 IAV infection (Supplementary Table 1, scaledRAW column). For the RIG-I negative regulator screen, the positive control siRNA (IAV NS1 siRNA) was also set at an arbitrary value of 0.1, and the negative control siRNAs (non-targeting siRNAs) at 1.0 (Supplementary Table 2, scaledRAW column). Wells with lower values corresponded to RIG-I signaling enhancement during wild type IAV infection. Each gene was targeted by at least two sets of independent siRNA pools and each siRNA set was run in duplicate. The reporter activity of the replicate wells were averaged to give an ISRE Activity Score (Fig. 1c; Supplementary Table 1 and 2, Score column). The ISRE-activity scores were then subjected to Redundant siRNA Activity (RSA) analysis, which ranks genes based on the activity of all individual siRNAs targeting a gene21. Genes were ranked based on LogP (RSA_LogP) values, where LogP of −2 is equivalent to P value of 0.01. Although the siRNAs were pooled to 2 siRNAs per well, we were able to apply RSA because each gene was targeted with a minimum of two pools and run in duplicate. See http://carrier.gnf.org/publications/RSA/ for the detailed RSA algorithm and procedures for analysis.
RIG-I negative regulator screen hit confirmation and triaging strategy
115 RSA-filtered hits (P<2×10−3) selected from the negative regulator screen based on pooled siRNAs (Supplementary Table 3, Tab 1 denoted as “RSA Hit” in Source column, LogP<−2.7) were supplemented with an additional 312 genes (Supplementary Table 3, Tab 1 denoted as “Konig” in Source column) from previously reported IAV host factors from our group26. We reasoned that a subset of these host factors might also function through antagonizing RIG-I-mediated innate immune signaling and thus included in the conformation step. From a combined 427 factors, we were able to confirm the activities of 134 factors, based on a criteria of 2 or more independent siRNAs displaying greater than 2.5-fold enhancement of ISRE-reporter activity over non-targeting controls (Supplementary Table 3, Tab 1).
We further selected 56 of these genes for additional reconfirmation assays including evaluation of endogenous ISG54 mRNA induction upon IAV infection (Supplementary Table 3, Tab 2, ISG54 column). Thirty of these factors showed substantially enhanced ISG54 expression greater than 2-fold (P<0.05) upon siRNA depletion compared to the negative control. Subsequently, these 30 factors were then confirmed in the cIRF9 knock out cells to identify IFN-independent regulators and 28 factors enhanced ISG54 mRNA at least 1.5-fold (Fig. 2a; Supplementary Table 3, Tab 2, ISG54_cIRF9 column).
Negative regulatory properties of these confirmed factors of RIG-I signaling were assessed by their ectopic expression. Twenty-five factors that enhanced ISG54 mRNA in both wild type and cIRF9 cells upon depletion (Fig. 2a; Supplementary Table 2, Tab 2, Selected for cDNA Inhibition column) were ectopically expressed in ISRE-luciferase reporter cells and subsequently infected with delNS1 IAV.
Thirteen factors reduced the reporter activity by at least 45% and were selected for RIG-I pathway mapping (Fig. 2b; Supplementary Table 2, Tab 3, delNS1 column). Briefly, overexpression of either truncated RIG-I containing only the caspase activation and recruitment domains (2CARD RIG-I), MAVS, or TBK1 can constitutively induce the ISRE-reporter, independent of ligand (Supplementary Fig. 4a). In order to assess inhibition at each RIG-I activation step, increasing amounts of each of these RIG-I signaling activators were co-transfected with a fixed amount of each candidate negative regulator. Statistical comparison (PRISM) between each candidate regulator and the RevGFP control indicated as a P value was used to determine whether the cDNA expression of the candidate blocked (blue) or did not affect (activity-yellow) ISRE-reporter activity induced by the RIG-I signaling member (P<0.05; Fig. 2c; Supplementary Fig. 4a; Supplementary Table 3 Tab 3, 2CARD_p-Val, MAVS_p-Val, TBK1_p-Val columns). Based on the inhibition profiles against the 4 different RIG-I signaling activators (including delNS1 IAV infection), the negative regulators were grouped into 4 distinctive categories (Fig. 2c; Supplementary Table 3 Tab 3, Location column).
