Abstract
Proteolytic processing of the dengue virus polyprotein is mediated by host cell proteases and the virus-encoded NS2B-NS3 two-component protease. The NS3 protease represents an attractive target for the development of antiviral inhibitors. The three-dimensional structure of the NS3 protease domain has been determined, but the structural determinants necessary for activation of the enzyme by the NS2B cofactor have been characterized only to a limited extent. To test a possible functional role of the recently proposed Φx3Φ motif in NS3 protease activation, we targeted six residues within the NS2B cofactor by site-specific mutagenesis. Residues Trp62, Ser71, Leu75, Ile77, Thr78, and Ile79 in NS2B were replaced with alanine, and in addition, an L75A/I79A double mutant was generated. The effects of these mutations on the activity of the NS2B(H)-NS3pro protease were analyzed in vitro by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of autoproteolytic cleavage at the NS2B/NS3 site and by assay of the enzyme with the fluorogenic peptide substrate GRR-AMC. Compared to the wild type, the L75A, I77A, and I79A mutants demonstrated inefficient autoproteolysis, whereas in the W62A and the L75A/I79A mutants self-cleavage appeared to be almost completely abolished. With exception of the S71A mutant, which had a kcat/Km value for the GRR-AMC peptide similar to that of the wild type, all other mutants exhibited drastically reduced kcat values. These results indicate a pivotal function of conserved residues Trp62, Leu75, and Ile79 in the NS2B cofactor in the structural activation of the dengue virus NS3 serine protease.
Infection by dengue viruses is now widely recognized as a major public health concern, with more than 1 million cases of dengue hemorrhagic fever per year and case fatality rates ranging from 1 to 10% (23). There are four serotypes of dengue virus, which cause dengue hemorrhagic fever and dengue shock syndrome (21, 22). Dengue viruses, members of the Flaviviridae family, are small, enveloped, positive-stranded RNA viruses which are transmitted by Aedes mosquitoes (7). At present, neither a commercial vaccine nor a causative treatment is available for the prevention or cure of acute dengue virus diseases.
The genomic RNA of dengue virus serotype 2 contains 10,723 nucleotides and encodes a large polyprotein precursor of 3,391 amino acid residues which consists of three structural proteins (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (26). The polyprotein is co- and posttranslationally processed by proteases of the host cell and the virus-encoded two-component protease NS2B-NS3 to generate individual viral proteins (11, 18). Optimal activity of the NS3 serine protease (flavivirin, EC 3.4.21.91) is an essential requirement for maturation of the virus, and inhibition of this enzyme offers the prospect of an effective antiviral chemotherapy for severe cases of dengue hemorrhagic fever and dengue shock syndrome (for review see references 6, 39, and 41 and references herein).
The NS2B-NS3 two-component protease mediates cleavage in the nonstructural region of the viral polyprotein at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 junctions. Additional cleavages within the C, NS2A, and NS4A proteins and within a C-terminal portion of NS3 itself were described in the literature (1, 34, 35, 46). With the exception of the NS2B/NS3 junction, which contains a glutamine residue at the P2 position, the cleavage sites for the NS3 protease consist of a pair of dibasic amino acids (Arg or Lys) at the P1 and P2 positions, followed by a short-chain amino acid (Gly, Ala, or Ser) at the P1′ site (12, 13, 40, 48, 51). The minimum domain size required for protease activity of the 69-kDa NS3 protein has been mapped to 167 residues at the N terminus (33). Based on sequence comparisons with known serine proteases, a catalytic triad comprised of residues His51, Asp75, and Ser135 was identified, and replacement of the catalytic Ser135 residue by alanine resulted in an enzymatically inactive NS3 protease (47). The C-terminal two-thirds of the dengue virus NS3 protein are associated with the enzymatic functions of a nucleoside triphosphatase and RNA helicase (20, 27). The three-dimensional structure of the NS3 protease domain (NS3pro) encompassing the N-terminal 185 amino acids has been resolved by X-ray crystallography, and the protein exhibits the six-stranded β-barrel conformation typical of chymotrypsin-like serine proteases (31).
The presence of a small activating protein or cofactor is a prerequisite for optimal catalytic activity of the flaviviral proteases with natural polyprotein substrates (4, 13). Although the dengue virus NS3 protease exhibits NS2B-independent activity with model substrates for serine proteases such as N-α-benzoyl-l-arginine-p-nitroanilide, enzymatic cleavage of dibasic peptides is markedly enhanced with the NS2B-NS3 cocomplex, and the presence of the NS2B cofactor was shown to be an absolute requirement for trans cleavage of a cloned polyprotein substrate (50). Intramolecular cleavage at the NS2B/NS3 site conducive to the formation of a noncovalent complex was observed with the NS2B(H)-NS3pro molecule after purification from overexpressing Escherichia coli and subsequent refolding (50).
A genetically engineered NS2B(H)-NS3pro protease containing a noncleavable nonamer glycine linker between the NS2B activation sequence and the protease moiety exhibited higher specific activity with para-nitroanilide peptide substrates than the NS2B(H)-NS3pro molecule (32). Recently we have shown that the NS2B-NS3pro protease incorporating a full-length NS2B cofactor sequence could catalyze the cleavage of 12-mer peptide substrates representing native polyprotein junctions (28, 29). However, this protein appeared to be completely resistant to proteolytic self-cleavage.
The initial characterization of the cofactor requirement for the dengue virus NS3 protease revealed that the minimal region required for protease activity was located in a 40-residue central hydrophilic segment of NS2B spanning residues Leu54 to Glu93 (19). Mutagenesis experiments with the yellow fever virus NS2B protein demonstrated that specific residues within this core sequence are critical for protease activation (14). Deletion of residues 51 to 55, 53 to 55, and 56 to 93 within the conserved central domain yielded no detectable processing of an NS2B-NS3pro polyprotein precursor, whereas a four-amino-acid deletion of the sequence 67ISGS70 generated a protease with significantly reduced cleavage efficiency. Directed mutagenesis within the yellow fever virus NS2B protein confirmed a structural role for the N-terminal region of the conserved cofactor segment (17). Mutations within a charged N-terminal cluster comprising residues 52ELKK55 impaired cis cleavage activity at the NS2B/NS3 site, and deletion analysis revealed that the conserved domain alone provided only basal cofactor activity, while the optimal function of the cofactor required both hydrophobic flanking regions of NS2B.
A significant reduction of NS3 cleavage activity was observed for the alanine substitutions at residues Val95 and Gln96 within the dengue virus NS3 protease sequence (37). It was proposed that these two residues are located at the C terminus of the NS2B binding cleft and that they are involved in precleavage association of NS2B with NS3 and proper processing at the NS2B/NS3 site.
