Abstract
The subgroup A to E avian sarcoma and leukosis viruses (ASLVs) are highly related and are thought to have evolved from a common ancestor. These viruses use distinct cell surface proteins as receptors to gain entry into avian cells. Chickens have evolved resistance to infection by the ASLVs. We have identified the mutations responsible for the block to virus entry in chicken lines resistant to infection by subgroup A ASLVs [ASLV(A)]. The tva genetic locus determines the susceptibility of chicken cells to ASLV(A) viruses. In quail, the ASLV(A) susceptibility allele tvas encodes two forms of the Tva receptor; these proteins are translated from alternatively spliced mRNAs. The normal cellular function of the Tva receptor is unknown; however, the extracellular domain contains a 40-amino-acid, cysteine-rich region that is homologous to the ligand binding region of the low-density lipoprotein receptor (LDLR) proteins. The chicken tvas cDNAs had not yet been fully characterized; we cloned the chicken tva cDNAs from two lines of subgroup A-susceptible chickens, line H6 and line 0. Two types of chicken tvas cDNAs were obtained. These cDNAs encode a longer and shorter form of the Tva receptor homologous to the Tva forms in quail. Two different defects were identified in cDNAs cloned from two different ASLV(A)-resistant inbred chickens, line C and line 72. Line C tvar contains a single base pair substitution, resulting in a cysteine-to-tryptophan change in the LDLR-like region of Tva. This mutation drastically reduces the binding affinity of TvaR for the ASLV(A) envelope glycoproteins. Line 72 tvar2 contains a 4-bp insertion in exon 1 that causes a change in the reading frame, which blocks expression of the Tva receptor.
Retroviruses share a common strategy for entry into cells (reviewed in references 27 and 51). The entry process is initiated by an interaction between the viral envelope glycoprotein and a specific cell surface protein that acts as a receptor. This interaction triggers a conformational change in the structure of the viral glycoprotein that leads to the fusion of the viral and cellular membranes. Despite the complexity of the interaction between the viral glycoprotein and the receptor, closely related retroviruses carry envelope glycoproteins that use different cellular proteins as receptors. In the avian sarcoma and leukosis viruses (ASLVs), there are five highly related envelope subgroups, subgroups A to E, that are thought to have evolved from a common ancestor (reviewed in references 7 and 50). The presence of viral subgroups that utilize distinct receptors helps the virus overcome host resistance and promotes coinfection. We and others have developed strategies that mimic resistance to ASLV entry in cell culture to study the evolution of the envelope glycoprotein. These studies demonstrated that blocking virus entry could select viral variants with mutations in the viral glycoproteins that altered receptor usage (24, 25, 31, 34, 43). The goal of the present study was to identify and characterize the mutations in the subgroup A receptor in lines of chickens that cause resistance to infection by ASLVs carrying the subgroup A envelope glycoproteins.
Three genetic loci in chicken cells determine the susceptibility and resistance to subgroup A to E ASLVs: tva (susceptibility to subgroup A viruses), tvb (susceptibility to subgroup B, D, and E viruses), and tvc (susceptibility to subgroup C viruses) (49, 50). Alleles that confer susceptibility to ASLV infection are dominant: two recessive resistance alleles are required at these loci to confer resistance. Because the tvar, tvbr, and tvcr resistance alleles are recessive, it is unlikely that these alleles encode dominant-negative forms of the receptor protein. The resistance alleles are likely to contain defects that either block receptor expression or prevent its use as an efficient ASLV receptor (29).
Several alleles of the tvb genetic locus and three related Tvb receptors have been identified. Two different susceptibility alleles have been defined at the chicken tvb locus. The tvbs1 allele confers susceptibility to subgroups B, D, and E; the tvbs3 allele confers susceptibility to only subgroups B and D (1, 3). These alleles encode the chicken TvbS1 (3) and TvbS3 (12) receptors, respectively. TvbS3 differs from TvbS1 by a single amino acid change, cysteine to serine at position 62, which presumably alters the structure of the TvbS1 protein so that it no longer functions as an ASLV(E) receptor. A third tvb receptor, the turkey TvbT receptor (2), which confers susceptibility to only subgroup E ASLV, has also been cloned. The Tvb proteins are members of the tumor necrosis factor receptor (TNFR) family. The recessive tvbr resistance allele does not support the entry of subgroup B, D, or E ASLVs. Recently, the tvbr allele from inbred chicken line 72 was cloned and shown to contain a mutation that introduces an in-frame stop codon (29). This molecular defect produces a severely truncated protein, abolishing its use as an ASLV receptor.
There appears to be only one susceptibility allele of tva and tvc. The subgroup A receptors are related to the low-density lipoprotein receptor (LDLR) family (9, 52). The tvc locus has not yet been cloned. The cDNAs obtained from the quail tvas allele are derived from two alternatively spliced mRNA products. These cDNAs produce two different proteins that differ in the C-terminal transmembrane and cytoplasmic regions of the protein (9, 52). The longer product, Tva950, is a typical type I single transmembrane glycoprotein, whereas the shorter Tva800 form lacks the full transmembrane region of Tva950 and is attached to the membrane via a glycosyl phosphatidylinositol anchor. Both forms of the quail Tva receptor have identical 83-amino-acid extracellular domains that contain a 40-amino-acid cysteine-rich region homologous to the LDLR ligand binding domain (LDL-A). LDL-A is required for efficient interaction with the ASLV(A) envelope glycoproteins (35, 55). There are three disulfide bonds in the LDL-A region, amino acid residues 11 to 50 in quail Tva (C1-C3, C2-C5, and C4-C6), that involve six cysteine residues (Cys-11, Cys-18, Cys-28, Cys-35, Cys-41, and Cys-50) (10). Extensive mutational analyses identified three amino acid residues (Asp-46, Glu-47, and Trp-48) (37, 56) in the protein loop formed by the C4-C6 disulfide bond and several other residues in the carboxy-terminal half of this region (Leu-34, His-38, and Gly-49) (36, 46, 48, 53) as critical components for efficient interaction between the quail Tva receptor and ASLV(A). Two somewhat different structures of the quail Tva LDL-A module have been solved using nuclear magnetic resonance (44, 47).
The chicken Tva receptor has not been extensively characterized in part because the complete coding region of the gene has not been fully identified and sequenced (9, 52). The sequence of the chicken Tva LDL-A domain was known from a cloned and partially sequenced 7-kb genomic DNA fragment (52). The carboxy-terminal halves of the LDL-A domains of the quail and chicken Tva receptors are identical; however, the N-terminal regions are only 50% identical. These differences in the N-terminal halves of the LDL-A modules presumably allow the envelope glycoproteins of some mutant ASLV(A) viruses to preferentially bind the chicken Tva receptor relative to the quail Tva receptor (25, 31). The cloned chicken ASLV(A) receptor gene fragment was genetically mapped to the chicken tva locus. A restriction fragment length polymorphism was used to screen the progeny of a cross between the inbred ASLV(A)-susceptible line 63 and the ASLV(A)-resistant line 72 (8). In this same study, the LDL-A region of exon 2 of the chicken tva gene was amplified by PCR and cloned from both chicken lines in an attempt to identify the molecular defect(s) in the tvar allele of line 72 (8). Although the analysis identified two putative amino acid differences between the line 72 tvar allele and the line 63 tvas allele, these differences did not correlate with the ASLV(A)-sensitive or -resistant phenotype.
The goals of our study were to completely define the chicken tva gene and identify and characterize the molecular defect(s) in the tvar allele that confers resistance to ASLV(A) infection in lines of chickens. Here we report the complete nucleotide sequence of the chicken tva gene and the cloning of chicken tva cDNAs from several ASLV(A)-susceptible and -resistant chicken lines. Two different ASLV(A)-resistant lines of inbred White Leghorn chickens contained different tvar mutations. One mutation is predicted to completely eliminate the expression of the Tva protein, while the other mutation changes a cysteine residue that apparently alters the structure of the Tva protein so that it no longer functions as an efficient receptor for ASLV(A).
