Abstract
At present, a large number of long noncoding RNAs (lncRNAs) from the human genome have been discovered. Meanwhile, emerging evidence has indicated that lncRNAs could play a critical role in the regulation of cellular processes such as cancer progression and metastasis. However, the functions of some new lncRNAs in the complex transcriptional process are mostly unknown at present. Existing studies suggest that loss of WW domain-containing oxidoreductase (WWOX) expression is linked with poor prognosis in numerous cancers, including epithelial ovarian cancer (EOC). However, the functional role of its antisense transcript RP11-190D6.2 is not clear to date. In this study, WWOX antisense transcript RP11-190D6.2 was analyzed specifically in EOC cells using real-time polymerase chain reaction and gain-/loss-of-function studies. We found that RP11-190D6.2 expression was positively correlated with WWOX expression. The RP11-190D6.2 expression was markedly downregulated in tumor tissues compared with normal tissues, but the RP11-190D6.2 expression was significantly downregu-lated in four EOC cell lines compared with human ovarian surface epithelial cell line. RP11-190D6.2 overexpression resulted in the increase of WWOX expression, whereas its knockdown led to the decrease of WWOX expression. We also found that RP11-190D6.2 was restored by 5-aza-2′-deoxycytidine treatment in EOC. In addition, the RP11-190D6.2 overexpression and knockdown experiments revealed that RP11-190D6.2 overexpression inhibited proliferation, migration, and invasion abilities in HO8910-PM cells, whereas RP11-190D6.2 knockdown in HEY-A8 cells had the opposite effect. The analyses in EOC implicate that RP11-190D6.2 may play a pivotal role in the regulation of tumor metastasis, suggesting that RP11-190D6.2 may serve as a potential biomarker and therapeutic target for EOC.
Keywords: epithelial ovarian cancer, lncRNA, WWOX, RP11-190D6.2, metastasis
Introduction
Epithelial ovarian cancer (EOC) is one of the most common genital system tumors that leads to cancer-related death among females.1 Recently, its high mortality rate worldwide among various types of gynecological malignancy has become a serious threat to female health due to the difficulties in early diagnosis and poor treatment.2 It has been proven that a large portion of the human genome can be transcribed into noncoding RNAs (ncRNAs).3 Long ncRNAs (lncRNAs) are one type of ncRNAs whose transcripts are longer than 200 nucleotides and have no protein-coding capacity.3 lncRNAs are dissimilar to miRNAs or else other smaller noncoding RNAs because they can regulate the downstream target gene at transcriptional and posttranscriptional levels by various means of cis and trans regulation.4,5 In recent years, many studies have demonstrated that lncRNAs play crucial roles in human diseases and various cancers.6–8 Particularly, research concerning the function of lncRNAs in EOC has also been reported.9 Moreover, what most cancer-related lncRNAs have in common is that they have an antisense partner of protein coding gene, such as ANRIL (CDKN2B-AS1),10,11 CTBP1-AS,12 HNF1A-AS1,13 and GAS6-AS1.14
From our early lncRNAs chip data (Jie Yao, Affymetrix Company, unpublished data, 2012), we noticed that lncRNA RP11-190D6.2 had differential expression between high and low metastatic cell lines of human tumor. Based on the data from USCS database website, we found that lncRNA RP11-190D6.2 is the antisense transcript of protein-coding gene WW domain-containing oxidoreductase (WWOX). WWOX is a type of tumor suppressor gene and is linked with the FRA16D common chromosomal fragile site.15 Furthermore, researchers found that it was associated with many cancers, including EOC,16 hepatocellular carcinoma,17 and colon cancer,18 bladder cancer,19 osteosarcoma,20 and other cancers. In EOC, WWOX plays significant roles in the regulation of the cell cycle and apoptosis.16 Mainly, loss of heterozygosity in WWOX and reduced expression of WWOX in many cancers have been reported.21,22 In addition, a recent study has identified that WWOX expression is largely regulated by DNA hypermethylation.23 Nevertheless, the evidence concerning the functional role of RP11-190D6.2 in EOC remains unclear, which prompted us to conduct this study.
