Abstract
Computational and biological approaches were undertaken to characterize the role of the human astrovirus nonstructural protein nsP1a/4, located at the C-terminal fragment of nsP1a. Computer analysis reveals sequence similarities to other nonstructural viral proteins involved in RNA replication and/or transcription and allows the identification of a glutamine- and proline-rich region, the prediction of many phosphorylation and O-glycosylation sites, and the occurrence of a KKXX-like endoplasmic reticulum retention signal. Immunoprecipitation analysis with an antibody against a synthetic peptide of the nsP1a/4 sequence detected polyprotein precursors of 160, 75, and 38 to 40 kDa as well as five smaller proteins in the range of 21 to 27 kDa. Immunofluorescence labeling showed that the nsP1a/4 protein is accumulated at the perinuclear region, in association with the endoplasmic reticulum and the viral RNA. These results suggest the involvement of nsP1a/4 protein in the RNA replication process in endoplasmic reticulum-derived intracellular membranes.
Human astroviruses (HAstV) are recognized as common viral pathogens causing gastroenteritis in children (13). Astroviruses are nonenveloped positive-stranded RNA viruses that belong to the family Astroviridae, which includes both mammalian and avian viruses. Their 6.8-kb polyadenylated genome contains three overlapping open reading frames (ORFs). ORF1a and ORF1b encode the viral protease and polymerase, respectively, and ORF2 encodes the capsid precursor (33). The nonstructural proteins (nsPs) are translated from the genomic RNA as two large polyproteins, nsP1a and nsP1a/1b, through a translational frameshifting mechanism (28, 29). Upon infection, nonstructural proteins participate in transcribing a full-length negative-strand RNA, which serves as the template for the transcription of new genomic and subgenomic RNAs. Large amounts of structural proteins are presumably synthesized from the polyadenylated subgenomic RNA that contains the complete ORF2 (34, 39).
While it is clear that the nsP1b corresponds to the RNA-dependent RNA polymerase (21), little is known about the role of most of the nsP1a mature products. Sequence analysis has predicted a 3C-like serine protease motif, a nuclear localization signal, as well as some transmembrane domains close to the N-terminal end (21, 22). Although most RNA viruses with a genome larger than 6 kb encode a viral helicase, no helicase domain has been identified in any part of the complete astrovirus genome (21). An immunoreactive epitope close to the C terminus of nsP1a was partially characterized some years ago (35). In addition, a hypervariable region has been identified close to this epitope by different authors after genetic characterization of HAstV-3 and HAstV-8 (37, 41).
Interestingly, a 45-nucleotide deletion within this region had been described and associated with the adaptation of astrovirus to HEK and LLCMK2 cell lines (59). Recently, computer analysis of nsP1a revealed the presence of two coiled-coil regions common to all known human astrovirus (22), and a putative death domain (15). In addition, a similarity between the putative astrovirus nuclear localization signal and calicivirus genome-linked proteins has also been described (22).
Data about HAstV nonstructural polyprotein processing are still controversial; at least four cleavage sites have been suggested within nsP1a, and it is assumed that both viral and cellular proteases are responsible for the process (11, 12, 24, 36, 60). Processing at the amino terminus of nsP1a and nsP1a/1b is likely to occur cotranslationally, probably at residue Ala174, giving rise to a ≈20-kDa N-terminal product (36). A ≈27-kDa protein containing the protease motif would be generated by cleavage at residues Val409 and Glu654 (11). The cleavage site reported by Kiang and Matsui (30) at Gln567 of nsP1a, which would produce two proteins of ≈64 and ≈38 kDa, does not seem to occur in HAstV-8-infected cells (36), but it is unclear whether it could be functional in other HAstV strains. With antibodies against approximately 30% of the nsP1a C-terminal region, different authors identified products of 34, 20, 6.5, and 5.5 kDa, but no other cleavage sites were suggested (36, 60). According to this information, nsP1a polyprotein would generate at least four products (see Fig. 1A), which in this work will be named nsP1a/1 (≈20-kDa N-terminal product), nsP1a/2, nsP1a/3 (protease), and nsP1a/4, which could be processed even further.
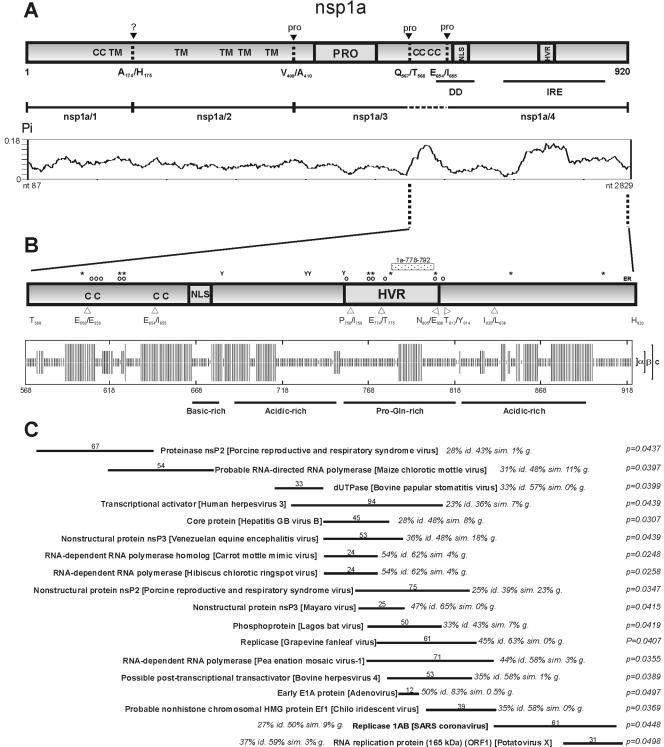
FIG. 1.
(A) Schematic depiction of HAstV nsP1a polyprotein. Predicted transmembrane helices (TM), protease motif (PRO), predicted nuclear localization signal (NLS), immunoreactive epitope (IRE), coiled-coil structures (CC), predicted death domain (DD), and hypervariable region (HVR) are shown. Black arrowheads refer to putative identified proteolytic cleavage sites, which seem to be dependent on both cellular proteases (?) and the viral protease (pro). Below the diagram, analysis of nucleotide variability (Pi) through the nsP1a coding region, with a sliding window size of nine nucleotides and a step size of three. (B) Schematic diagram of the computational analysis of the nsP1a/4 amino acid sequence (HAstV-4). The small dotted box indicates the relative position of the synthetic peptide used to generate the anti-1a778-792 antibody. Potential posttranslational modifications are indicated by symbols above the diagram (*, Ser/Thr phosphorylation sites; Y, Tyr phosphorylation sites; O, O-glycosylation sites), as well as the KKXX-like endoplasmic reticulum retention signal (ER). Open arrowheads below the diagram depict predicted foot-and-mouth disease virus 3C cleavage sites. Analysis of predicted nsP1a/4 secondary structure is shown within a square box which is drawn to scale. Both α-helix and β-strand conformations are indicated on the right of the diagram. Underlined fragments depict amino acidregions containing residues with shared characteristics. (C) Best-match results from the sequence similarity search analysis (BlastP) of nsP1a/4 protein against the viral database. Each match shows information on the length of the region, percentages of sequence identity (id.) and similarity (sim.), percentage of gaps (g.), and the Smith and Waterman statistics for the alignment. Nucleotide (nt) and amino acid (aa) positions are numbered according to HAstV-1 Oxford reference strain (accession no. L23513).
