Abstract
In latent infection, Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen 1 (LANA1)-specific binding to KSHV terminal repeat DNA mediates multicopy episome persistence. We now use electrophoretic mobility shift assays to investigate LANA1 binding to its 20-bp cognate sequence. Mutations at positions 6, 7, and 8 (6CCC8) severely reduced LANA1 binding, whereas mutations at other positions only modestly reduced binding. Since 6CCC8 is in the 5′ half of an inverted repeat sequence, these results are consistent with an asymmetric role for the inverted repeat in LANA1 binding.
Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8, is tightly associated with KS, primary effusion lymphoma, and multicentric Castleman's disease (8, 10, 28, 32). KSHV infection in tumor cells and in primary effusion lymphoma cell lines is predominantly latent. Latently infected cells have multiple copies of extrachromosomal, circular, KSHV episomes (plasmids) (9, 13). To persist in proliferating cells, episomes must replicate and efficiently segregate to progeny cells.
Latency-associated nuclear antigen 1 (LANA1) (21, 22, 29), encoded by one of a small number of viral genes expressed in latent infection, is necessary and sufficient for KSHV episome persistence (2, 3). Expression of LANA1 in uninfected B-lymphoma cells permits episome persistence of DNA containing specific KSHV terminal repeat (TR) sequence. Confocal microscopy demonstrates that LANA1 concentrates at sites of KSHV DNA along mitotic chromosomes (2, 11). LANA1 also mediates replication of TR-associated DNA in transient assays (16, 18, 25). Taken together, these data support a model in which LANA1 mediates KSHV TR DNA replication and tethers episomes to mitotic chromosomes to effect efficient partitioning to progeny nuclei. Similar roles in episome persistence have been proposed for the Epstein-Barr virus (EBV) EBNA1 and bovine papillomavirus (BPV) E2 proteins (5, 19, 24, 30, 31, 33, 34).
Consistent with this model, LANA1 binds specific TR sequence (3, 12, 15) with high affinity (14). Footprinting analyses indicate that LANA1 also binds to a low-affinity site which is adjacent to the high-affinity site (14). The crystal structures of the EBNA1 and E2 DNA-binding domains complexed with cognate sequence have been determined, revealing their structural homology (6, 17). Despite an absence of primary amino acid homology with EBNA1, the structure of the LANA1 C-terminal DNA-binding domain is predicted to be similar to that of EBNA1 (and thereby to that of E2) (16). EBNA1 and E2 each bind inverted repeat sequence as a dimer. There is an 8-bp inverted repeat within the LANA1 cognate sequence (Fig. 1A) (3, 12, 15), but its importance for LANA1 binding is unclear. The similarities between LANA1 and EBNA1 (and E2) suggest the possibility that these proteins recognize DNA through similar mechanisms.
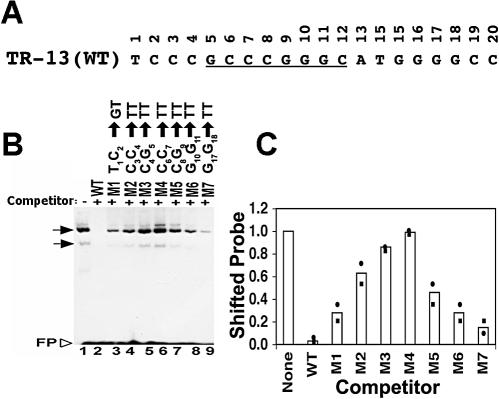
FIG. 1.
Mutation of adjacent base pairs in the LANA1 binding site compromises the ability of oligonucleotides to compete LANA1 in binding to the WT probe. (A) The WT TR-13 sequence is shown. The 8-bp inverted repeat is underlined. (B) In vitro-translated LANA1 was incubated with radiolabeled TR-13 probe, and EMSA was performed. A 50-fold excess of the indicated nonradiolabeled competitor was added to the incubation mixture for lanes 2 to 9. The substitutions in each mutant (M) are indicated above the lanes. Horizontal arrows indicate LANA1-specific complexes. The upper complex sometimes resolves into two bands (3). FP, free probe. The data shown are representative of more than five experiments. (C) Quantitation of EMSA results. Signal from two experiments was captured with a PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant software. The average values (bars) and individual values (circles and squares) are shown. Values were determined by dividing the signal in the shifted position after incubation with each competitor oligonucleotide by the signal in the absence of competitor.
