Abstract
Adeno-associated virus (AAV) is a unique gene transfer vector which takes approximately 4 to 6 weeks to reach its expression plateau. The mechanism for this slow-rise expression profile was proposed to be inefficient second-strand DNA synthesis from the input single-stranded (ss) DNA viral genome. In order to clarify the status of ss AAV genomes, we generated AAV vectors labeled with bromodeoxyuridine (BrdU), a nucleotide analog that can be incorporated into the AAV genome and packaged into infectious virions. Since BrdU-DNA can be detected only by an anti-BrdU antibody when DNA is in an ss form, not in a double-stranded (ds) form, ss AAV genomes with BrdU can be readily tracked in situ. Although ss AAV DNA was abundant by Southern blot analysis, free ss AAV genomes were not detectable after AAV transduction by this new detection method. Further Southern blot analysis of viral DNA and virions revealed that ss AAV DNA was protected within virions. Extracted cellular fractions demonstrated that viral particles in host cells remained infectious. In addition, a significant amount of AAV genomes was degraded after AAV transduction. Therefore, we conclude that the amount of free ss DNA is not abundant during AAV transduction. AAV transduction is limited by the steps that affect AAV ss DNA release (i.e., uncoating) before second-strand DNA synthesis can occur. AAV ss DNA released from viral uncoating is either converted into ds DNA efficiently or degraded by cellular DNA repair mechanisms as damaged DNA. This study elucidates a mechanism that can be exploited to develop new strategies to improve AAV vector transduction efficiency.
Adeno-associated virus (AAV) is a defective parvovirus which requires helper viruses such as adenovirus or herpesvirus to complete its productive replication (23). Its life cycle consists of both lytic and latent infection. During its lytic cycle, AAV replicates through a double-stranded (ds) DNA molecule intermediate but packages only plus or minus strands of AAV genomes into preassembled capsids (22, 29). Historically, AAV was thought to have a duplex DNA genome due to the confusion caused by Southern blot analysis of its genome size. This issue of an ss versus a ds DNA genome was eventually settled by experiments in which the AAV genome was labeled with bromodeoxyuridine (BrdU), which allowed the separation of the plus- and minus-stranded viruses in a cesium chloride (CsCl) density gradient (4). During latent infection, single-stranded (ss) AAV genomes in the virion are integrated into the host genome as a duplex molecule. For wild-type AAV or recombinant AAV supplied by Rep proteins, these integration events have been shown to occur primarily in human chromosome 19 with a frequency of more than 70% (16, 17, 19, 26, 32).
Gene transfer vectors based on AAV have shown great promise in directing long-term gene expression without eliciting destructive T-cell-mediated immune responses against the transduced target cells (12, 30). Unlike other gene transfer vectors such as adenovirus or retrovirus, transgene expression profiles from AAV vectors appear to be unique in that the expression levels increase gradually after vector administration and require approximately 4 to 6 weeks before a plateau is reached. Due to the ss DNA nature of AAV genomes, it has been proposed previously that the slow conversion of ss AAV DNA to a duplex form is the primary cause for the delayed expression profile (10, 11). Further studies focusing on AAV vector genomes revealed a variety of circular molecules converted from AAV single-stranded DNA genomes both in vitro and in vivo (7, 8, 28) which may eventually be converted into high-molecular-weight multimers (21, 30). Regarding the mechanism for the delayed transgene expression postadministration, the first hypothesis is that the rate-limiting step for AAV transduction is second-strand DNA syntheses. This hypothesis was supported by experiments showing that large amounts of single-stranded DNA could be detected by Southern blot analysis of low-molecular-weight DNA extracted from cells that were transduced by recombinant AAV vectors. In contrast, the amount of extracted double-stranded DNA is far less than that of ss AAV DNA. Additional evidence includes results from genotoxic agents, such as hydroxyurea, UV irradiation, and adenovirus E4orf6 protein, demonstrating that they could increase the amount of ds AAV DNA detected and improve recombinant AAV (rAAV) transduction efficiency (1, 2). However, in several recent studies, other experimental evidence suggested that additional steps are the main barriers for AAV transduction. In NIH 3T3 cells, Hansen et al. pointed out that hydroxyurea may actually affect intracellular endocytic processing of AAV (13, 14). Other groups proposed that intracellular trafficking and the ubiquitin-proteasome pathway are the barriers for AAV high transduction rates of airway epithelia or muscle (6, 9, 27, 28, 31). Using pseudotyped AAV serotype 6 (AAV6) and AAV8 vectors, Thomas et al. suggested that the uncoating of vector genomes is the primary step limiting the AAV transduction efficiency of liver (29a).
To clarify what the rate-limiting step for rAAV transduction is, we designed a new strategy to directly examine the status of AAV single-stranded DNA in situ. AAV genomes could exist in host cells in three major forms: (i) ss DNA in virions, (ii) free ss DNA in host cells, and (iii) ds DNA (circular, linear, or integrated). We took advantage of BrdU-labeled AAV vectors and used in situ hybridization and in situ cell enzyme-linked immunosorbent assay (ELISA) to discern these three forms which are otherwise difficult to distinguish by Southern blot analysis. To our surprise, free single-stranded AAV genomes were not detectable in all of our assays. Our results suggested that ss DNA AAV either was converted into ds DNA efficiently or degraded rapidly as a DNA-damaging signal. In turn, this supports the theory that the rate-limiting step(s) for rAAV transduction is the processes prior to the release of ss AAV genomes.