The objective of this study was to identify the expression of RP11-190D6.2 in EOC and evaluate its functional role in EOC cell lines as well as the study the potential modulating mechanism. More importantly, its potential roles in the diagnosis and treatment of EOC were also evaluated.
Materials and methods
Patient specimens
All specimens with EOC (n=33) were collected from patients who underwent initial surgery from 2011 to 2014 at the Chinese PLA General Hospital. All patients enrolled in this study had not received therapy before surgical resection. This study and the utilization of tumor materials for research was approved by the ethical committee of PLA General Hospital. Written informed consent was obtained from the patients or their families for this study. For International Federation of Gynecology and Obstetrics (FIGO) stage I (n=5), FIGO stage II (n=8), FIGO stage III (n=12), FIGO stage IV (n=8), all samples were immediately frozen at the time of resection until use.
Cancer cell lines
The human EOC cell lines HO8910-PM, SKOV3, HEY-A8, and OVCAR3, and normal cell line human ovarian surface epithelial (HOSE) were used in the in vitro studies. The specimens were purchased from American Type Culture Collection (Manassas, VA, USA). Cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and incubated at 37°C in a humidified atmosphere with 5% (v/v) CO2.
RNA extraction and real-time quantitative polymerase chain reaction
Total RNA was extracted using the Trizol Total RNA Reagent (Invitrogen) following the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized with 2 μg total RNAs using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Takara, Otsu, Japan). All primers were obtained from GenePharma (Shanghai, People’s Republic of China), and the sequences we used were listed in Table 1. Real-time quantitative polymerase chain reaction (PCR) was analyzed using the SYBR PrimeScript RT-PCR kit (Takara) in an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, and 60°C for 34 s.
Table 1.
Primers for real time PCR analysis
| Gene name | Forward | Reverse |
|---|---|---|
| β-actin | 5′-CCACTGGCATCGTGATGGA-3′ | 5′-CGCTCGGTGAGGATCTTCAT-3′ |
| WWOX | 5′-CAAGGGCGAGTGAAGCAGT-3′ | 5′-GGCGGAGGGTGGTATTTTGT-3 |
| RP11-190D6.2 | 5′-CAGAGTGAGAACCACTGGTGAT-3′ | 5′-CGAAGGATGAGTTAAAAAGTTT-3′ |
Abbreviations: PCR, polymerase chain reaction; WWOX, WW domain-containing oxidoreductase.
Western blot analysis
Cells were rinsed with ice-cold phosphate-buffered saline, harvested, and lysed in the lysis buffer (40 nM Tris [pH 7.4], 150 mM NaCl, 10 mM ethylenediamine tetraacetic acid, 10% glycerol, 1% Triton X-100, 10 mM glycerophosphate, 1 mM Na3VO4, and 1 mM phenyl-methylsulfonyl fluoride) containing protease inhibitor (Roche, Mannheim, Germany). Proteins were separated on a NuPAGE 4%–12% Bis-Tris Gel (Invitrogen) and then transferred onto a polyvinylidene difluoride membrane (Invitrogen). The membrane was blocked for 60 min with 5% skim milk at room temperature, incubated with the primary antibody overnight at 4°C, and then incubated with corresponding secondary antibody. Finally, the polyvinylidene fluoride membrane was developed using enhancing chemiluminescence solutions (Thermo Fisher Scientific, Waltham, MA, USA).
Overexpression of RP11-190D6.2 in EOC cells
The RP11-190D6.2 was expressed at a low level in HO8910-PM cells and these were selected to be transfected with plasmid cDNA (pcDNA)-RP11-190D6.2 by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. RP11-190D6.2 cDNA was cloned into BamHI-EcoRI sites of pcDNA3.1. Those transfected cells were harvested for RNA isolation, scratch wound-healing assay, and matrigel invasion assay.