Identification of the TR-13 sequence critical for LANA1 DNA binding.
Mutations were introduced into the 20-bp high-affinity LANA1 binding site, TR-13 (Fig. 1A), to identify nucleotides important for LANA1 binding. Previous electrophoretic mobility shift assays (EMSAs) had implicated the first seven TR-13 positions as having a role in LANA1 DNA binding (3). In these experiments, an excess of an unlabeled oligonucleotide which included TR-13 positions 1 through 7 weakly competed LANA1 in binding to the TR probe. In contrast, an excess of an unlabeled oligonucleotide which included positions 8 through 20 did not compete LANA1 in binding to the TR probe. Therefore, we initially focused our attention on the 5′ end of TR-13, including positions 1 through 7. The first 11 TR-13 nucleotides were mutated to T's in adjacent pairs except for at position 1, where the native T was mutated to a G. Positions 17 and 18 were also mutated to T's.
Each mutated oligonucleotide was assayed for the ability to compete LANA1 in binding to the TR-13 probe. In vitro-translated LANA1 was incubated with an end-radiolabeled TR-13 probe and a 50-fold excess of nonradiolabeled TR-13 or a mutant oligonucleotide prior to EMSA, which was performed as described previously (3, 23). In the absence of competitor, LANA1 formed two major complexes with the TR-13 probe (Fig. 1B, lane 1), similar to earlier results (3, 4, 23). An excess of unlabeled TR-13 efficiently competed the LANA1 gel-shifted complexes (Fig. 1B, lane 2, and C). In contrast, mutant 4 (6CC7 → TT) did not compete the LANA1 complexes (Fig. 1B, lane 6, and 1C). Therefore, the TR-13 6CC7 sequence is critical for LANA1 binding. The ability to compete LANA1 complexes was not abolished in any of the other mutated oligonucleotides. However, the ability of each oligonucleotide to compete the LANA1 complexes was reduced compared to that of wild-type (WT) TR-13 (Fig. 1B and C). Interestingly, the magnitude of the impairment of the ability to compete LANA1 complexes was related to the proximity of mutations to 6CC7. For instance, oligonucleotide mutant 3 (4CG5 → TT) only weakly competed LANA1 complexes (Fig. 1B, lane 5, and 1C), whereas mutant 2 (3CC4 → TT) (Fig. 1B, lane 4, and 1C) and mutant 1 (1TC2 → GT) (Fig. 1B, lane 3, and 1C) competed LANA1 complexes more efficiently. Therefore, these results indicate that the TR-13 6CC7 sequence is critical for LANA1 DNA binding and that other sequence is less important.
Transition mutations indicate that TR-13 positions 6, 7, and 8 (6CCC8) are each critical for LANA1 binding to DNA.
To more precisely investigate the role of individual TR-13 bases in LANA1 binding, scanning transition (purine to purine or pyrimidine to pyrimidine) substitutions were introduced into TR-13. EBNA1 binding to its cognate sequence is significantly less sensitive to transition mutations than to transversion (purine to pyrimidine or pyrimidine to purine) mutations (1). Due to similarities between EBV EBNA1 and LANA1 (16, 23), we therefore reasoned that transition mutations might be less disruptive to LANA1 binding.
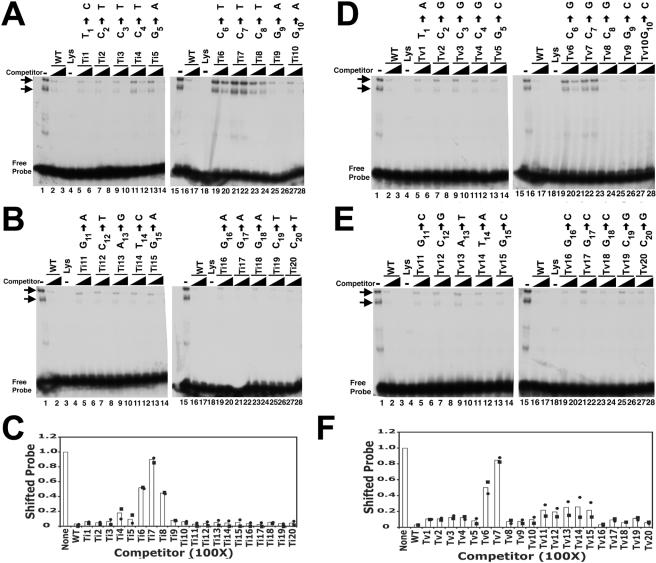
Each of the 20 TR-13 bases was individually mutated by transition substitution, and the mutated oligonucleotides were termed transition 1 (Ti1) through Ti20. Excess nonradiolabeled oligonucleotides with each transition mutation were then assayed for the ability to compete LANA1 in binding to the TR-13 probe (Fig. 2A to C). As expected, an excess of unlabeled WT TR-13 efficiently competed LANA1 in binding (Fig. 2A and B, lanes 2, 3, 16, and 17, and C). Excesses of nonradiolabeled Ti1 through Ti5 and Ti9 through Ti20 (Fig. 2A to C) also greatly diminished the shifted LANA1 complexes, although Ti4 and Ti5 competed slightly less efficiently.
