Abstract
An increasing number of microRNAs have been found to be involved in tumorigenesis, including melanoma tumorigenesis. miR-204-5p is down-regulated and functions as a tumor suppressor in many human malignant tumors. miR-204-5p expression is also decreased in melanoma tissues, but its biological roles and molecular mechanisms in malignant melanoma remain unclear. In this study, the aberrant down-regulation of miR-204-5p was detected in melanoma, especially in metastatic melanoma. miR-204-5p also served as a protective factor for the prognosis of melanoma patients. We determined that miR-204-5p suppresses cell proliferation, migration and invasion, and promotes cell apoptosis in melanoma. Matrix metalloproteinases-9 and B-cell lymphoma-2 are the functional targets of miR-204-5p, through which it plays an important biological role in malignant melanoma. The effect of miR-204-5p on malignant melanoma is verified using a xenograft model. We also determined that miR-204-5p increases 5-fluorouracil and cisplatin (DDP) chemosensitivity in malignant melanoma cells. This finding elucidates new functions and mechanisms for miR-204-5p in melanoma development, and provides potential therapeutic targets for the treatment of melanoma.
Keywords: miR-204-5p, MMP9, BCL2, melanoma, cell apoptosis, migration, invasion
Introduction
Malignant melanoma, a tumor derived from melanocytes, is the leading cause of skin cancer-related deaths, with an increased global incidence in the recent years.1–3 It is highly malignant, progresses rapidly and metastasizes early. There is no cure for patients with advanced melanomas.4 The main cause of melanoma-related deaths is the metastatic phenotype.5 The development of melanoma involves complex changes in multiple genes, multiple signaling pathways and gene regulation.6,7 Therefore, further study of the underlying molecular mechanisms involved in the progression of melanoma is crucial.
MicroRNAs (miRNAs) are approximately 22-nt long non-coding single-stranded RNA molecules, which are encoded by endogenous genes.8 miRNAs play a key role in gene regulation by cleaving or repressing the translation of their target mRNAs at the posttranscriptional level.9 Accumulating evidence indicates that miRNAs are involved in many biological processes, especially tumorigenesis. They can modulate cancer cell proliferation, invasion, metastasis and apoptosis.10–12 Dysregulation of miRNAs is related to initiation of various types of cancers, including melanoma.13 The function and role of several miRNAs in melanoma genesis and development have been identified.14,15
miR-204-5p is a member of the miRNA family. It is down-regulated and functions as a tumor suppressor in various types of human tumors, such as glioblastoma, prostate cancer and non-small-cell lung carcinoma (NSCLC).16–18 It has been reported that miR-204-5p is down-regulated in melanoma tissues.19 However, the function and molecular mechanism of this in melanoma remains unclear. In this study, we showed that the miR-204-5p expression was decreased in melanoma tissue, and its expression serves as a protective factor improving the prognosis of melanoma patients. We also demonstrated that miR-204-5p suppressed the proliferation, invasion and migration of melanoma cells, and promoted melanoma cell apoptosis. Moreover, matrix metalloproteinases-9 (MMP9) and B-cell lymphoma-2 (BCL2) are the direct functional targets of miR-204-5p in melanoma. The up-regulation of miR-204 could increase 5-fluorouracil (5-FU) and cisplatin (DDP) chemosensitivity in malignant melanoma cells. Based on these findings, we report a novel molecular mechanism of miR-204-5p as a tumor suppressor in malignant melanoma. miR-204-5p could serve as a potential therapeutic target and survival indicator for melanoma patients.
Materials and methods
Human tissues
Thirty malignant melanoma tissues (including nineteen primary melanoma tissues and eleven metastatic melanoma tissues) and benign nevi tissues were collected during surgery at the Affiliated People’s Hospital of Jiangsu University. None of the patients received chemotherapy or radiotherapy before surgical resection. Clinical pathological diagnosis of the tissues was performed independently by two professional pathologists. The samples were stored in liquid nitrogen. This study was approved by the Human Research Ethics Committee of the Affiliated People’s Hospital of Jiangsu University. A written informed consent was obtained for this study from human study participants or their next of kin if under 16 years of age.
Cell lines and cell culture
The human malignant melanoma cell line A375 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). The human malignant melanoma cell lines: WM35, SK-MEL-5 and SK-MEL-2, were provided by American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA USA) supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. The human epidermal melanocytes, adult, lightly pigmented (HEMa-LP) were purchased from Invitrogen, and the cells were maintained in medium 254 and human melanocyte growth supplement (Cascade Biologics). All cell lines were incubated in a humidified incubator containing 5% CO2 at 37°C.