An essential requirement for the activation of the protease is the presence of hydrophobic residues in the cofactor, which may act as an anchor in the enzyme-cofactor complex. Within the hepatitis C virus NS4A cofactor, two residues, Ile25 and Ile29, are critical for complete activation of the NS3 protease (9, 44, 45). For the NS4A cofactor from GB virus, a minimum region which supports NS3 protease activity was mapped to a sequence spanning residues Phe22 to Val36, and two central residues, Val27 and Trp31, were indispensable for maximal proteolytic activity (10).
A peptide comprising residues Ser69 to Glu81 of the dengue virus NS2B cofactor was recently proposed as a substitute for the cofactor (8), however, it failed to reconstitute catalytic activity of the NS3pro protease in vitro (32). Therefore, it seems likely that additional residues located further at the N terminus of the NS2B core sequence play a role in NS3 activation.
These findings were suggestive of a common structural motif involved in activation of the flaviviral proteases. The Φx3Φ motif is comprised of two bulky hydrophobic residues separated by three unspecified residues, and it was speculated that additional residues located outside this sequence motif would contribute to the stringent specificity of the protease for the corresponding polyprotein substrate (10).
Substantial biochemical data were accumulated for the hepatitis C virus protease which offer some structural and mechanistic explanations for the activation of this flaviviral protease by its cofactor. Interaction of the NS3 protease with the NS4A cofactor was shown to affect the folding of the NS3 protease, resulting in conformational rearrangements of the N-terminal 28 residues of the protease and a strand displacement conducive to the formation of a well-ordered array of three β-sheets in which the cofactor becomes an integral part of the protease fold (30, 49). The result of these conformational changes is a reorientation of the residues of the catalytic triad which is more favorable for proton shuttling during catalysis. A number of studies provided evidence that structural rearrangements leading to a fully activated protease are induced not only by binding of the cofactor but also by the substrate, as shown for the competitive inhibition of the NS3 protease by its cleavage products (2, 3, 24, 25, 38). Based on these findings, the hepatitis C virus enzyme has been described as an induced-fit protease (6, 39). However, analogies to the hepatitis C virus system should be treated with caution, since preliminary data obtained with the dengue virus and the GB virus enzymes are indicative of major structural differences in the activation process (10, 50).
We demonstrate in this report that alanine substitutions at residues Trp62, Leu75, and Ile79 in the dengue virus NS2B cofactor result in marked effects on autoprocessing at the NS2B/NS3 site and that activity of the mutant NS3 proteases with the synthetic peptide substrate is mainly affected by significantly reduced kcat values. To analyze the structure-activity relationships which we have observed experimentally, we generated a molecular model for the NS2B/NS3 cocomplex based on homology to the hepatitis C virus NS3/NS4A protease.
MATERIALS AND METHODS
Construction of pTH/NS2B(H)-NS3pro by SOE-PCR.
The recombinant plasmid encoding the dengue virus serotype 2 NS2B(H)-NS3pro protein was generated with the previously described plasmid pTH/NS2B-NS3 as a template for splicing by overlap extension (SOE)-PCR (15). The sequence for NS2B(H) was obtained with primers 5′-TGCTCACTGGAGGATCCGCCGATTTGGAACTGGAG-3′ (nucleotides 4259 to 4293 in dengue virus serotype 2) and 5′-CTTCACTTCCCACAGGTACCACAGTGTTTGTTCTTCCTC-3′ (nucleotides 4399 to 4416). For amplification of NS3pro, primers 5′-GAACAAACACTGTGGTACCTGTGGGAAGTGAAGAAAC-3′ (nucleotides 4492 to 4516) and 5′-CTTCTCTTTCAGGATCCCTAATCTTCGATCTCTGGGTTG-3′ (nucleotides 5043 to 5081) were used.
SOE-PCR was performed with a combination of both templates with the NS2B(H) forward and the NS3pro reverse primers incorporating an overlapping region of 33 nucleotides. The product of SOE-PCR comprises the sequence of NS2B from amino acid residues 48 to 95 followed by residues 121 to 130 and 180 N-terminal residues of the NS3 protease domain. The PCR product was cut with BamHI and cloned into the pTrcHisA expression vector (Invitrogen) to yield the polyhistidine-tagged fusion protein. The sequence of the resulting construct was verified by DNA sequencing with an ABI Prism model 377 sequencer with a dye terminator cycle sequencing reaction kit (Perkin Elmer).
Mutation constructs.
Alanine substitutions were introduced in the NS2B sequence at residues Trp62, Ser71, Leu75, Ile77, Thr78, and Ile79 with the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's directions. In addition, a double Leu75/Ile79 mutant was generated with the L75A mutant as the template for PCR with Ile79 mutagenic oligonucleotides. Additional base changes creating restriction sites suitable for screening of the resulting mutant constructs were introduced in the primer sequences.
The following pairs of forward and reverse primers were used for mutagenesis (bold letters indicate changed nucleotides, and italic letters represent restriction sites): W62A, 5′-CGATGTCAAAGCTGAAGACCAGGCAGAG-3′ and 5′-CTCTGCCTGGTCTTCAGCTTTGACATCG-3′; S71A, 5′-GAGATATCAGGAGCTAGTCCAATCC-3′ and 5′-GGATTGGACTAGCTCCTGATATCTC-3′; L75A, 5′-GCAGTCCGATCGCGTCAATAACAATATCAG-3′ and 5′-CTGATATTGTTATTGACGCGATCGGACTGC-3′ ; I77A, 5′-CCAATCCTGTCAGCTACAATCTCAGAAGATGG-3′ and 5′-CCATCTTCTGATATTGTAGCTGACAGGATTGG-3′ ; T78A, 5′-CCAATCCTGTCAATAGCAATCTCAGAAGATGG-3′ and 5′-CCATCTTCTGAGATTGCTATTGACAGGATTGG-3′ ; and I79A, 5′-TCAATAACAGCCTCAGAAGATGGTAGC-3′ and 5′-GCTACCATCTTCTGAGGCTGTTATTGAC-3′.
The catalytically inactive NS3 protease mutant with an S135A substitution was obtained as described earlier (28). Plasmid DNA from the mutants was analyzed by DNA sequencing to confirm that only the desired mutation was incorporated.
Expression and purification of protease constructs.
The pTrcHis plasmids containing the recombinant NS2B(H)-NS3pro sequences were transformed into Escherichia coli C41(DE3). Transformants were grown in Luria broth (LB) medium supplemented with ampicillin (100 μg/ml) at 37°C. At an A600 of 0.5, isopropyl-1-thio-β-d-galactopyranoside was added to 0.1 mM, and the culture was grown at 37°C for 8 h. Cells were harvested by centrifugation (5,000 × g, 10 min, 4°C), resuspended in 20 ml of lysis buffer A (100 mM Tris-HCl, pH 7.5, 300 mM NaCl), and lysed with a French pressure cell at 14.000 lb/in2. The lysate was clarified by centrifugation (10,000 × g, 30 min, 4°C), and the pellet fraction was washed two times with lysis buffer containing 1% Triton X-100.