MATERIALS AND METHODS
Plasmid and cell culture.
HeLa and 293 cells were obtained from the American Type Culture Collection and cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS; HyClone), penicillin (100 U/ml), and streptomycin at 37°C in a moisturized environment supplied with 5% CO2.
Antibodies.
Monoclonal anti-BrdU antibody recognizing BrdU (note that the antibody does not recognize thymidine in ss DNA, free BrdU, or BrdU coupled to a protein carrier) was purchased from Becton Dickinson. Additional antibodies against BrdU were part of the reagent kits from Roche and used for ELISA of ss DNA labeled with BrdU.
Recombinant AAV vector production, purification, and infection.
The vectors used in this study are all based on AAV serotype 2. Recombinant AAV vectors with different reporter genes were generated by a triple-transfection method which has been described previously (5). Briefly, a vector plasmid (with AAV inverted terminal repeats), a helper plasmid (with the AAV rep and cap genes), and a mini adenovirus helper plasmid (pFΔ6, with essential regions from the adenovirus genome) were cotransfected into 293 cells at a ratio of 1:1:2 by calcium phosphate precipitation. At 96 h after transfection, cells were harvested, and vectors were purified by two rounds of CsCl gradient centrifugation. Fractions in the density range of 1.38 to 1.42 g/ml were collected, dialyzed against phosphate-buffered saline (PBS), and stored in PBS with 3% glycerol. Genome titer was determined by using dot blot or quantitative PCR.
For AAV infection, 2.5 × 106 HeLa cells per well were seeded onto 6-well plates and allowed to grow for 24 h. The cells were approximately 50% confluent prior to infection. Before infection, cells were cooled to 4°C for 10 min. The desired amount of vectors was then diluted in 1 ml of cold DMEM without FBS and applied to each well. The vectors and the cells were incubated at 4°C for 1 h to allow sufficient binding. After incubation, the cells were then washed once with PBS and incubated in DMEM with 10% FBS at 37°C. After 48 h, the cells were close to full confluency.
DNA preparation and analysis.
Isolation of total cellular DNA from mammalian cells was carried out with an Easy DNA kit (Invitrogen). Extraction of low-molecular-weight DNA was performed by using modified procedures recommended by Roche for the High Pure plasmid purification kit. In detail, cells were resuspended in 320 μl of extraction buffer (10 mM Tris [pH 8.0], 10 mM EDTA, 50 μg of RNase/ml, 1% sodium dodecyl sulfate [SDS]) and incubated at 37°C for 30 min. Proteinase K (Roche) was then added to a final concentration of 40 μg/ml. After the proteinase K digestion was carried out at 37°C for 2 h, NaCl was then added to a final concentration of 1.1 M. The digested samples were incubated at 4°C overnight and centrifuged at 13,000 × g for 30 min at 4°C. Finally, the supernatant was collected and extracted with phenol-chloroform, and DNA was precipitated with ethanol (EtOH) before being resuspended in 10 mM Tris (pH 8.0).
Virion-protected AAV DNA was isolated by using a method described by Ferrari et al. (10). In detail, cells were resuspended in 500 μl of buffer containing 50 mM Tris (pH 8.0), 0.2% deoxycholate, and 10% EtOH. The cell suspension was incubated at room temperature for 1 h and then centrifuged at 13,000 × g for 10 min. The supernatant was then collected. In the supernatant, RNase was added to a final concentration of 20 μg/ml, DNase was added to a final concentration of 200 μg/ml, and CaCl2 and MgCl2 were added to a final concentration of 2 mM. The mixture was then incubated at 37°C for 90 min before EGTA and EDTA were added to a final concentration of 10 mM. Next, a 1/20 volume of 10% sarcosine was added. The sample was further incubated at 70°C for 10 min. Afterwards, the sample was cooled to 37°C, which was followed by proteinase K digestion (1 mg/ml) at 37°C for 2 h. The processed sample was then extracted once with phenol-chloroform. At the last step, DNA was precipitated with 2 volumes of ethanol, washed with 70% ethanol, and resuspended in 100 μl of Tris-EDTA buffer.
For Southern blotting, 10% of isolated DNA from 106 cells was applied to each lane. Southern blotting was performed according to standard procedures for DNA transfer. After the membrane was hybridized with a 32P-labeled green fluorescent protein (GFP) probe, it was washed under high-stringency conditions twice for 30 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 65°C and twice for 30 min with 0.1× SSC-0.1% SDS at 65°C. The membrane was then analyzed by both phosphorimaging and X-ray autoradiography.
Recovery of infectious AAV particles from AAV-transduced cells.
To obtain a cell lysate from cells infected by AAV vectors, 106 cells were resuspended in a 500-μl solution containing 10 mM Tris (pH 8.0) and 10 mM EDTA. The cells were subjected to multiple rounds of freezing-thawing cycles or sonication to break the cell membranes. The treated cells were then centrifuged at 5,000 × g for 5 min to remove debris. The supernatant was collected and analyzed for vectors.
Generation of BrdU-labeled rAAV vector.
Recombinant AAV vectors were produced by a modified triple-plasmid transfection of HEK293 cells based on calcium phosphate precipitation, as described above. To incorporate BrdU into the vector genome, we followed procedures that were initially described by Berns and Adler (4). In detail, 10 μg of BrdU (Sigma)/ml and 0.5 μg of 5-fluoro-2′-deoxyuridine (FrdU) (Sigma)/ml were added to the medium 8 h after transfection. Cells were harvested 96 h after transfection, and vectors were purified by two rounds of CsCl gradient centrifugation. Fractions in the density range of 1.38 to 1.48 g/ml were collected, dialyzed against PBS, and stored in PBS with 3% glycerol.
