Abstract
Four cyclopentenone-containing ansamycin polyketides (mccrearamycins A–D), and six new geldanamycins (Gdms B–G, including new linear and mycothiol conjugates), were characterized as metabolites of Streptomyces sp. AD-23-14 isolated from the Rock Creek underground coal mine acid drainage site. Biomimetic chemical conversion studies using both simple synthetic models and Gdm D confirmed that the mccrearamycin cyclopentenone derives from benzilic acid rearrangement of 19-hydroxy Gdm, and thereby provides a new synthetic derivatization strategy and implicates a potential unique biocatalyst in mccrearamycin cyclopentenone formation. In addition to standard Hsp90α binding and cell line cytotoxicity assays, this study also highlights the first assessment of Hsp90α modulators in a new axolotl embryo tail regeneration (ETR) assay as a potential new whole animal assay for Hsp90 modulator discovery.
Keywords: ansamycin, axolotl, biomimetic synthesis, Hsp90, regeneration
Mining for geldanamycins
Six new geldanamycins (Gdms) and four new ring-contracted cyclopentenone macrolactams are reported as metabolites of an abandoned Kentucky coal mine-associated microbe. A biosynthetic pathway via benzilic acid rearrangement is proposed and an axolotl embryo tail regeneration assay is utilized to assess the Hsp90 inhibitory activities of the metabolites.
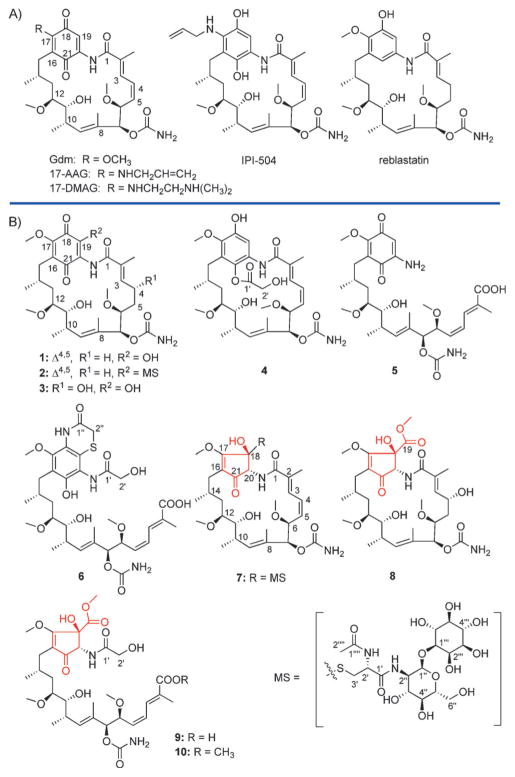
Geldanamycin (Gdm)-type polyketides are prototypical microbial benzoquinone ansamycin anticancer agents that target the N-terminal ATP-binding domain of heat shock protein 90 (Hsp90; Figure 1A).[1] While a number of elegant and efficient Gdm synthetic and biosynthetic production and derivatization strategies have been developed,[2] C-17 semi-synthetic Gdm modification was a key to both first (tanespimycin/17-AAG[3] and orally bioavailable alvespimicin/17-DMAG)[4] and second (retaspimycin hydrochloride/IPI-504)[5] generation analogues advanced to the clinic (Figure 1A), the latter of which displayed improved solubility and reduced hepatotoxicity.[6] More recent medicinal chemistry efforts have focused on C-19 substitution to prohibit non-specific alkylation (a putative contributor to non-selective toxicity), analogues of which were found to opportunistically favor the cis-amide conformer observed in the Gdm-Hsp90 ligand-bound complex.[7] As part of a microbial natural products discovery effort from coal-mining-associated environments in Kentucky, USA,[8] herein we describe the isolation and structure elucidation of six new Gdm analogues (1–6), and four unprecedented ring-contracted cyclopentenone macrolactams (mccrearamycins A–D, 7–10) from the Rock Creek (McCreary County) underground coal mine acid drainage isolate Streptomyces sp. AD-23-14 (Figure 1B). Biomimetic studies using both simple synthetic models and isolated Gdm analogues revealed the ortho-quinone to undergo a facile benzilic acid rearrangement to provide the ring-contracted cyclopentenone scaffold, presenting both a new synthetic strategy and implicating the role of a potential novel biocatalyst for ansamycin ring contraction. In addition to expanding Hsp90α inhibitor SAR, these studies also highlight the first assessment of Hsp90α modulators in a new axolotl (Ambystoma mexicanum) embryo tail regeneration (ETR) assay.[9]
Figure 1.