Oligonucleotides, plasmids and cell transfection
The oligonucleotides were chemically synthesized by GenePharma (Shanghai, People’s Republic of China), and the sequences were as follows: hsa-miR-204-5p mimic, 5′-UUCCCUUUGUCAUCCUAUGCCU-3′; negative control (NC), 5′-UUCUCCGAACGUGUCACGUTT-3′. For constructing MMP9 and BCL2 constructs, the full open reading frame complementary DNA (cDNA) clones for MMP9 and BCL2 were transcribed, and the products were amplified and inserted into pcDNA3.1. 5-FU and cisplatin were obtained from Sigma-Aldrich Co. (St Louis, MO, USA). The oligonucleotides and plasmids were transfected into melanoma cells with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen). To detect the levels of miR-204-5p, MMP9 and BCL2, reverse transcription (RT) was conducted with Fermentas reverse transcription reagents using an Applied Biosystems® TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). An ABI StepOne Plus system (Applied Biosystems) was used to perform the amplification reaction. The specific primers for miR-204-5p were purchased from Guangzhou RiboBio Co Ltd (Guangzhou, People’s Republic of China), and U6 was used for normalization. For the analysis of the levels of BCL2 and MMP9, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization. The following primers were used: BCL2 (forward) 5′-GGATTGTGGCCTTCTTTGAG-3′ and (reverse) 5′-CCAAACTGAGCAGAGTCTTC-3′; MMP9 (forward) 5′-CCTGGAGACCTGAGAACCAATC-3′ and MMP9 (reverse) 5′-CCACCCGAGTGTAACCATAGC-3′. The 2−ΔΔCt method was used to analyze the data. Each experiment was performed in triplicate.
CCK-8 assay
A cell-counting kit-8 (CCK-8; Beyotime, Shanghai, People’s Republic of China) assay was applied to evaluate the proliferative ability of the melanoma cells. The transfected A375 cells (5×103 cells) were added to 96-well plates. At 12, 24, 36 and 48 h, the medium of each well was replaced with 100 μL of fresh culture media that contained 10% CCK-8 solution. Subsequently, these cells were incubated for 2 h at 37°C, and the absorbance was measured using a microplate reader (Multiskan FC, Thermo Scientific) at a wavelength of 450 nm. Experiments were independently repeated in triplicate.
Apoptosis assay
The related oligonucleotides and plasmids were transfected into melanoma cells. After 48 h, an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) was used to analyze the apoptosis ratio according to the manufacturer’s instructions. The apoptosis percentages were evaluated using Annexin V-FITC and propidium iodide (PI) double staining; cells without Annexin V and PI staining were used as controls. The Annexin V+ and PI− cells were considered apoptotic. Experiments were repeated in triplicate.
Cell invasion and migration assays
A Transwell assay was used to evaluate the invasion ability of melanoma cells. Transfected A375 cells were digested and then resuspended in serum-free DMEM. Then, the cells were placed on the top of the matrigel-coated invasion chambers (BD Biosciences). We added 500 μL of DMEM containing 10% FBS to the lower chamber as a chemoattractant. Cotton swabs were used to remove the non-invasive cells after 24 h. The invading cells were fixed and stained with 0.1% crystal violet. The cells were counted and photographed using a microscope (×100). The migration ability of melanoma cells was determined using a scratch wound assay. After transfection for 24 h, a 200 μL pipette tip was used to scratch the cell layers to form wound gaps. The cell culture was continued in 10% FBS-supplemented DMEM at 37°C. At 0 and 24 h, the cells were photographed under a microscope, and the wound width was recorded. Experiments were repeated in triplicate.
Luciferase reporter assay
The 3′-untranslated region (UTR) fragments of BCL2 and MMP9 containing the putative binding sequences of miR-204-5p were cloned into pMIR-REPORT vectors, and a mutated plasmid was used as a control. The melanoma cells were co-transfected with a hsa-miR-204-5p mimic and related reporter plasmids. The luciferase activity was measured using a Dual Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA) after transfection for 48 h according to the manufacturer’s instructions.
Western blot
Radioimmunoprecipitation assay (RIPA) buffer (KenGEN, Nanjing, People’s Republic of China) was used to extract the total protein from cells after transfection, and the protein concentrations were determined using a bicinchoninic acid (BCA) Protein Assay Kit (Beyotime, People’s Republic of China). Equal amounts of protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk and incubated overnight with diluted antibodies against MMP9 (1:1,000, Abcam, UK) and BCL2 (1:1,000, Abcam), followed by incubation with horseradish peroxidase-conjugated secondary antibody (1:2,500, Santa Cruz, CA, USA). GAPDH was used as a control (1:2,500, Abcam).