MATERIALS AND METHODS
Chicken lines.
The inbred chicken line H6 was originally developed at the Northern Poultry Breeding Station (Reaseheath, Cheshire, United Kingdom) and imported to Prague in 1989. Line H6 is sensitive to ASLV(A) infection (21). Inbred chicken line C was initially developed in 1938 at the Northern Poultry Breeding Station. Lines CB and CC were derived from line C based on a heterozygosity in the major histocompatibility complex system (23) and bred at the Institute of Molecular Genetics (Prague, Czech Republic) by continuous brother × sister mating. Both lines are resistant to ASLV(A) and ASLV(E) infection and in this report are referred to as line C. Line 72 and line Rh-C chickens were from the Avian Disease and Oncology Laboratory (East Lansing, Mich.). Line 72 is resistant to ASLV(A), ASLV(B), ASLV(D), and ASLV(E) infection (5). Line Rh-C was bred at the Avian Disease and Oncology Laboratory from the original line C stock imported from the Northern Poultry Breeding Station.
Vector constructions.
The chicken Tva expression plasmids pTvaS and pTvaR were constructed in a similar way to the quail Tva expression plasmid described previously (10). In brief, the sequence encoding the chicken Tva receptor was truncated using PCR after the third codon (Lys114) of the intracellular tail and fused in frame with a sequence encoding the 9-amino-acid hemagglutinin (HA) epitope tag (YPYDVPDYA) followed by 10 histidine residues. The resulting fragment was cloned into the EcoRI site of the pSG5 eukaryotic expression vector under the control of the simian virus 40 early promoter (20). The plasmid pTvaS contains the sequence for the wild-type receptor, and pTvaR encodes the Cys40Trp mutation. The plasmid pKZ261 encodes the quail Tva 950 receptor protein under the control of the cytomegalovirus promoter in the expression plasmid pCB6. This plasmid was described previously (55).
Genes encoding soluble forms of the chicken TvaS (tvasstva-mIgG) and TvaR (tvarstva-mIgG) receptors were constructed as described previously for quail stva-mIgG (26) and chicken stva-mIgG (ckstva-mIgG) (25). The stva-mIgG genes encode the extracellular domain of the particular Tva receptor fused to the constant region of a mouse immunoglobulin G (IgG) heavy chain and are in the CLA12NCO adaptor plasmid (16). The stva-mIgG gene cassettes were isolated as ClaI fragments and subcloned into the ClaI site of the expression plasmid TFANEO to generate TFANEO/tvasstva-mIgG and TFANEO/tvarstva-mIgG plasmids. The expression cassette of TFANEO contains two long terminal repeats derived from the RCAS vector that provide strong promoter, enhancer, and polyadenylation site sequences. The long terminal repeats flank a unique ClaI insertion site (15, 16). The TFANEO plasmid also contains a neo resistance gene expressed under the control of the chicken β-actin promoter and an ampicillin resistance gene for selection in Escherichia coli.
The RCASBP(A)AP retroviral vector, an ASLV-based replication-competent vector with a subgroup A env gene that expresses the heat-stable human placental alkaline phosphatase (AP) gene, was described previously (16, 17, 19).
Cell culture and virus propagation.
Chicken embryo fibroblasts (CEFs) were isolated from 10-day-old embryos from lines H6, CC, CB, Rh-C, and 72 by standard procedures (16). The CEFs and DF-1 cells, a continuous fibroblastic cell line derived from line O CEFs (C/E) (22, 39), were grown in Dulbecco's modified Eagle's medium (GIBCO/BRL) supplemented with 10% fetal bovine serum (GIBCO/BRL), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Quality Biological, Inc., Gaithersburg, Md.) at 39°C and 5% CO2. Human 293 cells were grown in the same medium at 37°C. The clonal DF-1 cell lines expressing the soluble Tva-mIgG receptor proteins were grown in the above medium but supplemented with 250 μg of G418/ml.
The long forms of the TvaS and TvaR receptor proteins were expressed in 293 cells by transfection of eukaryotic expression plasmid DNAs (pTvaS and pTvaR). The eukaryotic expression plasmid pKZ261, which expresses the 950 form of the quail Tva receptor on 293 cells, was used as a positive control (55). As a control for transfection efficiency, an expression plasmid encoding the enhanced green fluorescent protein (EGFP), pCMS-EGFP (BD Biosciences Clontech, Palo Alto, Calif.), was included in each transfection mixture. In standard transfection mixtures, 8.8 μg of purified plasmid DNA was introduced into 3 × 106 293 cells by using the Polyfect transfection reagent following the manufacturer's protocol (QIAGEN Inc., Valencia, Calif.).
RCASBP(A)AP virus was generated by transfection of plasmid DNA that contained the retroviral vector in proviral form (16). In standard transfections, 5 μg of purified plasmid DNA was introduced into DF-1 cells by the calcium phosphate precipitation method (28). Viral spread was monitored by assaying culture supernatants for ASLV capsid protein (CA) by enzyme-linked immunosorbent assay (ELISA) (40). Virus stocks were generated from cell supernatants cleared of cellular debris by centrifugation at 2,000 × g for 10 min at 4°C and stored in aliquots at −80°C.
The TFANEO/tvasstva-mIgG and TFANEO/tvarstva-mIgG plasmids were used to generate clonal DF-1 cell lines as described previously for the generation of the TF/sTva-4 cell line, a clonal line of DF-1-derived cells expressing quail sTva-mIgG from the TFANEO expression plasmid (25), and the clonal TF/cksTva-15 DF-1-derived line expressing chicken sTva-mIgG (31). DF-1 cells transfected with TFANEO/tvasstva-mIgG or TFANEO/tvarstva-mIgG plasmid DNA were grown in 500 μg of G418 per ml and selected for neomycin-resistant cells. Clones were isolated using cloning cylinders (Bellco Glass Inc., Vineland, N.J.), expanded, and maintained with standard medium supplemented with 250 μg of G418/ml.
Reverse transcription-PCR (RT-PCR).
Total RNA was prepared from CEFs with the RNeasy total RNA isolation system (QIAGEN Inc.). RNA samples (1 μg) were reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) with an oligo(dT) primer. The cDNA was then PCR amplified with Taq DNA polymerase (Roche Diagnostics) and primers TVA4 (5′-GCATGGTGCGGTTGTTGGAG-3′) and TVA5 (5′-TCGTGTCCAAATTCAGCCAG-3′). Amplification was performed under the following conditions: 45 s at 98°C; 33 cycles of 20 s at 95°C, annealing for 50 s at 64°C, and 90 s at 72°C; and a final extension of 3 min at 72°C.
PCR.
Genomic DNA was isolated from DF-1 cells and CEFs by standard procedures (38). Each PCR mixture contained 1.25 μl of 10× PCR buffer (final concentration, 50 mM Tris · Cl [pH 8.3], 50 mM KCl, 7 mM MgCl2, 1.1 mM beta-mercaptoethanol), 1.25 μl of a 1.7-mg/ml solution of bovine serum albumin, 0.5 μl of each deoxynucleoside triphosphate at 25 mM, 0.5 μl of each primer (A260 = 5), 6.0 μl of H2O, and 1.0 μl of DNA (genomic DNA, ∼100 ng/μl; plasmid DNA, ∼2 ng/μl). The reaction mixtures were heated to 96°C for 1 min, and reactions were initiated by the addition of 1.5 μl of Taq DNA polymerase (Promega, Madison, Wis.) diluted 1:10 (vol/vol) (0.75 U). Thirty cycles of PCR were carried out as follows: 96°C for 40 s and then 63°C for 80 s. The diagnostic primers used to detect the line 72 4-nucleotide insertion were TVA10 (5′-TGCTGCGCGCCGTCCGCCCGCTCG-3′) in tva exon 1 and TVA9 (5′-CGCCCGTACCTGTGCCGCCGG-3′) in tva exon 2.