Transfection of siRNAs in EOC cells
The RP11-190D6.2 was expressed at a high level in HEY-A8 cells, and these were selected to carry out RP11-190D6.2 small interfering RNA (siRNA) transfection. siRNA sequences for target genes and negative-control (NC) siRNA were purchased from GenePharma. All sequences we used are listed in Table 2. In detail, ~5% cells were planted into 12-well plates and cultured for 24 h to obtain a concentration of 30%–50% before transfection. siRNA transfection was conducted with X-tremeGENE transfection reagent (Roche) according to the manufacturer’s recommendations. Scratch wound healing assay and matrigel invasion assay were performed 48 h posttransfection.
Table 2.
Sequences for small interfering (si) RNA analysis
| Gene name | Sense (5′–3′) | Antisense (5′–3′) |
|---|---|---|
| RP11-190D6.2-si1 | AUGGAUAUUUCCAAGCUGGAACCGA | UCGGUUCCAGCUUGGAAAUAUCCAUGU |
| RP11-190D6.2-si2 | AUUGAAAAUGUAAGAACUAUUCUGA | UCAGAAUAGUUCUUACAUUUUCAAUCA |
Treatment of HO8910-PM and HEY-A8 cells with 5-aza-2′-deoxycytidine (5-aza-dC)
To detect the RP11-190D6.2 expression, HO8910-PM and HEY-A8 cells were seeded into the plastic 6-well plates containing 5 μM 5-aza-dC (Sigma, St Louis, MO, USA) and then cultured for 3 days.
Cell proliferation assay
HO8910-PM or HEY-A8 cells were plated in 96-well plates at a density of 2,000 cells per well after transfection. The cells proliferation abilities were observed with MTS assay (Promega) following the manufacturer’s recommendation. Each well contained 100 μL culture medium, and to this was added 20 μL MTS reagent. Subsequently, the 96-well plate was incubated in a humidified atmosphere containing 5% CO2 at 37°C. After 2 h of incubation, the plate was measured at 490 nm using a plate reader.
Scratch wound-healing assay
HO8910-PM and HEY-A8 cells, grown on plastic 6-well plates, were scraped, to form uniform wounds, via a pipette tip before transfection, and the initial gap length (0 h) was measured from photomicrographs. After 48 h, the residual gap length was calculated.
Matrigel invasion assay
Transwell assay was performed to observe the cells’ invasion ability using modified Boyden Chambers (BD Biosciences, Bedford, MA, USA) that consist of transwell-precoated Matrigel membrane filter inserts with 8-mm pores in 24-well tissue culture plates. DMEM supplemented with 10% fetal bovine serum served as chemoattractant in the lower chamber. After incubation, invading cells on the lower chamber were calculated using photographic images, and the mean invasion rate was tested by measuring the increasing radius of the entire cell population.
Statistical analyses
Each mentioned experiment was performed at least three times. The differences between groups were performed using Student’s t-test or Chi-square test. The differences between patient groups were compared using the log-rank test. For correlation between genes expression, Pearson’s correlation was used. Statistical analysis was performed using SPSS version 18.0 (SPSS, Chicago, IL, USA), respectively. All analyses were considered statistically significant at P<0.05(*).
Results
Expression of RP11-190D6.2 was downregulated in EOC tissue samples
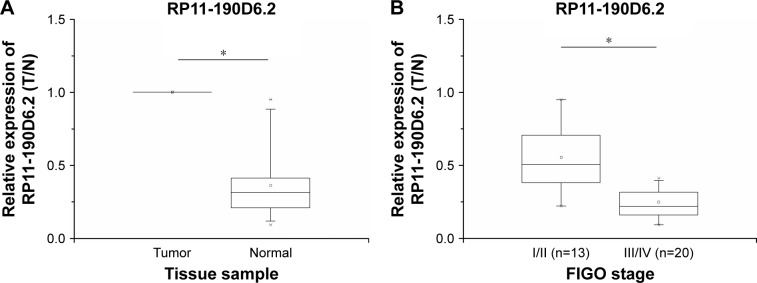
To examine the RP11-190D6.2 expression levels, panel-paired specimen assays were performed on the specimens that were obtained from the 33 patients suffering from EOC. Compared with the matched nontumor tissues, the expression level of RP11-190D6.2 in tumor tissues was markedly decreased (Figure 1A). Meanwhile, the expression of RP11-190D6.2 was remarkably downregulated in tumors with higher FIGO stage (III/IV) compared with tumors with lower FIGO stage (I/II) (Figure 1B).