Affinity-purified mass spectrometry (AP-MS)
cDNAs of RIG-I pathway members and reported negative regulators (RIG-I, MAVS, TRIM25, NLRX1, PCBP2, PSMA7, RNF5, RNF125, DHX58, A20, IKKε, STING, CYLD, TBK1, IRF3, EYA4, DDX3X, and PIN1) were cloned into pcDNA4/TO-3×FLAG-C (a gift from Nevan Krogan) and subjected to AP-MS as described in38,55,56. Briefly, HEK293T cells expressing each tagged protein were immunoprecipitated against FLAG tag, followed by mass-spectrometry analysis to identify co-immunoprecipitating proteins. Each affinity purification analysis was conducted in triplicate, and the data were analyzed and scored with both Mass spectrometry interaction Statistics (MiST) algorithm and CompPASS57,58. Using the MiST reproducibility (0.32), specificity (0.68) and abundance (0.01) weights were used as previously reported55,59. All bait-prey pairs with a MiST score greater than 0.70 or a top 5% CompPASS WD score were considered for further analysis.
Protein Network Data Analyses
Protein networks were constructed for the combination of identified RIG-I negative regulators, cofactors and canonical members. Two protein-protein binding databases were compared, an aggregated public database that was previously used and the commercial GeneGo MetaCore database (http://portal.genego.com/)26,60. A subset of GeneGo interactome (v3.3.1) consisting of 814,128 interactions was extracted using Metabase R-script Library, where we excluded low-confident interactions derived from data mining, or from publications with high-throughput screens, as well as those derived based on co-expression similarity calculations. GeneGo was later chosen for this study, because it led to a more complete network. The resultant network consists of 167 proteins and 402 interactions (P<0.001, estimated based on 1,000 permutations; Fig. 1d; Supplementary Table 6). A small sub-network was extracted based on AP-MS interactions with the RNAi hits and canonical RIG-I bait proteins (red edges). First neighbor RNAi hits that had direct interaction (one hop) with this sub-network was then utilized to expand this network using GeneGo interaction information (blue edges; Fig. 1d). Molecular Complex Detection (MCODE) analysis was applied to the 167-node RIG-I network to identify densely connected sub-networks (Supplementary Fig. 2a) and gene ontology analysis identified multiple enriched functional clusters (Supplementary Fig. 2b–e; see below). All network visualization was based on Cytoscape (http://www.cytoscape.org/, version 3.3.0)61.
Gene Function Enrichment Network
The 167 RIG-I network proteins were analyzed with Metascape (http://metascape.org) for functional enrichment using ontology terms collected from Gene Ontology (http://geneontology.org), GeneGo pathways and processes (http://portal.genego.com), KEGG pathways, and structural complexes (http://www.genome.jp/kegg), as well as multiple gene sets from MSigDB (http://www.broadinstitute.org/gsea/msigdb), including canonical pathways, hallmark gene sets, immunological signatures, and chemical and genetic perturbation sets. Among them, 163 statistically significant terms were extracted through manual curation. These terms were then clustered and rendered through Metascape into an enrichment network (Fig. 1e), where each enriched term is depicted as a pie chart, and terms with kappa similarity >0.3 were connected to form local functional clusters. The size of a pie is proportional to number of network proteins contained in the given ontology term. Pie sectors are sized by the portion of negative regulators versus positive regulators (red versus green, respectively). Edge width is proportional to kappa similarity and they are bundled for improved clarity. The functional network illustrator shows key biological processes that RIG-I proteins were involved in and their relationship to each other.
Antibodies
The anti-IRF3 antibody was a gift from Michael David62. The following antibodies were used in this study: anti-phospho-IRF3 (S386) (Genetex; GTX130422), anti-STAT1 (BD; 610115), anti-phospho-STAT1 (Y701) (Cell Signaling; 9171), anti-COX-IV (Cell Signaling; 4850), anti-β-Actin (Cell Signaling; 4970), anti-KHSRP (Cell Signaling; 13398), anti-RIG-I (EMD Millipore; MABF297), anti-RIG-I (Adipogen; AG-20B-0009-C100), anti-RIG-I (Sigma-Aldrich; SAB2104315), anti-FLAG (M2) (Sigma; F3165), anti-Flag (Sigma; F7425), anti-V5 (Sigma; V8137), anti-HA (Sigma; H6908), HA-probe (Santa Cruz Bio; sc-7392), and anti-phospho-RIG-I (Ser8) (Abnova; PAB15905).