Generation of single-stranded BrdU-labeled DNA.
Cloning vector pBluescriptII (Stratagene) was transformed into competent bacteria. To label the plasmid with BrdU, bacteria were grown overnight in Luria-Bertani medium containing 33 μg of BrdU (Sigma)/ml and 0.55 μg of FrdU (Sigma)/ml. DNA was isolated with a QIAGEN Plasmid Mega kit. Labeled DNA was digested with XbaI and KpnI, and ss DNA was generated by using ExoIII nuclease. In a typical reaction, 50 μg of restriction enzyme-digested DNA was incubated with 100 U of ExoIII (Promega) at 37°C for 30 min before the reaction was stopped by adding 2 μl of 0.5 M EDTA. Control single-stranded pBluescript DNA without BrdU labeling was generated in a similar way.
Transfection with BrdU-labeled DNA.
HeLa cells (105) were seeded onto culture slides (Falcon) 24 h before transfection. Each well was transfected overnight with 200 ng of total DNA by using Lipofectamin 2000 (Invitrogen). The negative control included only 200 ng of single-stranded unlabeled pBluescript DNA. In the test samples, 10 ng or 100 pg of BrdU-labeled single-stranded pBluescript DNA was transfected, and the total DNA was adjusted to 200 ng with unlabeled carrier DNA. After transfection, cells were washed with PBS and subjected to immunohistochemical analysis.
In situ cell-based ELISA for detecting BrdU-containing DNA.
Approximately 5 × 105 HeLa cells per well were seeded onto 96-well plates 24 h before infection. Cells were infected with BrdU-labeled vector at a multiplicity of infection (MOI) of 10,000. The cells were analyzed at 0, 6, 12, 24, and 48 h after infection with a cell proliferation ELISA kit (Roche). For staining, the cells were washed once with PBS and fixed with a nondenaturing fixation solution (70% EtOH) and/or the denaturing Fix/Denat solution supplied with the kits for 30 min at room temperature. The cells were then washed three times with wash buffer, and detection of BrdU was performed with an anti-BrdU antibody which is conjugated with peroxidase. The anti-BrdU peroxidase-conjugated antibody was diluted 1:100 in antibody dilution solution, and 100 μl was applied to each well for 90 min at room temperature. After the plate was washed three times with wash buffer, 100 μl of substrate was applied to each well, and the absorption was measured at an optical density of 405 nm (OD405). Samples were run in quadruplicate and repeated twice.
ELISA for detecting single-stranded DNA with BrdU.
To analyze BrdU-labeled DNA by ELISA, a cellular DNA fragmentation ELISA kit (Roche) was used with minor modifications. An anti-DNA antibody was diluted 1:50 in coating buffer, and 100 μl per well was applied to an ELISA plate. After incubating at 4°C overnight, the plate was washed three times with PBS-Tween and incubated with blocking buffer for 2 h at room temperature. Labeled and nonlabeled single-stranded pBluescript DNA or isolated vector DNA from BrdU-labeled rAAV vectors was diluted in 100 μl of incubation buffer and applied to the coated ELISA plate. After binding at room temperature for 90 min, the plate was washed again. Finally, the BrdU-labeled DNA was detected by using an anti-BrdU peroxidase-conjugated antibody. The anti-BrdU peroxidase-conjugated antibody was diluted 1:50 in incubation buffer and applied to each well. After a final incubation and washing step, 100 μl of substrate was applied to every well, and the absorption was measured at OD405. No denaturing steps were performed to detect accessible free labeled single-stranded DNA. Samples were performed in quadruplicate and repeated twice.
Immunofluorescence staining.
HeLa cells were seeded onto culture slides (Falcon) 24 h before infection. For infection with BrdU-labeled vector, cells were precooled for 10 min at 4°C. The vector was diluted in cold DMEM without FBS, and 250 μl was applied to each chamber. After incubation at 4°C for 1 h, cells were washed with cold medium and then shifted to incubation at 37°C. The cells were fixed under either denaturing or nondenaturing conditions. Under nondenaturing conditions, cells were fixed with 100% methanol for 30 min at −20°C and the slides were washed once with PBS and incubated with 3% H2O2 to destroy endogenous peroxidases. Under denaturing conditions, the cells were treated with 0.07 N NaOH at room temperature for 2 min and subsequently washed with PBS and PBT (PBS containing 0.1% Tween 20). After the samples had gone through the fixing step, a tyramide signal amplification kit from NEN Life Sciences (Perkin Elmer Life Science Inc.) was used to amplify the signal. The slides were blocked in tyramide signal amplification blocking buffer for 30 min at room temperature. Antibody against BrdU (mouse; Becton Dickinson) in a 1:10 dilution in blocking buffer was added to the samples and incubated in a humidified chamber at 4°C overnight. After the primary antibodies were washed by PBT, secondary anti-mouse antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology) in a 1:200 dilution was applied to the slides and incubated for 2 h at room temperature. Finally, slides were incubated with Texas Red-conjugated streptavidin molecules (NEN Life Sciences) at a 1:200 dilution in PBT at 37°C for 30 min. Nuclei were stained by using Hoechst 33342. Confocal microscopy was performed with a Leica DMIRE2 microscope and the Leica TCS SP2 confocal system. Images were processed and assembled by using Photoshop software (Adobe).