A) Chemical structures of representative Gdm-type ansamycins and B) new compounds isolated from Streptomyces sp. AD-23-14. The unique cyclopentenone ring structure of mccrearamycins A–D is highlighted in red.
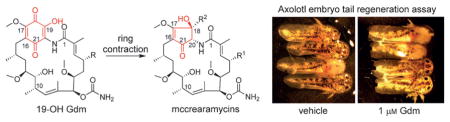
Gdms B–G (1–6) were characterized as new Gdm analogues (including mycothiol conjugate 2 and linear Gdms 5–6) based on NMR, MS, and comparison with literature precedent (see Figure 1 and the Supporting Information). While 7–10 also shared the signature spectral features of Gdm 19-membered macrolactams (Figures S2–S4), they notably lacked indicators of the corresponding Gdm 1,4-benzoquinone. Key HMBC correlations [for example, for 7, from a nitrogen-bearing CH (δH = 5.12 ppm, 20-H) to C-16 (δC = 116.2 ppm), C-17 (δC = 177.0 ppm) and C-18 (δC = 71.1 ppm), and from 18-OH (δH = 6.16 ppm) to C-17 and C-18] implicated an unprecedented alternative cyclopentenone ring (Figure 1B) in 7–10. Determination of C-18 substitution (MSH in 7; methyl formate in 8–10) relied on HMBC correlations (Figures S2 and S3). The relative configurations of 7–10 were established through NOESY (Figures S3 and S4) where many observed modifications paralleled those of corresponding Gdm analogues. Namely, like 3 (Gdm D), hydration of the 8 C-4/C-5 double bond was observed, and similar to 6 (Gdm G), 9 and 10 were also identified as N-20-acyl (2-hydroxy-acetate) linear metabolites (Tables S1 and S4, Figures S2–S4). These cumulative analyses established 7–10 as new ring-contracted cyclopentenone macrolactams and thus were named mccrearamycins A–D in reference to the structural novelty and the producing strain3s point of origin.
The similarities between Gdms and mccrearamycins from Streptomyces sp. AD-23-14 implicated Gdms as potential mccrearamycin progenitors (Scheme 1). In addition, while NOESY firmly established the cyclopentenone C-18/C-20 relative trans-configuration in 7, the key 1H NMR resonance for 18-OH was lacking for 8–10. For further validation, a model study was pursued to assess cyclopentenone formation via ring contraction of a 19-OH Gdm progenitor (Scheme 1) reminiscent of the classical cyclohexanone to cyclopentane-1-carboxylate benzilic acid rearrangement.[10] While the corresponding Gdm rearrangement is unprecedented, the analogous Hooker oxidation rearrangement of hydroxynaphthoquinones to indane carboxylic acids served as related precedent.[11] For this study, the synthesis of the Gdm model surrogate 2-hydroxyquinone 21 (Scheme 2) commenced with aryl lithiation–alkylation of benzyl methyl ether 12. DDQ-mediated oxidation of 13 followed by hydroxy-directed iodination provided phenol 15. The iodide was then treated with copper powder in basic medium to provide catechol 16, which was selectively methylated by Me2SO4. Methoxymethyl protection of the remaining phenolic hydroxyl followed by Baeyer–Villiger oxidation produced the key intermediate 18. Consistent with challenges associated with hexasubstituted benzene syntheses,[12] amination, amidation, and nitration of 18 directly, or of corresponding halogenated derivatives using transition-metal catalysts, failed to give desired aniline 19 or amide 20. However, azo coupling with sulfanilic acid,[13] followed by dithionite reduction, gave aniline 19 in 62% yield. Sequential acetylation, hydrolysis, oxidation, and deprotection furnished template 21 in 73% yield, and methylation of 21 further afforded the corresponding 2-methoxy quinone 22 as an additional comparator.