Inclusion bodies were suspended in 15 ml of denaturing buffer B (100 mM Tris-HCl, pH 8.0, 300 mM NaCl, 8 M urea) followed by sonication (10 bursts at power setting 3 for 15 s) with a ultrasonic processor (Misonix). The suspension was centrifuged (10,000 × g, 30 min, 4°C), and the supernatant was loaded on a Hitrap chelating column (Pharmacia) equilibrated with denaturing buffer. The column was washed with 10 column volumes of denaturing buffer containing 20 mM imidazole and eluted at a flow rate of 0.5 ml min−1 with denaturing buffer containing 50 mM imidazole. Fractions of 1 ml were collected, and aliquots were analyzed for the presence of NS2B(H)-NS3pro by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 15% polyacrylamide gels.
Peak fractions were pooled and loaded on a Superdex 200 HR 10/30 gel filtration column (Pharmacia). The column was eluted with denaturing buffer at a flow rate of 0.3 ml min−1 and the fractions containing NS2B(H)-NS3pro, as analyzed by SDS-PAGE, were pooled and diluted with the same buffer to 0.5 mg ml−1. Refolding of the protein was initiated by stepwise dialysis of 1-ml samples with a dialysis tubing (cutoff, 8 kDa) at 4°C against three changes of 100 mM Tris-HCl, pH 8.0-300 mM NaCl (200 ml) and one change against 200 ml of 100 mM Tris-HCl, pH 9.0-50 mM NaCl (buffer C). The dialysate was centrifuged (10,000 × g, 10 min, 4°C), and the protein concentration was determined with a Bradford protein assay kit (Bio-Rad). Preparations of the NS2B(H)-NS3pro protein were stored at −20°C in 100 mM Tris-HCl, pH 9.0-50 mM NaCl-50% glycerol.
Determination of NS3 protease activity.
Autocleavage of NS2B(H)-NS3pro at the NS2B/NS3 site was monitored by Tricine-SDS-PAGE (43). Samples containing 0.5 μg μl−1 of purified NS2B(H)-NS3pro in buffer C were incubated at 37°C, and aliquots of 20 μl were removed at fixed intervals. The reaction was quenched by the addition of 7 μl of SDS-PAGE sample buffer (200 mM Tris-HCl, pH 7.5, 4% [wt/vol] SDS, 40% glycerol, 0.1% bromophenol blue, 100 mM dithiothreitol), and precursor processing was quantitated by densitometric analysis of band intensities obtained from SDS-PAGE with the ONE-D scan program (Scanalytics). Cleavage at the NS2B/NS3 site was confirmed by automated Edman amino acid sequencing of the protease fragment NS3pro and by Western blot analysis with anti-Xpress antibodies (Invitrogen) of the N-terminal cleavage fragment (His)6NS2B(H).
The fluorogenic substrate GRR-AMC (Peptide International) was used for the in vitro assay of NS3. The assay was performed in 96-well microtiter plates with a Labsystems Fluoroscan II (Labsystems) at an excitation wavelength of 355 nm and emission wavelength of 460 nm at 37°C. Assay reactions contained in a 100-μl final volume NS2B(H)-NS3pro and the mutant proteins at a concentration of 0.15 μM in 100 mM Tris-HCl, pH 9.0. The substrate concentration was varied between 10 and 250 μM, and signals were converted to concentrations by comparison with standard amounts of free AMC. Km and Vmax values were obtained from measurements of initial velocities prior to 10% substrate depletion, assuming that 100% of the protease was enzymatically active. To determine Km and Vmax values, Michaelis-Menten kinetics, v = Vmax[S]/[S] + Km, were transformed into double reciprocal Lineweaver-Burk plots by nonlinear regression analysis with the GraphPad Prism software. Three independent experiments were carried out for each set of data points, and data are reported as mean ± standard error.
Coordinates and molecular modeling.
The crystal structures of the dengue virus type 2 NS3 serine protease (protein database identifier 1BEF) (31) and the hepatitis C virus NS3/NS4A complex (protein database identifier 1JXP) (49) were obtained from the protein database (Brookhaven National Laboratory). To obtain a structure for the dengue virus NS2B core segment in the complex with the NS3 protease, the NS2B peptide (residues 56 to 93) was initially aligned with the NS4A peptide (residues 21 to 32) according to the published sequence comparison (10). The structure for the NS2B peptide was generated with the Modeller 6v2 software (42). The dengue virus serotype 2 protease domain was superimposed on the hepatitis C virus protease domain, and the structures of the NS3 protease and the NS2B peptide were combined. Several 1-ns molecular dynamics trajectories of NS3pro in complex with NS2B were generated. The simulation of the resulting structure was performed with the Gromacs simulation package (http://www.gromacs.org) with GROMOS96 force field in a single-point charge water box. The space between protein and box walls was set to a minimum distance of 7 Å, and the system was energy minimized with the steepest descent. The simulated structures were visualized by Deepview Swiss-PdbViewer v3.5b4. The final model was evaluated by a Ramachandran plot, and 98.2% of the nonglycine residues were in allowed conformations.
RESULTS
Generation of mutants.
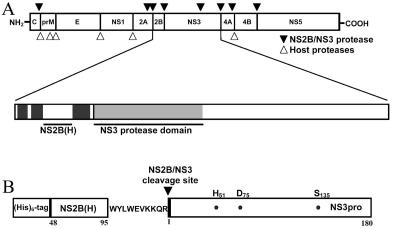
The sequence 75LSITI79 represents the Φx3Φ motif in the NS2B cofactor of dengue virus serotype 2 which was recently proposed to play a functional role in the association of the flaviviral proteases with their corresponding cofactors (10). To analyze the effects of amino acid substitutions in the NS2B cofactor on the enzymatic activity of the NS3 serine protease, we constructed the NS2B(H)-NS3pro polyprotein precursor by SOE-PCR (Fig. 1) (15). A sequence alignment of known flavivirus cofactor sequences and the location of the Φx3Φ motif are shown in Fig. 2.
FIG. 1.
Organization of the dengue virus polyprotein and the NS2B(H)-NS3(pro) construct. (A) Sites on the dengue virus polyprotein cleaved by host-encoded proteases (▿) and the virus-encoded two-component protease NS2B-NS3 (▾). The NS3 protease domain (NS3pro) is shaded, and the 40-residue minimum cofactor region which supports catalytic activity of the NS3 protease is indicated by a bar. (B) Structural organization of the expression construct NS2B(H)-NS3pro as generated by SOE-PCR. The construct contains the N-terminal polyhistidine tag for purification purposes, the central activation sequence of NS2B from residues 48 to 95 followed by residues 121 to 130, and 180 N-terminal residues of NS3 representing the protease domain. The residues of the catalytic triad, His51, Asp75, and Ser135, are indicated, and the sequence of the native NS2B/NS3 polyprotein cleavage junction is shown.
FIG. 2.