Xenograft tumor assay
Ten immunodeficient female nude mice were used to test the effects of miR-204-5p in malignant melanoma in vivo. All nude mice were obtained from the Beijing Laboratory Animal Center (Beijing, People’s Republic of China). These nude mice were divided into two groups (the miR-204-5p group and the NC group). About 2×106 logarithmically growing A375 cells (the miR-204-5p mimic-transfected cells and the NC-transfected cells) were subcutaneously injected into nude mice, respectively. Subsequently, the miR-204-5p mimic was locally injected into every xenograft tumor in the miR-204-5p group, and in the scramble group the tumor was injected with NC oligonucleotides. These injections were performed every 4 days. Tumor volume was measured after each injection and calculated according to the formula: V (mm3) =0.5 * a * b2 (a represents the longest axis and b the shortest axis). After 32 days, the tumors were stripped and weighed. Total proteins and RNA were extracted from tissues; the expressions of miR-204-5p, MMP9 and BCL2 were detected using western blot analysis or quantitative real-time polymerase chain reaction (qRT-PCR). The study was approved by the Experimental Animal Ethics Committee of the Affiliated People’s Hospital of Jiangsu University, and conducted according to the guidelines and regulations of the Experimental Animal Ethics Committee of the Affiliated People’s Hospital of Jiangsu University.
Statistical analysis
The data were presented as the mean ± standard deviation and were analyzed with an Statistical Package for the Social Sciences (SPSS) 13.0 software package (IBM Corp., Armonk, NY, USA). The data were analyzed by Student’s t-test or one-way analysis of variance. The Pearson correlation analysis was performed using MATLAB software. Log-rank tests were used to analyze the difference in the overall survival time. P<0.05 was considered to be statistically significant. All experiments were independently performed in triplicate.
Results
miR-204-5p is down-regulated in melanoma tissues and cells, and confers a protective effect that improves the prognosis of patients with melanoma.
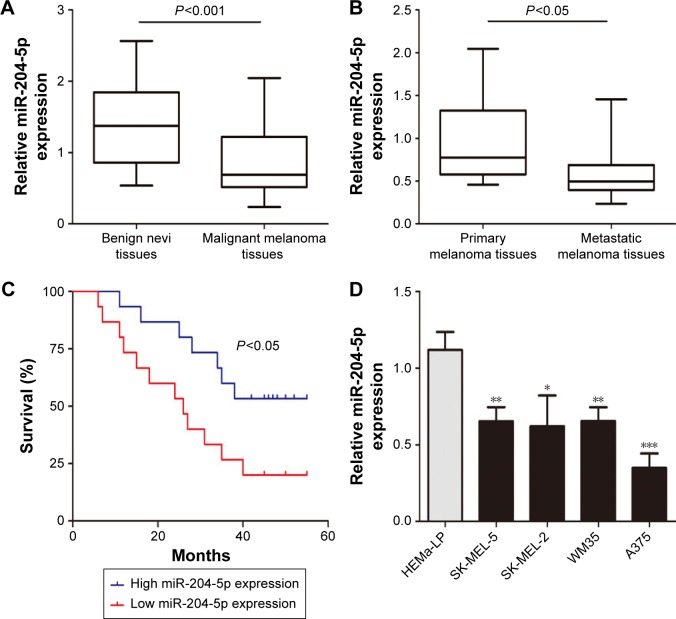
We first investigated miR-204-5p expression in thirty malignant melanoma tissues and twenty benign nevi tissues using RT-qPCR. As shown in Figure 1A, the miR-204-5p expression was markedly decreased in melanoma compared to benign nevi. In addition, miR-204-5p levels were more reduced in metastatic melanoma tissues relative to primary melanoma tissues (Figure 1B). We next examined the correlation between miR-204-5p expression and overall survival of melanoma patients. Low miR-204-5p expression in patients was associated with poor overall survival compared with high miR-204-5p expression (Figure 1C). We also detected the miR-204-5p levels in human melanoma cell lines (A375, WM35, SK-MEL-2 and SK-MEL-5) and HEMa-LP. miR-204-5p was considerably lower in melanoma cells than in HEMa-LP, especially in A375 cells (Figure 1D). These findings indicated that down-regulation of miR-204-5p may contribute to the malignant progression of melanoma.
Figure 1.
miR-204-5p is down-regulated in melanoma tissues and cells, and confers a protective effect that improves the prognosis of patients with melanoma. (A) miR-204-5p levels were analyzed in malignant melanoma tissues and benign nevi tissues. (B) The expression of miR-204-5p was analyzed in metastatic melanoma tissues and primary melanoma tissues. (C) The overall survival curves of melanoma patients with high miR-204-5p expression and low miR-204-5p expression. (D) The miR-204-5p expression profile in human melanoma cell lines (A375, WM35, SK-MEL-2 and SK-MEL-5) and human epidermal melanocytes (HEMa-LP). *P<0.05, **P<0.01, ***P<0.001.