Scheme 1.
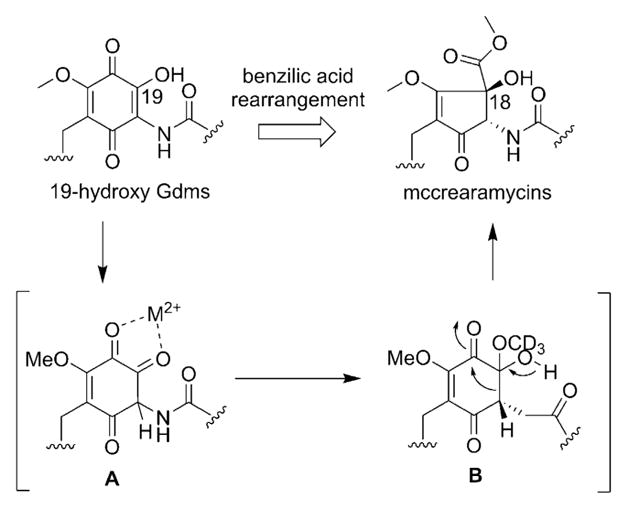
Proposed metal (M2+)-mediated benzilic acid rearrangement of the Gdm hydroxyquinone to afford the mccrearamycin cyclopentenone.
Scheme 2.
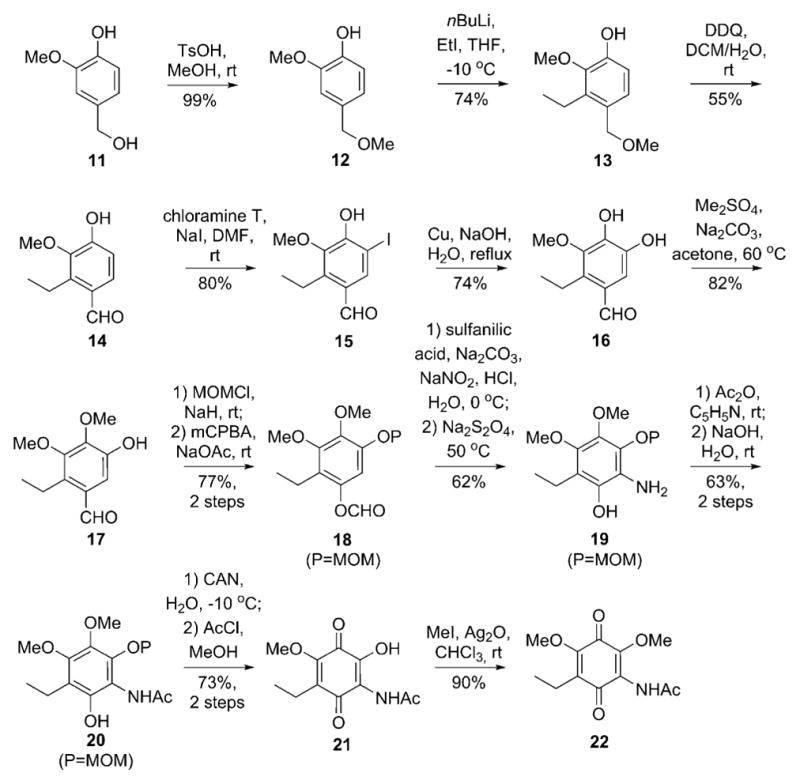
Synthesis of templates 21 and 22.
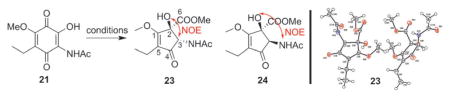
Consistent with the impact of CuCl2 on benzilic acid rearrangement stereoselectivity and yield,[14] evaluation of the putative 21 benzilic acid rearrangement in the presence of transition metal salts and various other known benzilic acid rearrangement promoters revealed CoCl2 to afford the best overall yield and stereoselectivity (Table 1). Single-crystal X-ray diffraction of the isolated product 23 further established the relative C-2/C-3 trans-configuration (Table 1 and S7, CCDC 1496415), consistent with the signature 23 2-OH to 3-CH NOE and corresponding 18-OH to 20-CH NOE of mccrearamycin A (7). A putative mechanism for Co2+-assisted benzilic acid rearrangement is depicted in Scheme 1. Consistent with this mechanism, the substitution of CH3OH with CD3OD as solvent led to selective isotopic label incorporation in 25 (entry 18, Table 1). Importantly, the 2-methoxy model 22 and the prototypical Hooker reaction substrate lawsone failed to give the desired benzilic acid rearrangement under the optimized conditions (Scheme S1).