Figure 1.
Relative expression in 33 EOC tissue samples were examined by real-time PCR and normalized to β-actin expression.
Notes: A fold change in tumor tissues relative to normal tissues is depicted. (A) Compared with the matched nontumor tissues, the expression level of RP11-190D6.2 in tumor tissues was evidently decreased (*P<0.05). (B) The expression of RP11-190D6.2 was remarkably downregulated in tumors with higher FIGO stage (III/IV) compared with tumors at lower FIGO stage (I/II) (*P<0.05).
Abbreviations: EOC, epithelial ovarian cancer; FIGO, International Federation of Gynecology and Obstetrics; PCR, polymerase chain reaction.
Expression of RP11-190D6.2 was correlated with expression of WWOX
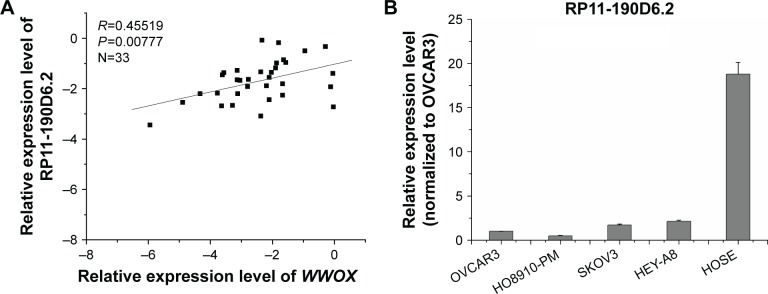
Real-time PCR was performed to evaluate the expression levels of RP11-190D6.2 and WWOX with an aim to investigate their potential association. As depicted in Figure 2A, the RP11-190D6.2 expression was positively correlated with WWOX expression (R=0.45519, P<0.00777).
Figure 2.
WWOX mRNA expression level was positively correlated with the RP11-190D6.2 lncRNA overexpression or knockdown in EOC cell lines.
Notes: Real-time PCR was performed to evaluate the expression levels of RP11-190D6.2 and WWOX, aiming to investigate their potential association. (A) The RP11-190D6.2 expression was positively correlated with WWOX expression (R=0.45519, P<0.00777). (B) The expression of RP11-190D6.2 was significantly lower in each EOC cell line compared with HOSE cell line. Additionally, the expression of RP11-190D6.2 was the highest in HEY-A8, but the lowest in H08910-PM. (C) The data showed that the lncRNA RP11-190D6.2 expression significantly increased. Meanwhile, using real-time PCR, we found that the mRNA expression level of WWOX was apparently upregulated by pcDNA RP11-190D62. (D) In RP11-190D6.2 siRNA groups, we found that RP11-190D6.2 expression was evidently decreased, as was WWOX expression. *P<0.05.
Abbreviations: EOC, epithelial ovarian cancer; HOSE, human ovarian surface epithelial; PCR, polymerase chain reaction; WWOX, WW domain-containing oxidoreductase; pcDNA control, pcDNA, plasmid complementary DNA; lncRNA, long noncoding RNA; mRNA, messenger RNA; siRNA, small interfering RNA; NC, negative control.
WWOX expression was coregulated together with the RP11-190D6.2 overexpression or knockdown in EOC cell lines
Real-time PCR was performed to observe the expression levels of RP11-190D6.2 in the four EOC cell lines and the HOSE normal cell line. The results showed that expression of RP11-190D6.2 was significantly lower in each EOC cell line compared with HOSE cell line (Figure 2B). Additionally, the expression of RP11-190D6.2 was the highest in HEY-A8 but the lowest in HO8910-PM among four human EOC cell lines (Figure 2B). Based on this result, the overexpression and knockdown experiments were conducted.