miR-204-5p acts as a tumor suppressor in melanoma
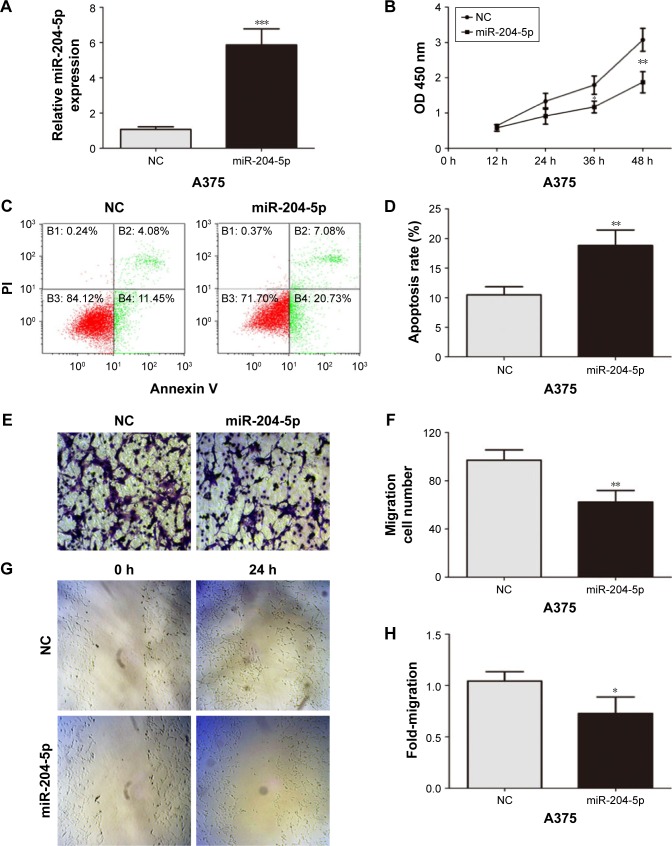
To further explore the biological function of miR-204-5p in melanoma cells, the commercially synthesized miR-204-5p mimic was transfected into A375 cells (Figure 2A). A CCK-8 assay showed that overexpression of miR-204-5p markedly repressed the proliferation of A375 cells (Figure 2B). In an apoptosis assay, the miR-204-5p mimic-transfected cells exhibited more apoptotic cells compared to the control group (Figure 2C and D). Transwell assays and scratch wound assays revealed that the invasive and migratory abilities of A375 cells were inhibited by the miR-204-5p mimic (Figure 2E–H). Taken together, these results suggest that miR-204-5p suppresses cell proliferation, migration and invasion, and promotes cell apoptosis in melanoma.
Figure 2.
miR-204-5p acts as a tumor suppressor in melanoma. (A) Transfection efficiency of the miR-204-5p mimic was determined by PCR. (B) The proliferative ability of A375 cells was measured by a CCK-8 assay. (C and D) Effect of miR-204-5p on apoptosis of A375 cells was assessed by flow cytometry. (E and F) The effect of miR-204-5p on the invasive capacity of melanoma cells was assessed by a Transwell assay. (G and H) The effect of miR-204-5p on the migratory ability of melanoma cells was assessed by the scratch wound assay. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: PCR, polymerase chain reaction; CCK-8, cell-counting kit-8; NC, negative control; PI, Propidium iodide; OD, optical density.
MMP9 and BCL2 are the direct functional targets of miR-204-5p
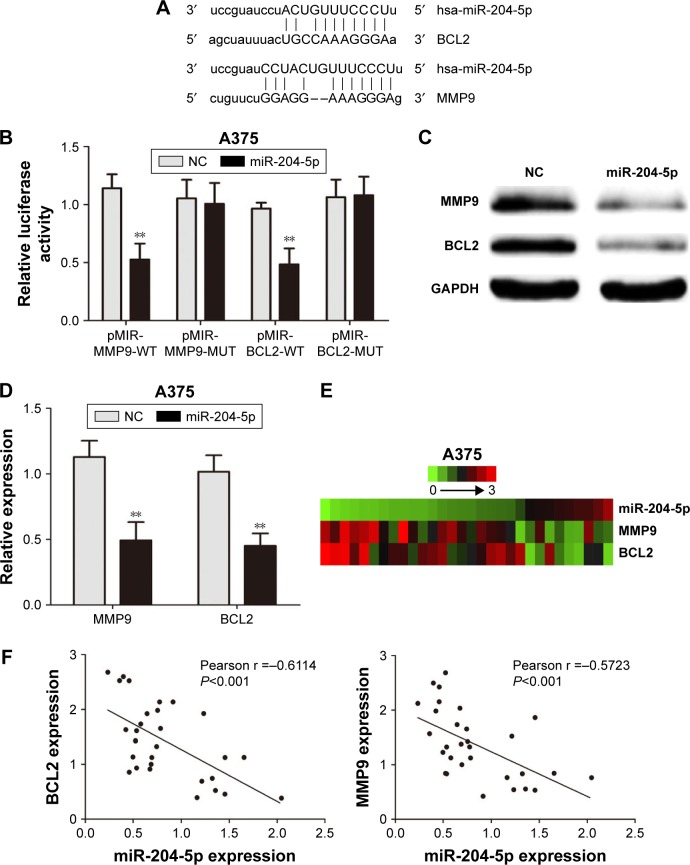
We further studied the mechanism of action of miR-204-5p in melanoma cells. miRanda (http://www.microrna.org/) was used to investigate the direct targets of miR-204-5p. The binding sites of miR-204-5p matched the 3′-UTR of MMP9 (associated with invasion) and BCL2 (associated with apoptosis), suggesting that MMP9 and BCL2 are potential targets of miR-204-5p (Figure 3A). We then constructed luciferase reporter plasmids containing the wild-type (WT; pMIR-MMP9-WT and pMIR-BCL2-WT) and mutant (Mut; pMIR-MMP9-MUT and pMIR-BCL2-MUT) binding sites. Dual luciferase reporter assays showed that overexpression of miR-204-5p suppressed luciferase activity in cells with the WT plasmid, though no significant change was detected in the A375 cells transfected with the Mut plasmid (Figure 3B). Meanwhile, western blotting