Abstract
Myeloid differentiation is controlled by a multilayered regulatory circuitry consisting of various elements, including histone modifications, transcription factors, and posttranscriptional regulators such as miRNAs, long noncoding RNAs, and circular RNAs. However, the molecular mechanism underlying this biological process remains unclear. In this study, through epigenetic profiling analysis using chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq), we identified an lncRNA, HOTAIRM1, with a critical role in myeloid development. Further ChIP-chip analysis showed obvious H3K4me3 and H3K27me3 histone modification peak changes in the promoter region of HOTAIRM1 during the process of monocyte to dendritic cell (DC) differentiation. In line with this observation, HOTAIRM1 RNA expression was downregulated when monocytes differentiated into DCs. Moreover, we found that HOTAIRM1 RNA was regulated by epigenetic factors such as RBBP4 and RBBP7. Mechanistically, we found that the silencing of HOTAIRM1 caused changes in the expression of several monocyte differentiation markers such as CD14 and B7H2. In addition, based on the “competing endogenous RNA” hypothesis, we discovered miR-3960 targeting both HOTAIRM1 and the DC differentiation repression gene, HOXA1, by most possibly constructing a potential competing endogenous RNA network. Increased miR-3960 expression could downregulate both of these two long RNAs and finally lead peripheral blood cells to differentiate into DCs. In summary, our study demonstrates that HOTAIRM1 competitively binds to miR-3960 and finally regulates the process of hematopoiesis, which reveals a novel regulatory mechanism of lncRNA function.
Keywords: myeloid cells, lncRNA, HOTAIRM1, miR-3960, myeloid differentiation
Introduction
Cellular immunity is a process involved in the activation of multiple types of cells such as neutrophils, macrophages, and dendritic cells (DCs), all of which originate from the same progenitor cell, bone marrow stem cell. These stem cells can differentiate into granulocyte–macrophage progenitor cells, which then further convert to monocytes, and finally differentiate into DCs.1 DCs are potent professional antigen-presenting cells (APCs) serving as a bridge linking the innate and adaptive immune systems.2 Human DCs have many reported markers such as CD40, CD80, and CD86. In addition, the B7:CD28 family of costimulatory molecules also play a key role in triggering T-cell activity.3 In the DC microenvironment, cytokines, chemokines, and other stimulatory factors have a great influence on DC differentiation and maturation.4 For instance, under lipopolysaccharide (LPS) stimulation, monocytes could differentiate into mature DCs, while transforming growth factor beta (TGF-β) could inhibit their maturation and further interrupt the activation of T cells.5
Recently, long noncoding RNAs (lncRNAs), a new class of transcripts, have attracted increasing attention for their contributions to different kinds of human diseases and genetic disorders. lncRNAs are RNAs longer than 200 nucleotides (nt) and lack protein-coding function. Although not encoding protein products, they play critical functional roles in regulating gene expression by linking different gene regulatory elements together, including miRNAs, histone modifications, and transcription factors.6,7 Extensive evidences have shown that lncRNA HOTAIR1, a gene transcribed antisense to the HOXA1/2 intergenic region, can influence myeloid cell development through interaction with various chromatin modifiers to regulate myelopoiesis by modulation of epigenetic state of corresponding genes at the HoxA locus.8,9 HOTAIR expression levels are closely associated with diverse types of cancer, including breast cancer, colon cancer, liver cancer, and glioma, and it is also a critical regulator of cell cycle progression.10–13 HOTAIRM1 could be induced by retinoic acid.14,15 It is