ELISA.
The ASLV CA protein was detected in culture supernatants by ELISA as described previously (40). The levels of sTva-mIgG proteins were quantitated in culture supernatants by ELISA for the mouse IgG tag as previously described (26). The linear range for a standard experiment was between 0.5 and 50 ng of ImmunoPure mouse IgG Fc fragment per ml.
ASLV AP assay.
For AP assays, DF-1 or 293 cell cultures (∼30% confluent) were incubated with 10-fold serial dilutions of the appropriate RCASBP/AP virus stocks for 36 to 48 h. The assay for AP activity was described previously (26).
Western immunoblot analysis.
Supernatants from confluent DF-1 cultures expressing the soluble Tva-mIgG proteins were cleared of cellular debris by centrifugation at 2,000 × g for 10 min at 4°C. The sTva-mIgG proteins were immunoprecipitated with anti-mouse IgG-agarose beads (Sigma), and the precipitated proteins were denatured, separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a nitrocellulose membrane as described in reference 26. Cell lysates were prepared from transfected 293 cells by treating the cells with lysis buffer (50 mM Tris [pH 8.0], 62.5 mM EDTA, 0.4% deoxycholate, and 1% Igepal [Sigma] supplemented with Complete protease inhibitor [Roche; 1 tablet per 10 ml]) for 1 to 5 min at room temperature, removing the cell debris by centrifugation at 4,000 × g for 10 min, and mixing the recovered supernatant with an equal part of 2× Laemmli buffer (125 mM Tris [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.2% bromophenol blue). The proteins were denatured, separated by SDS-12% PAGE, and transferred to a nitrocellulose membrane.
The Western transfer filters were blocked and washed as described previously (26). The immunoblots were probed with either peroxidase-conjugated goat anti-mouse IgG (heavy and light [H+L]; 50 ng/ml; Kirkegaard & Perry Laboratories, Gaithersburg, Md.) directly, or first the 12CA5 monoclonal antibody (18, 45) against the influenza virus HA (1:3,000 dilution) followed by the peroxidase-conjugated goat anti-mouse IgG (H+L). After extensive final washing, immunodetection of the protein-antibody-peroxidase complexes was performed with the Western blot chemiluminescence reagent (DuPont NEN, Boston, Mass.). The immunoblots were then exposed to Kodak X-Omat film.
FACS analysis.
Transfected 293 cells or RCASBP(A)AP-infected DF-1 cells were removed from culture with trypsin de Larco (Quality Biological, Inc.) and washed with Dulbecco's phosphate-buffered saline (PBS). The cells were fixed with 4% paraformaldehyde in PBS at room temperature for 15 min and then washed with PBS. The EGFP-positive cells in the 293 transfected cell populations were quantitated directly by fluorescence-activated cell sorting (FACS) with a Becton Dickinson FACSCalibur apparatus and CellQuest 3.1 software.
Approximately 106 DF-1 cells in PBS supplemented with 1% calf serum (PBS-CS) were incubated with supernatant containing one of the four sTva-mIgG proteins on ice for 30 min. The stable DF-1 cell lines TF/cksTva-15 (chicken sTva-mIgG), TF/sTva-4 (quail sTva-mIgG), TF/sTvaS-18, and TF/sTvaR-8 were the sources of the sTva-mIgG proteins. The cells were then washed with PBS-CS and incubated with 5 μl of goat anti-mouse IgG (H+L) linked to phycoerythrin (Kirkegaard & Perry Laboratories) in PBS-CS (1 ml total volume) on ice for 30 min. The cell/soluble receptor-mIgG/Ig-phycoerythrin complexes were washed with PBS-CS, resuspended in 0.5 ml of PBS-CS, and analyzed with a Becton Dickinson FACSCalibur apparatus and CellQuest 3.1 software.
Apparent dissociation constant (Kd) calculations.
The maximum possible fluorescence and Kd values for each data set obtained from the FACS binding assays were estimated by fitting the data via nonlinear least squares to a log logistic growth curve function as follows: f(y) = M/[1 + e−r(logx− logKd)], where y is the mean fluorescence, M is the maximum fluorescence, r is the rate, x is the concentration of sTva-mIgG, and Kd is the dissociation constant defined as the concentration of sTva-mIgG at half-maximal binding (25).
Nucleotide sequence accession number.
The sequences reported in this study have been submitted to GenBank and assigned accession numbers as follows: AY531258, tvas longer form; AY531259, tvas shorter form; AY531260, tvar longer form; AY531261, tvar shorter form; AY531262, tva genomic sequence.
RESULTS
Cloning chicken tva cDNAs and identification of the chicken tva coding region.
The complete sequence of the chicken tvas receptor gene had not been reported; the tva mRNAs and the Tva protein are expressed at very low levels (9, 52). The recent availability of the chicken EST databases (11, 13) combined with the partial sequence from the chicken Tva coding region enabled us to assemble a tentative nucleotide sequence of the complete chicken tva receptor gene. Using RT-PCR and primers designed based on the predicted coding region of this assembled tva sequence, two cDNA products were amplified from total RNA isolated from both ASLV(A)-sensitive (line H6) and ASLV(A)-resistant (line C) CEFs (Fig. 1). The RT-PCR products were cloned, and the nucleotide sequences were determined from both representative clones and directly from the RT-PCR products. The deduced amino acid sequences of line H6 tva long cDNA and tva short cDNA (Fig. 1) are homologous to the two alternatively spliced quail tva mRNAs (9, 52). The chicken tva cDNA sequences were used to prepare additional PCR primers, and the line H6 tva genomic region was amplified and sequenced (Fig. 2).
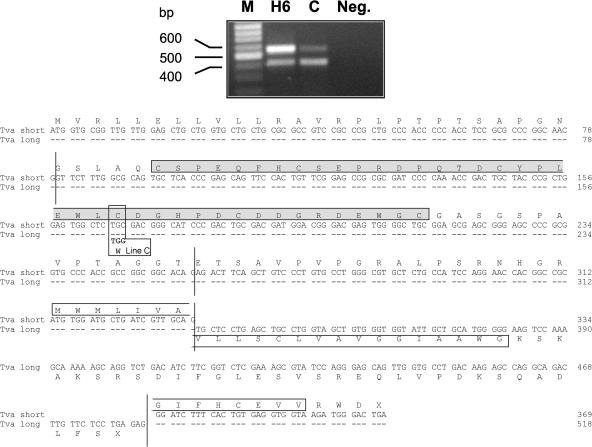
FIG. 1.
Two alternatively spliced Tva transcripts are present in ASLV(A)-sensitive and -resistant chicken cells. RNA samples were prepared from CEFs of inbred lines H6 (sensitive) and C (resistant) and subjected to RT and PCR amplification. The primers used span the entire predicted Tva coding region and yield cDNA products from the longer and shorter alternatively spliced RNAs of 569 and 420 bp, respectively. The nucleotide sequences of the gel-purified cDNA products were determined directly. The nucleotide and deduced amino acid sequences of the coding regions of the long and short chicken Tva cDNAs are shown. Identical nucleotides are represented by dashes, and vertical lines mark the exon boundaries. The 40-amino-acid LDLR homologous cysteine-rich sequence (LDL-A) is highlighted in a shaded box. The putative transmembrane segments are highlighted in open boxes. We assume that the shorter form of Tva is attached to the cell surface via a glycosyl phosphatidylinositol linkage. The codon containing the single nucleotide difference between the line H6 and line C Tva cDNAs that results in the Cys40Trp mutation is labeled line C.