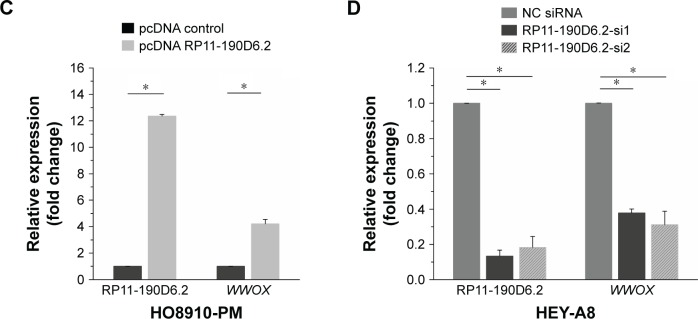
HO8910-PM cells transfected with RP11-190D6.2 cDNA plasmid were prepared for the overexpression experiment. The data showed that the lncRNA RP11-190D6.2 expression was significantly increased (Figure 2C). Meanwhile, using real-time PCR, we found that the mRNA expression level of WWOX was apparently upregulated by pcDNA RP11-190D6.2 (Figure 2C).
HEY-A8 cells transfected with RP11-190D6.2 siRNAs were prepared for the knockdown experiment. The mRNA expression levels of RP11-190D6.2 and WWOX were assessed using real-time PCR analysis. In RP11-190D6.2 siRNA groups, we found that RP11-190D6.2 expression was evidently decreased as well as WWOX expression (Figure 2D). Taken together, these results suggested that WWOX expression may be regulated by lncRNA RP11-190D6.2.
RP11-190D6.2 expression was restored by 5-aza-dC treatment in ovarian cancer cell lines
According to the UCSC database (http://genome.ucsc.edu/), we found that extensive hypermethylation could be found at the RP11-190D6.2 promoter region. To investigate the functional role of DNA methylation in lncRNA RP11-190D6.2 inactivation in tumor, we examined the influence of DNA demethylating agent (5-aza-dC) on RP11-190D6.2 expression. The results suggested that RP11-190D6.2 expression was significantly higher in HEY-A8 and HO8910-PM cells exposed to 5-aza-dC than that in control group (Figure 3).
Figure 3.
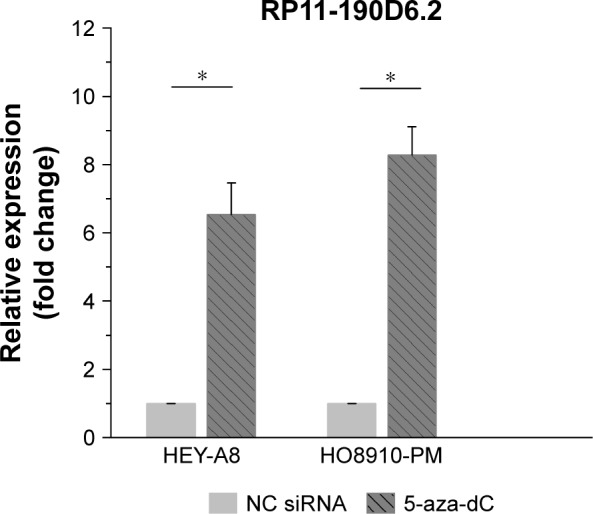
Influence of DNA demethylating agent (5-aza-dC) on RP11-190D6.2 expression.
Notes: The results suggested that RP11-190D6.2 expression was significantly higher in HEY-A8 and H08910-PM cells exposed to 5-aza-dC than that in NC group (*P<0.05).
Abbreviations: 5-aza-dC, 5-aza-2′-deoxycytidine; NC, negative control; siRNA, small interfering RNA.