specifically expressed in the myeloid lineage and found most highly in the terminal stage of monocyte differentiation.16 Meanwhile, HOTAIRM1 is a positive regulator of these monocyte-related HOXA genes, especially HOXA1,17 by which it acquires a function in further regulating this development process and interfering with monocyte to DC differentiation. Indeed, knockdown of HOTAIRM1 in mice could disturb the transcription of CD11b and CD18.18 In this context, recent emerging studies suggested that HOTAIRM1 regulates HOXA1 gene by epigenetic modification of interaction with the PRC2 complex in mouse embryonic stem cells.18,19
In addition to the lncRNA of HOTAIRM1, miRNAs mediate the regulation of HOXA genes, which has been identified as another complex molecular mechanism. miRNAs are short noncoding RNAs with a standard size of 20–22 nt, which could finally repress target gene expression by leading to mRNA degradation through binding to the untranslated regions (3′UTRs) of mRNA. Along with this, miR-3960 that is mainly expressed in bone and liver could target HOXA2 and block its function, which finally affects osteoblast differentiation.20 However, besides the direct interaction with target mRNAs, recent studies illustrated that miRNAs can also influence protein-coding gene expression by binding noncoding RNAs, such as circular RNAs (circRNAs), pseudogene, and lncRNAs, to act as competing endogenous RNAs (ceRNAs).21,22 These ceRNAs function as molecular decoys or “sponges” of miRNAs to impair the activity of miRNAs through sequestration, thereby upregulating miRNA target gene expression.23,24
Since the expression of myeloid differentiation-related HOXA genes can be influenced by both lncRNA HOTAIRM1 and miRNA miR-3960, we decided to investigate whether there is a potential interaction among HOTAIRM1, miR-3960, and HOXA genes based on the “ceRNA theory”. In this study, we show that miR-3960 targets both HOTAIRM1 and the DC differentiation repression gene, HOXA1, by most possibly constructing a potential ceRNA network. Increased miR-3960 expression could downregulate both of these two long RNAs and finally lead peripheral blood cells to differentiate into DCs. In summary, our study demonstrates that HOTAIRM1 competitively binds to miR-3960 and finally regulates the process of hematopoiesis, which reveals a novel regulatory mechanism of lncRNA function.
Materials and methods
Human peripheral blood-derived DCs and cell lines
Human peripheral blood monocytes (HPBMs) were provided by Tianjin Blood Center (Tianjin, People’s Republic of China). Mononuclear cells were obtained from blood sample using ficoll. Briefly, the cells were washed in RPMI 1640 medium supplemented with 1% antibiotic for 1.5 h to separate monocytes and T and B lymphocytes. Subsequently, cells were cultured in complete RPMI 1640 medium supplemented with GM-CSF (40 ng/mL) and IL-4 (40 ng/mL). After 3 days, the cells were harvested by centrifugation at 1,600 rpm for 8 min and then plated into a 24-well plate. Two days later, the cells were divided into the following three groups: control, LPS (20 ng/mL) stimulation group, and TGF-β (20 ng/mL) stimulation group. Cells were used for follow-up experiments after 24 h stimulation. Human 293T cells were cultured in dulbecco's modified eagle medium (DMEM) with 10% fetal bovine serum (FBS) (HyClone, Logan, UT, USA) at 37°C in a humidified 5% CO2 atmosphere. Ethical permission for the study was obtained from Nankai University and Tianjin Blood Center.
Reagent and antibodies
RPMI 1640, DMEM, and FBS were obtained from Gibco (Grand Land, NY, USA). Recombinant human LPS, TGF-β, GM-CSF, and IL-4 were purchased from Sigma-Aldrich (St Louis, MI, USA). Anti-CD14-FITC and anti-B7H2-PE antibodies were purchased from BD Biosciences (San Diego, CA, USA). Small interfering RNA (siRNA), microRNA (miRNA) mimics, and control oligonucleotides were purchased from Riobio (Guangzhou, China).