FIG. 2.
Competition of LANA1 complexes with oligonucleotides containing point mutations. (A to C) EMSA results after competition with oligonucleotides containing point transition mutations. (A and B) An EMSA was performed after the TR-13 radiolabeled probe was incubated with in vitro-translated LANA1 (lanes 1 and 15) or reticulocyte lysate (lanes 4 and 18). Unlabeled TR-13 (WT) or the indicated transition mutant (Ti1 through Ti20) at a 25- or 100-fold excess was included in the incubation mixture prior to the EMSA as indicated. The transition substitution in each mutant is indicated above the lanes. Horizontal arrows identify LANA1 complexes. Data shown are representative of three experiments. (C) Quantitation of EMSA results. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). Data are from competition with oligonucleotides at a 100-fold excess. (D to F) EMSA results after competition with oligonucleotides containing point transversion mutations. (D and E) EMSA with the TR-13 radiolabeled probe after incubation with in vitro-translated LANA1 (lanes 1 and 15) or reticulocyte lysate (lanes 4 and 18). Unlabeled TR-13 (WT) or the indicated transversion mutant (Tv1 through Tv20) at a 25- or 100-fold excess was included in the incubation mixture prior to the EMSA. The transversion substitution for each mutant is indicated above the lanes. Arrows indicate LANA1 complexes. The data shown are representative of three experiments. (F) Quantitation of EMSA results. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). Data are from competition with oligonucleotides at a 100-fold excess. (C and F) Values were determined by dividing the shifted signal after incubation with competitor oligonucleotide by the shifted signal in the absence of competitor.
In contrast, oligonucleotides with transitions at positions 6 through 8 were each significantly impaired in their ability to compete the LANA1 complexes. Ti7 (7C → T) was the most severely impaired. Unlabeled Ti7 at a 25- or even 100-fold excess only minimally competed LANA1 complexes (Fig. 2A, lanes 21 and 22, and 1C). The ability of Ti6 (6C → T) and Ti8 (8C → T) to compete LANA1 complexes was also significantly compromised (Fig. 2A and C). Therefore, these data indicate that transitions at positions 6, 7, and 8 each greatly decrease the ability of oligonucleotide to compete LANA1 complexes with WT TR-13. A transition at position 7 had the most dramatic effect.
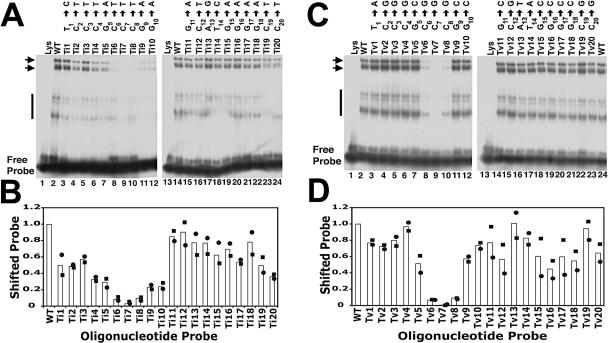
To directly assay the ability of LANA1 to complex with each transition mutant, EMSA was performed using radiolabeled transition mutants or WT TR-13 (Fig. 3A and B). As expected, LANA1 efficiently formed complexes with WT TR-13 (Fig. 3A, lanes 2 and 14). Less-intense, higher-mobility LANA1 complexes were also present (Fig. 3A, lanes 2 and 14). These complexes are often present with various intensities and are LANA1 specific since they are not present with reticulocyte lysate alone (Fig. 3A, lanes 1 and 13), can be competed with unlabeled competitor oligonucleotide, and are supershifted with anti-LANA1 antibody (data not shown). Ti1 through Ti3 (Fig. 3A, lanes 3 to 5, respectively) and Ti11 through Ti20 (Fig. 3A, lanes 15 to 24, respectively) bound LANA1 in complexes that were easily detectable. These oligonucleotides formed complexes at mildly reduced levels of ∼50 to 90% relative to the level of the WT, although Ti20 formed complexes at a level of ∼36% of that of the WT (Fig. 3B). Ti4 (Fig. 3A, lane 6), Ti5 (Fig. 3A, lane 7), Ti9 (Fig. 3A, lane 11), and Ti10 (Fig. 3A, lane 12) complexed with LANA1 at moderately reduced levels of ∼23 to 33% of the WT level (Fig. 3B). Therefore, Ti1 through Ti3 and Ti11 through Ti20 were mildly impaired in LANA1 binding, while oligonucleotides Ti4, Ti5, Ti9, and Ti10, whose mutations are immediately adjacent to 6CCC8, were moderately impaired.