Table 1.
Optimization of benzilic acid rearrangement and biomimetic conversion of 3 to 8.
| ||||||
|---|---|---|---|---|---|---|
| Entry[a] | Additives | Temp. [°C] | Solvent | Time [h] | Ratio[b] (23/24) | Yield[c] of 23 |
| 1 | none | 50 | MeOH/CH2Cl2 | 16 | –[d] | |
| 2 | KOtBu | 50 | MeOH/CH2Cl2 | 16 | –[d] | |
| 3 | DBU | 50 | MeOH/CH2Cl2 | 16 | 2:1 | 51% |
| 4 | TEA | 50 | MeOH/CH2Cl2 | 16 | 5:1 | 60% |
| 5 | DABCO | 50 | MeOH/CH2Cl2 | 16 | 1:1 | |
| 6 | DIPEA | 50 | MeOH/CH2Cl2 | 16 | 4:1 | 55% |
| 7 | CoCl2 | 50 | MeOH/CH2Cl2 | 16 | >10:1 | 73% |
| 8 | NiCl2 | 50 | MeOH/CH2Cl2 | 16 | –[e] | |
| 9 | CuCl2 | 50 | MeOH/CH2Cl2 | 16 | –[e] | |
| 10 | AgOTf | 50 | MeOH/CH2Cl2 | 16 | 1:5 | 39%[f] |
| 11 | Au(PPh3)Cl | 50 | MeOH/CH2Cl2 | 16 | –[d] | |
| 12 | Co(OAc)2 | 50 | MeOH/CH2Cl2 | 16 | ND[g] | 24% |
| 13 | Co(acac)2 | 50 | MeOH/CH2Cl2 | 16 | ND[g] | 15% |
| 14 | CoCl2 | 50 | MeOH/CHCl3 | 16 | ND[g] | 29% |
| 15 | CoCl2 | 50 | MeOH/DCE | 16 | >10:1 | 31% |
| 16 | CoCl2 | 50 | MeOH | 40 | >10:1 | 82% |
| 17 | CoCl2 | 80 | MeOH | 16 | >10:1 | 85% |
| 18 |

|
|||||
| 19 |
|
|||||
Reactions contained substrate (0.05 mmol) and additives (0.1 mmol) in 0.5 mL solvent under specified conditions with product formation subsequently assessed by analytical HPLC and NMR (key NOE signatures are highlighted, see the Supporting Information for experimental details).
Based on analytical HPLC peak integration.
Yields of isolated products.
No reaction.
Undefined mixture.
Yield of isolated product 24.
Not determined.
To probe the relevance to mccrearamycins, this biomimetic model study was subsequently extended to the corresponding 19-hydroxy-substituted Gdm D (3). Remarkably, reaction of 3 under the same optimized conditions led to 50% conversion to mccrearamycin B (8; entry 19, Table 1 and Figures S6–S8). The established stereoselectivity of the model reaction implicates an 8 cyclopentenone C-18/C-20 trans-configuration identical to that of 7 and 23. Comparison of select 13C NMR chemical shifts in mccrearamycins B–D (8–10) to that of the trans- and cis-configured models (23 and 24, respectively) provide further support of a common benzilic acid rearrangement-derived C-18/C-20 trans-configuration in all of the mccrearamycins (Table S5). Subsequent indirect mccrearamycin absolute configuration assignment was accomplished through electronic circular dichroism (ECD) analysis. Specifically, comparison of the ECD spectra of 8 in MeOH to the theoretical ECD spectra [generated using time-dependent density functional theory (TDDFT)][8a,15] for two possible isomers of 8 (8a: 4R, 6S, 7S, 10S, 11R, 12S, 14R, 18S, 20S and 8b: 4R, 6S, 7S, 10S, 11R, 12S, 14R, 18R, 20R), revealed that of 8a as providing the best spectral match (Figure S9).