Quantitative real-time polymerase chain reaction (PCR)
Total RNA was isolated from cells using TRIzol reagent (Carlsbad, CA, Invitrogen, USA). One micro-gram of RNA was reverse transcribed to cDNA using the M-MLV Reverse Transcriptase (Tiangen, Beijing, China) according to the manufacturer’s instructions. Quantitative PCR was performed using SYBR Mixture (CWBIO, Beijing, China), and the fluorescence signal was detected by iQ5 Multicolor real-time (RT)-PCR system (Bio-Rad, Hercules, CA, USA). PCR primer for human was as follows: glyceraldehyde-3-phosphate dehydroge-nase (GAPDH), 5′-GAAGGTGAAGGTCGGAGTC-3′ (sense), 5′-GAAGATGGTGATGGGATTTC-3′ (antisense); miR-3960 stem-loop, 5′-GTATCCAG TGCAGGGTCCGAGGTATTCG CACTGGATACGACCCCCCG-3′; miR-3960 forward, 5′-ATATATAGGCGGCGGCGGA-3′; universal reverse for miRNA, 5′-GTGCAGGGTCCGAGGT-3′; U6, 5′-CTCGCTTCGGCAGCACA-3′ (sense), 5′-AACGCTTCACGAATTTGCGT-3′ (antisense); HOTAIRM1, 5′-CCCACCGTTCAATGAAAGATG-3′ (sense), 5′-TCAAACACCCACATTTCAACC-3′ (antisense); HOXA1, 5′-CAAAAGAAACCCTCCCAAAAC-3′ (sense), 5′-CGTCAGGTACTTGTTGAAG-3′ (antisense). GAPDH was used as an endogenous control. Fold changes were calculated using the 2−ΔΔCt method. PCRs were conducted in triplicate for each sample. All the reactions were performed in triplicate.
Transfection
The cells were cultured in a 24-well plate for the process of monocyte to DC differentiation and then transfected with either HOTAIRM1 or negative control siRNA (100 nM) by using the HiPerFect Transfection Reagent (Qiagen, Alencia, CA, USA) according to the manufacturer’s instructions.
Cell surface staining
Cells transfected with either control or experimental siRNA were collected and maintained in ice-cold PBS with 1% FBS. The expression of CD14 or B7H2 was detected by incubating with antibodies for 15–30 min. After washing twice using PBS, cells were fixed in 1% paraformaldehyde and analyzed using FACScan flow cytometer (BD Biosciences).
Prediction of binding sites
The binding site of miR-3960 on HOTAIRM1 lncRNA was predicted using the RNAfold webserver (http://nhjy.hzau.edu.cn/kech/swxxx/jakj/dianzi/Bioinf4/miRNA/miRNA1.htm).
Results
HOTAIRM1 is downregulated during the process of monocyte to DC differentiation
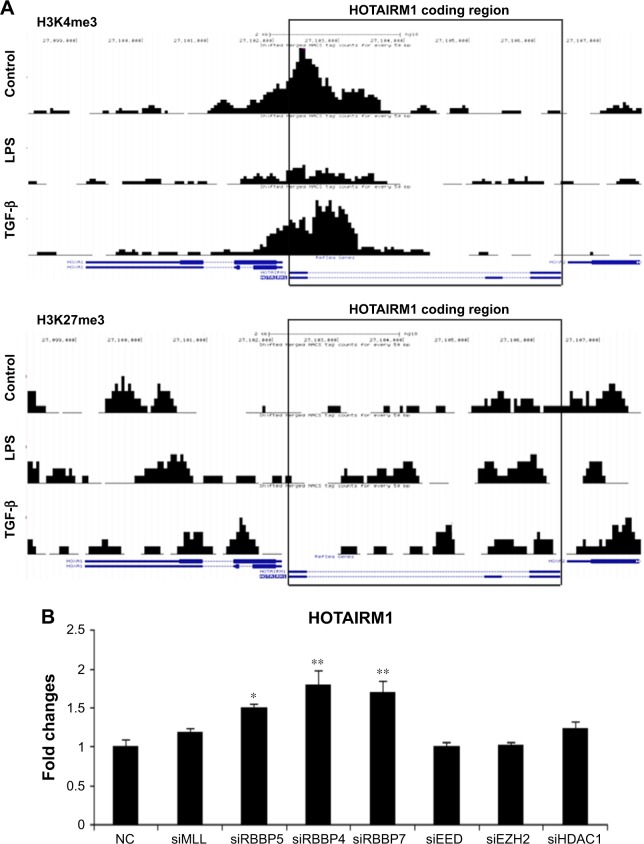
Previous studies have suggested that HOTAIRM1 played a key role in myeloid cell differentiation.16 To explore the potential mechanisms of HOTAIRM1 involved in DC differentiation, an epigenetic modification study was performed using the H3K4me3 and H3K27me3 methylation ChIP-chip map analyses. Indeed, H3K4me3 and H3K27me3 peaks were found in HOTAIRM1-related regions and were highly dynamic along with the process of DC differentiation. Moreover, stimulation with LPS to promote the maturation of HPBMs to DCs led to the downregulation of the transcription activation marker H3K4me3, while the inhibition marker H3K27me3 was increased. In contrast, TGF-β treatment resulted in