FIG. 3.
EMSA with oligonucleotides containing point substitutions. (A) Point transition mutations. The TR-13 (WT) probe (0.5 ng) was incubated with reticulocyte lysate (lanes 1 and 13) or in vitro-translated LANA1 (lanes 2 and 14). Transition mutants (Ti1 through Ti20) were each radiolabeled and 0.5 ng was incubated with an equivalent amount of LANA1 prior to the EMSA. The transition substitution for each position is indicated above each lane. Horizontal arrows and the vertical line indicate specific LANA1 complexes. The data shown are representative of more than six experiments. (B) Quantitation of EMSA results with oligonucleotides containing point transition mutations. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). (C) EMSA with oligonucleotides containing point transversion substitutions. The TR-13 (WT) probe (0.5 ng) was incubated with reticulocyte lysate (lanes 1 and 13) or in vitro-translated LANA1 (lanes 2 and 24). Transversion mutants (Tv1 through Tv20) were each radiolabeled and 0.5 ng was incubated with an equivalent amount of LANA1 prior to EMSA. The transversion substitution at each position is indicated above each lane. Horizontal arrows and the vertical line indicate specific LANA1 complexes. The data shown are representative of more than six experiments. (D) Quantitation of EMSA results with oligonucleotides containing point transversion mutations. The signals from two experiments (shown as circles and squares) were captured and averaged (bars). (B and D) The fraction of shifted signal was determined by dividing the shifted signal by the sum of the signals from shifted and free probes. The fraction of the probe in the shifted position was then compared to that of TR-13 (WT), which was normalized to 1.
In contrast, and consistent with the competition experiments, Ti6 through Ti8 (Fig. 3A, lanes 8 to 10, respectively) were greatly compromised in their ability to bind LANA1. LANA1 complexes were barely detectable for these mutants. Ti6 and Ti8 bound LANA1 at ∼8.5 and 9.5% of WT levels, respectively (Fig. 3B). Consistent with the competition data, Ti7 was most affected and bound LANA1 at a 3% level (Fig. 3B). Therefore, a transition mutation at position 6, 7, or 8 exerted severe effects on LANA1 binding to DNA.
Transversion mutations indicate that TR-13 positions 6, 7, and 8 (6CCC8) are each critical for LANA1 binding to DNA.
Experiments with scanning transversion mutations were also performed. Each of the 20 TR-13 bases was individually replaced by transversion, and the oligonucleotides with replacements were termed transversion 1 (Tv1) through Tv20. Excess nonradiolabeled oligonucleotides with each transversion mutation were then assayed for the ability to compete LANA1 in binding to the TR-13 probe (Fig. 2D to F). An excess of unlabeled TR-13 (WT) again efficiently competed the LANA1 complexes (Fig. 2D and E, lanes 2, 3, 16, and 17). Excesses of nonradiolabeled Tv1 through Tv5 and Tv8 through Tv20 (Fig. 2D to F) also significantly competed the LANA1 complexes. The results with Tv1 through Tv5 and Tv9 through Tv20 generally paralleled those with the corresponding transition mutants (Fig. 2A to C).
However, results with Tv8 (8C → G) greatly differed from those with a transition at position 8. In contrast to Ti8 (8C → T), Tv8 efficiently competed the LANA1 complexes at both 25- and 100-fold excesses (Fig. 2D, lanes 23 and 24, and 2F). This result indicates that a transition at position 8 has a greater impact on the ability to compete LANA1 complexes than a transversion at position 8.