RESULTS
Existence of abundant amounts of single-stranded AAV genomes after recombinant AAV vector infection.
During transduction, rAAV vectors based on AAV serotype 2 bind to their receptors on the host cell surface (heparan sulfate, integrins, and fibroblast growth factor receptor) and enter the host cells by clathrin-dependent or -independent internalization. Subsequently, AAV particles undergo an intracellular trafficking process and escape from the endosomal compartment to the nucleus. Single-stranded AAV genomes are then released from the virion through an uncoating process, which can be processed to the following possible forms: free single-stranded AAV genome, annealed plus and minus genome, double-stranded DNA converted from free single-stranded AAV genome, circular double-stranded AAV genome, and integrated AAV genome. Since a duplex DNA template is essential for RNA transcription and subsequent protein expression and ss DNA is the immediate precursor for ds molecules, the status of ss DNA is critical for elucidating the mechanism of rAAV transduction. As shown in Fig. 1, we and others have observed an enormous amount of ss AAV DNA postinfection by Southern blot analysis of low-molecular-weight DNA extracted by using the Hirt DNA protocol (10, 21). The data also revealed that the total amount of vector DNA detected decreased over the first 48 h. There was only approximately 20% of original AAV DNA detected at 48 h postinfection, which suggests that there is a considerable amount of viral DNA being degraded in the process.
FIG. 1.
Single-stranded AAV genomes are abundant after rAAV infection. HeLa cells were transduced with an AAV2-CMV-EGFP vector. Low-molecular-weight DNA was isolated at 0, 1.5, 4, 24, and 48 h postinfection and probed with a Southern blot using a fragment that specifically hybridizes to GFP genes. The DNA standard is indicated in kilobases. SS, single-strand DNA.
Generation of AAV vectors with BrdU-labeled genomes.
The Hirt DNA extraction procedure doesn't selectively isolate free single-stranded AAV genomes from the transduced cells. Both ss DNA encapsidated in and protected by AAV capsids and free ss DNA can be extracted by this method. Since the intact virions do not contribute to the expression level of transgene product, the ratio of free single-stranded DNA to intact virus is a key issue that has implications in transduction efficiency. In order to selectively monitor the free single-stranded AAV genomes in host cells, we took advantage of AAV vectors with BrdU-labeled genomes. BrdU is a nucleotide analog that can be incorporated into the AAV genome without interfering with AAV packaging (4). The anti-BrdU antibody used in this study distinguishes incorporated BrdU from thymidine in ss DNA, free BrdU, and BrdU coupled to a protein carrier. In addition, it will not recognize BrdU in double-stranded DNA since the BrdU epitope is buried beneath the duplex groove. When the AAV-BrdU vector is present in the host cells, an antibody recognizing the BrdU epitopes can therefore be utilized to follow the process of conversion of AAV genomes from ss to ds AAV.
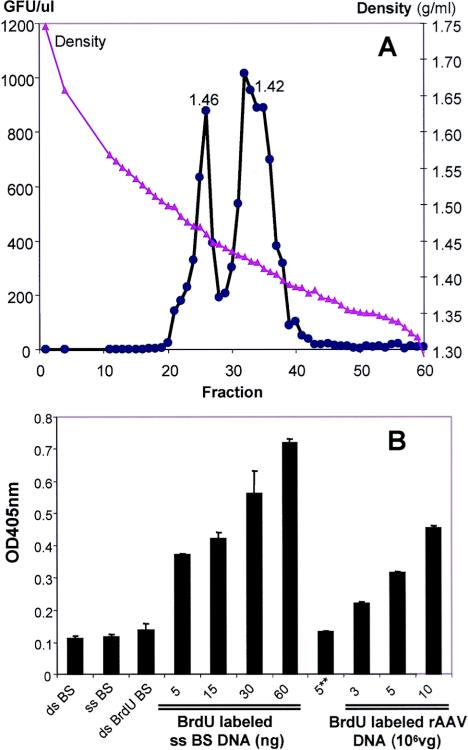
The standard buoyant density of AAV virus is approximately 1.40 to 1.42 g/ml in a CsCl gradient. As shown in Fig. 2A, we also observed that AAV vectors produced in cells cultured in medium supplied with BrdU shifted to a higher density. There are two distinct peaks at 1.46 and 1.42 g/ml which represent the minus- and plus-stranded AAV vectors, respectively. To confirm that BrdU incorporated into AAV genomes can be recognized by the anti-BrdU antibody, we designed a DNA ELISA based on a cellular DNA fragmentation ELISA kit (Roche). Under nondenaturing conditions, BrdU-labeled single-stranded pBluescript DNA and the extracted BrdU-labeled rAAV genome were captured by an anti-DNA antibody on an ELISA plate, which was followed by detection with an anti-BrdU peroxidase-conjugated antibody. As shown in Fig. 2B, we observed a dose-dependent OD405 reading with increased amounts of BrdU-labeled substrate, ss plasmid, or ss AAV genome. The controls with non-BrdU-labeled substrates produced only a basal level signal. This study demonstrated that BrdU molecules had been incorporated into AAV genomes and can be detected by using a monoclonal anti-BrdU antibody.
FIG. 2.