In vitro-transcribed SeVDI vRNA production and transfection
The Sendai virus defective interfering RNA (SeVDI) construct (a gift from the Garcia-Sastre lab) was subjected to in vitro transcription reaction to generate SeVDI RNAs as described before36.
Co-immunoprecipitation
Cells were lysed in the lysing buffer (300 mM KCl, 12.5 mM MgCl2, 1% Triton X-100, 10% glycerol, 20 mM HEPES, 2 mM EDTA) supplemented with protease inhibitors (Sigma) and subjected to immunoprecipitation with indicated antibodies, followed by conventional western blotting. Typically, 100ug of total proteins were immunoprecipiated with 2 ug of antibody in 300ul of binding buffer (100 mM KCl, 0.5% Triton X-100, 10% glycerol, 20 mM HEPES pH 7.4, 2 mM EDTA) over night, followed by three times washing in the binding buffer supplemented with 300 mM KCl.
Crosslinking and immunoprecipitation (CLIP) of RIG-I-associated Sendai virus RNA
The CLIP assay was modified from Runge and Baum, et al51,63. Briefly, cells replaced with cold PBS were UV irradiated with a UV Stratalinker 2400 (Stratagene) at 254 nm with 150mJ/cm2 and lysed in a buffer (20 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 2.5 mM EDTA, 0.5% NP-40, 0.1% SDS), supplemented with 2 mM DTT, 50 U/mL RNAseOUT (Invitrogen), 0.2 mM PMSF, and 1× protease inhibitor, for 15 mins on ice. Lysates were clarified by centrifugation. Typically, 100 ug of total protein in 300 ul in the above buffer supplemented with 1 mg/mL Heparin (binding buffer) was subjected to immunoprecipitation with anti-RIG-I antibodies (EMD Millipore; MABF297 or Adipogen; AG-20B-0009-C100) or normal IgG control overnight. The IPs were washed with binding buffer supplemented with 500 mM KCl and TE buffer (0.1% SDS), sequentially. The final immunoprecipitants were subjected to proteinase K (Ambion) digestion for 2 hour at 55°C, followed by RNA extraction using Trizol LS (Life Technologies) and RT-qPCR analysis.
Quantitative real-time PCR
Quantitative real-time PCR were performed using high capacity RNA-to-cDNA kit (ABI) and using PowerSybr (ABI) on the Viia7 Real-Time PCR System. Quantification of ISG54, IFNβ, KHSRP, and PIN1 mRNA are relative to TATA binding protein (TBP) as the reference gene unless otherwise specified. The following primers were used: IFNβ: 5’-CATTACCTGAAGGCCAAGGA-3’ and 5’-CAATTGTCCAGTCCCAGAGG-3’, ISG54: 5’-CACGCTGTGGCTCATCTGA-3’ and 5’-GGCTGGCAAGAATGGAACA-3’, TBP: 5’-CCACTCACAGACTCTCACAAC-3’ and 5’-CTGCGGTACAATCCCAGAACT-3’, SeV 5’ DI: 5’-TCCAAGACTATCTTTATCTATGTCCA-3’ and 5’-GCTTCAAACTTCTGGTCAGG-3’, SeV 3’ DI: 5’-GACAGCTCGTAATAATTAG-3’ and 5’-GTCCAAGACTTCCAGGTACCGCGGAG-3’, KHSRP: 5’-ATCCGCAAGGACGCTTTCG-3’ and 5’-GGAGTGCTGTTATTCACTGTCG-3’, PIN1: 5’-GCCTCACAGTTCAGCGACT-3’ and 5’-ACTCAGTGCGGAGGATGATGT-3’.