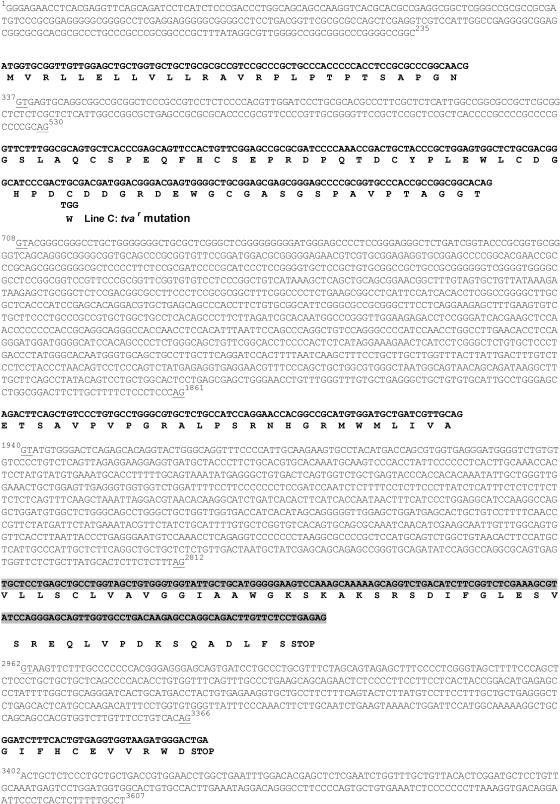
FIG. 2.
The nucleotide and putative amino acid sequences of the chicken tva locus of line H6. The genomic DNA region of tva was amplified by PCR, and the nucleotide sequence was determined. Each exon is shown in bold with the deduced amino acid sequence. The alternate exon of the long cDNA is highlighted with a shaded box. The nucleotide sequence of the tva introns and flanking regions are shown, and their boundaries are underlined and numbered. The codon containing the single nucleotide difference between the tva cDNAs from line H6 and line C that results in the Cys40Trp mutation is labeled line C.
A comparison of the deduced sequence of the chicken and quail immature Tva long proteins is shown in Fig. 3A with the predicted signal peptidase cleavage sites, extracellular, transmembrane, and intracellular regions marked. The predicted sequence of the immature chicken Tva protein contains two additional amino acids compared to the quail Tva protein. In addition, the location of the predicted signal peptidase cleavage site is different between the chicken and quail receptors. As a consequence, the mature chicken Tva protein contains five additional amino acids compared to the mature quail Tva protein. This changes the numbering of the amino acids: for example, the fourth cysteine in the LDL-A module is residue 40 in the mature chicken TvaS protein but residue 35 in the mature quail Tva protein (Fig. 3A).
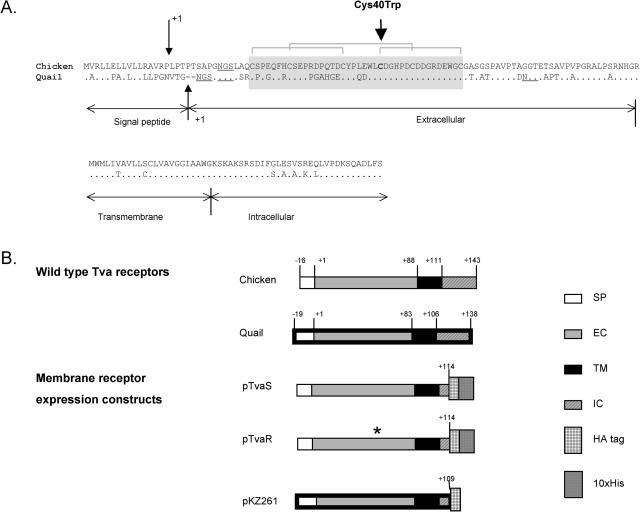
FIG. 3.
The chicken and quail Tva receptors and the expression constructs. (A) Comparison of the deduced amino acid sequences of the chicken and quail Tva receptors. Identical amino acids are denoted by dots, and gaps are indicated by dashes. Only the longer forms of both receptors are shown. The quail receptor is predicted to have a signal peptide of 19 residues, followed by an extracellular domain of 83 residues which includes the LDL-A module, marked by a shaded box (9). The chicken Tva receptor is predicted to have a leader peptide of 16 amino acids (32) followed by an extracellular domain of 88 residues. The predicted cleavage sites of the leader peptidase are marked by small vertical arrows, and the amino acid numbering starts with the first residue (+1) of the mature chicken and quail proteins generated by the cleavage. There is considerable similarity in the sequences of the Tva proteins, and the extracellular, transmembrane, and intracellular regions of the chicken receptor are assumed to correspond to the regions in the quail Tva protein. The transmembrane and cytoplasmic tail regions of both proteins consist of 23 and 32 amino acids, respectively. The three disulfide bonds in the LDL-A module are indicated by brackets, and the potential N-linked glycosylation sites are underlined. The large vertical arrow marks the site of the mutation in line C, which involves Cys40 in chicken Tva (corresponding to Cys35 in quail Tva). (B) Schematic diagram of the Tva receptors and expression constructs used in these studies. Only the longer forms of the chicken and quail Tva receptors are shown; their extracellular, transmembrane, and cytoplasmic regions were included in the expression constructs. The construct pKZ261 was described previously (55). The constructs pTvaR and pTvaS were generated specifically for these experiments and are described in Materials and Methods. The horizontal bars with thin and thick black boundaries denote sequences of chicken and quail origin, respectively. The numbers indicate the position of amino acid residues in mature proteins; signal peptide residues have negative numbers. The asterisk indicates the presence of the Cys40Trp mutation in the pTvaR construct. SP, signal peptide; EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain; HA, epitope tag from influenza virus HA protein; His, histidine residue tag.
Two tva cDNAs were also amplified from the ASLV(A)-resistant line C RNA and had a single nucleotide difference from the line H6 tva cDNAs (Fig. 1). Comparison of the deduced sequence of the Tva receptors from line H6 and line C showed that this single nucleotide change caused a single amino acid substitution in line C, changing the fourth conserved cysteine residue in the LDL-A module to tryptophan (codon 40 [TGC to TGG]). This mutation was also present in the line C tva genomic sequence (Fig. 2). The Cys40Trp mutation would prevent the formation of the disulfide bond between the fourth and sixth cysteines in the LDL-A module, which is critical for the proper folding of Tva and ASLV(A) receptor function (10). An adenine mutation introduced into the quail Tva protein at the fourth cysteine position of the LDL-A module (Cys35Ala) reduced receptor function to ∼3% of wild-type levels when the mutant receptor protein was transiently expressed in COS-7 cells (10). Although this is a significant reduction in receptor function, ASLV(A) infection was relatively efficient compared to the ∼106-fold resistance of line C CEFs to ASLV(A) infection (data not shown).
The Cys40Trp mutation significantly reduces the ability of the line C Tva receptor to mediate ASLV(A) infection compared to that of the line H6 Tva receptor.
To determine if the Cys40Trp mutation in the chicken Tva receptor accounts for the ASLV(A)-resistant phenotype of line C cells, expression plasmids encoding the long form of the line H6 TvaS receptor (pTvaS) or the long form of the line C TvaR receptor (pTvaR) were constructed. The coding regions were truncated at the C terminus after the codon for the third amino acid (Lys114) of the intracellular domain and fused to an HA epitope tag and a histidine tag (Fig. 3B). Expression plasmids encoding TvaS, TvaR, quail Tva950 receptor, or an empty plasmid vector were transfected into a mammalian cell line, human 293 cells (Fig. 4A). Mammalian cells do not normally express a functional form of any of the ASLV receptors. Each transient-transfection mixture contained 8.8 μg of plasmid DNA, including 0.8 μg of plasmid DNA encoding EGFP. The efficiency of the transfections was estimated by counting the number of green cells by FACS. Two days after transfection, the cells were challenged with RCASBP(A)AP, an ASLV(A) virus also encoding human placental AP.