Sequence alignment of the conserved central domain within the NS2B cofactors of members of the Flaviviridae family. Numbers on the left indicate positions of amino acid residues in the polyprotein sequence. Residues which are identical in all sequences are shaded in dark grey, and conserved residues are shaded in light grey. The location of the Φx3Φ motif is indicated above the alignment, and residues within the dengue virus NS2B cofactor that were changed to alanine are labeled by dots. Abbreviations: DEN, dengue virus; JEV, Japanese encephalitis virus; KUNJIN, Kunjin virus; MVE, Murray valley encephalitis virus; TBE, tick-borne encephalitis virus; WNV, West Nile virus; YFV, yellow fever virus.
Previous work had shown that the NS2B(H)-NS3pro protein undergoes proteolytic self-cleavage at the NS2B/NS3 site that is conducive to the formation of a noncovalent complex (50). Alanine substitutions were introduced at residues Trp62, Ser71, Leu75, Ile77, Thr78, Ile79, and Leu75 plus Ile79 in the NS2B sequence. An enzymatically inactive NS2B(H)-NS3pro protein was obtained by replacing the catalytic Ser135 residue with alanine and used as negative control for the activity assays as described previously (28). All recombinant plasmids were subjected to DNA sequencing, and no mutations were found at nontargeted sites. Expression of the mutant derivatives as inclusion bodies in E. coli and purification under denaturing conditions by immobilized metal chelate chromatography and gel filtration yielded homogeneous products, as determined by SDS-PAGE analysis. The 29.8-kDa (His)6-NS2B(H)-NS3pro molecule displayed anomalous migration in gel electrophoresis with a higher apparent molecular mass of approximately 37 kDa. Subsequent refolding was performed by stepwise dialysis, and correct cleavage at the NS2B/NS3 junction was confirmed for the wild-type protein by N-terminal amino acid sequencing of the 20-kDa cleavage product, which yielded the sequence AGVLW, identical to the first five residues of the NS3 protein.
Effect of alanine substitutions on self-cleavage efficiency.
Protein samples purified by metal affinity chromatography and gel filtration were analyzed by SDS-PAGE for autocleavage at the NS2B/NS3 site after various periods of incubation (Fig. 3). After extensive dialysis, wild-type NS2B(H)-NS3pro exhibited complete autoproteolytic cleavage, which resulted in two protein products of 20 kDa and 10 kDa, whereas the S135A mutant was completely inactive in the self-cleavage assay. In Western blot analysis, only the 10-kDa protein reacted with anti-Xpress antibodies directed against the polyhistidine tag, which confirmed that this protein represents the N-terminal cleavage fragment (His)6-NS2B(H) (data not shown).
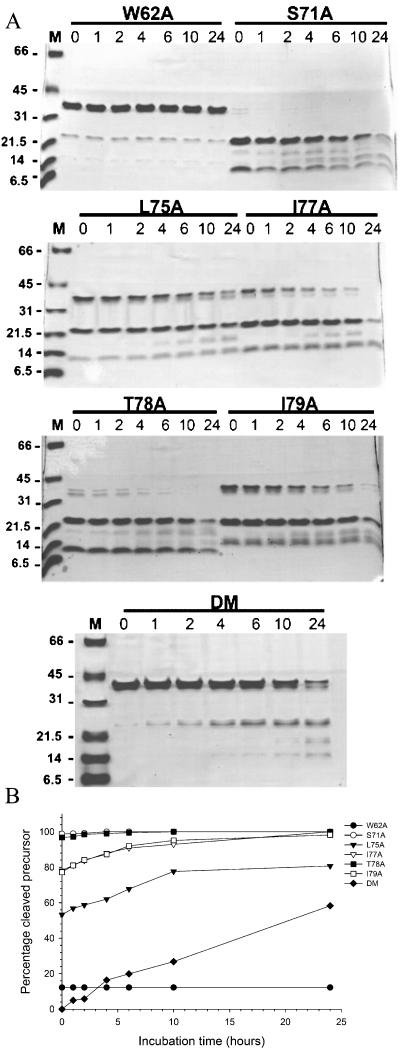
FIG. 3.
Kinetics of proteolytic autoprocessing of the NS2B(H)-NS3(pro) mutant derivatives. (A) Samples of wild-type NS2B(H)-NS3pro and the mutant proteins were refolded by successive dialysis, incubated at 37°C, and analyzed on Coomassie blue-stained Tricine-SDS-PAGE gels as described in the text. Lane M, protein molecular size markers. Numbers above the lanes indicate incubation times ranging from 0 to 24 h. (B) Band intensities of wild-type NS2B(H)-NS3pro and the mutant polypeptides as observed on Tricine-SDS-PAGE gels were quantitated by densitometric analysis with the ONE-D scan program, and the fraction of cleaved NS2B(H)-NS3(pro) precursor was plotted as a function of incubation time.
The NS2B mutants displayed different levels of proteolytic processing when analyzed by SDS-PAGE immediately after refolding. A densitometric analysis based on the amount of the NS2B(H)-NS3pro precursor remaining revealed that self-cleavage was almost completely abolished in the L75A/I79A double mutant. Autoprocessing was markedly reduced in the W62A mutant, which gave approximately 10% of wild-type activity. In contrast, the cleavage efficiencies of the S71A and the T78A mutants were not significantly affected by the substitutions and were comparable to that of the wild type. A sequence alignment of known flaviviral cofactor sequences shows that a serine at position 71 is preferred in most viruses of the Flaviviridae family, and substitution of this residue with alanine did not have a marked effect on proteolytic self-cleavage.
Thr78 is part of the Φx3Φ motif, but this residue is not very well conserved among the Flaviviridae and we expected that one alanine residue in the context of the serotype 2 sequence would have only a marginal effect on the activity of the NS3 protease. In accordance with this prediction, at this position the presence of the hydrophobic alanine residue is well tolerated. In contrast, alanine replacements at the hydrophobic residues Leu75, Ile77, and Ile79 caused a reduction in autocleavage activity of the NS2B(H)-NS3pro protein. Substitutions at Leu75 and Ile79 reduced autoprocessing to approximately 55 and 75% of the wild-type value, and the L75A mutation had a greater effect on cleavage efficiency than the I77A substitution, which still allowed for approximately 80% of wild-type precursor cleavage.
The data presented here support an important function for the Φx3Φ motif in activation of the NS3 protease. In agreement with a critical role for this hydrophobic sequence element, we found that self-cleavage was substantially decreased for the L75A/I79A double mutant, which gave only 2% of wild-type cleavage.
We also examined the effect of an alanine replacement at the W62 residue. This position is strictly conserved among the members of the Flaviviridae family (Fig. 2) and is located in the N-terminal region of the NS2B activation sequence. Alanine substitution at this position had a dramatic effect on protease activity, and self-cleavage was markedly reduced with this mutant protein; a finding which suggests a pivotal function for this invariant residue in protease activation.
Delayed processing kinetics of the mutants.