This work characterizes a putative biological role of the nonstructural protein encoded by astrovirus ORF1a at its C terminus (nsP1a/4). The results of computational and biological characterization of the nsP1a/4 protein show colocalization of this protein with viral RNA and host cellular membranes, suggesting its role in viral RNA replication.
MATERIALS AND METHODS
nsP1a/4 gene sequencing.
Primers A1739 (5′-GGTGCACCAGTTTGTGACA-3′; positions 1739 to 1758 from accession no. L23513) and RFS (5′-TCTGGGGCCCTTTGTAGTTT-3′; positions 2797 to 2816 from accession no. L23513) were used to amplify and sequence the ORF1a C terminus of the HAstV-4 strain used throughout this work (GenBank accession no. AY257977).
Proteomic computational analysis.
Together with the HAstV-4 cell-adapted strain sequence, five HAstV complete genome sequences available at GenBank were included in the analysis (accession nos.: L23513 for HAstV-1 Oxford reference strain; Z25771 for HAstV-1 Newcastle strain; L13745 for HAstV-2 Oxford reference strain; AF141381 for HAstV-3 Rostock strain; and AF260508 for HAstV-8 Yuc-8 strain). A computer analysis in search of protein homologues to the ORF1a-encoded nsP1a/4 protein was performed with different available servers and the amino acid sequence that spans Thr568 and His920 (positions according to accession no. L23513). The consensus amino acid secondary structure prediction was performed with Network Protein Server Analysis (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) (7). Coiled coils were predicted with COILS version 2.2 at the EMB net (http://www.ch.embnet.org/software/COILS_form.html), according to Lupas (32). Phosphorylation sites were predicted with the NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/) (2), N-glycosylation sites were predicted with the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/), and O-β-GlcNAc O-linked glycosylation sites were predicted with the YinOYang 1.2 Server (http://www.cbs.dtu.dk/services/YinOYang/) (17).
The SAPS program was used to evaluate a wide variety of protein sequence properties by statistical criteria (http://www.isrec.isb-sib.ch/software/SAPS_form.html) (4). Properties considered include compositional biases, clusters and runs of charged and other amino acid types, and different kinds of repetitive structures. The protein-protein BlastP from the Basic Local Alignment Search Tool at the (http://www.ncbi.nlm.nih.gov/BLAST/) was performed, increasing the threshold for less stringent matches. The Superfamily program was used to provide structural assignments to protein sequence at the superfamily level (http://supfam.org/SUPERFAMILY/) (14). Protein fold recognition with one-dimensional and three-dimensional sequence profiles coupled with secondary structure and solving potential information was studied with the web server 3D-PSSM V2.6.0 (http://www.sbg.bio.ic.ac.uk/~3dpssm/) (23). Functional motifs were scanned against the Prosite database (19), with the PredictProtein server at http://cubic.bioc.columbia.edu/predictprotein. PRSS3, a sequence alignment algorithm, was used to determine the significance (P < 0.05) of protein alignments (53). The NetPicoRNA program V1.0, which identifies targets for foot-and-mouth disease virus 3C protease, was used to describe potential cleavage sites (3).
Analysis of genetic variability.
Sequence alignments were performed with Clustal W (18). Nucleotide and amino acid distances were calculated as the number of substitutions per site (p-dist) with pairwise deletion for treating indels with the MEGA2.1 program (25). Ks and Ka values (where Ks is the frequency of synonymous substitutions per synonymous site and Ka is the frequency of nonsynonymous substitutions per nonsynonymous site) and Ks/Ka ratios were calculated based on the Nei-Gojobori method (40) with the DnaSP 3.0 software (47). The Pi parameter was used as a measure of nucleotide diversity (p-dist) and it was analyzed through the total nsP1a coding region with a sliding window size of nine nucleotides and a step size of three, with the DnaSP 3.0 software (47).
Antibody production.
The highest peaks of hydrophilicity, potential indicators of antigenic sites of a protein, were predicted for the total nsP1a, with the method described by Hopp and Woods, and a window size of 9 (20) from the ProtScale program available at http://ca.expasy.org/cgi-bin/protscale.pl. A polyclonal ascite antibody (anti-1a778-792) was obtained after immunization of mice with a synthetic peptide belonging to amino acid positions 778 to 792 from accession no. L23513 (QPLDLSQKKEKQPEH). The peptide was coupled to keyhole limpet hemocyanin before being administered to 8-week-old female Swiss mice in the presence of Freund's complete adjuvant. The peptide preparation was diluted 1:10 in complete adjuvant and administered in five doses of 25 μg of peptide each at weeks 0, 2, 3, 4, and 5.
Cells and virus.
The human colon adenocarcinoma cell line CaCo-2 was grown in Eagle's minimum essential medium supplemented with 10% fetal calf serum. A cell culture-adapted strain of HAstV-4 (p23795, kindly provided by W. D. Cubitt from the Great Ormond Street Hospital for Children, London), was used in this study. CaCo-2 cells were infected as previously described (44) with some modifications. Briefly, cell monolayers were washed twice with phosphate-buffered saline (PBS) and inoculated with viral stocks pretreated with 10 μg of trypsin (GIX Sigma) per ml for 30 min at 37°C. After a 1-h adsorption at 37°C, minimal essential medium supplemented with 2% fetal calf serum was added.
Immunoprecipitation analysis.
Infections were carried out with 2 × 106 CaCo-2 cells at a multiplicity of infection of 5. Trypsin (5 μg/ml) was added to the serum-free overlay medium. In the indicated experiments, protease inhibitors were added to the postinfection medium (1 μg of aprotinin and 10 μg of leupeptin per ml). Cell lysates were prepared at different times postinfection in 0.2 ml of TNE buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM EDTA) containing 1% NP-40, 1 μg of aprotinin per ml, and 10 μg of leupeptin per ml. After a 1-h incubation at 4°C, homogenized suspensions were centrifuged for 5 min at 10,000 × g at 4°C to remove cell debris and immunoprecipitated overnight at 4°C with either the anti-1a778-792 mouse polyclonal antibody diluted 1:75 or a nonimmune ascitic fluid. Immune complexes were harvested by the addition of protein A-agarose and a 3-h incubation at 4°C, followed by centrifugation at 10,000 × g for 1 min. Pellets were washed twice for 1 h at 4°C in TNE buffer supplemented with 0.1% NP-40 and resuspended in 40 μl of the same buffer. After adding 10 μl of Laemmli buffer and boiling the samples for 10 min, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12 to 15% polyacrylamide gels and examined by Western blot analysis as previously described (15), with a 1:10 dilution of the same anti-1a778-792 antibody. An alkaline phosphatase-conjugated anti-mouse immunoglobulin antibody (BD Biosciences Pharmingen) was used as the detecting antibody and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) (Roche) were used as substrates.