(A) Generation and purification of BrdU-labeled rAAV vector. Vectors were produced by using a standard triple-transfection method in the presence of BrdU in the medium (see Materials and Methods) and subjected to two rounds of CsCl gradient centrifugation. After the second round of CsCl gradient centrifugation, the whole volume was collected as 200-μl fractions. One microliter of solution from each fraction was used to infect HeLa cells which had been seeded onto a 24-well plate. The number of GFP-positive cells was counted 24 h later. The number of GFP-positive cells (GFU/μl) is shown on the left y axis, and the density of each fraction is shown on the right y axis. The x axis stands for each fraction. The two peaks indicating the separation of sense and antisense strand, which incorporated different amounts of BrdU, are identified. (B) Validation of BrdU-labeled rAAV vector. Linear ds pBluescript plasmid (ds BS), ss pBluescript plasmid (ss BS), BrdU-labeled ss pBluescript plasmid (ss BrdU BS), and isolated viral genomes from a BrdU-labeled AAV vector in the indicated amounts were used in a DNA ELISA under nondenaturing conditions. 5**, 5 × 108 vector genomes from a nonlabeled AAV2 vector; vg, vector genomes. The OD405 is shown on the y axis.
In situ analysis of the AAV single-stranded DNA genome.
As shown in Fig. 1, a large amount of single-stranded DNA genome can be detected by Southern blot analysis of isolated low-molecular-weight cellular DNA following AAV infection. We then designed an in situ cell-based ELISA to monitor and track ss AAV genomes. This assay was carried out in situ in order to avoid the potential annealing of AAV plus strands and minus strands during DNA manipulation procedures. We utilized most components from a cell proliferation ELISA kit (Roche) to perform this assay. The cells infected with AAV vectors were fixed with either 70% EtOH or Fix/Denat solution provided with the cell proliferation ELISA kit (Roche), which represents nondenaturing or denaturing conditions, respectively. In the cells treated with the Fix/Denat solution, there were positive signals for the BrdU-labeled genome (Fig. 3B). However, under nondenaturing conditions which are specific for free AAV ss DNA detection, no BrdU signals were found (Fig. 3A), even with an increased MOI of vectors or prolonged incubation time (Fig. 3A and C). A signal could be detected only under denaturing conditions (Fig. 3B and D). Our results suggested that the amount of free single-stranded DNA found in the cells is rare. Free single-stranded DNA can be available only under denaturing conditions which can cause either annealed or converted ds DNA dissociation.
FIG. 3.
Detection of BrdU-labeled viral genomes by ELISA. (A and B) HeLa cells were cultured on a 96-well plate and transduced with a BrdU-labeled AAV2 (AAVBrdU) vector at MOI of 10,000 (10k) and 20,000 (20k), transduced with a nonlabeled AAV2 vector at an MOI of 10,000, or not infected (Mock). Cells were then fixed 24 h postinfection under different conditions, and BrdU-labeled genomes were monitored by ELISA using procedures described in Materials and Methods. (A) ELISA was carried out under nondenaturing conditions. (B) ELISA was performed under denaturing conditions. (C and D) HeLa cells were seeded onto 96-well plates and infected with a BrdU-labeled AAV2 vector (AAVBrdU) or a normal AAV2 vector (AAV) at an MOI of 10,000. At various time points postinfection, cells were washed and assayed for the presence of BrdU by ELISA. (C) ELISA under nondenaturing conditions. (D) ELISA under denaturing conditions.
The ELISA readout reflects an overall estimation of the total amount of free ss DNA in the transduced cells. To examine the levels of free ss AAV genomes at the individual cell level, we performed in situ immunofluorescence staining of cells infected with BrdU-labeled vector (Fig. 4). Under denaturing conditions in which cells are treated with 0.07 N NaOH, AAV ss DNA genomes were detectable in the majority of the cells by as early as 4 h in the cell nuclei. In contrast, there was no detectable BrdU signal in the cells that were fixed with nondenaturing agents such as 70% ethanol or methanol-acetone (data not shown). In ds DNA, the bromine atom is hidden in the phosphodiester backbone of the double helix rendering the accessibility of anti-BrdU antibodies. The BrdU epitope becomes available only when either the ds DNA is converted into ss DNA or the ss DNA in the AAV virion becomes accessible under denaturing conditions. As a positive control, cells were transfected with BrdU-labeled ss plasmid DNA, and immunofluorescence staining was performed under nondenaturing conditions (Fig. 4G to I). We were able to detect 100 pg of BrdU-labeled ss DNA in these cells (Fig. 4H), indicating that our immunofluorescence analysis is sensitive enough to detect free ss DNA in these cells. This assay demonstrated that there were few AAV-transduced cells with free single-stranded AAV genomes in the nuclei.
FIG. 4.
Detection of BrdU-labeled viral ss DNA by immunofluorescence staining. HeLa cells were infected with BrdU-labeled AAV2 vector at an MOI of 10,000, and the incorporated BrdU in the viral genome was detected at (A) 10 min, (B) 6 h, (C) 12 h, (D) 24 h, and (E) 48 h postinfection by using a BrdU antibody. Under nondenaturing (Non D) conditions, no signal is detectable. Under denaturing (Denatured) conditions, BrdU signal (red) can be detected in the nucleus, which is stained with Hoechst 33342 (blue) at 6 h postinfection or later on. The strongest signal is detected 24 h postinfection, which is reduced following that time point. As a negative (neg) control, the same time course experiment was performed by using a nonlabeled AAV2-CMV-GFP vector which is shown at 12 h postinfection (F). (G, H, and I) SS plasmid DNA was prepared from bacteria that were grown in Luria-Bertani medium supplemented with BrdU. HeLa cells were transfected with 10 ng (G) or 100 pg (H) of labeled ss DNA or 200 ng of unlabeled ss DNA (I). Cells were fixed at 6 h posttransfection and stained with BrdU antibody (red) under nondenaturing conditions. The nucleus was stained with Hoechst 33342 (blue). The scale bar is 20 μm for all panels.