and real-time PCR showed that the up-regulation of miR-204-5p led to a decrease in the expression of endogenous MMP9 and BCL2 (Figure 3C and D). We then investigated the possible correlations among miR-204-5p, MMP9 and BCL2 expression in melanoma tissues. The heat map data of thirty malignant melanoma tissues revealed a negative correlation between miR-204-5p, and MMP9 and BCL2 (Figure 3E). The Pearson correlation of miR-204-5p and MMP9 expression levels was negative, while the correlation between miR-204-5p and BCL2 was the same (Figure 3F). Overall, we demonstrated that MMP9 and BCL2 are the functional targets of miR-204-5p in melanoma.
Figure 3.
MMP9 and BCL2 are the direct functional targets of miR-204-5p. (A) The binding sites of miR-204-5p within the 3′-UTR of MMP9 and BCL2 were predicted by miRanda. (B) Overexpression of miR-204-5p suppressed luciferase activity in cells with the WT plasmid but did not cause a significant change in A375 cells with the Mut plasmid. (C) Western blots identified MMP9 and BCL2 protein expression changes following transfection with a miR-204-5p mimic or NC; GAPDH was used as a control. (D) PCR to assess the mRNA level of MMP9 and BCL2 in A375 cells following transfection with a miR-204-5p mimic. (E) The heat map shows that MMP9 and BCL2 are negatively correlated with miR-204-5p in thirty malignant melanoma tissues. (F) The Pearson correlation of miR-204-5p and MMP9 and BCL2 expression in thirty malignant melanoma tissues. **P<0.01.
Abbreviations: MMP9, matrix metalloproteinases-9; BCL2, B-cell lymphoma-2; Mut, mutant type; PCR, polymerase chain reaction; UTR, untranslated region; WT, wild type; NC, negative control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Expression of MMP9 and BCL2 attenuates the effects of miR-204-5p in melanoma
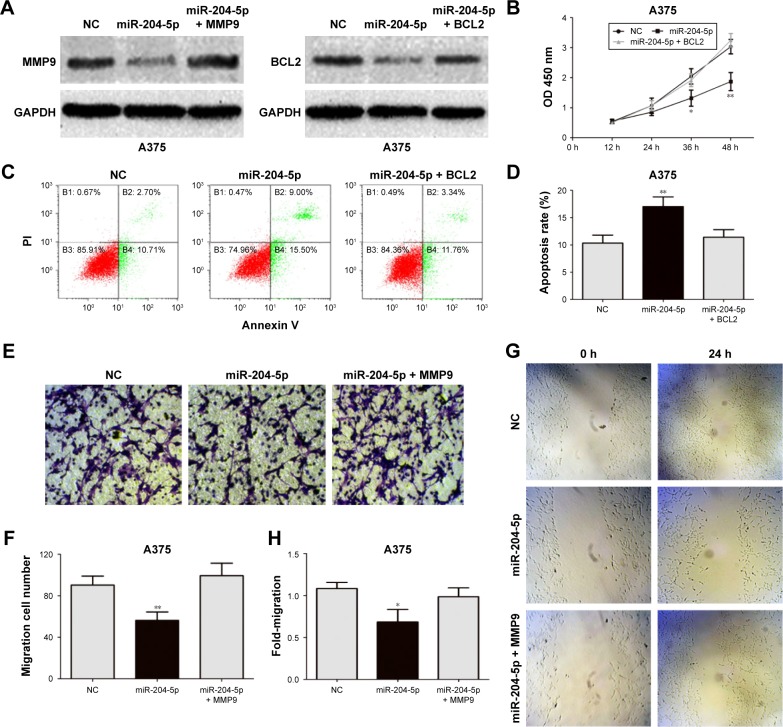
To elucidate the relationship between miR-204-5p and MMP9 and BCL2, we transfected A375 cells with the miR-204-5p mimic together with the MMP9 and BCL2 plasmids, and performed a series of functional assays. Western blotting was used to confirm MMP9 and BCL2 protein expression (Figure 4A). Importantly, the effect of miR-204-5p on the proliferation, migration, invasion and apoptosis of A375 melanoma cells was rescued by the MMP9 and BCL2 plasmids (Figure 4B–H). In summary, these results suggested that MMP9 and BCL2 are the critical targets of miR-204-5p in regulating the biological function of melanoma cells.