To answer the question of whether the alanine substitutions at critical positions within the NS2B cofactor resulted in a catalytically inefficient NS3 serine protease, we analyzed the levels of self-cleavage after various periods of incubation ranging from 1 to 24 h (Fig. 3).
The wild-type NS2B(H)-NS3pro molecule and the S71A mutant underwent complete autoproteolytic cleavage during the refolding process, whereas progressive proteolysis leading to complete cleavage of the precursor was observed for the I77A, T78A, and I79A mutants. Continued proteolysis of the S71A mutant protein resulted in additional cleavage fragments in SDS-PAGE at molecular masses of approximately 16 and 12 kDa. These proteins could represent fragments generated by cleavage at internal sequences within the NS3pro molecule. The NS3pro sequence contains paired basic residues at positions 63KRI65 and 142KKG144 and a monobasic site resembling the NS2B/NS3 junction at 27QRG29 which could serve as additional substrates for the protease, and cleavage at these sites would generate products of the observed sizes. Whether the additional cleavage products are formed by internal cleavage at these sites remains to be investigated. For the L75A mutant, approximately 75% cleaved precursor was observed at 24 h of incubation. The L75A/I79A double mutant showed weak cleavage activity and yielded only 50% precursor cleavage after 24 h. The W62A mutant did not display a significant increase in the amount of autoprocessing products at 24 h. Therefore, it is likely that the W62A substitution within the NS2B cofactor results in a significant inactivation of the NS2B-NS3 protease.
Reactivity with small substrate peptides.
The NS3 protease reacts with small model substrates for serine proteases in the absence of the NS2B cofactor, but cleavage efficiency of the protease towards synthetic tripeptide substrates is significantly stimulated in the presence of the 40-residue NS2B activation sequence (50). Cleavage at the NS2B/NS3 site is not a prerequisite for the reaction with small substrates, as shown by the activity of the NS2B-NS3pro precursor with 12-mer peptides (29). This protein did not display significant levels of autocleavage.
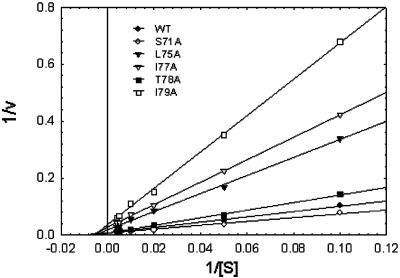
Comparison of activities with the synthetic peptide GRR-AMC between wild-type NS2B(H)-NS3pro and the mutant derivatives of NS2B revealed that the alanine substitutions affected the rate of substrate hydrolysis. Data for the kinetic parameters are presented in Table 1. The alanine substitutions mainly affected the kcat values of the recombinant proteases, whereas Michaelis-Menten equilibrium constants appeared to be less affected (Fig. 4). For the T78A mutant, only a 1.5-fold increase in Km was observed, whereas the largest changes in kcat occurred in the L75A and I79A mutants, which had kcat values that were 24- and 40-fold lower, respectively, than that of the wild type. In the case of the S71A mutant, the replacement resulted in a slightly more active enzyme, with a kcat/Km value that was 1.2-fold higher than that of the wild type. Catalytic efficiencies expressed as kcat/Km values were substantially reduced for the L75A, I77A, and the I79A mutants, which had 30-, 12-, and 52-fold-lower efficiencies, respectively, compared to the wild type. The activity of the L75A/I79A double mutant and the W62A mutant with GRR-AMC was negligible under the conditions of the assay, and a 10-fold increase in enzyme concentration did not result in detectable conversion of the substrate.
TABLE 1.
Kinetic parameters of the NS2B(H)-NS3pro mutant proteasesa
| Construct | Activity (% of wild-type activity) | Km (μM) | kcat (min−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Wild type | 100 | 146 ± 5.4 | 1.2 ± 0.07 | 137 ± 3.0 |
| S135A | ND | ND | ND | ND |
| W62A | ND | ND | ND | ND |
| S71A | 118 | 133 ± 7.0 | 1.3 ± 0.03 | 163 ± 10.0 |
| L75A | 3.4 | 180 ± 4.9 | 0.05 ± 0.007 | 4.7 ± 0.5 |
| I77A | 8.4 | 171 ± 15.3 | 0.12 ± 0.01 | 11.5 ± 0.13 |
| T78A | 65 | 225 ± 5.4 | 1.2 ± 0.09 | 89.0 ± 9.0 |
| I79A | 1.9 | 181 ± 8.0 | 0.03 ± 0.004 | 2.6 ± 0.2 |
| L75A/I79A | ND | ND | ND | ND |
Protease activity was assayed in 0.1 M Tris-HCl (pH 9.0) for 60 min at 37°C with the fluorogenic peptide GRR-AMC at concentrations ranging from 10 to 250 μM. Standard reactions contained protease at a concentration of 0.15 μM. No activity was observed with the W62A mutant and the L75A/I79A double mutant at a 1.5 μM enzyme concentration. The activity of the wild-type enzyme NS2B(H)-NS3pro with GRR-AMC was taken as 100%. ND, not detectable.
FIG. 4.
Steady-state cleavage kinetics of the GRR-AMC substrate by NS2B(H)-NS3pro derivatives in vitro. Reactions were performed in triplicate at a 0.15 μM protein concentration over a range of substrate concentrations from 10 to 250 μM. Assays were performed with 60-min incubation periods, and kinetic parameters were obtained by nonlinear regression analysis of initial velocities prior to 10% substrate depletion. The L75A/I79A double mutant and the W62A mutant had no measurable activity at a 10-fold-higher enzyme concentration (1.5 μM). Data were plotted in a double reciprocal format.
It is unlikely that the differences in catalytic efficiency which we observed between the wild-type and mutant NS2B(H)-NS3pro proteins result simply from a distortion of the NS2B/NS3 cleavage site, since the mutations seem to affect autocleavage and reactivity with small peptides as well. Since the gel electrophoresis assay does not allow detection of trans cleavage activity, we cannot exclude the possibility that some of the proteolysis products shown in Fig. 3 are generated by trans cleavage of the NS2B(H)-NS3pro precursor. Autoprocessing at the NS2B/NS3 site is not strictly required for trans cleavage activity, and for mutants displaying intermediate levels of autoproteolysis, the unprocessed precursor NS2B(H)-NS3pro may also contribute to enzymatic conversion of the synthetic substrate (29). However, mutations in the NS2B/NS3 cleavage site sequence which abolish autoprocessing result in catalytically poor NS3 proteases with inefficient trans cleavage of less than 10% of the wild-type activity (R. Khumthong, unpublished data).
In summary, the alanine substitutions in the cofactor had greater effects on the reaction rates of the NS3 protease than on substrate binding. This would imply a model for the activation of the NS3 protease in which the cofactor contributes mainly to an arrangement of the residues of the catalytic triad that is optimized for proton transfer during substrate cleavage.