The majority of AAV single-stranded DNA exists in intact virions at the early stage of infection.
There is a substantial amount of single-stranded AAV genomes detected in the Southern blot analysis (Fig. 1). However, the BrdU tracking experiments showed few free single-stranded AAV genomes. Therefore, it is likely that the ss AAV genomes detected by Southern blot analysis were from intact AAV virions, where AAV single-stranded DNA remained protected and was inaccessible to the BrdU antibodies under nondenaturing conditions. To test this hypothesis, we compared the total cellular DNA and protected AAV virion DNA after AAV infection. The protected AAV virion DNA was obtained by extensive DNase treatment before proteinase K digestion of the AAV virion. As presented in Fig. 5A, the protected AAV DNA was at a level similar to that of AAV DNA detected in the total cellular DNA extracted at 0, 4, 24, and 48 h after AAV infection. This result is consistent with previous observations that showed that there is a limited amount of AAV vector DNA converted as double-stranded DNA (10, 11). To further confirm that the majority of AAV vector DNA is still associated with AAV capsid proteins, we carried out a Southern blot of vector DNA by directly using cell lysate from rAAV infection (Fig. 5B). The cell lysate was loaded onto gel for electrophoreses without further treatment. If AAV DNA was associated with capsid proteins, the DNA should have been detected in a very-high-molecular-weight region by Southern blot. Our result confirmed this hypothesis. There was no single-stranded DNA visible in the blot, and the pattern of vector DNA in cell lysate is similar to that of purified intact AAV vectors. The result again supports the hypothesis that the majority of vectors were still not uncoated.
FIG. 5.
Southern analysis of viral genomes. HeLa cells were transduced with an AAV2-CMV-GFP vector. DNA or cell lysate was isolated at different time points postinfection. The Southern blot was done with a GFP fragment as the probe. (A) Total cellular DNA and protected virus DNA were extracted at 0, 4, 24, and 48 h postinfection. (B) Southern blot of cell lysate of transduced HeLa cells without additional procedures. Mock, not infected. The DNA standard is indicated in kilobases.
We speculated that if the vectors in the cells still remain as intact particles, they could still be infectious. To test this hypothesis, we extracted cell lysates from cells that were previously infected with AAV-cytomegalovirus (CMV)-enhanced GFP (EGFP) vectors. The crude cell lysates were then used to infect fresh HeLa cells. This result is presented in Fig. 6. As expected, a significant number of positive cells with GFP expression were observed. The number of green cells decreased over the testing period from 4 to 48 h, which is consistent with results of the Southern blot analysis shown in Fig. 1, indicating a significant vector DNA lost in that period.
FIG. 6.
Infectious AAV particles in transduced cells. HeLa cells were transduced with an AAV2-GFP vector, and a cell lysate was extracted at different time points postinfection. The cells were infected with equal amounts of crude lysates, and GFP expression was visualized with a UV fluorescence microscope. Mock, cell lysate from noninfected cells.
Both the annealing of plus- and minus-stranded AAV genomes and second-strand DNA synthesis are involved in AAV transduction.
The plus- and minus-stranded AAV vectors can be separated by BrdU labeling as shown in Fig. 2. To test the infectivity of BrdU-labeled virus, we infected HeLa cells with rAAV-CMV-GFP from fractions with a density of 1.42 or 1.46 g/ml, which represents two different polarities of AAV vectors. As shown in Fig. 7, 1 μl of either fraction yielded a similar amount of GFP-positive cells, suggesting that both polarities are equally infectious. This result is consistent with the results of Fig. 2B showing that both peak fractions contain a similar amount of GFP-forming units (GFU). However, a mix of both fractions (2 μl) yielded not only more GFP-positive cells but also stronger green fluorescence in the cells. The brighter gene fluorescence is an indication of more ds AAV genomes. Therefore, DNA annealing is another mechanism, in addition to AAV second-strand DNA synthesis, that is involved in AAV transduction.
FIG. 7.
Infectivity of BrdU-labeled AAV-CMV-EGFP vectors. After plus- and minus-stranded BrdU-labeled vectors were separated by CsCl gradient, 1-μl (containing approximately 900 GFUs) or 2-μl peak fractions or the mix of 1-μl each was used for infecting HeLa cells. The fluorescence images were taken at 48 h posttransduction.
DISCUSSION
The single-stranded AAV genome is a critical form in the AAV life cycle which consists of both lytic and latent infection. In lytic infection, AAV ss DNA is sequestered into the protected AAV virion, while in latent infection, the ss DNA genome is converted by host enzymes into ds DNA in either circular or linear form, which may be integrated into the host chromosome (8, 21). Intact AAV virions could be detected by immunofluorescence staining using antibodies that specifically recognize intact AAV virion or label the capsid proteins with fluorescent dyes (3, 18). Previously, there were no special experimental techniques to specifically track the fate of the ss AAV genome, which has been difficult to discern from its ds forms. The coexistence of the plus- and the minus-stranded viral DNA also added more complexity because of their potential for annealing. In this study, we successfully established an assay that detects AAV ss DNA at the molecular level by using in situ cell ELISA and immunofluorescence staining. The in situ analysis also eliminated the possibility of generating artifacts in DNA isolation procedures. To visualize ss AAV DNA in situ, we took advantage of the property of the base analog BrdU which, when incorporated into DNA, can still get packaged and interact with anti-BrdU antibody but only if the DNA is in the single-stranded form (25). In ds DNA, the bromine atom is hidden in the phosphodiester backbone of the double helix and is therefore not accessible to the antibody. This method could be applied to study the biology of all single-stranded DNA virus, with some minor modifications.