Figure 4.
Expression of MMP9 and BCL2 attenuates the effects of miR-204-5p in melanoma. (A) Western blots identified MMP9 and BCL2 protein expression changes following transfection with miR-204-5p alone or in combination with MMP9 and BCL2. (B) BCL2 plasmid reversed the effect of miR-204-5p on the proliferative ability of A375 cells. (C and D) Expression of BCL2 reversed miR-204-5p-induced apoptosis as assessed by flow cytometry. (E and F) The effect of miR-204-5p on the invasive ability of A375 cells was largely abrogated by the MMP9 plasmid. (G and H) The MMP9 plasmid reversed the effect of miR-204-5p on the migratory ability of A375 cells. *P<0.05, **P<0.01.
Abbreviations: MMP9, matrix metalloproteinases-9; BCL2, B-cell lymphoma-2; NC, negative control; PI, Propidium iodide; OD, optical density.
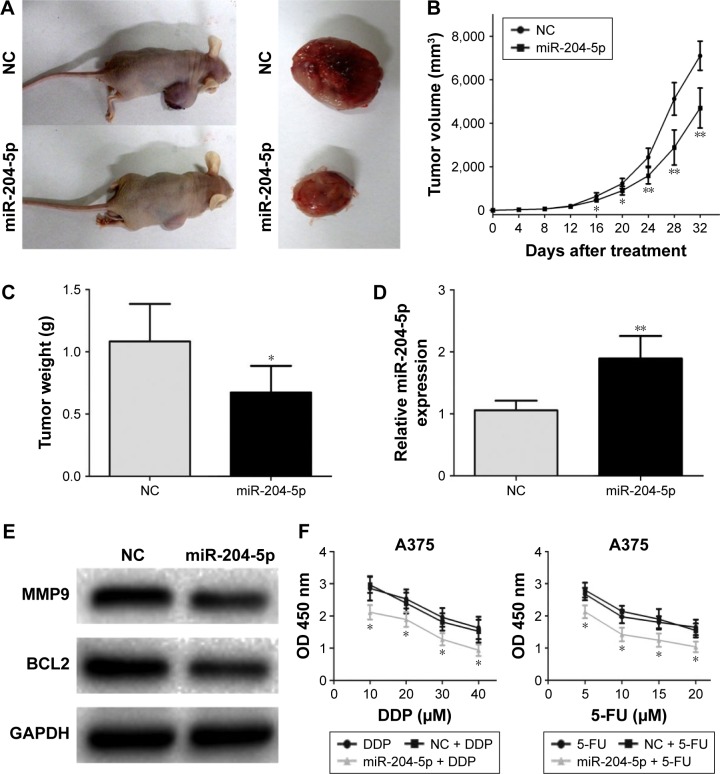
miR-204-5p inhibits melanoma growth in vivo and increases sensitivity of melanoma cells to chemotherapy
To test the function of miR-204-5p in vivo, we generated the melanoma xenograft model by subcutaneously inoculating the miR-204-5p mimic- or NC-transfected A375 cells (Figure 5A). As shown in Figure 5B, the tumor volume in the NC group was significantly bigger compared with the miR-204-5p overexpression group between 16 and 32 days post-injection. After 32 days, the mice were sacrificed, and tumors were stripped and weighed. The tumor weight in the miR-204-5p overexpression group was significantly less than that in the NC group (Figure 5C). Finally, western blotting revealed that the expression of BCL2 and MMP9 was decreased in miR-204-5p mimics-treated group (Figure 5D and E). These results indicated that miR-204-5p inhibits melanoma growth in vivo. Finally, a dose-dependent 5-FU and DDP treatment at different concentrations was conducted on miR-204-5p mimic- or NC-transfected melanoma cells. After 48 h, a CCK-8 assay showed that the inhibition of cell proliferation increased with 5-FU and DDP dose, and the cell proliferation was more inhibited in miR-204-5p-transfected group (Figure 5F). These results suggested that miR-204-5p increases the sensitivity of melanoma cells to chemotherapy.
Figure 5.
miR-204-5p inhibits melanoma growth in vivo and increases sensitivity of melanoma cells to chemotherapy. (A) Tumor formation in nude mice and the excision tumor of A375 xenografts. (B) Difference in tumor volume between the NC group and the miR-204-5p group. (C) The tumor weight of excision tumor. (D) PCR identified miR-204-5p expression changes. (E) Western blots identified MMP9 and BCL2 protein expression changes. (F) A dose-dependent 5-FU and DDP treatment was conducted on miR-204-5p mimic- or NC-transfected melanoma cells. The proliferative ability of A375 cells was measured by CCK-8 assay. *P<0.05, **P<0.01.