repressing their DC conversion, whereas H3K4me3 and H3K27me3 expression showed an opposite effect (Figure 1A). Moreover, the effect of two pivotal epigenetic-associated factors RBBP4 and RBBP7 on the regulation of HOTAIRM1 was further investigated. As shown in Figure 1B, silencing of these two factors significantly increased the mRNA level of HOTAIRM1 (Figure 1B). RBBP4 and RBBP7 were found to repress transcription by promoting histone deacetylation and nucleosome remodeling. In addition, RBBP4 and RBBP7 were found to be the components of multiple epigenetic function-associated complexes such as histone deacetylase (HDAC) complex, the nucleosome remodeling, HDAC complex (the NuRD complex), and the PRC2/EED–EZH2 complex.25,26 These findings indicated that HOTAIRM1 was tightly regulated during DC maturation and its downregulation was closely associated with this process.
Figure 1.
Epigenetic screening reveals that lncRNA HOTAIRM1 is associated with histone methylation.
Notes: (A) Monocytes were isolated from human peripheral blood and stimulated with LPS or TGF-β. Then, the prepared cells were detected by ChIP-chip technology. The H3K4me3 and H3K27me3 methylation states around HOTAIRM1 gene regions were analyzed by IGV genome explorer. (B) RT-PCR result of HOTAIRM1 RNA level in DCs transfected with various epigenetic-associated factor siRNAs. *P<0.05; **P<0.01.
Abbreviations: DCs, dendritic cells; lncRNA, long noncoding RNA; RT-PCR, real-time polymerase chain reaction; NC, negative control.
HOTAIRM1 is tightly regulated by histone modifications during DC differentiation
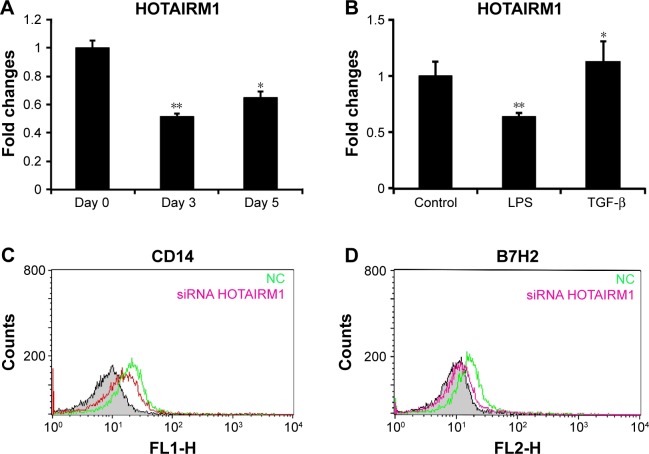
In addition to the research mentioned earlier on histone modifications of HOTAIRM1 gene, we also detected its RNA level directly and the result was consistent with the H3K4me3 and H3K27me3 changes. Stimulation of HPBMs with granulocyte-macrophage colony stimulating factor (GM-CSF) and interleukin-4 (IL-4) for the induction of differentiation led to a downregulation of HOTAIRM1 as detected by reverse transcription polymerase chain reaction (RT-PCR) analysis. Although HOTAIRM1 expression on day 5 was slightly higher than that on day 3, it still maintained a relatively low level in comparison to control level as seen on day 0. This suggested an opposite relationship between HOTAIRM1 expression and DC differentiation (Figure 2A). This conclusion was supported by the fact that the addition of DC differentiation-promoting agent LPS was accompanied by decline in this RNA level, while the inhibitory factor TGF-β could upregulate HOTAIRM1 (Figure 2B). In addition, flow cytometry results visually showed that, compared with the control group, downregulation of HOTAIRM1 using its siRNA had the effect of reducing the monocyte markers CD14 and B7H2 more severely (Figure 2C and D), implying that this lncRNA had the ability of keeping monocyte markers and inhibiting them from developing into DCs.