HO8910-PM cells’ proliferation, migration, and invasion abilities were inhibited by RP11-190D6.2 overexpression
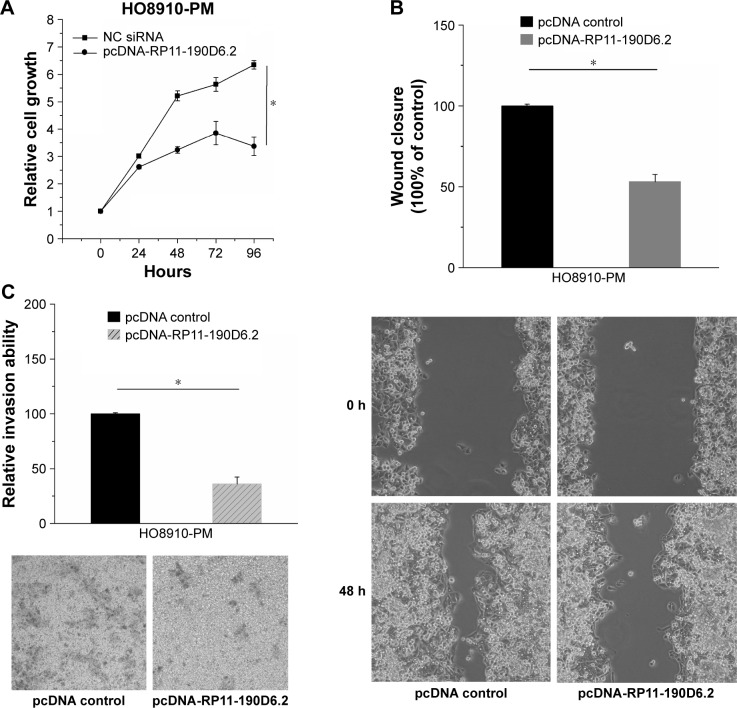
RP11-190D6.2 overexpression assay was performed in HO8910-PM cells transfected with pcDNA RP11-190D6.2 or pcDNA control, aiming to further investigate the functional role of RP11-190D6.2. As shown in Figure 4A, we found that the growth of cells was slower in pcDNA-RP11-190D6.2 group than that in pcDNA control group. The wound healing assay and matrigel invasion assay revealed that the migration and invasion abilities of cells were remarkably suppressed in pcDNA-RP11-190D6.2 group than that in pcDNA control group, which are respectively depicted in Figure 4B and C.
Figure 4.
Cell proliferative, migratory, and invasive properties were repressed by pcDNA-RP11-190D6.2 in H08910-PM cells.
Notes: (A) We found that the growth of cells was not evident in pcDNA-RP11-190D6.2 group compared with NC group. (B, C) The wound healing assay and matrigel invasion assay revealed that the migration and invasion abilities of cells were remarkably suppressed in pcDNA-RP11-190136.2 group than that in pcDNA control group (*P<0.05). Each image was taken at 40× magnification (objective: 4×, eyepiece: 10×).
Abbreviations: NC, negative control; pcDNA, plasmid complementary DNA; siRNA, small interfering RNA.
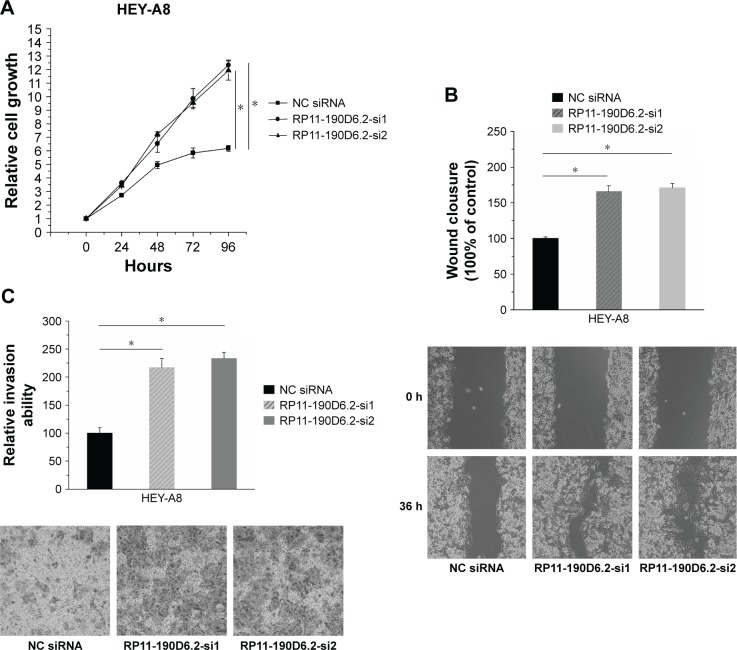
HEY-A8 cells proliferation, migration, and invasion abilities were enhanced by RP11-190D6.2 knockdown
RP11-190D6.2 knockdown assay was carried out in HEY-A8 cells transfected with RP11-190D6.2 siRNA or NC siRNA. The growth rate of cells transfected with RP11-190D6.2 siRNA showed great elevation compared with the NC siRNA group (Figure 5A). As seen from Figure 5B and C, the scratch wound-healing assay and matrigel invasion assay showed a significant elevation of cell migration and invasion in RP11-190D6.2 siRNA group compared with the NC siRNA group.