Based on the established mechanism of Gdm and related analogues, all of the isolated compounds were evaluated in standard Hsp90α inhibition[8b] and cancer cell line (human non-small cell lung A549) cytotoxicity assays (Table S7). This cumulative analysis revealed the parental prototypes (Gdm, reblastatin, and 17-O-demethyl-reblastatin) to afford greatest Hsp90α inhibition (IC50s of 5–30 nM) with notably divergent corresponding cytotoxicities (Gdm IC50 1 nM, reblastatin IC50 0.7 μM, and 17-O-demethyl-reblastatin IC50 > 50 μM), suggesting the oxidation state and substitution pattern contribute to differences in cellular uptake and/or alternative cytotoxicity mechanisms consistent with prior Gdm SAR studies.[7a,16] Similar to that of the parental prototypes, the corresponding cytotoxicity of the new Streptomyces sp. AD-23-14 metabolites did not correlate with Hsp90α inhibitory potential in some cases.
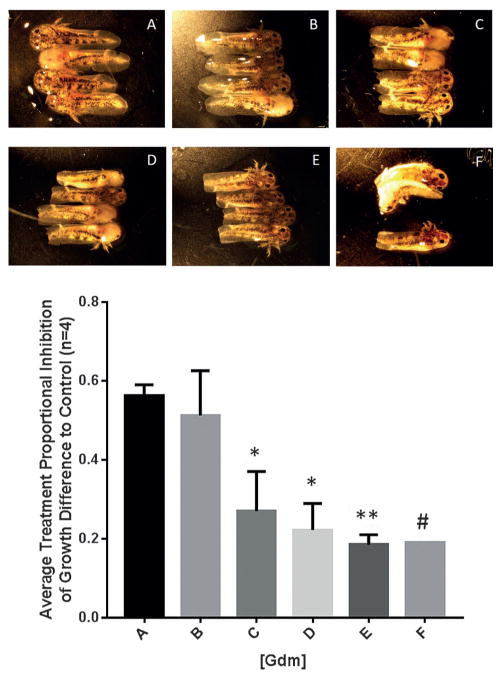
Streptomyces sp. AD-23-14 metabolites were also evaluated using a highly regenerative salamander model, the Mexican axolotl (Ambystoma mexicanum).[17] Previous transcriptional studies found hsp90aa1 to be significantly upregulated 12 hours after axolotl limb and tail amputation, suggesting a role for Hsp90 in tissue regeneration.[9] To investigate this further, we used the axolotl embryo tail regeneration (ETR) assay[9b] to test Gdm for an inhibitory effect on tail regeneration. Tail-amputated axolotl embryos were incubated in microtiter plates in the absence (vehicle control, DMSO) or presence of 10 μM agent (Gdm, reblastatin, 7-O-demethyl-reblastatin, and 1–10) and imaged on day 1 (pre-treatment) and day 7. An initial single dose screen revealed Gdm to completely inhibit tail regeneration with no effect observed for all of the other test agents. Subsequent studies revealed a clear dose-response for Gdm, with developmental abnormalities and toxicity observed at the highest dose (10 μM), inhibition of regeneration at intermediate doses, and no effect on regeneration at the lowest dose (0.1 μM; Figure 2).
Figure 2.
The impact of [Gdm] on axolotl embryo tail regeneration as determined by the ETR assay (*p <0.005, **p <0.0001, n =4; #: 3 axolotls were dead at day 7; A: DMSO control; B: 0.1 μM; C: 1 μM; D: 2.5 μM; E: 5 μM; F: 10 μM).