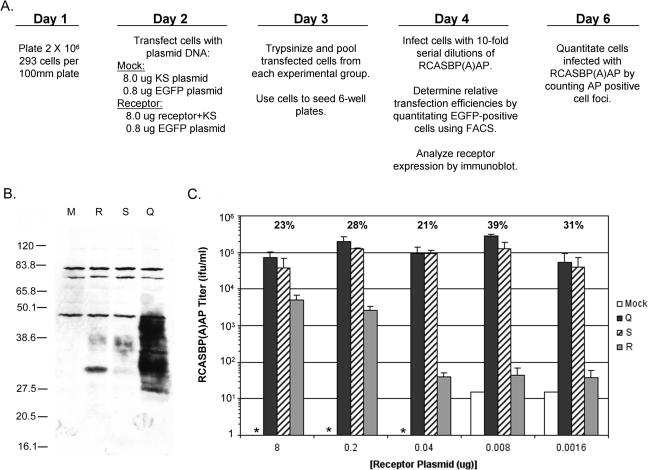
FIG. 4.
The different Tva receptors confer different susceptibilities to ASLV(A) infection. (A) Schematic representation of the experimental approach. (B) Western immunoblot analysis of the levels of Tva receptor expressed by 293 cells transfected with 8 μg of receptor plasmid DNAs. Clarified cell lysates were made, the proteins were separated by SDS-12% PAGE and transferred to nitrocellulose, the filter was probed with anti-HA monoclonal antibody 12CA5, and the bound protein complexes were visualized by chemiluminescence. Molecular sizes (in kilodaltons) are given on the left. Lanes: M, mock; Q, quail Tva950; S, chicken TvaS; R, chicken TvaR. (C) 293 cells were transfected with 8.8 μg of plasmid DNA that contained different amounts of a plasmid encoding a Tva receptor. Transfected 293 cells expressing the different Tva receptors, quail Tva950 (Q), chicken TvaS (S), chicken TvaR (R), or mock (M), were challenged with 10-fold serial dilutions of RCASBP(A)AP and the titers were determined by AP assay. The asterisk indicates that the titer was below the limit of detection, 1 IFU/ml. The transfection efficiencies for each experimental group are shown above the titers as an average percentage. The results are averages of triplicate experiments. Error bars show standard deviations.
When the maximum amount of each Tva receptor expression plasmid (8 μg) was transfected into 293 cells, detectable levels of the Tva receptor proteins were expressed as shown by Western immunoblot analysis (Fig. 4B). The expression levels of the chicken TvaS and TvaR receptor proteins were similar but appeared to be lower than the expression level of the quail Tva950 protein, even though the transfection efficiencies were similar (21 to 25%). This difference may be due in part to differences in the promoters used to express the Tva proteins: the simian virus 40 early promoter was used to express the chicken TvaS and TvaR receptors, and the cytomegalovirus promoter was used to express the quail Tva receptor. The chicken Tva receptor proteins also displayed a lesser amount of glycosylation (ranging from ∼30 to 40 kDa) compared to the quail Tva receptor (ranging from ∼25 to 50 kDa) (Fig. 4B). The quail Tva receptor contains three potential N-linked glycosylation sites; the chicken Tva receptor contains only one (Fig. 3A). The fastest-migrating chicken TvaS and TvaR receptor proteins, which are presumably the unglycosylated forms of the receptors (30 kDa), contain the histidine tag and are larger than the unglycosylated form of quail Tva (Fig. 3B). The expression of each Tva receptor from 8 μg of plasmid DNA transfected in 293 cells conferred susceptibility to ASLV(A) infection (Fig. 4C) but did not confer susceptibility to ASLV(B) or ASLV(C) infection (data not shown). However, ASLV(A) infection was ∼20-fold less efficient with the TvaR receptor that contained the Cys40Trp mutation compared to the TvaS and quail Tva receptors. The reduction in ASLV(A) receptor activity when the chicken TvaR was expressed in 293 cells was similar to the loss of receptor function observed when the quail Tva Cys35Ala receptor mutant was expressed in COS-7 cells (10), but it was still relatively efficient when compared to the observed ∼106-fold resistance of line C.
Since the process of transfecting plasmid DNA into eukaryotic cells results in multiple copies of the plasmid in each transfected cell, it is likely that the transfected cells express much higher levels of the Tva receptor proteins relative to the normal expression level on avian cells. Normally, the Tva receptor proteins are expressed at very low levels on avian cells. Tva cannot be detected by Western immunoblotting in extracts of avian cells; however, this low level of expression allows it to function efficiently as the receptor for ASLV(A) viruses (9, 52). To test this hypothesis, wild-type Tva and Tva carrying the Cys40Trp mutation were expressed at lower levels by transfecting smaller amounts of the Tva expression plasmids (8 to 0.0016 μg) into 293 cells. A total of 8.8 μg of plasmid DNA was used in each transfection mixture. This reduced the amount of Tva expressed in the 293 cells; the Western immunoblotting procedure could not detect Tva proteins when <1 μg of the receptor plasmid DNA was used in the transfection (data not shown). Both of the wild-type Tva receptors, chicken TvaS and quail Tva, conferred similar levels of susceptibility to ASLV(A) infection in 293 cells transfected using a wide range of receptor plasmid DNA concentrations, 8 μg to 8 ng (Fig. 4C). However, transfection of lower amounts of the pTvaR plasmid, which would result in lower amounts of the mutant TvaR receptor, resulted in up to a 2,500-fold loss in susceptibility to ASLV(A) infection (Fig. 4C). These data imply that the Cys40Trp mutation and the low level of expression of the Tva receptor in chicken cells account for the resistance of line C cells to ASLV(A) infection.
The Cys40Trp mutation significantly lowers the binding affinity of the Tva receptor for the ASLV(A) envelope glycoproteins.
The six conserved cysteines in the LDL-A module are critical for the proper folding of the Tva protein. In TvaR, the Cys40Trp mutation eliminates a critical disulfide linkage and introduces a large aromatic amino acid that may further distort the structure of the protein. Despite these defects, the TvaR protein can still function as a receptor for ASLV(A), albeit at a much lower efficiency. We hypothesized that the Cys40Trp mutation lowers the binding affinity of the TvaR receptor for the ASLV(A) glycoproteins. A lower affinity between TvaR and ASLV(A) would be partially offset if the TvaR receptor were expressed at high levels on the surface of the cell.
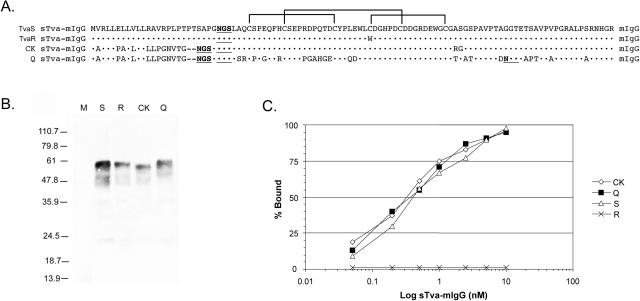
To test this hypothesis, we estimated the binding affinities of the wild-type and Cys40Trp mutant Tva receptors for subgroup A envelope glycoproteins expressed on the surface of ASLV(A)-infected chicken cells. To perform this analysis, we prepared soluble forms of the TvaS and TvaR receptors fused to a mouse IgG domain: TvaSsTva-mIgG and TvaRsTva-mIgG (Fig. 5A). The production of soluble forms of the quail Tva receptor, QsTva-mIgG, and a partial chicken Tva receptor, CKsTva-mIgG, was described previously (25, 26). The sTva-mIgG proteins used in these experiments were analyzed by immunoprecipitation and Western immunoblotting from culture supernatants from DF-1 producer cells (Fig. 5B). The fusion proteins migrate as broad bands (45 to 60 kDa) due to glycosylation. The soluble Tva-mIgG proteins form dimers due to interaction of the mIgG domains. Different amounts of each sTva-mIgG receptor protein were assayed for ability to bind to ASLV(A) glycoproteins expressed on the surface of ASLV(A)-infected DF-1 cells by FACS as described previously (25). The TvaS, Q, and CK sTva-mIgG receptors bound the subgroup A glycoproteins with high binding affinities, ∼0.3 nM (Fig. 5C), in agreement with previous reports (6, 25, 31, 56). However, binding of the TvaR sTva-mIgG receptor to subgroup A glycoproteins was not detected over the protein concentrations tested, demonstrating that the Cys40Trp mutation in Tva causes a significant loss in the binding affinity of the TvaR receptor for the ASLV(A) glycoproteins (Fig. 5C).