Glycoprotein detection.
The presence of carbohydrates was studied on infected and mock-infected cell extracts previously immunoprecipitated with the anti-1a778-792 antibody, as mentioned above. After immunoprecipitation, proteins were separated on sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Carbohydrate moieties were oxidized with 10 mM sodium periodate, followed by incorporation of biotin hydrazide. Biotinylated complexes were detected by streptavidin-alkaline phosphatase (Sigma) and NBT-BCIP (Roche) color development reagents.
Detection of phosphorylated proteins.
A monoclonal anti-phosphoserine/threonine antibody (anti-PSPT) (BD Biosciences Pharmingen) was used at a 1:100 dilution to immunoprecipitate phosphorylated proteins from infected and mock-infected CaCo-2 cell extracts, essentially as described above, but incubating samples overnight at 37°C. Immunoprecipitated proteins were resolved by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis, and the presence of nsP1a/4 protein was analyzed by Western blot with the anti-1a778-792 antibody, as previously described.
Immunofluorescence studies.
At 48 h postinfection, CaCo-2 cells grown on glass coverslips in 24-well plates and infected at a multiplicity of infection of 0.5 were rinsed twice with PBS and fixed with 3% paraformaldehyde in PBS for 30 min at room temperature. Permeabilization was performed for 15 min at room temperature in PBS containing 0.5% Triton X-100. After washing, cells were blocked for 30 min at room temperature in PBS containing 10% fetal calf serum. Cells were incubated with a mixture of an anti-astrovirus rabbit polyclonal antibody (kindly provided by D. M. Bass, Division of Pediatric Gastroenterology, Stanford University) diluted 1:2,000 and the anti-1a778-792 mouse antibody diluted 1:1,000. An anti-rabbit fluorescein isothiocyanate-conjugated antibody (Sigma) and an anti-mouse immunoglobulin indocarbocyanine 3-conjugated antibody were used as secondary antibodies. After immunofluorescence labeling, cells were stained with 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI) in PBS. Mounting was done in Fluoromont G mounting medium (Southern Biotechnologies), and samples were analyzed under a fluorescence microscope (Leica DMRB FLUO).
Electron microscopy.
For transmission electron microscopy studies, approximately 107 infected (multiplicity of infection of 5) and mock-infected CaCo-2 cells were processed at 48 h postinfection as previously described (15). For immunoelectron microscopy, samples were fixed with 4% (vol/vol) paraformaldehyde and 0.2% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. After fixation, samples were embedded in 10% gelatin and immersed in a cryoprotection buffer. The immunostaining was performed on ultrathin cryosections with the anti-1a778-792 mouse antibody at a dilution of 1:100 as the primary antibody, and a 1:25 dilution of a goat anti-mouse immunoglobulin G serum conjugated with 10-nm colloidal gold particles as the secondary antibody. Antibody incubations were done in saline solution containing 2% fetal calf serum, 1% bovine serum albumin, and 0.1% Tween for 1.5 h at room temperature. Extensive washing and staining with uranyl acetate and lead citrate were performed before observation (Hitachi HT-800).
Double labeling of HAstV proteins and viral RNA.
nsP1a/4 detection was performed by immunofluorescence, and viral RNA was detected by fluorescence in situ hybridization with a digoxigenin-labeled DNA probe. A double-stranded DNA probe of 194 bp was generated by reverse transcription-PCR amplification with primers A1 and A2 as previously described (16, 59), with 100 μM digoxigenin-11-dUTP (Roche) in the PCR mixture. CaCo-2 cells grown on glass coverslips in 24-well plates were infected at a multiplicity of infection of 0.5 and fixed at 24 h postinfection as specified above. After two washes in PBS, cells were permeabilized by an overnight 4°C treatment with 70% ethanol and stored air-dried. For hybridization, cells were rehydrated for 5 min at room temperature in 2× (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-50% formamide and hybridized overnight at 37°C in 25 μl of a mixture containing 1X SSC, 10% dextran sulfate, 50% formamide, 200 μg of herring sperm DNA per ml and 2 ng of DNA probe/μl. The hybridization mixture was heated at 100°C for 10 min to denature the probe. Following hybridization, coverslips were washed twice in 50% formamide-2X SSC for 15 min at room temperature and then in PBS for 15 min at room temperature.
For immunological detection of nsP1a/4, coverslips were blocked in 10% fetal calf serum in PBS and incubated with either a 1:1,000 dilution of anti-1a778-792 or a 1:10,000 dilution of the anti-astrovirus monoclonal antibody 8E7 (kindly provided by J. Herrmann, Department of Medicine/Infectious Diseases, University of Massachusetts Medical School). After three washes with PBS, samples were incubated with a mixture of anti-mouse indocarbocyanine 3-conjugated secondary antibody and an antidigoxigenin fluorescein isothiocyanate-conjugated antibody (Roche). All antibodies were diluted in 10% fetal calf serum in PBS, and incubations were performed for 2 h at room temperature. After immunofluorescence labeling, cells were washed three times in PBS and observed under a fluorescence microscope for colocalization analysis (Leica DMRB FLUO). Controls included identically treated mock-infected cells and infected cells incubated in the absence of probe.
Double labeling of HAstV proteins and the endoplasmic reticulum.
At 24 h postinfection, CaCo-2 cells grown on coverslips in 24-well plates and infected with a multiplicity of infection of 0.5 were washed with PBS and incubated in 200 μl of postinfection medium supplemented with 2% fetal calf serum and 1 μM RE-Tracker Blue-White DPX (Molecular Probes), for 1 h at 37°C. After washing the cell monolayers twice in PBS for 10 min, cells were fixed with 3% paraformaldehyde in PBS for 30 min at room temperature and processed for immunofluorescence as described above, with either the anti-1a778-792 antibody or the 8E7 monoclonal antibody as the primary antibody. Permeabilization was performed for 10 min at room temperature in PBS containing only 0.2% Triton X-100. ER-Tracker labeling was visualized with a UV long-pass filter under a fluorescence microscope (Leica DMRB FLUO).
RESULTS
Proteomic computational analysis of HAstV nsP1a/4.
To gather information on the features of the nsP1a/4 protein, proteomic computational predictions were performed based on the five published complete genome sequences of four serotypes, HAstV-1, HAstV-2, HAstV-3, and HAstV-8, and the complete nsP1a/4 sequence of our HAstV-4 cell-adapted strain. The results are summarized in Fig. 1B. Protein structure prediction revealed the presence of many α-helix secondary conformations and several coiled-coil domains. The proportion of amino acid residues that were not assigned to a particular secondary structure was higher within this region of nsP1a than in other regions of the genome, revealing the high degree of structural flexibility that this region may have. Computational analysis of HAstV nsP1a/4 also suggested the existence of many posttranslational modification motifs, such as protein kinase C, casein kinase II tyrosine phosphorylation sites, and serine and threonine targets for O-glycosylation.