RIG-I conformational studies
Limited protease digestion was used to determine RIG-I conformational changes as described64. RIG-I Flag-tagged cDNAs were ectopically expressed in wild type or cKHSRP cells in 10cm dish format for 24 hours. Cells were harvested and immediate lysed in 100uL PBS-0.5% Triton-X-100 solution for 10 mins and lysates were clarified by centrifugation and snap frozen for −80C storage. For protease assays, 2.5ug/ul of the lysates in PBS + 0.15% Triton-X-100 digested with trypsin (0.4ug/uL final) for 15 mins at 37C. Digestion was stopped by addition of SDS sample buffer and immediately boiled at 95C for 10 mins. Samples were analyzed by commassie staining and western blotting with an anti-RIG-I (Alme-1) antibody.
Ex vivo replication
MHV-68 viral replication was measured using a MHV-68 luciferase reporter virus using BriteGlo luciferase assay (Promega) 48 hours post infection. For measuring IAV and VSV viral replication, supernatant aliquots of triplicates taken at the indicated time points were titrated on MDCK or BHK-21 cells respectively, and using a plaque assay as described65. Briefly, MDCK or BHK-21 cells were plated in 12 well plates the day before the plaque assay was performed. Ten-fold serial dilutions of supernatants were adsorbed onto cells for 1 hour at RT. The inoculum was removed, then washed with dPBS, and the cells were then covered with MEM – 1.25% Avicel RC581 solution (FMC Biopolymer, Philadelphia, PA) supplemented with TPCK-treated trypsin (2.5 µg/ml) for IAV titration. After 72 hours (IAV) or 36 hrs (VSV), cells were fixed and stained for 20 minutes at RT in one step by using a solution of 0.1% crystal violet + 10% formaldehyde and visible plaques were counted.
Animal experiments
For in vivo influenza A virus infection studies, 6 week old C57BL/6 male mice were purchased from Jackson laboratories. Mice were anesthetized by intraperitoneal injection of a mixture of Ketamine (100 µg per gram of body weight) and Xylazine (5 µg per gram) and inoculated via the intranasal route (i.n.) with the indicated 100 µg of PPMOs or PR8 influenza virus (500 pfu) in 40µl of phosphate-buffered saline (PBS). Mice were monitored daily for weight loss and clinical signs. For measuring the virus titers, lung homogenates were prepared using a FastPrep24 system (MP Biomedicals). After addition of 800 µl of PBS containing 0.3% BSA, lungs were subjected to two rounds of mechanical treatment for 10 s each at 6.5 m/s. Tissue debris was removed by low-speed centrifugation, and virus titers in supernatants were determined by performing 10-fold serial dilutions in PBS with 0.3% BSA followed by plaque assay on MDCKII cells. For this study, mice for each experimental group (n=5) were randomly selected and kept in separate cages throughout the experiment. The investigator was blinded to the experimental group code for measuring mouse body weight and viral titer in lungs. All work with animals conformed to guidelines approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai.
Supplementary Material
Acknowledgments
The authors thank the Krogan laboratory for running and analysis of the AP-MS experiments, Monika Schneider for help in editing the manuscript, and the Chanda laboratory for discussions and advice. We thank Randy Albrecht for providing the H1N1 PR8/34 IAV stocks used in the in vivo mouse experiments. We thank Charlotte Lässig and Karl-Peter Hopfner for providing recombinant RIG-I protein for in vitro assays. We acknowledge support from the National Institutes of Health (U19 AI106754 and P50 GM085764). This work was also supported by a generous grant from the James B. Pendleton Charitable Trust and by CRIP (Center for Research on Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C).