Figure 5.
Cell proliferation, migration, and invasion were elevated by RP11-190D6.2 siRNA in HEY-A8 cells.
Notes: (A) The growth rate of cells transfected with RP11-190D6.2 siRNA showed great elevation compared with the NC group. (B, C) The scratch wound-healing assay and matrigel invasion assay showed a significant elevation of cell migration and invasion in RP11-190D6.2 siRNA group compared with the NC siRNA group (*P<0.05). Each image was taken at 40× magnification (objective: 4×, eyepiece: 10×).
Abbreviations: NC, negative control; siRNA, small interfering RNA.
Discussion
Nowadays, a large number of lncRNAs have been found to be linked with diagnosis, staging, and progression in cancer.6–8 lncRNA RP11-190D6.2 is the antisense partner of the protein coding gene WWOX. WWOX is an important tumor suppressor in various human malignancies, and its expression is mainly controlled by promoter hypermethylation.16–20
In the present study, our data demonstrated that the deregulation of lncRNA RP11-190D6.2 was associated with the tumor occurrence in EOC, and the reduced expression of RP11-190D6.2 was linked with increased FIGO stages. In in vitro experiments, the expression of RP11-190D6.2 was remarkably lower in the four EOC cell lines compared with the HOSE cell line. Additionally, the results also showed that overexpression of RP11-190D6.2 suppressed proliferation, migration, and invasion abilities of the HO8910-PM cells, but the knockdown of RP11-190D6.2 showed the opposite effect. It is probable that the expression level and functional role of lncRNA RP11-190D6.2 in cancer was analyzed for the first time.
In a previous study, researchers found that lncRNA HOTAIR could promote the progression of ovarian cancer cells by regulating the cell cycle and apoptosis.24 CCAT2 is also associated with EOC metastasis, and its expression is upregulated in EOC.25 It is worth noting that our study provides the first evidence that lncRNA RP11-190D6.2 might be negatively linked with tumor progression and inhibit cell proliferation, migration, and invasion abilities in EOC.
It has been demonstrated that the antisense lncRNAs often regulate their sense mRNA partners through positive or negative expression relationship.26–28 Thus far, multiple antisense lncRNAs have been found to be associated with various malignant tumors.6–8 Our results showed that RP11-190D6.2 expression level was positively correlated with its sense partner WWOX. To further investigate the expression relationship between lncRNA RP11-190D6.2 and its protein-coding gene WWOX, overexpression and knockdown experiments were conducted, separately. Our data suggested that the WWOX expression concurred with the up- and downregulation of RP11-190D6.2 expression in EOC cells. According to these results, we speculated that lncRNA RP11-190D6.2 may function as a tumor suppressor by regulating the WWOX expression.