Figure 2.
HOTAIRM1 is involved in DC differentiation.
Notes: (A) The HOTAIRM1 RNA concentration was analyzed by RT-PCR in the process of monocyte differentiation to DCs. (B) HOTAIRM1 lncRNA level in monocytes stimulated with LPS or TGF-β. Flow cytometry to detect CD14 (C) and B7H2 (D) in cells transfected with HOTAIRM1 or NC siRNA. Each experiment was independently performed three times. *P<0.05; **P<0.01.
Abbreviations: DCs, dendritic cells; lncRNA, long noncoding RNA; RT-PCR, real-time polymerase chain reaction; NC, negative control.
The expression of HOTAIRM1 shows negative correlation with miRNA-3960
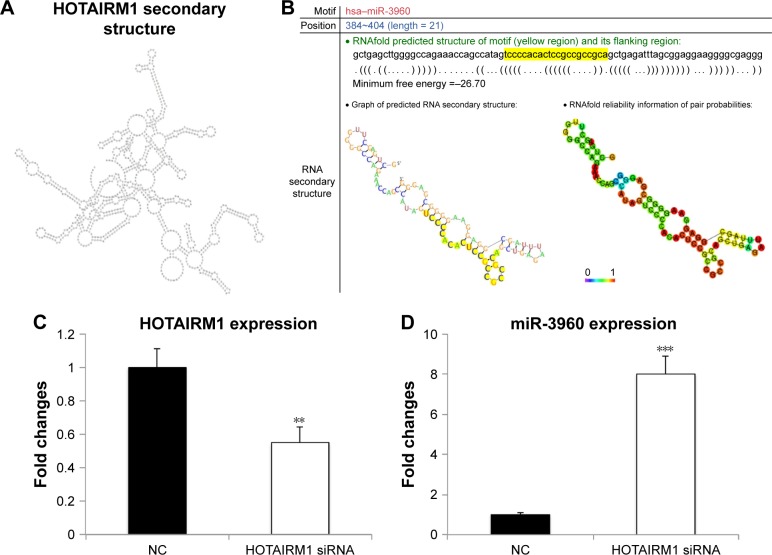
lncRNAs function as ceRNAs to bind to miRNAs, which has recently been identified as a novel posttranscriptional regulatory mechanism associated with multiple tumorigenesis. To elucidate whether HOTAIRM1 performed its function through this mechanism in myeloid cells, we analyzed the secondary structure of HOTAIRM1 and then searched the potential miRNAs binding to HOTAIRM1 using RNAfold webserver. Finally, we identified miR-3960 as a promising putative interaction partner of HOTAIRM1. The seed sequence of this lncRNA to interact with miR-3960 located at position 384–404 with a length of 21 nt, which is much longer than the usual miRNA-binding site in mammalian cells with a length of 6–8 nt. Finally, the interaction of miR-3960 with HOTAIRM1 would result in the degradation of these two noncoding RNAs, which needed a long seed region to mediate this effect, rather than just inhibiting the usual activity of target RNA without the occurrence of any decay event (Figure 3A and B).