BrdU labeling is a method that has been commonly used for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays. In a standard TUNEL assay, terminal deoxyribonucleotidyl transferase adds approximately 20 to 100 nucleotides to the DNA breaks. In BrdU-labeled AAV genomes, each genome may get a maximum of 1,000 BrdU molecules. Therefore, this assay could be used to identify BrdU molecules in ss AAV genomes. This is confirmed by the data shown in Fig. 4, in which 100 pg of ss BrdU-labeled plasmid DNA can be detected under nondenaturing conditions. In the ELISA assay shown in Fig. 3, 3 × 106 vector genomes of labeled AAV genomes can give rise to a detectable signal. In a typical in situ ELISA assay, 5 × 105 cells are used. Thus, this assay can detect an average of less than 10 free ss DNA molecules per cell. Therefore, this assay is sensitive enough to address the question of whether there is significant ss DNA accumulation for an AAV administration dose of 20,000 MOI.
It has been accepted that second-strand DNA synthesis is a rate-limiting step for rAAV transduction, which was based on the existence of abundant amounts of ss DNA after AAV infection and few double-stranded forms. However, the state of these single-stranded DNA molecules is so far unclear. A common misconception in the field is that these molecules exist as free ss DNA that is available to the cellular enzymatic machinery. The inefficiency with regard to rAAV transduction is therefore caused by the deficiency of host enzymes to process the vast pool of AAV single-stranded DNA genomes. In this study, we clearly demonstrate for the first time that free single-stranded DNA is rare during AAV infection. First, no free BrdU-labeled single-stranded AAV genomes could be detected by in situ cell-based ELISA and immunohistochemistry staining with anti-BrdU antibodies under nondenaturing conditions. Such single-stranded DNA can be detected only under denaturing conditions, which either partially damaged the protected AAV virions and exposed the ss AAV genomes or allowed the BrdU epitopes of ds DNA to be accessible to anti-BrdU antibody. Second, we also showed that the protected AAV virion is a major contribution to the abundant single-stranded DNA previously detected (Fig. 5). Furthermore, the vectors extracted from previously infected cells were still infectious (Fig. 6). Since these particles are still infectious, it rules out the possibility that the main source of released ss DNA is from special cellular compartments which are not accessible to BrdU antibody under nondenaturing conditions.
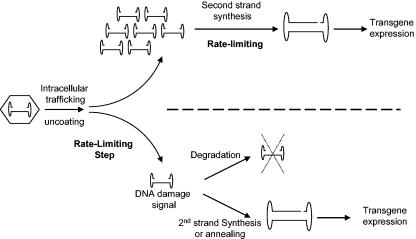
Therefore, a clearer picture of what happens after AAV infection appears to be that viral particle processing, an event prior to the uncoating process, is the rate-limiting step. The amount of ds DNA available for mRNA transcription is still limited because only a few free ss DNA molecules are available for second-strand DNA synthesis. However, free AAV ss DNA may have been either converted or annealed into ds DNA genomes almost instantly after uncoating or being degraded by host enzymes. Previous studies have suggested that ss DNA can serve as a DNA damage signal which can cause cell death and stimulate the cellular repair mechanism (15). Cells are extremely sensitive to the presence of ss DNA; a few ss DNA molecules are sufficient to cause cell arrest in the G1 phase. Therefore, our result is consistent with theories established in DNA repair studies. A summary of this pathway is illustrated in Fig. 8. Although we rule out the existence of abundant free single-stranded AAV DNA, our results still support earlier studies that found that the annealed or fully synthesized ds AAV genomes are rather limited, which ultimately determines the expression levels of transgene. As shown in Fig. 7, a mix of 1 μl each of both polarities is far better than a 2-μl fraction of either polarity. This result suggests that in addition to second-strand DNA synthesis, the annealing of both polarities is part of the mechanism that occurs and improves the transduction efficacy. Since this result is observed in a cell line, we do not rule out the possibility that there may be only one dominant mechanism that determines rAAV transduction in other situations (24, 33).
FIG. 8.
Illustration of potential rate-limiting steps for rAAV transduction. In the top part of the scheme, a large amount of free single-stranded DNA is shown. In bottom part of the scheme, degradation of ss viral DNA and complementary-strand synthesis are proposed to be a fast reaction.
We clearly observed that a significant amount of viral DNA got lost in the first 48 h, presumably degraded by cellular nucleases (Fig. 1). This finding suggests that the cellular mechanism may prefer degrading AAV ss DNA instead of converting it into double-stranded DNA, which is essential for AAV transduction. The enhanced AAV transduction by genotoxic agents may actually help shift the balance of degradation to conversion. This hypothesis can be supported by previously published studies (10, 11). The most direct supporting evidence comes from self-complementary AAV (scAAV) vector studies (6, 20). scAAV does not need second-strand DNA synthesis for RNA transcription. However, the onset of scAAV expression is similar to that of normal AAV vectors. The high efficiency of scAAV expression may also be partly due to the fact that there is no ss DNA-induced viral genome degradation since a double-stranded DNA genome is more stable than the ss AAV genome. Our study further suggests that it is critical to improve the stability of AAV genomes and vector trafficking or uncoating to improve rAAV transduction efficiency.