Based on the comparative analysis of the amino acid composition in nsP1a/4 and other parts of the HAstV genome, different domains could be identified: a positively charged cluster that spans amino acids 665 to 686 and corresponds to the previously identified nuclear localization signal (12 Lys and Arg residues in 22 amino acids, 54.5%); a negatively charged acidic domain that spans amino acids 690 to 749 of nsP1a (25 Asp and Glu residues in 60 amino acids, 41.6%); a proline- and glutamine-rich region that spans amino acids 755 to 818 (11 Pro and 9 Gln residues in 64 amino acids, 17.2% and 14.1%, respectively); and one more acidic domain that spans amino acids 821 to 890 (15 Asp and Glu residues in 70 amino acids, 21.4%) (amino acid positions according to accession no. L23513). Additionally, computational analysis led to the detection of a large number of acidic clusters consisting of stretches of four acidic residues, seven potential protease cleavage sites, and an endoplasmic reticulum membrane retention signal (KKXX-like motif) at the C terminus of nsP1a/4 (amino acid positions 916 to 919 according to accession no. L23513).
An analysis of the genetic variability within nsP1a/4 was performed at the nucleotide and amino acid level. The nsP1a/4 protein was the most variable region of the whole nonstructural region of HAstV (Fig. 1A and Table 1). The percentage of mutations that resulted in an amino acid change was higher within the nsP1a/4 coding region than within the nsP1a/123 coding region (17.4% versus 13.1%). Moreover, while the percentage of nonconservative amino acid substitutions within nsP1a/4 was 52.2%, within the nsP1a/123 region this percentage was only 20.6%. At the same time, all identified motifs were found to be highly conserved among different HAstV serotypes through all nsP1a/4 protein, with the exception of those in the hypervariable region that spans 50 amino acids at positions 758 to 807 of nsP1a (accession no. L23513).
TABLE 1.
Analysis of genetic variability
| Parameter | Genomic region
|
|||
|---|---|---|---|---|
| nsP1a/123 | nsP1a/4 | ORF1b | ORF2 | |
| No. tested | 5 | 6 | 5 | 5 |
| Length (bp) | 1704 | 1027-1080 | 1557 | 2346-2388 |
| No. of insertions or deletions | 0 | 2 | 0 | 7 |
| p-dist (nt) | 0.076 | 0.077 | 0.100 | 0.275 |
| p-dist (aa) | 0.027 | 0.042 | 0.037 | 0.252 |
| Ks | 0.280 | 0.273 | 0.378 | 0.549 |
| Ka | 0.012 | 0.020 | 0.019 | 0.175 |
| Ks/Ka | 22.75 | 13.61 | 20.21 | 3.14 |
Among the different serotypes of HAstV, this nsP1a/4 hypervariable region differs enormously in both length and amino acid composition, and the number and position of the predicted phosphorylation/glycosylation sites are also highly variable. In all sequenced serotypes, between 60.0% and 65.2% of all predicted phosphorylation sites of nsP1a were consistently accumulated within the nsP1a/4 coding region, which represents only between 38.4 and 39.4% of the whole nsP1a polyprotein, depending on the serotype. Moreover, 20.0 to 40.0% of the nsP1a/4 phosphorylation sites were located within the hypervariable region, which represents 14.2 to 17.7% of the total nsP1a/4 protein. Regarding O-glycosylation sites, between 71.4 and 85.7% of all predicted sites of nsP1a were again accumulated within the nsP1a/4 coding region, and 20 to 50% of these were located within the hypervariable region. Among all the nonconserved amino acid positions that were present in the nsP1a/4 protein alignment, 15.4% and 26.1% of the substitutions affected predicted phosphorylation and O-glycosylation sites, respectively. For the hypervariable region, these percentages were 25.0% and 18.7, respectively, and interestingly, they were only 0 and 0.03%, respectively, for the nsP1a/123 region. Assuming that phosphorylation and/or O-glycosylation plays a key role on the regulation of nsP1a/4 function, and taking into account the presence of the hypervariable region within this protein, some biological differences could be postulated between distinct protein variants.
With the aim of identifying proteins that show a significant degree of sequence similarity with HAstV nsP1a/4, a BlastP computer search analysis was performed against both viral and cellular databases. Unlike other regions of the astrovirus genome, which show extensive homologies with nonstructural proteins from other positive-strand RNA virus families, all the homologous proteins identified for HAstV nsP1a/4 displayed a moderate degree of similarity. Restricting the Blast database to viruses, the most significant results involved nonstructural proteins from single-stranded RNA viral families, although some matches also belonged to DNA viruses. The results are summarized in Fig. 1C. Interestingly, a common feature shared by all these proteins is that they are involved in the formation of the RNA replicase complexes and/or in the regulation of both viral and cellular transcription processes. Computer BlastP homology searches against cellular proteins did not reveal any close similarity to published protein sequences in the database. With the Superfamily HMM (hidden Markov model) library and genome assignment server, in search of structural and functional identities, nsP1a/4 was assigned to the “winged-helix” DNA-binding domain superfamily (SSF46785), which includes proteins with diverse biological functions such as transcription factors, helicases, endonucleases, and histones. However, the nsP1a/4 protein did not show exact consensus motifs for leucine zippers, nucleoside triphospate-binding domains, HMG box domains, zinc fingers, or homeodomains.
Biological characterization of HAstV nsP1a/4: colocalization with viral RNA and the endoplasmic reticulum.
The molecular weight of the nsP1a/4 mature protein and its protein intermediates was evaluated through an immunoprecipitation Western blotting technique with the anti-1a778-792 antibody. A kinetic analysis at 12, 24, 36, and 48 h postinfection showed that the anti-1a778-792 antibody recognized proteins of 160 kDa, 75 kDa, 38 to 40 kDa, and 21 to 27 kDa (Fig. 2A). The identification of at least five electrophoretically distinct similarly sized bands (27, 25, 24, 22, and 21 kDa) could either indicate differential cleavages during the proteolytic processing of the precursor protein or reflect posttranslational modifications, such as phosphorylation and/or glycosylation.
FIG. 2.