DISCUSSION
Previous studies have shown that the activity of the dengue virus NS3 serine protease depends critically on the presence of the small NS2B cofactor protein (32, 50). In this report, we further investigated the structural determinants for the interaction of the NS3 protease with the NS2B cofactor by generating alanine substitutions at selected positions possibly involved in association of the protease-cofactor complex. Effects on the enzymatic activity of the NS3 protease were determined by analysis of autoprocessing at the NS2B/NS3 site and by reaction with the synthetic peptide GRR-AMC. The enzymatic data which we obtained for both types of reactions indicate an extreme sensitivity of NS3 cleavage activity to the correct conformation of the NS2B cofactor.
First, we examined the functional relevance of the Φx3Φ motif for cofactor-induced activation of the NS3 protease. Based on structural and mutational evidence obtained for the GB virus and hepatitis C virus NS3 proteases, a consensus sequence element involved in flaviviral protease activation was recently identified (10). By comparison between the structures of the hepatitis C virus NS3 protease and the NS3/NS4A cocomplex, it was hypothesized that the binding pocket for the first Φ residue (Leu75 in dengue virus type 2) undergoes a substantial conformational change, whereas the pocket for the second Φ residue (Ile79) remains largely unchanged upon complexation with the cofactor (30, 36). The second hydrophobic amino acid was proposed to occupy the hydrophobic pocket between the two β-barrel subdomains and to contribute to stabilization of the relative orientation of these subdomains. Our data show that the alanine substitution at Ile79 had a greater effect on NS3 autocleavage activity than the substitution at Leu75, whereas the catalytic efficiency was approximately 1.8-fold lower in the I79A mutant. A drastic effect on protease activation was observed with the L75A/I79A double mutant, in which autocleavage was almost completely eliminated and enzymatic activity with the GRR-AMC peptide was not detectable under the conditions of our assay.
In addition, we examined two noncritical residues, Ile77 and Thr78, which are located within the Φx3Φ motif. Although these two mutations had only marginal effects on autocleavage, the alanine substitution at Ile77 resulted in reduced enzyme activity, as demonstrated by a 12-fold-lower kcat/Km value compared to the wild-type enzyme. The T78A substitution yielded a less active enzyme with a 1.5-fold higher Km value for the GRR-AMC substrate than the wild type. These results support a role for the Φx3Φ motif in the activation mechanism of the dengue virus NS2B cofactor in which the unspecified residues also contribute to the interactions which are necessary for protease activation, a feature which discriminates the dengue virus protease from the recently analyzed GB virus NS3 protease (10). It appears that the association between the cofactor and the protease is mainly directed by hydrophobic interactions and that the mutations introduced in this region had greater effects on the catalytic efficiency of the protease than on the Km values, suggesting that perturbation of the hydrophobic interactions in the Φx3Φ motif may primarily affect the geometry of the catalytic triad.
Intermediate effects on enzymatic activity, as observed for the I77A, I79A, and T78A mutants, may reflect a role of these residues not only in the conformational activation of the catalytic apparatus of the enzyme but also for the stabilization of a ternary substrate-cofactor-protease complex. For the hepatitis C virus protease, a synergistic cooperation between the cofactor and the substrate was proposed to form an induced-fit protease with optimal catalytic activity and high specificity for the polyprotein substrate (5). Therefore, in the dengue virus NS2B/NS3 protease, additional residues located outside the Φx3Φ motif may contribute to the structural rearrangements induced by cofactor binding.
Substitution of Trp62 with alanine had the strongest effect on the activity of the NS3 protease. This residue is located outside the proposed short activation sequence which is homologous to the hepatitis C virus NS4A peptide (8). A deletion construct lacking the Trp62 residue was previously shown to be inactive in cleavage assays, and the sequence comprising residues 58 to 62 was implicated in the conformational stabilization of the protease (19). The finding that Trp62 is of paramount importance for the activation mechanism also provides an explanation for the inability of the peptide Gly69-Glu81 to replace the NS2B core sequence in vitro (32); however, it does not exclude the possibility that additional interactions at the N terminus of the NS2B(H) sequence are necessary for optimal activity, as was shown earlier for the yellow fever virus NS3 protease (17).
With the exception of the Trp62 substitution, the mutations which we introduced in the NS2B sequence of dengue virus type 2 do not simply abolish the enzymatic activity of the NS3 protease. Instead, we observed slow kinetics for autoprocessing at the NS2B/NS3 site upon prolonged incubation of the mutant enzymes. The kinetic analysis of GRR-AMC cleavage and the self-cleavage reaction indicates the existence of an inefficient catalytic machinery for both types of substrate conversion in the NS2B mutants.
In the absence of a crystallographic structure for the dengue virus NS2B-NS3 complex, we generated a model based on homology with the hepatitis C virus NS4A peptide (49). In analogy to the structure of the hepatitis C virus NS3/NS4A complex, the model predicts a threading of the cofactor in an extended conformation on a large and mainly hydrophobic surface groove of the NS3 protease formed by the N- and C-terminal domains (Fig. 5). Both hydrophobic residues Leu75 and Ile79 of the Φx3Φ motif occupy a hydrophobic pocket at the domain interface. The interactions observed in this model also predict a crucial role for the Gln96 residue in the NS3 sequence by formation of a hydrogen bond to Ile79 in the NS2B main chain, an observation which would be consistent with the weak protease activity of a recently described V95A/Q96A NS3 mutant (37).
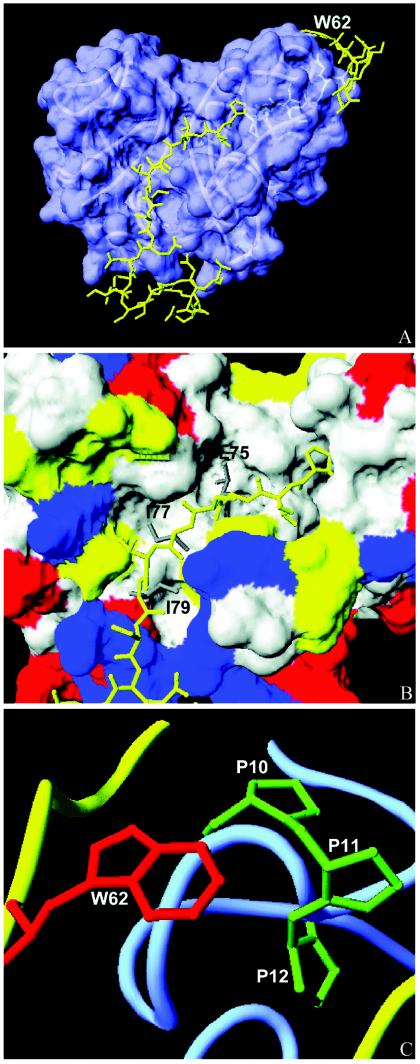
FIG. 5.