Figure 3.
HOTAIRM1 interacts with miR-3960.
Notes: (A and B) HOTAIRM1 secondary structure was stimulated, and its potential binding site for miR-3960 was predicted by the RNAfold webserver. (C) HOTAIRM1 concentration was detected in peripheral blood cells loaded with HOTAIRM1 siRNA or negative control siRNA using RT-PCR. (D) The corresponding miR-3960 concentrations after HOTAIRM1 siRNA or NC siRNA transfection were also detected. **P<0.01; ***P<0.005.
Abbreviations: RT-PCR, real-time polymerase chain reaction; NC, negative control.
Previous studies have shown that miR-3960 affected osteoblast differentiation by targeting HOXA215 and, coincidently, HOTAIRM1 located at the region between HOXA1 and HOXA2. This gave us a hint that there may be a potential correlation between HOTAIRM1 and miR-3960 in the regulation of gene expression. To test the relationship between HOTAIRM1 and miR-3960, HOTAIRM1 siRNA was transfected in peripheral blood cells. Interestingly, downregulation of HOTAIRM1 by siRNA led to an increased miR-3960 expression (Figure 3C and D), suggesting a negative correlation between these two noncoding RNAs. Under normal condition, HOTAIRM1 binds miR-3960 with its seed sequence. This binding made both of them enter the pathway of degradation. Therefore, miR-3960 could not be detected at a high concentration. However, when HOTAIRM1 was decayed by its siRNA in advance, a large amount of miR-3960 could escape from this lncRNA-mediated degradation and be detected at a higher level. Taken together, all these results displayed that there was a reciprocal inhibition mechanism between HOTAIRM1 and miR-3960.
HOTAIRM1 promoted the expression of HOXA1 through downregulation of miR-3960
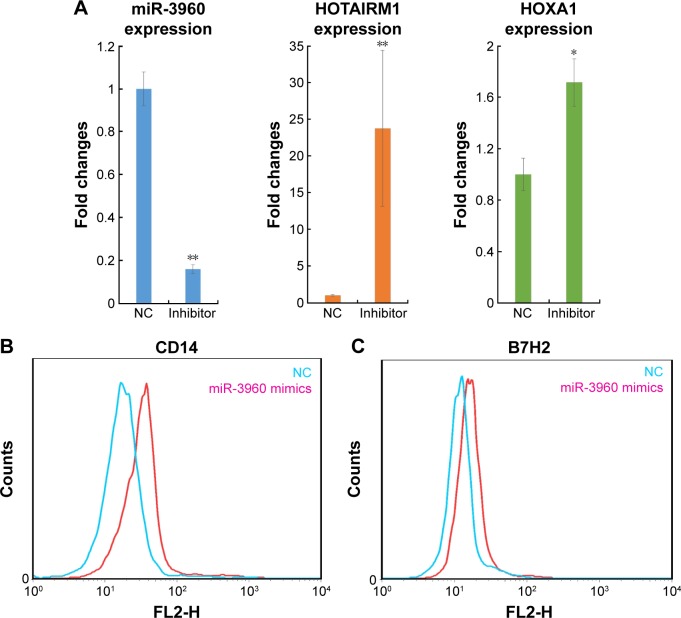
To determine whether the regulatory correlation between HOTAIRM1 and miR-3960 has functional consequences for the regulation of gene expression, the well-established target genes HOXA1 and HOXA2, were investigated. A previous study showed that HOXA1 was positively regulated by HOTAIRM1 and HOXA2 was a target of miR-3960.15 Although these two HOXA genes are different, in view of their homology, we hypothesized that one of these genes, HOXA1, could be regulated by those noncoding RNA interactions. Indeed, miR-3960 not only had an effect on HOTAIRM1 RNA level, but it could also influence HOXA1, similar to HOXA2, as demonstrated by the experiment on 293T cells. Depletion of miR-3960 by transfection of miR-3960 inhibitor resulted in a significant increase in both HOTAIRM1 and HOXA1 mRNA expressions (Figure 4A), which further influenced the process of monocyte to DC differentiation. As shown in Figure 4B and C, decreased miR-3960 could keep cells in a monocytic phenotype with high expression levels of CD14 and B7H2. Previous studies had described that HOXA genes accounted for the same function.15 In contrast, when peripheral blood monocytes were transfected with miR-3960 to consequently downregu-late HOTAIRM1 and HOXA1 levels, these cells exhibited a phenotype with low CD14 and B7H2 expression levels, indicating that the cells were liberated from the inhibitory effects of the two RNAs on DC differentiation. Based on these findings, we conclude the existence of the ceRNA network of HOTAIRM1–miR-3960–HOXA1, which acted as a regulatory module during myeloid development.