FIG. 5.
Binding affinity of the Tva receptors for ASLV(A) envelope glycoproteins. (A) Comparison of the signal peptides and extracellular domains of the quail Tva receptor (Q), the previously published quail/chicken Tva receptor (CK), the chicken TvaS receptor, and the chicken TvaR receptor used in the soluble Tva receptor constructs. Tva amino acids identical to the TvaS receptor are indicated (•). The three disulfide bonds are indicated by brackets. (B) Western immunoblot analysis of the sTva-mIgG proteins. The sTva-mIgG proteins were immunoprecipitated with goat anti-mouse IgG-agarose beads from clarified supernatants collected from the clonal DF-1 producer lines, denatured, separated by SDS-12% PAGE, and transferred to nitrocellulose. The filter was probed with peroxidase-conjugated goat anti-mouse IgG, and the bound protein-antibody complexes were visualized by chemiluminescence using Kodak X-Omat film. M, mock supernatant; S, chicken TvaS sTva-mIgG; R, chicken TvaR sTva-mIgG; CK, chicken sTva-mIgG; Q, quail sTva-mIgG. Molecular sizes (in kilodaltons) are given on the left. (C) Binding of the sTva-mIgG proteins to ASLV(A) glycoproteins. Uninfected DF-1 cells and DF-1 cells infected with wild-type RCASBP(A)AP virus were fixed with paraformaldehyde and incubated with different amounts of a sTva-mIgG protein and the envelope glycoprotein/sTva-mIgG complexes bound to goat anti-mouse Ig antibody linked to phycoerythrin. The amount of phycoerythrin bound to the cells was determined by FACS, and the maximum fluorescence was estimated (see Materials and Methods). The data were plotted as percent maximum fluorescence bound versus sTva-mIgG concentration. The values shown are averages of duplicate experiments.
A different mutation is present in ASLV(A)-resistant line 72.
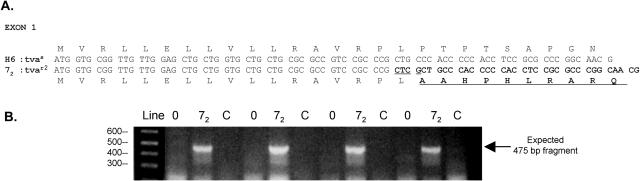
While the Cys40Trp mutation and the low level of TvaR expression appear to explain the ASLV(A) resistance of line C, a previous study did not find this mutation in another inbred line of chickens resistant to ASLV(A) infection, line 72 (8). To identify the possible defect(s) in the tva gene in line 72, tva cDNAs were amplified by RT-PCR and cloned from RNA prepared from line 72 CEFs. As controls, tva cDNAs were also amplified and cloned from different ASLV(A)-susceptible CEFs, line 0, and a parental stock of the ASLV(A)-resistant line C CEFs, line Rh-C. Long and short forms of tva cDNAs were identified in all three lines (data not shown). The tva cDNAs isolated from line 0 were identical to line H6 cDNAs. The tva cDNAs isolated from the parental line Rh-C were identical to the line C cDNAs and, as expected, contained the Cys40Trp mutation (data not shown). However, the nucleotide sequence of the line 72 tva cDNAs showed one difference from the line H6 and line 0 cDNAs: a 4-nucleotide insertion in exon 1 (Fig. 6A). This insertion causes a change in the predicted translational reading frame of both forms of the tva mRNAs, presumably preventing expression of the Tva proteins. The same 4-nucleotide insertion was found in exon 1 of line 72 genomic DNA amplified by PCR (data not shown). We have named this ASLV resistance allele tvar2. We also developed a PCR assay that specifically detects the line 72 tvar2 gene but not the tva or tvar genes by using a 5′ primer that includes the 4-nucleotide insertion at the 3′ end of the primer (Fig. 6B). This assay can be used to identify genomic DNAs that carry the tvar2 gene.
FIG. 6.
The tvar2 gene of chicken Line 72 contains a 4-nucleotide insertion. (A) The nucleotide sequence and deduced amino acid sequence of exon 1 of the tvas gene of the ASLV(A)-susceptible line H6 (H6:tvas) and the tvar2 gene of ASLV(A)-resistant line 72 (72:tvar2). The 4-nucleotide insertion and altered amino acid sequence of line 72 are highlighted in bold. (B) A PCR assay was developed to specifically detect the 4-nucleotide insertion in the tvar2 gene of line 72 genomic DNA. The picture shows a gel electrophoresis of DNAs produced by PCR amplification of four samples each of line 0 (0), line 72 (72), and line Rh-C (C) genomic DNA isolated from CEFs separated on a 2% agarose gel and visualized with ethidium bromide. The sizes of the DNA markers are shown on the left in base pairs.
DISCUSSION
ASLV infection of chickens leads to a variety of serious diseases and can cause death. Thus, populations of chickens exposed to ASLV would be under selective pressure to develop resistance to ASLV. So why haven't lines of chickens been naturally selected that are resistant to all subgroups of ASLVs? For example, the exogenous ASLV(A) viruses are a significant threat to flocks in poultry houses, yet there are very few lines of chickens that carry a tvar allele. Most likely, the susceptibility of chickens to the ASLVs is the result of breeding patterns and a balance of several different evolutionary forces. Most lines of chickens that have been extensively studied are inbred lines that have been selected for traits that have nothing to do with resistance to viruses, for example, egg production, growth rates, or plumage colors. In these cases, there may be little or no selective pressure for ASLV resistance. In addition, the selection of ASLV resistance alleles would not be favorable if the normal cellular proteins used as ASLV receptors were important for the normal growth and development of the bird. Unfortunately, the normal cellular functions of Tva and Tvb are not yet known. However, it is likely that these proteins are not absolutely required, since there are tvar and tvbr alleles that appear to severely disrupt these cellular proteins without apparently harming chickens homozygous for these ASLV resistance alleles. Since the Tva and Tvb proteins are members of large multimember families of receptors, possibly other members of the LDLR- and TNFR-related receptors may compensate for their loss. It is also possible that these genes may be more important for the survival of the bird in the wild than in captivity. The selection of resistance alleles to ASLVs may also be hampered by the ability of retroviruses to evolve the structure of their envelope glycoproteins to alter their receptor usage. A variety of studies have demonstrated that ASLV can acquire mutations that alter the envelope and the viral host range (24, 25, 31, 34, 43). Subgroups of highly related viruses that use different proteins as receptors may vary the selective pressure to make it difficult for chickens to develop complete resistance to ASLV entry.
One ASLV resistance allele that is prevalent in inbred chicken lines is the tvbs3 allele, which confers resistance to subgroup E ASLVs. Almost all lines of chickens contain endogenous subgroup E ASLV loci; a number of loci encode infectious virus and/or subgroup E viral glycoproteins (50). Adkins et al. proposed that the evolution of the tvbs3 allele is a result of the selective pressure of exposure to endogenous ASLV(E)s on chickens, implying that a mutant TvbS1 protein that does not bind ASLV(E) provides a selective advantage (3). While ASLV(E) viremia and/or the expression of subgroup E glycoproteins has much more modest effects on the health of chickens than infection with exogenous ASLVs, expression of endogenous ASLVs does reduce the immunological response of birds to exogenous ASLV infection. It is also possible there are other unknown detrimental effects that are produced by exposure to endogenous ASLV(E) viremia and subgroup E glycoproteins in chickens. For example, the interaction of ASLV(E) glycoproteins with the TvbS1 receptor may cause the death of certain cell types in the bird by subgroup E glycoprotein receptor-mediated apoptosis. In addition, since other members of the TNFR family play important roles in immune responses against microbial pathogens, the Tvb proteins may also be involved in immune responses. In this scenario, it may be advantageous to have a Tvb protein expressed on the cell surface that cannot be blocked or down regulated by interactions with the subgroup E glycoprotein.