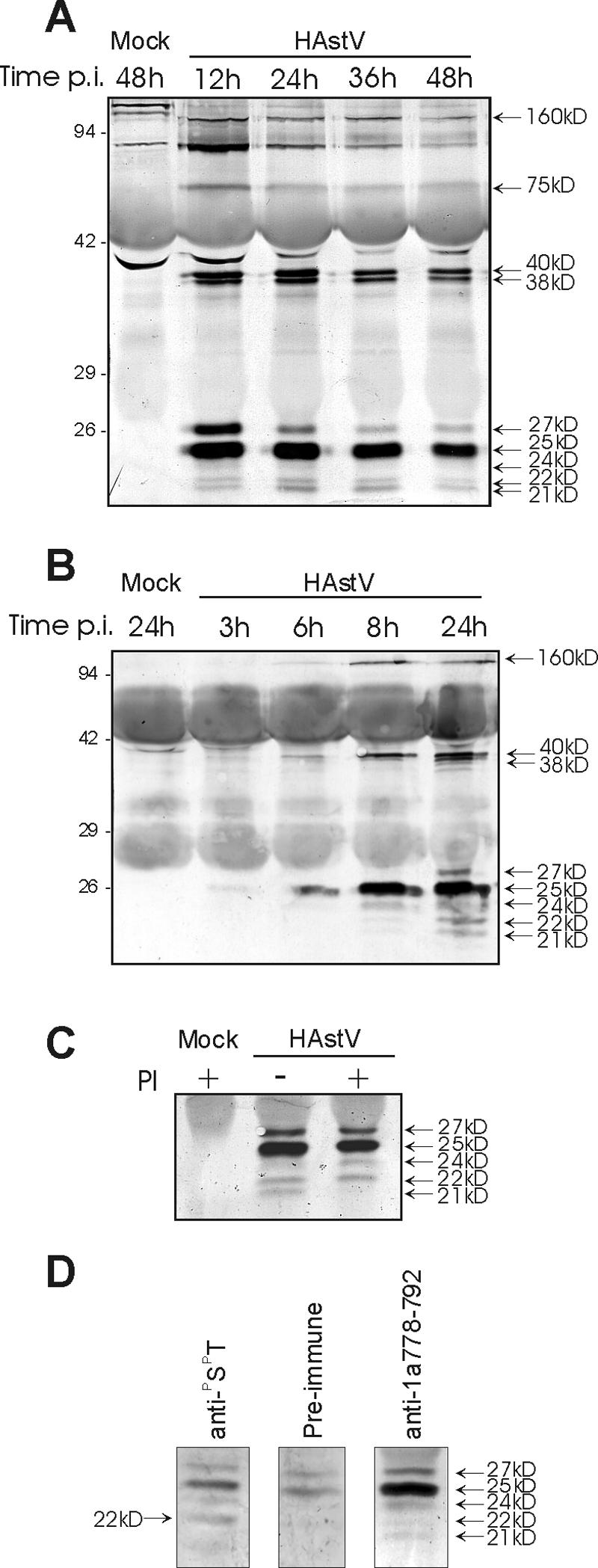
(A and B) Viral proteins immunoprecipitated with the nsP1a/4-specific antibody anti-1a778-792 and detected by Western blot with the same antibody. HAstV-infected and mock-infected CaCo-2 cell extracts were harvested at the indicated times postinfection (hours). The nonspecific staining observed in the mock-infected lane corresponds to mouse immunoglobulin chains of the antibodies used to immunoprecipitate, which are detected by the anti-mouse immunoglobulin-alkaline phosphatase conjugate antibody. (C) Immunoprecipitation experiment with the anti-1a778-792 antibody at 36 h postinfection, in the presence (+) or absence (−) of aprotinin and leupeptin protease inhibitors (PI) in the supernatant of infected cultures. Immunodetection was performed with the anti-1a778-792 antibody. (D) Analysis of phosphorylated proteins at 24 h postinfection, after immunoprecipitation with a specific anti-PSPT monoclonal antibody. A preimmune serum and the anti-1a778-792 antibody were used as negative and positive controls, respectively. In all three cases,immunodetection was performed with the anti-1a-778-792 antibody. Proteins were separated in 12% (A) or 15% (B, C, and D) polyacrylamide gels. The positions of molecular size markers are indicated on the left (in kilodaltons), and viral proteins are indicated on the right.
In order to elucidate the relationship between the different smallest nsP1a/4 forms, shorter kinetic experiments and analysis of the presence of posttranslational modifications were carried out. Since all proteins were detected at 12 h postinfection and remained stable up to at least 48 h postinfection, shorter postinfection periods were analyzed (Fig. 2B). A protein of ≈25 kDa could be detected as soon as 3 h after infection, indicating that the synthesis and processing of the polyproteins had already taken place and suggesting the occurrence of very early efficient proteolytic processing. Interestingly, the kinetics of the ≈27-kDa protein was slightly different compared to the other four proteins of similar size. While the first detectable protein was the one of ≈25 kDa, being the smaller bands detected later on in a progressive form up to 12 h postinfection, the ≈27-kDa protein was never detected before 12 h postinfection These results suggested that the ≈27-kDa protein would not be a precursor of the smaller proteins.
The addition of serine and cysteine protease inhibitors to the culture medium (Fig. 2C) did not prevent the generation of the 27-, 25-, and 22-kDa proteins, which were likewise detected, but it did affect the amount of 24- and 21-kDa proteins, indicating that the 21-kDa protein could correspond to a processed form of the 24-kDa protein. In contrast, unlike the 21-kDa polypeptide, the 22-kDa protein does not seem to result from the proteolytic processing of the 24-kDa protein; neither cleavage site that generates the 25- and 27-kDa proteins would be sensitive to protease inhibition. These data suggest that these cleavages could be produced by a cis-acting protease, which in general are more resistant to inhibitors, or that they are produced by a protease whose action is not inhibited by aprotinin and/or leupeptin. No modifications were observed on the larger intermediate forms.
To evaluate whether some of the 21- to 27-kDa nsP1a/4 forms were glycosylated, cell extracts immunoprecipitated with the anti-1a778-792 antibody were analyzed by periodate oxidation and biotin-hydrazide modification on a Western blot. However, none of the proteins obtained by immunoprecipitation were labeled. Finally, immunoprecipitation experiments with a monoclonal antibody against phosphorylated serine and threonine residues revealed that the 22-kDa protein was phosphorylated. Figure 2D shows immunoprecipitation results obtained from infected cell extracts at 24 h postinfection with a specific anti-PSPT monoclonal antibody. A preimmune serum and the anti-1a778-792 antibody were used as negative and positive controls, respectively. While the 27- and 25-kDa proteins were detected even in the negative control as a background level due to the high sensitivity of the anti-1a778-792 antibody used in the Western immunoblotting, the 22-kDa protein was specifically immunoprecipitated by the anti-PSPT antibody, indicating that this protein contains phosphorylated serine and/or threonine residues.
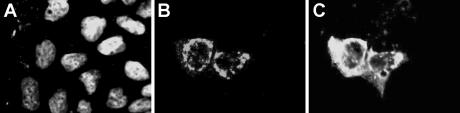
The subcellular localization of the predicted nsP1a C-terminal nsP1a/4 protein and the structural viral proteins were studied by double immunofluorescence staining at 48 h postinfection with the mouse anti-1a778-792 and a rabbit polyclonal antibody against purified astrovirus particles. Both nsP1a/4 and structural proteins were mostly detected in the cytoplasm of infected cells (Fig. 3), although some weak nuclear staining could also be observed. While expression of structural proteins resulted in a fluorescent signal widely distributed throughout the cytoplasm, nsP1a/4 staining showed prominent bright accumulations of stain concentrated along the perinuclear region, resulting in a punctate granular appearance, typical of membrane-derived aggregates. A kinetic analysis of expression over time indicated that while high levels of both nsP1a/4 and structural protein expression were evident at 24, 48, and 72 h postinfection, they could not be detected by immunofluorescence as early as 6 h postinfection (data not shown), indicating a lower sensitivity of immunofluorescence compared to immunoprecipitation to detect protein expression.