Figure 4.
HOTAIRM1 and miR-3960 play important functional roles in the regulation of target gene expression and cell differentiation.
Notes: (A) The RNA level of miR-3960, HOTAIRM1, and HOXA1 in 293T cells transfected with miR-3960 inhibitor or NC siRNA. Flow cytometry to detect the level of CD14 (B) and B7H2 (C) in peripheral blood cells transfected with miR-3960 mimics or NC siRNA. Each experiment was independently performed three times. *P<0.05; **P<0.01.
Abbreviation: NC, negative control.
Discussion
HOTAIRM1, a long intergenic noncoding RNA located between human HOXA1 and HOXA2 genes, is expressed specifically in the myeloid lineage.18 Our results have shown that HOTAIRM1 could hamper miR-3960 from functioning to repress monocyte-related HOXA1 mRNA expression and finally result in amplified HOXA1 expression in monocytes, while overexpression of miR-3960 could in turn downregulate both of these two long RNAs and liberate cells to enter into the DC differentiation pathway.
Extensive evidence had implied that lncRNAs play important roles in multiple biological processes. Our study focused on the regulatory mechanism of HOTAIRM1 in myeloid cell development. In view of the fact that several lncRNAs are controlled tightly by chromatin-modifying enzymes regulating the transcriptional activation or silencing of some genes,27 we investigated the factors associated with HOTAIRM1 expression using our H3K4me3 and H3K27me3 ChIP-chip map analyses. We found that during myeloid differentiation, these two histone modifications displayed significant changes in HOTAIRM1 gene-related regions, indicating the interaction between this lncRNA and DC development.
Furthermore, we showed that HOTAIRM1 silencing led to the downregulation of the two markers CD14 and B7H2 in peripheral blood cells. CD14 serves as a monocyte marker, and B7H2 is expressed in activated monocytes. Hence, the decline in their level is an evidence of the number of monocytes decreased. In contrast, most cells have transdifferentiated into DCs, which indicates that HOTAIRM1 is a negative regulator of monocyte–DC differentiation.28,29
To explore the mechanism of how HOTAIRM1 functions to repress this process, we have adopted the ceRNA theory here, which emphasizes that lncRNAs act as ceRNAs to impair miRNA activity by sequestering paired miRNAs and facilitate corresponding mRNAs to function without miRNA interference.30,31 In the light of this thought, we have found that miR-3960 and its target mRNA HOXA1 are able to build a ceRNA network with lncRNA HOTAIRM1, which functionally influences DC differentiation. The negative correlation between the expressions of HOTAIRM1/HOXA1 and miR-3960 and the subsequent downregulation of the monocyte markers CD14 and B7H2 demonstrate the potential regulatory mechanism of this ceRNA network.