To date, four different mutations that cause resistance to ASLV envelope subgroups have been described in inbred White Leghorn chickens (Table 1). The molecular defects encoded by these mutations either eliminate the expression of the receptor or alter the structure of the receptor protein to lower the binding affinity for the viral glycoproteins. These mechanisms are consistent with the recessive nature of the ASLV-resistant phenotypes. The tvar2 (this study) and tvbr (29) alleles apparently eliminate the production of the receptor proteins by altering the translational reading frame. In contrast, the tvar (this study) and tvbs3 (3) alleles encode proteins but with mutations in cysteine residues that presumably alter the shape of the proteins, significantly reducing their binding affinities for ASLV envelope glycoproteins. While these mutations alter the use of these proteins as ASLV receptors, it is not known if the changes in structure of the TvaR and TvbS3 proteins also affect their natural cellular function(s).
TABLE 1.
The characterized chicken ASLV receptor alleles, including the genetic defects and phenotypes of the known ASLV receptor resistance alleles
| Allele | Chicken line | Mutation | Phenotype | Ref. |
|---|---|---|---|---|
| tvas | H6 | Wild type | Susceptible to subgroup A ASLVs | This study |
| 0 | ||||
| tvar | C | Single-nucleotide mutation in tva resulting in the Cys40Trp change | Resistant to subgroup A ASLVs due to a drastically lower binding affinity for ASLV(A) envelope glycoprotein | This study |
| Rh-C | ||||
| tvar2 | 72 | 4-nucleotide insertion in exon 1 of tva resulting in a change in reading frame | Resistant to subgroup A ASLVs due to the absence of the Tva protein | This study |
| tvbs1 | 15B1 | Wild type | Susceptible to subgroup B, D, and E ASLVs | 3, 29 |
| tvbs3 | 0 | Single-nucleotide mutation in tvbs1 resulting in the Cys62Ser change | Susceptible to subgroup B and D ASLVs; resistant to subgroup E ASLVs due to a drastically lower binding affinity for only ASLV(E) envelope glycoprotein | 3, 29 |
| tvbr | 72 | Single-nucleotide mutation in tvbs1 resulting in a premature stop codon | Resistant to subgroup B, D, and E ASLVs due to truncation of the TvbS1 protein at amino acid 57 | 29 |
Another significant determinant of susceptibility to virus infection is the endogenous expression levels of cell surface proteins used as receptors by retroviruses. For example, Chinese hamster ovary (CHO) cells are relatively resistant to several retroviruses due to low-level expression of the wild-type cellular proteins used as receptors by these viruses (42). Overexpression of these same functional protein receptors in CHO cells makes them susceptible to viral infection. In our study, we found that the level of expression of TvaR carrying the Cys40Trp mutation is important for the level of susceptibility to ASLV(A) infection. Whereas the wild-type chicken TvaS and quail Tva proteins were efficient ASLV(A) receptors when the proteins were expressed at levels comparable to protein levels normally found on chicken cells, the mutant TvaR protein was not an efficient ASLV(A) receptor. However, expression of TvaR at high levels permits ASLV(A) infection at a level of 5% of wild-type Tva. High levels of the TvaR mutant receptor may increase the avidity of the interaction between the subgroup A envelope glycoproteins on a virion and the TvaR mutant receptors on the cell surface to a level that permits ASLV(A) infection. Alternatively, despite the loss of a disulfide bond from the Cys40Trp mutation, a small percentage of the synthesized TvaR mutant protein may fold into a structure similar to that of wild-type Tva and function efficiently as an ASLV(A) receptor; high levels of TvaR would be required to produce low levels of functional receptor.
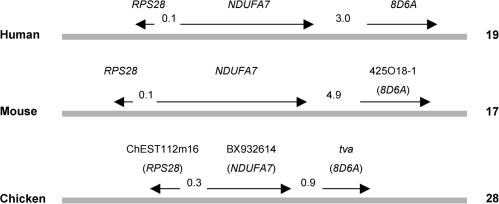
As discussed above, knowing the normal cellular functions of the Tva and Tvb proteins in chicken growth and development would help to identify the evolutionary forces involved in ASLV receptor choice and developing host resistance to ASLV infection. Recent data from genome and proteome projects may provide clues to a possible function for the Tva protein. The tva locus has recently been mapped to the end of chromosome 28 in chickens, which has conserved synteny with human chromosome 19 and mouse chromosome 17 (14, 41). Three chicken genes, RPS28, NDUFA7, and tva, are tightly linked on chromosome 28. The similar regions of human chromosome 19 and mouse chromosome 17 carry RPS28, NDUFA7, and a tva orthologue, 8D6A (GenBank accession no. NC000019), which are tightly linked (Fig. 7). The 8D6A gene encodes a 282-amino-acid protein, named the 8D6 antigen, that is abundantly expressed on follicular dendritic cells (30). It contains two LDL-A modules and a single transmembrane region. The 8D6 antigen appears to be a signaling molecule for the follicular dendritic cell stimulation of B-cell growth in germinal centers (54). Although the normal function of Tva is not known, it is interesting to speculate that it may also function in B-cell development in a way similar to the 8D6 antigen. The principal target organ for ASLV disease is the bursa of Fabricius; ASLV infection produces B-cell lymphomas (33). Tva may be one of the signaling molecules in the bursa and has a role in B-cell development. As discussed above, Tva does not appear to be absolutely required for the development of healthy chickens, and it is possible that related signaling molecules may compensate for the loss of Tva.
FIG. 7.
Maps of the regions encompassing syntenic genes in human, mouse, and chicken. Horizontal bars symbolize chromosomes; the organism name is given on the left, the chromosome number is on the right, and the gene symbols are on the top. The symbols of orthologous human genes are indicated in parentheses. The chicken genes represented by database entries ChEST112m16 and BX932614 showed the highest homology on a whole-genome level to human genes RPS28 and NDUFA7, respectively, as determined by the BLAST program (4). The directions of transcription of the genes are shown with arrows; the numbers between arrows indicate intergenic distances (in kilobases). The drawing is not to scale.
The interaction between the retroviral envelope glycoproteins and the cellular protein receptor is complex, involving multiple determinants in both proteins that specify receptor choice, binding affinity, and the ability to trigger the fusion mechanism of the viral glycoprotein. Despite the complexity of this interaction, retroviruses have the ability to evolve to use different cellular proteins as receptors. This ability may be critical for retroviruses to overcome host resistance and/or for coinfection of the host by several viral subgroups. Understanding the evolutionary mechanisms whereby hosts can develop resistance to retroviral infection and how the virus responds will provide valuable information for antiviral intervention and targeted gene delivery.
Acknowledgments
We thank Ali Fadly at the USDA Avian Disease and Oncology Laboratory (East Lansing, Mich.) for providing CEFs from several of their inbred lines of chickens. We also thank Steve Hughes and Linda Gregory for helpful discussions and critical reading of the manuscript.
This work was supported in part by grant no. 523/04/0489 from the Grant Agency of the Czech Republic (J.S.), by project no. K5011112 awarded by the Academy of Sciences of the Czech Republic (J.S.), and by National Institutes of Health grant AI48682 and the Mayo Foundation (M.J.F.).