FIG. 3.
Immunolocalization analysis of HAstV proteins within infected CaCo-2 cells at 48 h postinfection (A) Nuclear staining with 4′,6′-diamidino-2-phenylindole. (B) Immunofluorescence to detect nsP1a/4 protein. (C) Immunofluorescence to detect viral structural proteins.
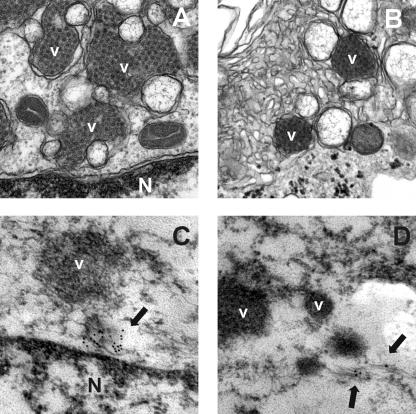
The ultrastructural analysis at 48 h postinfection revealed the presence of viral aggregates close to the nuclear periphery surrounded by a large number of double-membrane vacuoles (Fig. 4A and B). The fact that such a large amount of vacuoles was not observed in uninfected CaCo-2 cells indicates that these vesicles may be induced by astrovirus infection. Cryoimmunoelectron analysis after mild fixation showed immunostaining with the mouse anti-1a778-792 antibody, associated with small filamentous membranes accumulated at the perinuclear region (Fig. 4C and D).
FIG. 4.
Electron microscopy studies. Ultrastructural analysis was performed on HAstV-infected cells at 48 h postinfection (A and B). nsP1a/4 protein was detected by immunogold localization during infection (C and D). N, nucleus; V, viral aggregates. Arrows indicate areas positive for immunostaining.
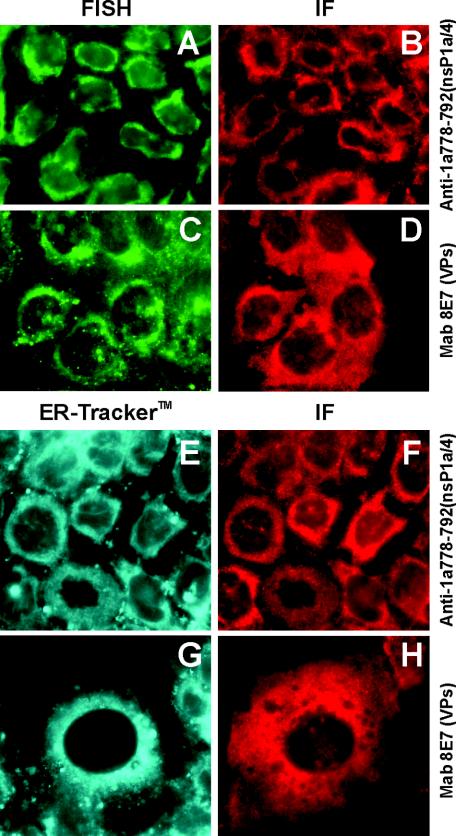
Viral RNA localization within infected cells was studied by fluorescence in situ hybridization with a digoxigenin-labeled DNA probe. Viral structural or nsP1a/4 proteins were concomitantly detected by immunofluorescence. Interestingly, it could be observed that nsP1a/4 protein completely colocalized with viral RNA (Fig. 5A and B), while structural proteins were widely dispersed along the entire cytoplasm (Fig. 5C and D).
FIG. 5.
Double-labeling analysis of viral RNA and viral proteins (A, B, C, and D), and double-labeling analysis of the endoplasmic reticulum and viral proteins (E, F, G, and H). RNA was detected by fluorescence in situ hybridization (FISH, A and C), the intracellular distribution of the endoplasmic reticulum was analyzed with the ER-Tracker (E and G), and viral proteins were concomitantly detected by immunofluorescence (IF) with either anti-1a-778-792 antibody for nsP1a/4 (B and F) or monoclonal antibody 8E7 for the structural proteins (VPs) (D and H). Photographs were generated with the same exposure times.
In order to identify the cellular compartment with which both nsP1a/4 and viral RNA were associated, and having in mind the occurrence of an endoplasmic reticulum retention signal, a series of colocalization studies with an endoplasmic reticulum tracker were undertaken. A precise localization of the nsP1a/4 protein at the endoplasmic reticulum around the perinuclear region was observed (Fig. 5E and F). In contrast, the structural protein staining was located not only at the perinuclear region but also through the complete cytoplasmic space (Fig. 5G and H).
DISCUSSION
This work deals with the molecular characterization of the putative HAstV nonstructural protein at the C terminus of nsP1a, which contains the hypervariable region and which in this work has been named nsP1a/4. Our data provide clear evidence that nsP1a/4 protein is localized close to the endoplasmic reticulum and viral RNA. Analysis of the amino acid composition of the nsP1a/4 protein revealed the presence of acidic regions and of glutamine- and proline-rich regions which have been generally related to motifs for transcriptional activation domains (38). Many putative glycosylation and phosphorylation sites and a KKXX-like endoplasmic reticulum retention signal were also identified. KKXX-like motifs (KKXX or XKXX, with lysine residues at the −3 or −3 and −4 positions from the C terminus) are present in type I integral membrane proteins, and they function as an efficient endoplasmic reticulum retrieval and retention signal (56).
At present, different available studies on HAstV nonstructural polyprotein processing suggest two possible protease cleavage sites between the protease domain and the end of the nsP1a polyprotein (11, 24). Our immunoprecipitation studies with the anti-1a778-792 nsP1a/4-specific antibody revealed the presence of polyprotein precursors of 160, 75, and 38 to 40 kDa, as well as five smaller proteins in the range of 21 to 27 kDa. Two cleavage sites yielding the nsP1a/4 have been proposed at positions Gln-567/Thr-568 (24) and Glu-654/Ile-655 (11), which would render products of 40.1 and 30.6 kDa, respectively. Our findings indicate that the C-terminal end of nsP1a must be further processed. With antibodies directed against a broader region of the nsP1a C terminus in a transient expression system on BHK-21 cells, other authors detected a 20-kDa protein, which was not observed with CaCo-2-infected cells at 12 h postinfection, probably due to further proteolytic processing of the protein (36). In addition, with similar approaches and longer incubation times, Willcocks et al. reported proteins of 34, 20, 6.5, and 5.5 kDa (60).