In summary, metabolic profiling led to the discovery of new Gdm analogues and a set of cyclopentenone macrolactams. The development and implementation of a cobalt-mediated benzilic acid rearrangement served as a key feature in mccrearamycin structure validation and highlights the potential synthetic utility in the context of 2-hydroxyquinone-containing complex natural products. That cyclopentenone formation requires distinct conditions may also implicate a unique biosynthetic pathway. These metabolites, together with the parental prototypes, also served as a test set to assess the impact of Hsp90 inhibitors in vivo using an axolotl ETR assay. While developmental abnormalities have been observed in many organisms (including zebrafish administered Gdm[18]) when Hsp90 activity is reduced below critical levels,[19] our results demonstrate that Gdm can be administered at a dose that blocks regeneration without overtly affecting development. This study implicates Gdm as a useful reagent to probe the role of Hsp90 in axolotl tail regeneration and suggests low dose Gdm could be used in a sensitized, ETR chemical genetic screen to identify new Hsp90 modulators.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R24 OD21479 (SRV, JST), T32 DA016176 (YZ), the University of Kentucky College of Pharmacy, the University of Kentucky Markey Cancer Center, and the National Center for Advancing Translational Sciences (UL1TR001998).
Footnotes
Conflict of interest
J.S.T. is a co-founder of Centrose (Madison, WI).
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: http://dx.doi.org/10.1002/anie.201612447.
Contributor Information
Dr. Xiachang Wang, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA).
Dr. Yinan Zhang, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA).
Dr. Larissa V. Ponomareva, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
Qingchao Qiu, Department of Biology, University of Kentucky, Lexington, KY 40506 (USA).
Dr. Ryan Woodcock, Department of Biology, University of Kentucky, Lexington, KY 40506 (USA)
Dr. Sherif I. Elshahawi, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
Dr. Xiabin Chen, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
Ziyuan Zhou, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA).
Bruce E. Hatcher, Kentucky Division of Abandoned Mine Lands, 300 Sower Blvd, Frankfort, KY 40601 (USA)
Prof. James C. Hower, Center for Applied Energy Research, University of Kentucky, Lexington, KY, 40511 (USA)
Prof. Chang-Guo Zhan, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
Dr. Sean Parkin, Department of Chemistry, University of Kentucky, Lexington, KY, 40506 (USA)
Prof. Madan K. Kharel, School of Pharmacy, University of Maryland Eastern Shore Princess Anne, Maryland 21853 (USA)
Prof. S. Randal Voss, Department of Biology, University of Kentucky, Lexington, KY 40506 (USA)
Dr. Khaled A. Shaaban, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
Prof. Jon S. Thorson, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, Kentucky 40536 (USA)
References
- 1.a) Rinehart KL, Jr, Sasaki K, Slomp G, Grostic MF, Olson EC. J Am Chem Soc. 1970;92:7591. doi: 10.1021/ja00729a018. [DOI] [PubMed] [Google Scholar]; b) DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. J Antibiot. 1970;23:442. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]; c) Whitesell L, Cook P. Mol Endocrinol. 1996;10:705. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]; d) Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Cell. 1997;90:65. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]; e) Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Proc Natl Acad Sci USA. 1994;91:8324. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Neckers L, Neckers K. Expert Opin Emerging Drugs. 