Conclusion
We have found HOTAIRM1 as an lncRNA to regulate myeloid development, which is strictly regulated by epigenetic factors such as histone modifiers H3K4me3 and H3K27me3. Furthermore, HOTAIRM1 can form a ceRNA network together with miR-3960 and HOXA1, which acts as a negative regulator of DC differentiation. Pairing miR-3960 by HOTAIRM1 can finally result in the maintenance of monocyte phenotype without converting to DCs. In conclusion, our findings provide a new target in the immune therapy against tumor and reveal a novel strategy for improving the antitumor responses. Thus, these results also offer valuable reference for the research on targeted drug.
Acknowledgments
This study was supported by NSFC grants 31470876, 91029736, and 91442111, and the Joint NSFC-ISF Research Program, jointly funded by the National Natural Science Foundation of China and the Israel Science Foundation (ISF-NSFC program 31461143010); a Ministry of Science and Technology grant (863 program, 2008AA02Z129); and the National Key Scientific Program (2011CB964902); the Program for Changjiang Scholars and Innovative Research Team in University (no IRT13023) and State Key Laboratory of Medicinal Chemical Biology. All authors concur with the submission.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Palucka KA, Taquet N, Sanchezchapuis F, et al. Dendritic cells as the terminal stage of monocyte differentiation. J Immunol. 1998;160(9):4587–4595. [PubMed] [Google Scholar]
- 2.Banchereau J, Steinman RM. Dendritic cells and the control immunity. Nature. 1998;392(6673):245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 3.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2(2):116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 4.Morelli AE, Larregina AT, Shufesky WJ, et al. Internalization of circulating apoptotic cells by splenic marginal zone dendritic cells: dependence on complement receptors and effect on cytokine production. Blood. 2003;101(2):611–620. doi: 10.1182/blood-2002-06-1769. [DOI] [PubMed] [Google Scholar]
- 5.Lyakh LA, Sanford M, Chekol S, Young HA, Roberts AB. TGF-beta and vitamin D3 utilize distinct pathways to suppress IL-12 production and modulate rapid differentiation of human monocytes into CD83+ dendritic cells. J Immunol. 2005;174(4):2061–2070. doi: 10.4049/jimmunol.174.4.2061. [DOI] [PubMed] [Google Scholar]
- 6.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibcus JH, Dekker J. The context of gene expression regulation. F1000 Biol Rep. 2011;4(1):8. doi: 10.3410/B4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lempradl A, Ringrose L. How does noncoding transcription regulate Hox genes? Bioessays. 2008;30(2):110–121. doi: 10.1002/bies.20704. [DOI] [PubMed] [Google Scholar]
- 9.Bramhill D, Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 10.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 12.Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–1515. doi: 10.1093/carcin/bgu055. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JX, Han L, Bao ZS, et al. Chinese Glioma Cooperative Group HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15(2):1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemons D, Mcginnis W. Genomic evolution of Hox gene clusters. Science. 2006;313(5795):1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- 15.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrea T, Amemiya CT, Chang-Bae K, et al. Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol. 2005;304(1):75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 17.Lin M, Pedrosa E, Shah A, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6(9):e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Lian Z, Padden C, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 25.Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015;72(13):2491–2507. doi: 10.1007/s00018-015-1880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millard CJ, Varma N, Saleh A, et al. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Elife. 2016;5:e13941. doi: 10.7554/eLife.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang SY, Wei L, Di G, et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11(12):2172–2183. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambrecht BN, Hammad H. The role of dendritic and epithelial cells as master regulators of allergic airway inflammation. Lancet. 2010;376(9743):835–843. doi: 10.1016/S0140-6736(10)61226-3. [DOI] [PubMed] [Google Scholar]
- 29.Yoshinaga SK, Whoriskey JS, Khare SD, et al. T-cell co-stimulation through B7RP-1 and ICOS. Nature. 1999;402(6763):827–832. doi: 10.1038/45582. [DOI] [PubMed] [Google Scholar]
- 30.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 31.Bian EB, Ma CC, He XJ, et al. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget. 2016;7(21):30610–30625. doi: 10.18632/oncotarget.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]