In our study, the most abundant proteins detected were in the range of 21 to 27 kDa. Occasionally, in some gels we also observed a smaller protein of ≈15 kDa (data not shown). Protein precursors of 160 and 75 kDa, which could correspond to the nsP1a/1b and to an N-terminally cleaved form of nsP1a polyprotein, respectively, have also been reported by other authors (24, 36, 60). The 38- to 40-kDa intermediates could correspond to intermediates covering amino acids between the end of the protease and the end of nsP1a (11). Different explanations could account for the five proteins in the range of 21 to 27 kDa detected. These polypeptides could correspond to posttranslationally modified forms of the same protein, or they could also be the result of differential cleavage during the proteolytic processing of the precursor protein. Our results suggest that a combination of both phenomena may occur during astrovirus infection. While only some of these proteins are indeed proteolytically related, such as the 24- and the 21-kDa proteins, the amounts of the other three proteins did not vary in the presence of protease inhibitors. Additionally, the present study also showed that the 22-kDa protein contains phosphorylated serine and threonine residues. In contrast, it was not possible to detect carbohydrates in any of the viral proteins obtained after immunoprecipitation, but this option cannot be totally ruled out due to the insufficient sensitivity of the glycoprotein staining method used. Further studies are required to ascertain the specific functional effect of posttranslational modifications on nsP1a/4 protein biological activity.
The results produced by the use of proteomics computational tools, together with the intracellular distribution of the nsP1a/4 protein in association with endoplasmic reticulum-derived membranous structures close to the perinuclear region, suggest the involvement of this protein in viral RNA replication through specific RNA-protein and/or protein-protein interactions. In fact, in other RNA viruses such as picornaviruses and several plant viruses (6, 48), RNA replication is associated with membranes derived from the endoplasmic reticulum. However, since the double-stranded DNA probe used in our fluorescence in situ hybridization analysis could detect negative-stranded replication intermediate RNA as well as assembled genomic RNA, the possibility that nsP1a/4 protein is also involved in virus morphogenesis or could form part of the assembled virions cannot be completely ruled out.
Most of the proteins from other viral families that showed sequence similarity to HAstV nsP1a/4 belonged to single-stranded RNA viral families and were related to the formation of the viral replication complex, to the regulation of viral replication, and/or to transcription (8, 9, 10, 26, 30, 31, 42, 43, 45, 46, 49, 50, 51, 52, 54, 55, 57, 58, 61). This observation provides additional support to the idea that nsP1a/4 protein may be involved in RNA replication. Of note, added to sequence similarity, HAstV nsP1a/4 protein shared some other features with all these proteins, such as, for example, a high degree of tolerance to insertions and deletions (51, 54), the occurrence of phosphorylation sites (30, 46, 52, 55) and immunogenic sequences (42), and intracellular colocalization with viral replication sites (31, 49).
Replication of the genomes of all well-characterized positive-strand RNA viruses, including plant, animal, and insect viruses, occurs in large replication complexes associated with intracellular membranes (5), and an increasing number of host cell proteins have been reported to be associated with these complexes (1), involving the establishment of specific interactions between host and viral proteins. The RNA replication complex of HAstV has been suggested to be anchored to the membrane through the transmembrane domains at the N-terminal region of the nsP1a product (21). The KKXX-like motif described in this report at the end of nsP1a could be responsible for retention of the entire polyprotein at endoplasmic reticulum-derived membranes, although it might be functional only after processing of the polyprotein. However, it is still unclear whether this signal would remain included as part of the nsP1a/4 protein after its processing, since any of the cleavages required to generate the 21- to 27-kDa forms could be located between amino acids corresponding to the synthetic peptide used to produce the antibody and the endoplasmic reticulum retention signal.
Moreover, it is also unclear whether the nsP1a-derived proteins would remain anchored to membranes after proteolytic processing of the nsP1a polyprotein. In our double labeling experiments, which were performed at 48 h postinfection, when the nsP1a/4 protein seems to be mainly in its 21- to 27-kDa processed forms, colocalization was also observed with the endoplasmic reticulum, suggesting either that these proteins keep the endoplasmic reticulum signal or that they are retained at the membrane complex even after proteolytic processing, by interaction with other nonstructural viral proteins, particularly those derived from the N-terminal region of nsP1a, where hydrophobic transmembrane regions have been identified. The elucidation of the interplay between polyprotein processing, phosphorylation and/or glycosylation, and membrane association to regulate the main functions of nsP1a/1b-encoded proteins will be an important issue in future studies of astrovirus replication. In other RNA viruses, the temporary regulation of protein processing of the replicase complex is necessary to regulate minus- and plus-strand RNA synthesis processes (27).
Since nsP1a/4 could be necessary for the efficient formation of the replication complex through a direct interaction with the viral RNA or with other proteins of the complex, mutations in this protein, specifically within the hypervariable region, could easily affect either virual replicative efficiency or the molecular mechanisms of pathogenicity. Work is currently in progress to elucidate the effect of nsP1a/4 genetic variability on the replicative properties of different human astrovirus mutants.
Acknowledgments
S. Guix was the recipient of an FI fellowship from the Generalitat de Catalunya. This work was supported in part by grant QLRT-1999-0594 from the European Union, 1997/SGR/00224 and 2001/SGR/00098 from the Generalitat de Catalunya, and the Centre de Referència de Biotecnología de Catalunya (CeRBa), Generalitat de Catalunya.
We acknowledge the technical expertise of the Serveis Científic-Tècnics of the University of Barcelona.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 3.Blom, N., J. Hansen, D. Blaas, and S. Brunak. 1996. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 5:2203-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brendel, V., P. Bucher, I. Nourbakhsh, B. E. Blaisdell, and S. Karlin. 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci. USA 89:2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucks, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plant and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carette, J. E., M. Stuiver, van Lent, J., J. Wellink, and A. van Kammen. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NSP@: network protein synthesis analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 8.Cotillon, A. C., M. Girard, and S. Ducouret. 2002. Complete nucleotide sequence of the genomic RNA of a French isolate of Pepino mosaic virus (PepMV). Arch. Virol. 147:2231-2238. [DOI] [PubMed] [Google Scholar]
- 9.Dé, I., C., Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 77:13106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaire, F., Schmitt, C., Stussi-Garaud, C., L. Pinck, and C. Ritzenthaler. 1999. Protein 2A of Grapevine Fanleaf Nepovirus is implicated in RNA2 replication and colocalizes to the replication site. Virology 264:25-36. [DOI] [PubMed] [Google Scholar]
- 11.Geigenmüller, U., T. Chew, N. Ginzton, and S. M. Matsui. 2002. Processing of nonstructural protein 1a of human astrovirus. J. Virol. 76:2003-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson, C. A., J. Chen, S. A. Monroe, and M. R. Denison. 1998. Expression and processing of nonstructural proteins of the human astroviruses. Adv. Exp. Med. Biol. 440:387-391. [DOI] [PubMed] [Google Scholar]
- 13.Glass, R. I., J. Noel, D. Mitchell, J. E. Herrmann, N. R. Blacklow, L. K. Pickering, P. Dennehy, G. Ruiz-Palacios, M. L. de Guerrero, and S. S. Monroe. 1996. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch. Virol. (Suppl.) 12:287-300. [DOI] [PubMed] [Google Scholar]
- 14.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 15.Guix, S., A. Bosch, E. Ribes, Dora Martinez, L., and R. M. Pintó. 2004. Apoptosis in astrovirus-infected CaCo-2 cells. Virology 319:249-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guix, S., S. Caballero, C. Villena, R. Bartolomé, C. Latorre, N. Rabella, M. Simó, A. Bosch, and R. M. Pintó. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta, R., and S. Brunak. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pacific Symposium on Biocomputing 7:310-322. [PubMed] [Google Scholar]
- 18.Higgins, D., J. Thompson, J. Gibson, D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA. 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonassen, C. M., T. O. Jonassen, T. M. Sveen, and B. Grinde. 2003. Complete genomic sequences of astroviruses from sheep and turkey: comparison with related viruses. Virus Res. 91:195-201. [DOI] [PubMed] [Google Scholar]
- 23.Kelley, L. A., R. M. MacCallum, and M. J. E. Sternberg. 2000. Enhanced Genome Annotation using Structural Profiles in the Program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 24.Kiang, D., and S. M. Matsui. 2002. Proteolytic processing of a human astrovirus nonstructural protein. J. Gen. Virol. 83:25-34. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetics analysis. Pennsylvania State University, University Park, Pa.