2005;10:137. doi: 10.1517/14728214.10.1.137. [DOI] [PubMed] [Google Scholar]; g) Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 2.a) Andrus MB, Meredith EL, Simmons BL, Soma Sekhar BB, Hicken EJ. Org Lett. 2002;4:3549. doi: 10.1021/ol0267432. [DOI] [PubMed] [Google Scholar]; b) Andrus MB, Meredith EL, Hicken EJ, Simmons BL, Glancey RR, Ma W. J Org Chem. 2003;68:8162. doi: 10.1021/jo034870l. [DOI] [PubMed] [Google Scholar]; c) Qin HL, Panek JS. Org Lett. 2008;10:2477. doi: 10.1021/ol800749w. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Eichner S, Eichner T, Floss HG, Fohrer J, Hofer E, Sasse F, Zeilinger C, Kirschning A. J Am Chem Soc. 2012;134:1673. doi: 10.1021/ja2087147. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Patel K, Piagentini M, Rascher A, Tian ZQ, Buchanan GO, Regentin R, Hu Z, Hutchinson CR, McDaniel R. Chem Biol. 2004;11:1625. doi: 10.1016/j.chembiol.2004.09.012. [DOI] [PubMed] [Google Scholar]; f) Kim W, Lee D, Hong SS, Na Z, Shin JC, Roh SH, Wu CZ, Choi O, Lee K, Shen YM, Paik SG, Lee JJ, Hong YS. ChemBioChem. 2009;10:1243. doi: 10.1002/cbic.200800763. [DOI] [PubMed] [Google Scholar]; g) Eichner S, Floss HG, Sasse F, Kirschning A. ChemBioChem. 2009;10:1801. doi: 10.1002/cbic.200900246. [DOI] [PubMed] [Google Scholar]; h) Lee K, Ryu JS, Jin Y, Kim W, Kaur N, Chung SJ, Jeon YJ, Park JT, Bang JS, Lee HS, Kim TY, Lee JJ, Hong YS. Org Biomol Chem. 2008;6:340. doi: 10.1039/b713407j. [DOI] [PubMed] [Google Scholar]
- 3.Le Brazidec JY, Kamal A, Busch D, Thao L, Zhang L, Timony G, Grecko R, Trent K, Lough R, Salazar T, Khan S, Burrows F, Boehm MF. J Med Chem. 2004;47:3865. doi: 10.1021/jm0306125. [DOI] [PubMed] [Google Scholar]
- 4.a) Tian ZQ, Liu Y, Zhang D, Wang Z, Dong SD, Carreras CW, Zhou Y, Rastelli G, Santi DV, Myles DC. Bioorg Med Chem. 2004;12:5317. doi: 10.1016/j.bmc.2004.07.053. [DOI] [PubMed] [Google Scholar]; b) Jez JM, Chen JC, Rastelli G, Stroud RM, Santi DV. Chem Biol. 2003;10:361. doi: 10.1016/s1074-5521(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.Hanson BE, Vesole DH. Expert Opin Invest Drugs. 2009;18:1375. doi: 10.1517/13543780903158934. [DOI] [PubMed] [Google Scholar]
- 6.a) Wagner AJ, Chugh R, Rosen LS, Morgan JA, George S, Gordon M, Dunbar J, Normant E, Grayzel D, Demetri GD. Clin Cancer Res. 2013;19:6020. doi: 10.1158/1078-0432.CCR-13-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Khandelwal A, Crowley VM, Blagg BS. Med Res Rev. 2016;36:92. doi: 10.1002/med.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Jhaveri K, Ochiana SO, Dunphy MP, Gerecitano JF, Corben AD, Peter RI, Janjigian YY, Gomes-DaGama EM, Koren J, III, Modi S, Chiosis G. Expert Opin Invest Drugs. 2014;23:611. doi: 10.1517/13543784.2014.902442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Kitson RR, Chang CH, Xiong R, Williams HE, Davis AL, Lewis W, Dehn DL, Siegel D, Roe SM, Prodromou C, Ross D, Moody CJ. Nat Chem. 2013;5:307. doi: 10.1038/nchem.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Cell. 1997;89:239. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 8.a) Wang X, Elshahawi SI, Shaaban KA, Fang L, Ponomareva LV, Zhang Y, Copley GC, Hower JC, Zhan CG, Kharel MK, Thorson JS. Org Lett. 2014;16:456. doi: 10.1021/ol4033418. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shaaban KA, Wang X, Elshahawi SI, Ponomareva LV, Sunkara M, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J Nat Prod. 2013;76:1619. doi: 10.1021/np400308w. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang X, Shaaban KA, Elshahawi SI, Ponomareva LV, Sunkara M, Zhang Y, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J Nat Prod. 2013;76:1441. doi: 10.1021/np400231r. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang X, Shaaban KA, Elshahawi SI, Ponomareva LV, Sunkara M, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J Antibiot. 