Footnotes
Data Availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Author contributions
S.S., S.Y., and S.K.C. wrote the manuscript. S.S., R.K., S.Y., S.K.C., designed experiments and interpreted data. Y.Z., and A.G.S., interpreted data. S.S., A.R.F., S.Y., F.G., N.J.H., S.T., V.R.B., A.I., M.T.S.A., E.D.C., P.D.D., Q.N., and N.K., performed experiments. H.M., D.S., provided reagents.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Tisoncik JR, et al. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. doi:1132998 [pii] 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. doi:10.1038/ni1087 ni1087 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 9.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 11.Zhong B, et al. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. Journal of immunology. 2010;184:6249–6255. doi: 10.4049/jimmunol.0903748. [DOI] [PubMed] [Google Scholar]
- 12.Lei Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–946. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitoh T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nature immunology. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 14.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. Journal of immunology. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 15.Choi SJ, et al. HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I. Embo J. 2016 doi: 10.15252/embj.201592586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow YJ. Type I interferonopathies: mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 18.Mibayashi M, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. Journal of virology. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gack MU, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. doi:S1931-3128(09)00107-3 [pii] 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman CS, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Rep. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konig R, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. doi:nmeth1089 [pii] 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- 22.Soulat D, et al. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. Embo J. 2008;27:2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 24.You F, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- 25.Jia Y, et al. Negative regulation of MAVS-mediated innate immune response by PSMA7. J Immunol. 2009;183:4241–4248. doi: 10.4049/jimmunol.0901646. [DOI] [PubMed] [Google Scholar]
- 26.Konig R, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. doi:nature08699 [pii] 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Yasukawa H, Sasaki A, Yoshimura A. Negative regulation of cytokine signaling pathways. Annu Rev Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 29.Komuro A, Bamming D, Horvath CM. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- 32.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 33.Lin R, et al. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. The Journal of biological chemistry. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 34.King PH, Chen CY. Role of KSRP in control of type I interferon and cytokine expression. J Interferon Cytokine Res. 2014;34:267–274. doi: 10.1089/jir.2013.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin WJ, et al. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wies E, et al. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi S, et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickup JC. Inflammation and activated innate immunity in the pathogenesis of type 2 diabetes. Diabetes Care. 2004;27:813–823. doi: 10.2337/diacare.27.3.813. [DOI] [PubMed] [Google Scholar]
- 40.Allam R, Anders HJ. The role of innate immunity in autoimmune tissue injury. Curr Opin Rheumatol. 2008;20:538–544. doi: 10.1097/BOR.0b013e3283025ed4. [DOI] [PubMed] [Google Scholar]
- 41.Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hampton RY. ER stress response: getting the UPR hand on misfolded proteins. Curr Biol. 2000;10:R518–R521. doi: 10.1016/s0960-9822(00)00583-2. [DOI] [PubMed] [Google Scholar]
- 43.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 45.Cho JA, et al. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13:558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Eckard SC, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol. 2014;15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 48.Reina CP, Nabet BY, Young PD, Pittman RN. Basal and stress-induced Hsp70 are modulated by ataxin-3. Cell Stress Chaperones. 2012;17:729–742. doi: 10.1007/s12192-012-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabral Miranda F, et al. CHIP, a carboxy terminus HSP-70 interacting protein, prevents cell death induced by endoplasmic reticulum stress in the central nervous system. Front Cell Neurosci. 2014;8:438. doi: 10.3389/fncel.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui S, et al. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 51.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alamares-Sapuay JG, et al. Serum- and glucocorticoid-regulated kinase 1 is required for nuclear export of the ribonucleoprotein of influenza A virus. J Virol. 2013;87:6020–6026. doi: 10.1128/JVI.01258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jager S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jager S, et al. Purification and characterization of HIV-human protein complexes. Methods. 2011;53:13–19. doi: 10.1016/j.ymeth.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verschueren E, et al. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics. 2015;49 doi: 10.1002/0471250953.bi0819s49. 8 19 11-18 19 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verschueren E, et al. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics. 2015;49 doi: 10.1002/0471250953.bi0819s49. 8 19 11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang CY, et al. Cofactors required for TLR7- and TLR9-dependent innate immune responses. Cell host & microbe. 2012;11:306–318. doi: 10.1016/j.chom.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dang O, Navarro L, Anderson K, David M. Cutting edge: anthrax lethal toxin inhibits activation of IFN-regulatory factor 3 by lipopolysaccharide. J Immunol. 2004;172:747–751. doi: 10.4049/jimmunol.172.2.747. [DOI] [PubMed] [Google Scholar]
- 63.Runge S, et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber M, Weber F. Monitoring activation of the antiviral pattern recognition receptors RIG-I and PKR by limited protease digestion and native PAGE. J Vis Exp. 2014:e51415. doi: 10.3791/51415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matrosovich M, Matrosovich T, Garten W, Klenk HD. New low-viscosity overlay medium for viral plaque assays. Virol J. 2006;3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.