Abbreviations: NC, negative control; 5-FU, 5-fluorouracil; CCK-8, cell-counting kit-8; PCR, polymerase chain reaction; MMP9, matrix metalloproteinases-9; BCL2, B-cell lymphoma-2; DDP, cisplatin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
miRNA, a non-coding RNA of about twenty-two nucleotides in length, plays an important role in several biological processes by regulating gene expression at the posttranscriptional level through binding of the 3′-UTR of the target mRNA.20,21 Extensive reports have shown that many miRNAs act as oncomiRNAs or tumor suppressors in diverse human tumors, including in melanoma.22–24 The role of miR-204-5p in tumor development has recently received much attention. Several researchers have reported that miR-204-5p is down-regulated and functions as a tumor suppressor in various types of human tumors. For example, miR-204-5p is down-regulated in glioblastoma, and suppresses the development and progression of glioblastoma by targeting ATF2.16 miR-204-5p suppresses NSCLC invasion and migration by targeting JAK2.17 Similarly, miR-204-5p acts as a tumor suppressor in prostate cancer and cutaneous squamous cell carcinoma.18,25 Some researchers have discovered that miR-204-5p expression is also decreased in melanoma tissues.19 However, the function and molecular mechanism of miR-204-5p in melanoma genesis and development remain unclear.
In this study, we report that miR-204-5p expression was decreased in melanoma tissues and cells, especially in metastatic melanoma. miR-204-5p may serve as a protective factor that predicts the prognosis of melanoma patients. These results indicate that low expression of miR-204-5p may play an important role in the progression of malignant melanoma. Furthermore, we investigated the biological function of miR-204-5p in melanoma cells. It inhibited proliferation, migration and invasion, and promoted apoptosis of melanoma cells. Then, we further examined the molecular mechanism of miR-204-5p in melanoma. miRanda was used to investigate the direct targets of miR-204-5p. We discovered that MMP9 and BCL2 may be the functional targets of miR-204-5p in melanoma.
BCL2, the most important antiapoptotic oncogene, inhibits apoptosis by interacting with proapoptotic members of the BCL2 family (BAD, BAX and BID).26,27 The imbalance of antiapoptotic and proapoptotic proteins plays an important role in tumor development, and a decrease in BCL2 expression disrupts the balance of antiapoptotic and proapoptotic signaling and activates the apoptotic cascade.28 MMPs drive metastasis by degrading the extracellular matrix (ECM), and type I and IV collagens are the major ECM components.29 MMP9, a member of the MMP family, degrades these collagens, thereby expediting and facilitating cancer cell invasion and metastasis.30 MMP9 is overexpressed in various human cancer types.31,32 A large number of studies have verified that many oncogenes and tumor suppressor genes influence cancer progression through regulation of BCL2 and MMP9, including melanoma.33,34 In this study, our data reveal a negative correlation between miR-204-5p, and MMP9 and BCL2 in melanoma tissues. We demonstrated that MMP9 and BCL2 are the functional targets of miR-204-5p in melanoma by luciferase reporter assays and a series of functional assays. Finally, we found that miR-204-5p inhibits melanoma growth in vivo by regulating the expression of BCL2 and MMP9.
Conclusion
We demonstrated that miR-204-5p acts as a tumor suppressor in melanoma by targeting MMP9 and BCL2, thus regulating cell proliferation, invasion, migration and apoptosis. Low levels of miR-204-5p participate in melanoma progression and are associated with poor overall survival of melanoma patients. We also determined that miR-204-5p increases the sensitivity of melanoma cells to chemotherapy. Thus, miR-204-5p may be a potential therapeutic target for melanoma.