Acknowledgments
We thank Valder Arruda, Kenneth Berns, Roland Herzog, Jude Samulski, and Xiao Xiao for helpful comments and scientific discussions. We also thank Marlene Webber for assistance in manuscript preparation.
This work was supported by grant HL69051 from the National Institutes of Health and a research grant from the Muscular Dystrophy Association.
REFERENCES
- 1.Alexander, I. E., D. W. Russell, and A. D. Miller. 1994. DNA-damaging agents greatly increase the transduction of nondividing cells by adeno-associated virus vectors. J. Virol. 68:8282-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, I. E., D. W. Russell, A. M. Spence, and A. D. Miller. 1996. Effects of gamma irradiation on the transduction of dividing and nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum. Gene Ther. 7:841-850. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns, K. I., and S. Adler. 1972. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J. Virol. 9:394-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, L., Y. Liu, M. J. During, and W. Xiao. 2000. High-titer, wild-type free recombinant adeno-associated virus vector production using intron-containing helper plasmids. J. Virol. 74:11456-11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, W., Z. Yan, R. Zak, M. Saavedra, D. M. Rodman, and J. F. Engelhardt. 2003. Second-strand genome conversion of adeno-associated virus type 2 (AAV-2) and AAV-5 is not rate limiting following apical infection of polarized human airway epithelia. J. Virol. 77:7361-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 72:8568-8577. (Erratum, 73:861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan, D., Y. Yue, Z. Yan, J. Yang, and J. F. Engelhardt. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Investig. 105:1573-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, K. J., K. Jooss, J. Alston, Y. Yang, S. E. Haecker, K. High, R. Pathak, S. E. Raper, and J. M. Wilson. 1997. Recombinant adeno-associated virus for muscle directed gene therapy. Nat. Med. 3:306-312. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J., K. Qing, H. J. Kwon, C. Mah, and A. Srivastava. 2000. Impaired intracellular trafficking of adeno-associated virus type 2 vectors limits efficient transduction of murine fibroblasts. J. Virol. 74:992-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, J., K. Qing, and A. Srivastava. 2001. Infection of purified nuclei by adeno-associated virus 2. Mol. Ther. 4:289-296. [DOI] [PubMed] [Google Scholar]
- 15.Huang, L.-C., K. C. Clarkin, and G. M. Wahl. 1996. Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc. Natl. Acad. Sci. USA 93:4827-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotin, R. M., J. C. Menninger, D. C. Ward, and K. I. Berns. 1991. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10:831-834. [DOI] [PubMed] [Google Scholar]
- 18.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linden, R. M., and K. I. Berns. 1997. Site-specific integration by adeno-associated virus: a basis for a potential gene therapy vector. Gene Ther. 4:4-5. [DOI] [PubMed] [Google Scholar]
- 20.McCarty, D. M., P. E. Monahan, and R. J. Samulski. 2001. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8:1248-1254. [DOI] [PubMed] [Google Scholar]
- 21.Miao, C. H., R. O. Snyder, D. B. Schowalter, G. A. Patijn, B. Donahue, B. Winther, and M. A. Kay. 1998. The kinetics of rAAV integration in the liver. Nat. Genet. 19:13-15. [DOI] [PubMed] [Google Scholar]
- 22.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 23.Muzyczka, N., and K. I. Berns. 2002. Parvoviridae: the viruses and their replication, p. 2327-2360. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samulski, R. J. 1993. Adeno-associated virus: integration at a specific chromosomal locus. Curr. Opin. Genet. Dev. 3:74-80. [DOI] [PubMed] [Google Scholar]
- 27.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by Rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanlioglu, S., D. Duan, and J. F. Engelhardt. 1999. Two independent molecular pathways for recombinant adeno-associated virus genome conversion occur after UV-C and E4orf6 augmentation of transduction. Hum. Gene Ther. 10:591-602. [DOI] [PubMed] [Google Scholar]
- 29.Steinbach, S., A. Wistuba, T. Bock, and J. A. Kleinschmidt. 1997. Assembly of adeno-associated virus type 2 capsids in vitro. J. Gen. Virol. 78:1453-1462. [DOI] [PubMed] [Google Scholar]
- 29a.Thomas, C. E., T. A. Storm, Z. Huang, and M. A. Kay. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 78:3110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao, X., J. Li, and R. J. Samulski. 1996. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 70:8098-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young, S. M., Jr., W. Xiao, and R. J. Samulski. 2000. Site-specific targeting of DNA plasmids to chromosome 19 using AAV cis and trans sequences. Methods Mol. Biol. 133:111-126. [DOI] [PubMed] [Google Scholar]
- 33.Zhong, L., W. Li, Z. Yang, L. Chen, Y. Li, K. Qing, K. A. Weigel-Kelley, M. C. Yoder, W. Shou, and A. Srivastava. 2004. Improved transduction of primary murine hepatocytes by recombinant adeno-associated virus 2 vectors in vivo. Gene Ther. 11:1165-1169. [DOI] [PubMed] [Google Scholar]