Both transversions at positions 6 and 7 significantly reduced the ability of oligonucleotides to compete LANA1 complexes, which was similar to the effects of transitions at these positions. Tv7 (7C → G) was particularly impaired (Fig. 2D, lanes 21 and 22, and 2F), while Tv6 (6C → G) was slightly less compromised than Tv7 (Fig. 2D, lanes 19 and 20, and 2F). Therefore, transversions at both positions 6 and 7 greatly impaired the competition of LANA1 complexes and the mutation at position 7 had the more severe effect.
To directly assay the effects of each transversion mutation on LANA1 DNA binding, an EMSA was performed after each mutated oligonucleotide or WT TR-13 was radiolabeled. Tv1 through Tv5 (Fig. 3C, lanes 3 to 7, respectively) and Tv9 through Tv20 (Fig. 3C, lanes 11, 12, and 14 to 23, respectively) each formed easily detectable LANA1 complexes. Binding of these mutants was generally mildly reduced, and all had at least ∼50% of WT TR-13 binding activity (Fig. 3D).
In contrast to the results with the competition data, but consistent with the results from the transition mutation experiments, Tv8 (Fig. 3C, lane 10) was also significantly impaired in its ability to complex LANA1 and formed complexes at a level of only ∼9% of the WT TR-13 level (Fig. 3D). Therefore, it appears that a transversion at position 8 produces an intermediate level of binding. A significant loss of binding is seen with direct labeling of a mutated probe, but a ≥25-fold excess of unlabeled Tv8 effectively competes LANA1 complexes.
Consistent with both the competition results and the data from the transition mutation experiments, Tv6 (Fig. 3C, lane 8) and Tv7 (Fig. 3C, lane 9) were greatly compromised in their ability to bind LANA1. LANA1 complexes were barely detectable for these mutants, and quantitation revealed an ∼6.5% binding efficiency for Tv6 and a less than 1% binding efficiency for Tv7 (Fig. 3D). Therefore, transversions at both position 6 and position 7 severely affect LANA1 binding.
This work identifies a core sequence that is critical for LANA1 binding to its high-affinity TR site. Each position of 6CCC8 within the 20-bp site TR-13 was critical for LANA1 binding. Changes at position 7 had particularly severe effects. Interestingly, mutations immediately proximal to 6CCC8 sometimes had a greater impact on LANA1 binding than more-distal changes. For instance, oligonucleotides with mutations at two adjacent bases were more impaired for competition of LANA1 complexes when the changes were proximal to 6CCC8 (Fig. 1). Further, radiolabeled oligonucleotides containing single-base transition substitutions immediately upstream (Ti4 and Ti5) or downstream (Ti8 and Ti9) of 6CCC8 bound LANA1 somewhat less efficiently than did transitions at other positions (Fig. 3A and B). One possibility is that these proximal mutations affect LANA1's interactions with 6CCC8. Overall, LANA1 binding did not appear to be more sensitive to transversion mutations than to transition mutations.
Although the core 6CCC8 sequence is critical for efficient LANA1 binding, the context of surrounding sequence is also important. For instance, the CCC sequence occurs at over 40 sites within the KSHV TR, yet LANA1 binds to only one of these sites with high affinity (3, 12, 14, 15). The point transition and transversion substitutions in sequences other than 6CCC8 also supported a role for adjacent sequence in LANA1 binding. For instance, single-nucleotide transition or transversion mutations at any non-6CCC8 TR-13 position, other than a transversion at position 13, reduced LANA1 binding to a mild or moderate degree (Fig. 3). Other results also demonstrate a role for neighboring sequence. For instance, LANA1 efficiently bound the TR-13 3CCGCCCGGGCATGGGGC19 sequence. However, removal of any additional bases from the 5′ or 3′ end significantly reduced the affinity of LANA1 binding (14). Further, LANA1 did not bind to TR-13 1TCCCGCCCGGGCA13 or 4CGCCCGGGCA13 sequences, although LANA1 did bind to the TR-13 4CGCCCGGGCATGG16 sequence (12). LANA1 also binds to a site in its own promoter that contains the TR-13 6CCCGGGC12 sequence but with lower affinity than it does to TR-13 itself (20). LANA1 was also reported to bind to a site in the TR that contains the CCCCCCC sequence but with a lower affinity than it does to the TR-13 1TCCCGCCCGGGCATGGG16 sequence (12). However, LANA1 did not bind to a 266-bp TR probe containing the CCCCCCC site but did bind to a 535-bp TR fragment containing TR-13 (3). These differences may have been due to different sensitivities of the assays used. LANA1 was also reported to bind to a 64-bp sequence containing multiple CC pairs with lower affinity than it does to the TR-13 1TCCCGCCCGGGCATGGG16 sequence (12).