Molecular model of the interaction between the NS2B core segment and the NS3 protease. The model was generated by Deepview Swiss-Pdbviewer. (A) NS2B(H) (residues 56 to 93) mapped onto the X-ray crystal structure of NS3. The surface of NS3pro is shown in blue, and NS2B(H) is shown as a ribbon in yellow. The location of Trp62 at the N terminus of NS2B(H) is shown. (B) Interaction of residues Leu75, Ile77, and Ile79 with the hydrophobic pocket of NS3. The surface of NS3 is colored by the residue type (yellow, polar; blue, basic; red, acidic; white, nonpolar); the NS2B segment is shown as a yellow stick with the side chains of Leu75, Ile77, and Ile79 in grey. (C) Association of the Trp62 residue (red) of NS2B(H) (yellow ribbon) with the N-terminal proline cluster of NS3 (side chains in green and NS3 in blue).
In the model presented here, the critical residue Trp62 is located in close proximity to an N-terminal cluster of proline residues, Pro10, -11, and -12, in the NS3 sequence, and the NS2B peptide is attached to a surface-exposed structure of the NS3 protease. This interaction, which is reminiscent of the N-terminal clamping observed with the hepatitis C virus NS4A peptide, may govern the correct association of the cofactor with the NS3 surface (49). Alternatively, it is conceivable that the binding of the cofactor contributes to a structural organization of the N-terminal region of the NS3 protein, since the N-terminal residues of NS3 display a high degree of conformational flexibility (31). For the hepatitis C virus NS3 protease, folding of the N-terminal 28 residues into a β-strand and an α-helix was observed as a result of NS4A binding (49).
Further predictions based on this model, especially changes in the geometry of the active site associated with NS2B binding, would be too speculative at the moment. Elucidation of the precise mechanism of cofactor-dependent activation of the dengue virus NS3 protease has to await the resolution of the three-dimensional structure of the NS2B-NS3 cocomplex.
The cofactor-induced activation process is comparatively well characterized for the hepatitis C virus NS3 protease by a number of structural and spectroscopic studies. Nuclear magnetic resonance experiments revealed large nonlocal structural changes leading to a catalytic triad which is better ordered in the presence of NS4A (38). Solution structures obtained with a covalently bound ketoacid inhibitor disclosed a hitherto unrecognized role for the substrate in stabilization of the catalytic machinery by the formation of hydrogen bonds within the S′ subsite of the enzyme (2). According to this model, complexation of the protease with the NS4A cofactor leads to indirect activation of NS3 and induces a conformation which is preorganized for substrate binding (5). It remains to be investigated whether similar enzyme-substrate interactions and induced-fit mechanisms contribute to active-site stabilization of the dengue virus NS2B-NS3 protease.
Taken together, our results support a model for the activation of the dengue virus protease by the NS2B cofactor which depends critically on the presence of specific residues in the cofactor core sequence rather than on the overall conformation. The residues located within the structural Φx3Φ motif play an important role in this activation process, an observation which confirms earlier findings for related flaviviral enzymes. In addition, we have shown that a single residue, Trp62, located in the N-terminal region of the NS2B core segment is of high relevance for conformational activation. The structural reasons for this unusual requirement are not entirely clear at the moment, and further studies are required to investigate the complex structure-activity relationships of the dengue virus two-component protease. These investigations are not only useful for the understanding of the cofactor-induced activation of proteolytic enzymes, but may also facilitate the development of inhibitors which interfere with the formation of viral protease complexes.
Acknowledgments
We thank P. Wilairat for helpful discussions, A. Ketterman for critical review of the manuscript, C. Maharat, Prince of Songkhla University, Thailand, for N-terminal amino acid sequencing, and A. Nirachanon for excellent secretarial assistance.
This work was supported by a Senior Researcher Fellowship (to S.P.) from the Thailand Research Fund and Basic Science Grant BRG4680006 (to G.K.) from the Thailand Research Fund. Royal Golden Jubilee Ph.D. research scholarships from the Thailand Research Fund to P.N., P.W., and S.C. are gratefully acknowledged.
REFERENCES
- 1.Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. [DOI] [PubMed] [Google Scholar]
- 2.Barbato, G., D. O. Cicero, F. Cordier, F. Narjes, B. Gerlach, S. Sambucini, S. Grzesiek, V. G. Matassa, R. De Francesco, and R. Bazzo. 2000. Inhibitor binding induces active site stabilization of the HCV protein serine protease domain. EMBO J. 19:1105-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbato, G., D. O. Cicero, M. C. Nardi, C. Steinkuehler, R. Cortese, R. De Francesco, and R. Bazzo. 1999. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol. 289:371-384. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., V. Lohman, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi, E., S. Orru, F. Dal Piaz, R. Ingenito, A. Casbarra, G. Biasiol, U. Koch, P. Pucci, and A. Pessi. 1999. Conformational changes in human hepatitis C virus NS3 protease upon binding of product-based inhibitors. Biochemistry 38:13844-13852. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi, E., and A. Pessi. 2002. Inhibiting viral proteases: challenges and opportunities. Biopolymers 66:101-114. [DOI] [PubMed] [Google Scholar]
- 7.Billoir, F., R. De Chesse, H. Tolou, P. De Micco, E. A. Gould, and X. De Lamballerie. 2000. Phylogeny of the genus flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J. Gen. Virol. 81:781-790. [DOI] [PubMed] [Google Scholar]
- 8.Brinkworth, R. I., D. P. Fairlie, D. Leung, and P. R. Young. 1999. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. J. Gen. Virol. 80:1167-1177. [DOI] [PubMed] [Google Scholar]
- 9.Butkiewicz, N., M. Wendel, R. Zhang, R. Jubin, J. Pichardo, E. B. Smith, A. M. Hart, R. Ingram, J. Durkin, P. Mui, M. G. Murray, L. Ramanathan, and B. Dasmahapatra. 1996. Enhancement of hepatitis C virus NS3 proteinase activity by association with NS4A-specific synthetic peptides: identification of sequence and critical residues of NS4A for the cofactor activity. Virology 225:328-338. [DOI] [PubMed] [Google Scholar]
- 10.Butkiewicz, N., N. Yao, W. Zhong, J. Wright-Minogue, P. Ingravallo, R. Zhang, J. Durkin, D. N. Standring, B. M. Baroudy, D. V. Sangar, S. M. Lemon, J. Y. Lau, and Z. Hong. 2000. Virus-specific cofactor requirement and chimeric hepatitis c virus/GB virus B nonstructural protein 3. J. Virol. 74:4291-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 12.Chambers, T. J., R. C. Weir, A. Grakoui, D. W. McCourt, J. F. Bazan, R. J. Fletterick, and C. M. Rice. 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc. Natl. Acad. Sci. USA 87:8898-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers, T. J., A. Grakoui, and C. M. Rice. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65:6042-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers, T. J., A. Nestorowitz, S. M. Amberg, and C. M. Rice. 1993. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J. Virol. 67:6797-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champreda, V., R. Khumthong, B. Subsin, C. Angsuthanasombat, S. Panyim, and G. Katzenmeier. 2000. The two-component protease NS2B-NS3 of dengue virus type 2: cloning, expression and Escherichia coli and purification of the NS2B, NS3(pro) and NS2B-NS3 proteins. J. Biochem. Mol. Biol. 33:294-299. [Google Scholar]
- 16.Clum, S., K. E. Ebner, and R. Padmanabhan. 1997. Cotranslational membrane insertion of the serine protease precursor NS2B-NS3(pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 272:30715-30723. [DOI] [PubMed] [Google Scholar]
- 17.Droll, D. A., H. M. Murthy, and T. J. Chambers. 2000. Yellow fever virus NS2B-NS3 protease: charged-to-alanine mutagenesis and deletion analysis define regions important for protease complex formation and function. Virology 275:335-347. [DOI] [PubMed] [Google Scholar]
- 18.Falgout, B., M. Pethel, Y. M. Zhang, and C. J. Lai. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falgout, B., R. H. Miller, and C. J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbalenya, A. E., A. P. Donchenko, E. V. Koonin, and V. Blinov. 1989. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 17:3889-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gubler, D. J. 2002. Epidemic dengue/dengue haemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100-103. [DOI] [PubMed] [Google Scholar]
- 22.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 23.Halstead, S. B. 1997. Epidemiology of dengue and dengue hemorrhagic fever, p. 23-44. In D. J. Gubler and G. Kuno (ed.), Dengue and dengue hemorrhagic fever. CAB International, New York, N.Y.