Acknowledgments
This work was supported by grants from the Social Development and Technology Support Foundation of Zhenjiang city (SH2011057 to Bin Xu).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Millet A, Martin AR, Ronco C, Rocchi S, Benhida R. Metastatic melanoma: insights into the evolution of the treatments and future challenges. Med Res Rev. 2017;37(1):98–148. doi: 10.1002/med.21404. [DOI] [PubMed] [Google Scholar]
- 2.Little EG, Eide MJ. Update on the current state of melanoma incidence. Dermatol Clin. 2012;30(3):355–361. doi: 10.1016/j.det.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118(9):1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 4.Haass NK, Schumacher U. Melanoma never says die. Exp Dermatol. 2014;23(7):471–472. doi: 10.1111/exd.12400. [DOI] [PubMed] [Google Scholar]
- 5.Russo AE, Ferraù F, Antonelli G, Priolo D, McCubrey JA, Libra M. Malignant melanoma in elderly patients: biological, surgical and medical issues. Expert Rev Anticancer Ther. 2015;15(1):101–108. doi: 10.1586/14737140.2015.961426. [DOI] [PubMed] [Google Scholar]
- 6.Nikolaou VA, Stratigos AJ, Flaherty KT, Tsao H. Melanoma: new insights and new therapies. J Invest Dermatol. 2012;132(3 Pt 2):854–863. doi: 10.1038/jid.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzdensky AB, Demyanenko SV, Bibov MY. Signal transduction in human cutaneous melanoma and target drugs. Curr Cancer Drug Targets. 2013;13(8):843–866. doi: 10.2174/1568009611313080004. [DOI] [PubMed] [Google Scholar]
- 8.Galasso M, Sandhu SK, Volinia S. MicroRNA expression signatures in solid malignancies. Cancer J. 2012;18(3):238–243. doi: 10.1097/PPO.0b013e318258b5f4. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Wang H, Tao K, et al. miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp Cell Res. 2013;319(8):1094–1101. doi: 10.1016/j.yexcr.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Luna-Aguirre CM, de la Luz Martinez-Fierro M, Mar-Aguilar F, et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark. 2015;15(3):299–310. doi: 10.3233/CBM-150465. [DOI] [PubMed] [Google Scholar]
- 11.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol. 2015;21(34):9838–9852. doi: 10.3748/wjg.v21.i34.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segura MF, Greenwald HS, Hanniford D, Osman I, Hernando E. MicroRNA and cutaneous melanoma: from discovery to prognosis and therapy. Carcinogenesis. 2012;33(10):1823–1832. doi: 10.1093/carcin/bgs205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Xu D, Xie H, et al. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1α. Cancer Biol Ther. 2015;16(6):846–855. doi: 10.1080/15384047.2015.1030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan W, Wang Y, Chen X, et al. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6(15):13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song S, Fajol A, Tu X, Ren B, Shi S. miR-204 suppresses the development and progression of human glioblastoma by targeting ATF2. Oncotarget. 2016;7(43):70058–70065. doi: 10.18632/oncotarget.11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P, Lv HY, Zhou DM, Zhang EN. miR-204 suppresses non-small-cell lung carcinoma (NSCLC) invasion and migration by targeting JAK2. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15026415. [DOI] [PubMed] [Google Scholar]
- 18.Lin YC, Lin JF, Tsai TF, Chou KY, Chen HE, Hwang TI. Tumor suppressor miRNA-204-5p promotes apoptosis by targeting BCL2 in prostate cancer cells. Asian J Surg. 2016;(16):30173–30177. doi: 10.1016/j.asjsur.2016.07.001. pii:S1015–9584. [DOI] [PubMed] [Google Scholar]
- 19.Kozubek J, Ma Z, Fleming E, et al. In-depth characterization of microRNA transcriptome in melanoma. PLoS One. 2013;8(9):e72699. doi: 10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9(1):e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun X, Li J, Sun Y, et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget. 2016;7(33):53558–53570. doi: 10.18632/oncotarget.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang CH, Wang Y, Sims M, et al. MiRNA203 suppresses the expression of protumorigenic STAT1 in glioblastoma to inhibit tumorigenesis. Oncotarget. 2016;7(51):84017–84029. doi: 10.18632/oncotarget.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toll A, Salgado R, Espinet B, et al. MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol Cancer. 2016;15(1):53. doi: 10.1186/s12943-016-0537-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Packham G. Mutation of BCL-2 family proteins in cancer. Apoptosis. 1998;3(2):75–82. doi: 10.1023/a:1009688706783. [DOI] [PubMed] [Google Scholar]
- 27.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27(50):6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 28.Humlová Z. Protooncogene bcl-2 in process of apoptosis. Review article. Sb Lek. 2002;103(4):419–425. [PubMed] [Google Scholar]
- 29.Zhou B, Wu Q, Chen G, Zhang TP, Zhao YP. NOP14 promotes proliferation and metastasis of pancreatic cancer cells. Cancer Lett. 2012;322(2):195–203. doi: 10.1016/j.canlet.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Poudel B, Kim DK, Ki HH, Kwon YB, Lee YM, Kim DK. Downregulation of ERK signaling impairs U2OS osteosarcoma cell migration in collagen matrix by suppressing MMP9 production. Oncol Lett. 2014;7(1):215–218. doi: 10.3892/ol.2013.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 32.Comincini S, Paolillo M, Barbieri G, et al. Gene expression analysis of an EGFR indirectly related pathway identified PTEN and MMP9 as reliable diagnostic markers for human glial tumor specimens. J Biomed Biotechnol. 2009;2009:924565. doi: 10.1155/2009/924565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt K, Joyce CE, Buquicchio F, et al. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 2016;15(9):2025–2037. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei Y, Zhang B, Zhang Y, et al. Lentivirus-mediated downregulation of MAT2B inhibits cell proliferation and induces apoptosis in melanoma. Int J Oncol. 2016;49(3):981–990. doi: 10.3892/ijo.2016.3603. [DOI] [PubMed] [Google Scholar]