These results are consistent with LANA1's low-affinity binding to the site adjacent to TR-13 in the TR. The low-affinity site contains a transversion at C8 to G within the critical core 6CCC8 sequence (14). This change significantly reduced LANA1 binding in our work. However, the low-affinity site also contains transversions at TR-13 positions C2, A13, G16, and C20 and a transition at position T1. Changes at all of these positions, except A13, had mild to moderate effects on LANA1 binding to the radiolabeled mutant probe. Therefore, it is not surprising that the sum of these changes greatly reduces LANA1's affinity to this site. Interestingly, the adjacent high-affinity TR-13 site allows cooperative binding of LANA1 to the low-affinity site (14).
These results indicate an asymmetric role for the 8-bp inverted repeat, 5GCCCGGGC12 (Fig. 1A) (12, 15), in the LANA binding site. Of note, both EBV EBNA1 and BPV E2 recognize inverted repeats. Whereas the 6CCC8 sequence in one half of the inverted repeat was critical for LANA1 binding, mutations in the other half of the inverted repeat (9GGGC12) did not exert severe effects on binding. These results are intriguing, since several lines of evidence suggest similarities between the EBNA1 and LANA1 DNA-binding domains. First, there is predicted structural similarity between the LANA1 and EBNA1 DNA-binding domains (16). The structure has been determined for the EBNA1 DNA-binding domain complexed with cognate, palindromic DNA (6). EBNA1 binds DNA as a dimer, and each monomer recognizes an equal half-site of the palindrome. Second, consistent with the predicted structural similarity, LANA1 self-associates to bind DNA, and LANA mutants which cannot self-associate do not bind DNA, similar to EBNA1. Third, the deletion of LANA1 residues 1007 through 1021 abolishes LANA1 binding to DNA but not LANA1 self-association. These residues are predicted to correspond to an EBNA1 sequence which directly contacts DNA but is not involved in dimerization (23). The asymmetric role of the inverted repeats raises the possibility that LANA1 may not recognize two equal half-sites and instead may bind DNA in a manner distinctly different from that of EBNA1. Of note, the stoichiometry of LANA1 bound to DNA is not known, and LANA1 might bind DNA as a dimer or higher-order oligomer. For instance, it is possible that LANA1 binds DNA as a higher-order structure, such as a hexamer, like the simian virus 40 large T antigen (27). Interestingly, the simian virus 40 large-T-antigen DNA-binding domain shares structural homology with BPV E2 and EBNA1 (7, 26). Alternatively, it is possible that a LANA1 dimer does recognize two equal half-sites in an inverted repeat sequence and that it compensates for certain mutations, perhaps by repositioning critical amino acid side chains. Further work should elucidate the mechanisms by which LANA1 binds DNA.
Acknowledgments
We thank Elliott Kieff for helpful discussions.
This work was supported by grants CA85751 (to M.E.B.) and CA82036 (to K.M.K.) from the National Cancer Institute.
REFERENCES
- 1.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency- associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 7.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 10.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 12.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 13.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 15.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegde, R. S., S. R. Grossman, L. A. Laimins, and P. B. Sigler. 1992. Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359:505-512. [DOI] [PubMed] [Google Scholar]
- 18.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong, J. H., J. Orvis, J. W. Kim, C. P. McMurtry, R. Renne, and D. P. Dittmer. 2004. Regulation and auto-regulation of the promoter for the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 279:16822-16831. [DOI] [PubMed] [Google Scholar]
- 21.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 23.Komatsu, T., M. E. Ballestas, A. J. Barbera, B. Kelley-Clarke, and K. M. Kaye. 2004. KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319:225-236. [DOI] [PubMed] [Google Scholar]
- 24.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 27.Mastrangelo, I. A., P. V. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 28.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 29.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 31.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M.-F. d'Agay, J.-P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 33.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]