In some other studies, many cancer-related lncRNAs have been discovered to be involved in epigenetic regulating network.6,7 DNA hypermethylation in the regulatory region has been found to be the cause of downmodulation of WWOX expression in cancers.21–23 Based on the UCSC data, extensive DNA methylation and histone modification markers can be found at the promoter of RP11-190D6.2 and WWOX (http://genome.ucsc.edu/). In our study, we found that the RP11-190D6.2 expression was markedly restored when cells were treated with 5-aza-dC. This result made us speculate that the DNA demethylation may contribute to the increase of RP11-190D6.2 expression in EOC cells.
In conclusion, this study provides the first evidence that the deregulation of lncRNA RP11-190D6.2 expression is linked with the diagnosis of EOC. The functional mechanism of RP11-190D6.2 is closely associated with the inhibition of cell proliferation, migration, and invasion abilities in EOC partly by regulating protein-coding gene WWOX. Tumor suppressor lncRNA RP11-190D6.2 may be regarded as a novel biomarker and therapeutic target for EOC treatment in future.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen Y, Tan X, Ding Y, et al. WWOX CNV-67048 functions as a risk factor for epithelial ovarian cancer in Chinese women by negatively interacting with oral contraceptive use. Biomed Res Int. 2016;2016:6594039. doi: 10.1155/2016/6594039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim W, Song G. Discovery of prognostic factors for diagnosis and treatment of epithelial-derived ovarian cancer from laying hens. J Cancer Prev. 2013;18:209–220. doi: 10.15430/JCP.2013.18.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 5.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren C, Li X, Wang T, et al. Functions and mechanisms of long non-coding RNAs in ovarian cancer. Int J Gynecol Cancer. 2015;25(4):566–569. doi: 10.1097/IGC.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 10.Pasmant E, Laurendeau I, Heron D, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 11.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayama K, Horie-Inoue K, Katayama S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Song JH, Cheng YL, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han L, Kong R, Yin DD, et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30:694. doi: 10.1007/s12032-013-0694-5. [DOI] [PubMed] [Google Scholar]
- 15.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 16.Yan H, Tong J, Lin X, Han Q, Huang H. Effect of the WWOX gene on the regulation of the cell cycle and apoptosis in human ovarian cancer stem cells. Mol Med Rep. 2015;12(2):1783–1788. doi: 10.3892/mmr.2015.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie B, Zen Q, Wang X, et al. ACK1 promotes hepatocellular carcinoma progression via downregulating WWOX and activating AKT signaling. Int J Cancer Oncol. 2015;46(5):2057–2066. doi: 10.3892/ijo.2015.2910. [DOI] [PubMed] [Google Scholar]
- 18.Nowakowska M, Pospiech K, Lewandowska U, Piastowska-Ciesielska AW, Bednarek AK. Diverse effect of WWOX overexpression in HT29 and SW480 colon cancer cell lines. Tumour Biol. 2014;35(9):9291–9301. doi: 10.1007/s13277-014-2196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Sun L, Mu Z, Huang Y, Fu C, Hu B. Ectopic WWOX expression inhibits growth of 5637 bladder cancer cell in vitro and in vivo. Biochem Biophys Rep. 2015;73(2):417–425. doi: 10.1007/s12013-015-0654-0. [DOI] [PubMed] [Google Scholar]
- 20.Del Mare S, Aqeilan RI. Tumor suppressor WWOX inhibits osteosarcoma metastasis by modulating RUNX2 function. Sci Rep. 2015;5:12959. doi: 10.1038/srep12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Liu J, Ren Y, Yang J, Liu P. Common chromosomal fragile site gene WWOX in metabolic disorders and tumors. Int J Biol Sci. 2014;10:142–148. doi: 10.7150/ijbs.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: clinical significance and future implications. Exp Biol Med (Maywood) 2014;239:253–263. doi: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- 23.Baryla I, Styczen-Binkowska E, Bednarek AK. Alteration of WWOX in human cancer: a clinical view. Exp Biol Med (Maywood) 2015;240(3):305–314. doi: 10.1177/1535370214561953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333(2):238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Huang S, Qing C, Huang Z, Zhu Y. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11(1):49. doi: 10.1186/s13000-016-0499-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;1:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yelin R, Dahary D, Sorek R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]