- 26.LaStarza, M. W., J. A. Lemm, and C. M. Rice. 1994. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 68:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis, T. L., H. B. Greenberg, J. E. Herrmann, L. X. Smith, and S. M. Matsui. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 68:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, T. L., and S. M. Matsui. 1996. Astrovirus ribosomal frameshifting in a infection-transfection transient expression system. J. Virol. 70:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, G., LaStarza, M. W., W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 33.Matsui, S. M., and H. B. Greenberg. 2001. Astroviruses, p. 875-893. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Matsui, S. M., D. Kiang, N. Ginzton, T. Chew, and U. Geigenmüller-Gnirke. 2001. Molecular biology of astroviruses: selected highlights. Novartis Found. Symp. 238:219-233. [DOI] [PubMed] [Google Scholar]
- 35.Matsui, S. M., J. P. Kim, H. B. L. Greenberg, M. L. Young, S. Smith, T. L. Lewis, J. E. Herrmann, N. R. Blacklow, K. Kupuis, and G. R. Reyes. 1993. Cloning and characterization of human astrovirus immunoreactive epitopes. J. Virol. 67:1712-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Méndez, E., Salas-Ocampo, M. P., M. E. Munguía, and C. F. Arias. 2003. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J. Virol. 77:11378-11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Méndez-Toss, M., P. M. Romero-Guido, E. Munguia, E. Méndez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 38.Mermod, N., E. A. O'Neil, T. J. Kelly, and R. Tjian. 1989. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell 58:741-753. [DOI] [PubMed] [Google Scholar]
- 39.Monroe, S. S., Stine, S. E., L. Gorelkin, J. E. Herrmann, N. R. Blacklow, and R. I. Glass. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J. Virol. 65:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 41.Oh, D., and E. Schreier. 2001. Molecular characterization of human astroviruses in Germany. Arch. Virol. 146:443-455. [DOI] [PubMed] [Google Scholar]
- 42.Oleksiewicz, M. B., A. Botner, T. P. Normann, and T. Storgaard. 2001. Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsP2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 75:3277-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panaviené Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191-205. [DOI] [PubMed] [Google Scholar]
- 44.Pintó, R. M., J. M. Díez, and A. Bosch. 1994. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J. Med. Virol. 44:310-315. [DOI] [PubMed] [Google Scholar]
- 45.Realdon, S., M. Gerotto, Dal Pero, F., O. Marin, A. Granato, G. Basso, M. Muraca, and A. Alberti. 2004. Proapoptotic effect of hepatitis C virus CORE protein in transiently transfected cells is enhanced by nuclear localization and is dependent of PKR activation. J. Hepatol. 40:77-85. [DOI] [PubMed] [Google Scholar]
- 46.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 47.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 48.Rust, R. C., L. Landman, R. Cosert, B. L. Tang, W. Hong, and H.-P. Hauri. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnitzler, P., M. Hug, M. Handermann, W. Janssen, E. V. Koonin, H. Delius, and G. Darai. 1994. Identification of genes encoding zinc finger proteins, non-histone chromosomal HMG protein homologue, and a putative GTP phosphohydrolase in the genome of Chilo iridescent virus. Nucleic Acids Res. 22:158-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, S., K. W. Liu, and D. X. Liu. 2000. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the nsP2 gene with a unique insertion. Arch. Virol. 145:871-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shih, C. M., C. M. Chen, S. Y. Chen, and Y. H. Wu Lee. 1995. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J. Virol. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, T. F., and M. W. Waterman. 1981. Identification of common molecular sequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 54.Tuttila, M., and A. E. Hinkkanen. 2003. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neuvovirulence. J. Gen. Virol. 84:1525-1533. [DOI] [PubMed] [Google Scholar]
- 55.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kääriäinen. 2001. Elimination of phosphorylation sites of Semliki Forest Virus replicase protein nsP3. J. Biol. Chem. 276:5747-5752. [DOI] [PubMed] [Google Scholar]
- 56.Vincent, M. J., A. S. Martin, and R. W. Compans. 1998. Function of the KKXX motif in endoplasmic reticulum retrieval of a transmembrane protein depends on the length and structure of the cytoplasmic domain. J. Biol. Chem. 273:950-956. [DOI] [PubMed] [Google Scholar]
- 57.Wassenaar, A. L. M., Spaan, W. J. M., A. E. Gorbalenya, and E. J. Snijder. 1997. Alternative proteolytic processing of the arteirivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Viol. 71:9313-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webster, L. C., K. Zhang, B. Chance, I. Ayene, J. S. Culp, W. J. Huang, Y. Wu, and R. P. Ricciardi. 1991. Conversion of the E1A Cys4 zinc finger to a nonfunctional His2,Cys2 zinc finger by a single point mutation. Proc. Natl. Acad. Sci. USA 88:9989-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willcocks, M. M., N. Ashton, J. B. Kurtz, W. D. Cubitt, and M. J. Carter. 1994. Cell culture adaptation of astrovirus involves a deletion. J. Virol. 68:6057-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willcocks, M. M., A. S. Boxall, and M. J. Carter. 1999. Processing and intracellular location of human astrovirus non-structural proteins. J. Gen. Virol. 80:2607-2611. [DOI] [PubMed] [Google Scholar]
- 61.Yan, L., M. Velikanov, P. Flook, W. Zheng, S. Szalma, and S. Kahn. 2003. Assessment of putative protein targets derived from the SARS genome. FEBS Lett. 554:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]