- 24.Ingallinella, P., S. Altamura, E. Bianchi, M. Taliani, R. Ingenito, R. Cortese, R. De Francesco, C. Steinkuehler, and A. Pessi. 1998. Potent peptide inhibitors of the human hepatitis C virus NS3 protease are obtained by optimizing the cleavage products. Biochemistry 37:8906-8914. [DOI] [PubMed] [Google Scholar]
- 25.Ingallinella, P., D. Fattori, S. Altamura, C. Steinkuehler, U. Koch, D. Cicero, R. Bazzo, R. Cortese, E. Bianchi, and A. Pessi. 2002. Prime site binding inhibitors of a serine protease: NS3/4A of hepatitis C virus. Biochemistry 41:5483-5492. [DOI] [PubMed] [Google Scholar]
- 26.Irie, K., P. M. Mohan, Y. Sasaguri, R. Putnak, and R. Padmanabhan. 1989. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain). Gene 75:197-211. [DOI] [PubMed] [Google Scholar]
- 27.Kadare, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khumthong, R., C. Angsuthanasombat, S. Panyim, and G. Katzenmeier. 2002. In vitro determination of dengue virus type 2 NS2B-NS3 protease activity with fluorescent peptide substrates. J. Biochem. Mol. Biol. 35:206-212. [DOI] [PubMed] [Google Scholar]
- 29.Khumthong, R., P. Niyomrattanakit, S. Chanprapaph, C. Angsuthanasombat, S. Panyim, and G. Katzenmeier. 2003. Steady-state cleavage kinetics for dengue virus type 2 NS2B-NS3(pro) serine protease with synthetic peptides. Protein Peptide Lett. 10:19-26. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343-355. [DOI] [PubMed] [Google Scholar]
- 31.Krishna Murthy, H. M., S. Clum, and R. Padmanabhan. 1999. Dengue virus NS3 serine protease: crystal structure and insights into interaction of the active site with substrates by molecular modeling and structural analysis of mutational effects. J. Biol. Chem. 274:5573-5580. [DOI] [PubMed] [Google Scholar]
- 32.Leung, D., K. Schroeder, H. White, N. X. Fang, M. J. Stoermer, G. Abbenante, J. L. Martin, P. R. Young, and D. P. Fairlie. 2001. Activity of recombinant dengue virus NS3 protease in the presence of a truncated NS2B cofactor, small peptide substrates and inhibitors. J. Biol. Chem. 276:45762-45771. [DOI] [PubMed] [Google Scholar]
- 33.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, C., S. M. Amberg, T. J. Chambers, and C. M. Rice. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 90:6218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love, R. A., H. E. Parge, J. A. Wickersham, Z. Hostomsky, N. Habuka, E. W. Moomaw, T. Adachi, and Z. Hostomska. 1996. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331-342. [DOI] [PubMed] [Google Scholar]
- 37.Matusan, A. E., P. G. Kelley, M. J. Pryor, J. C. Whisstock, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J. Gen. Virol. 82:1647-1656. [DOI] [PubMed] [Google Scholar]
- 38.McCoy, M. A., M. M. Senior, J. J. Gesell, L. Ramanathan, and D. F. Wyss. 2001. Solution structure and dynamics of the single-chain hepatitis C virus NS3 protease NS4A cofactor complex. J. Mol. Biol. 305:1099-1110. [DOI] [PubMed] [Google Scholar]
- 39.Pessi, A. 2001. A personal account of the role of peptide research in drug discovery: the case of hepatitis C. J. Peptide Sci. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 40.Preugschat, F., C. W. Yao, and J. H. Strauss. 1990. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J. Virol. 64:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan, M. D., S. Monaghan, and M. Flint. 1998. Virus-encoded proteinases of the Flaviviridae. J. Gen. Virol. 79:947-959. [DOI] [PubMed] [Google Scholar]
- 42.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 43.Schaegger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu, Y., K. Yamaji, Y. Masuho, T. Yokota, H. Inoue, K. Sudo, S. Satoh, and K. Shimotohno. 1996. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J. Virol. 70:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinkuehler, C., A. Urbani, L. Tomei, G. Biasiol, M. Sardana, E. Bianchi, A. Pessi, and R. De Francesco. 1996. Activity of purified hepatitis C virus protease NS3 on peptide substrates. J. Virol. 70:6694-6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teo, K. F., and P. J. Wright. 1997. Internal proteolysis of the NS3 protein specified by dengue virus 2. J. Gen. Virol. 78:337-341. [DOI] [PubMed] [Google Scholar]
- 47.Valle, R. P. C., and B. Falgout. 1998. Mutagenesis of the NS3 protease of dengue virus type 2. J. Virol. 72:624-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wengler, G., G. Czaya, P. M. Farber, and J. H. Hegemann. 1991. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 72:851-858. [DOI] [PubMed] [Google Scholar]
- 49.Yan, Y., Y. Li, S. Munshi, V. Sardana, J. L. Cole, M. Sardana, C. Steinkuehler, L. Tomei, R. De Francesco, L. C. Kuo, and Z. Chen. 1998. Complex of NS3 protease and NS4a peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 7:837-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yusof, R., S. Clum, M. Wetzel, H. M. Murthy, and R. Padmanabhan. 2000. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids. J. Biol. Chem. 275:9963-9969. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., P. M. Mohan, and R. Padmanabhan. 1992. Processing and localization of dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J. Virol. 66:7549-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]