2014;67:571. doi: 10.1038/ja.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wang X, Reynolds AR, Elshahawi SI, Shaaban KA, Ponomareva LV, Saunders MA, Elgumati IS, Zhang Y, Copley GC, Hower JC, Sunkara M, Morris AJ, Kharel MK, Van Lanen SG, Prendergast MA, Thorson JS. Org Lett. 2015;17:2796. doi: 10.1021/acs.orglett.5b01203. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Shaaban KA, Singh S, Elshahawi SI, Wang X, Ponomareva LV, Sunkara M, Copley GC, Hower JC, Morris AJ, Kharel MK, Thorson JS. J Antibiot. 2014;67:223. doi: 10.1038/ja.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Shaaban KA, Saunders MA, Zhang Y, Tran T, Elshahawi SI, Ponomareva LV, Wang X, Zhang J, Copley GC, Sunkara M, Kharel MK, Morris AJ, Hower JC, Tremblay MS, Prendergast MA, Thorson JS. J Nat Prod. 2017;80:2. doi: 10.1021/acs.jnatprod.6b00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Voss SR, Palumbo A, Nagarajan R, Gardiner DM, Muneoka K, Stromberg AJ, Athippozhy AT. Regeneration. 2015;2:120. doi: 10.1002/reg2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ponomareva LV, Athippozhy A, Thorson JS, Voss SR. Comp Biochem Physiol Part C. 2015;178:128. doi: 10.1016/j.cbpc.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Screttas CG, Micha-Screttas M, Cazianis CT. Tetrahedron Lett. 1983;24:3287. [Google Scholar]; b) Patra A, Ghorai SK, De SR, Mal D. Synthesis. 2006:2556. [Google Scholar]; c) Yamabe S, Tsuchida N, Yamazaki S. J Org Chem. 2006;71:1777. doi: 10.1021/jo051862r. [DOI] [PubMed] [Google Scholar]; d) Rozhko E, Raabova K, Macchia F, Malmusi A, Righi P, Accorinti P, Alini S, Babini P, Cerrato G, Manzoli M, Cavani F. ChemCatChem. 2013;5:1998. [Google Scholar]
- 11.a) Cunningham ID, Danks TN, O’Connell KTA, Scott PW. J Org Chem. 1999;64:7330. [Google Scholar]; b) Eyong KO, Puppala M, Kumar PS, Lamshoft M, Folefoc GN, Spiteller M, Baskaran S. Org Biomol Chem. 2013;11:459. doi: 10.1039/c2ob26737c. [DOI] [PubMed] [Google Scholar]
- 12.a) Snieckus V. Chem Rev. 1990;90:879. [Google Scholar]; b) Parsons PJ, Jones DR, Padgham AC, Allen LA, Penkett CS, Green RA, White AJ. Chem Eur J. 2016;22:3981. doi: 10.1002/chem.201504421. [DOI] [PubMed] [Google Scholar]
- 13.Shibuya K, Kawamine K, Sata Y, Miura T, Ozaki C, Edano T, Hirata M, Ohgiya T. 20040038987 A1. US. 2004
- 14.a) Stoltz BM, Wood JL. Tetrahedron Lett. 1996;37:3929. [Google Scholar]; b) Umland KD, Palisse A, Haug TT, Kirsch SF. Angew Chem Int Ed. 2011;50:9965. doi: 10.1002/anie.201103961. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2011;123:10140. [Google Scholar]
- 15.Abdel-Mageed WM, Bayoumi SA, Al-Wahaibi LH, Li L, Sayed HM, Abdelkader MS, El-Gamal AA, Liu M, Zhang J, Zhang L, Liu X. Org Lett. 2016;18:1728. doi: 10.1021/acs.orglett.6b00206. [DOI] [PubMed] [Google Scholar]
- 16.Onodera H, Kaneko M, Takahashi Y, Uochi Y, Funahashi J, Nakashima T, Soga S, Suzuki M, Ikeda S, Yamashita Y, Rahayu ES, Kanda Y, Ichimura M. Bioorg Med Chem Lett. 2008;18:1588. doi: 10.1016/j.bmcl.2008.01.072. [DOI] [PubMed] [Google Scholar]
- 17.a) Voss SR, Epperlein HH, Tanaka EM. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.emo128. [DOI] [PubMed] [Google Scholar]; b) Dall’Agnese A, Puri PL. Bioessays. 2016;38:917. doi: 10.1002/bies.201600015. [DOI] [PubMed] [Google Scholar]
- 18.a) Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lele Z, Hartson SD, Martin CC, Whitesell L, Matts RL, Krone PH. Dev Biol. 1999;210:56. doi: 10.1006/dbio.1999.9262. [DOI] [PubMed] [Google Scholar]
- 19.a) Rutherford SL, Lindquist S. Nature. 1998;396:336. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]; b) Schell R, Mullis M, Ehrenreich IM. PLoS Biol. 2016;14:e2001015. doi: 10.1371/journal.pbio.2001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.