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. T. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. T. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 94:11617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins, H. B., J. Brojatsch, and J. A. T. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74:3572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon, L. D., H. D. Hunt, and H. H. Cheng. 2000. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poultry Sci. 79:1082-1093. [DOI] [PubMed] [Google Scholar]
- 6.Balliet, J. W., J. Berson, C. M. D'Cruz, J. Huang, J. Crane, J. M. Gilbert, and P. Bates. 1999. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 73:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnard, R. J., and J. A. T. Young. 2003. Alpharetrovirus envelope-receptor interactions. Curr. Top. Microbiol. Immunol. 281:107-136. [DOI] [PubMed] [Google Scholar]
- 8.Bates, P., L. Rong, H. E. Varmus, J. A. T. Young, and L. B. Crittenden. 1998. Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus. J. Virol. 72:2505-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates, P., J. A. T. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 10.Belanger, C., K. Zingler, and J. A. Young. 1995. Importance of cysteines in the LDLR-related domain of the subgroup A avian leukosis and sarcoma virus receptor for viral entry. J. Virol. 69:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boardman, P. E., J. Sanz-Ezquerro, I. M. Overton, D. W. Burt, E. Bosch, W. T. Fong, C. Tickle, W. R. Brown, S. A. Wilson, and S. J. Hibbard. 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12:1965-1969. [DOI] [PubMed] [Google Scholar]
- 12.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. T. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 13.Cogburn, L. A., R. Morgan, and J. Burnside. 2003. Expressed sequence tags, DNA chip technology and gene expression profiling, p. 629-645. In W. M. Muir and S. E. Aggrey (ed.), Poultry genetics, breeding and biotechnology. CABI Publishing, Wallingford, United Kingdom.
- 14.Elleder, D., J. Plachy, J. Hejnar, J. Geryk, and J. Svoboda. 2004. Close linkage of genes encoding receptors for subgroups A and C of avian sarcoma/leucosis virus on chicken chromosome 28. Anim. Genet. 35:176-181. [DOI] [PubMed] [Google Scholar]
- 15.Federspiel, M. J., L. B. Crittenden, and S. H. Hughes. 1989. Expression of avian reticuloendotheliosis virus envelope confers host resistance. Virology 173:167-177. [DOI] [PubMed] [Google Scholar]
- 16.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 17.Fekete, D. M., and C. L. Cepko. 1993. Retroviral infection coupled with tissue transplantation limits gene transfer in the chicken embryo. Proc. Natl. Acad. Sci. USA 90:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field, J., J. I. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields-Berry, S. C., A. L. Halliday, and C. L. Cepko. 1992. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green, S., I. Issemann, and E. Sheer. 1988. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 16:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hala, K., and J. Plachy. 1997. Inbred strains of chickens, p. 2286-2292. In I. Lefkovitz (ed.), Immunology methods manual. Academic Press, London, England.
- 22.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 23.Hlozanek, I. 1974. Biology of avian RNA tumour viruses, p. 229-296. In F. J. Cleton, D. Crowther, and J. S. Malpas (ed.), Advances in acute leukemia. ASP Biological and Medical Press B.V., Amsterdam, The Netherlands.
- 24.Holmen, S. L., and M. J. Federspiel. 2000. Selection of a subgroup A avian leukosis virus [ALV(A)] envelope resistant to soluble ALV(A) surface glycoprotein. Virology 273:364-373. [DOI] [PubMed] [Google Scholar]
- 25.Holmen, S. L., D. C. Melder, and M. J. Federspiel. 2001. Identification of key residues in subgroup A avian leukosis virus envelope determining receptor binding affinity and infectivity of cells expressing chicken or quail Tva receptor. J. Virol. 75:726-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmen, S. L., D. W. Salter, W. S. Payne, J. B. Dodgson, S. H. Hughes, and M. J. Federspiel. 1999. Soluble forms of the subgroup A avian leukosis virus [ALV(A)] receptor Tva significantly inhibit ALV(A) infection in vitro and in vivo. J. Virol. 73:10051-10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 28.Kingston, R. E., C. A. Chen, and H. Okayama. 1989. Introduction of DNA into eukaryotic cells, p. 911-919. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 29.Klucking, S., H. B. Adkins, and J. A. T. Young. 2002. Resistance of infection by subgroups B, D, and E avian sarcoma and leukosis viruses is explained by a premature stop codon within a resistance allele of the tvb receptor gene. J. Virol. 76:7918-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, L., X. Zhang, S. Kovacic, A. J. Long, K. Bourque, C. R. Wood, and Y. S. Choi. 2000. Identification of a human follicular dendritic cell molecule that stimulates germinal center B cell growth. J. Exp. Med. 191:1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melder, D. C., V. S. Pankratz, and M. J. Federspiel. 2003. Evolutionary pressure of a receptor competitor selects different subgroup A avian leukosis virus escape variants with altered receptor interactions. J. Virol. 77:10504-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 33.Payne, L. N. 1992. Biology of avian retroviruses, p. 299-404. In J. A. Levy (ed.), The Retroviridae, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 34.Rainey, G. J. A., A. Natonson, L. F. Maxfield, and J. M. Coffin. 2003. Mechanisms of avian retroviral host range extension. J. Virol. 77:6709-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rong, L., and P. Bates. 1995. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J. Virol. 69:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong, L., K. Gendron, and P. Bates. 1998. Conversion of a human low-density lipoprotein receptor ligand-binding repeat to a virus receptor: identification of residues important for ligand specificity. Proc. Natl. Acad. Sci. USA 95:8467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong, L., K. Gendron, B. Strohl, R. Shenoy, R. J. Wool-Lewis, and P. Bates. 1998. Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 72:4552-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-0-derived cell line DF-1 supports efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 40.Smith, E. J., A. M. Fadly, and W. Okazaki. 1979. An enzyme-linked immunoabsorbant assay for detecting avian leukosis sarcoma viruses. Avian Dis. 23:698-707. [PubMed] [Google Scholar]
- 41.Smith, J., I. R. Paton, F. Murray, R. P. Crooijmans, M. A. Groenen, and D. W. Burt. 2002. Comparative mapping of human chromosome 19 and the chicken shows conserved synteny and gives an insight into chromosomal evolution. Mamm. Genome 13:310-315. [DOI] [PubMed] [Google Scholar]
- 42.Tailor, C. S., A. Nouri, and D. Kabat. 2000. Cellular and species resistance to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J. Virol. 74:9797-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taplitz, R. A., and J. M. Coffin. 1997. Selection of an avian retrovirus mutant with extended receptor usage. J. Virol. 71:7814-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonelli, M., R. J. Peters, T. L. James, and D. A. Agard. 2001. The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus. FEBS Lett. 509:161-168. [DOI] [PubMed] [Google Scholar]
- 45.Tyers, M., G. Tokiwa, R. Nash, and B. Futcher. 1992. The Cln3-cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 11:1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Q.-Y., K. Dolmer, W. Huang, P. G. W. Gettins, and L. Rong. 2001. Role of calcium in protein folding and function of Tva, the receptor of subgroup A avian sarcoma and leukosis virus. J. Virol. 75:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Q.-Y., W. Huang, K. Dolmer, P. G. W. Gettins, and L. Rong. 2002. Solution structure of the viral receptor domain of Tva and its implications in viral entry. J. Virol. 76:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Q.-Y., B. Manicassamy, X. Yu, K. Dolmer, P. G. W. Gettins, and L. Rong. 2002. Characterization of the LDL-A module mutants of Tva, the subgroup A Rous sarcoma virus receptor, and the implications in protein folding. Protein Sci. 11:2596-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, R. 1982. Experimental biology and assay of RNA tumor viruses, p. 209-260. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumor viruses, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 50.Weiss, R. A. 1992. Cellular receptors and viral glycoproteins involved in retrovirus entry, p. 1-108. In J. A. Levy (ed.), The retroviruses, vol. 2. Plenum Press, New York, N.Y. [Google Scholar]
- 51.Young, J. A. T. 2001. Virus entry and uncoating, p. 87-103. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 52.Young, J. A. T., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, X., Q.-Y. Wang, Y. Guo, K. Dolmer, J. A. T. Young, P. G. W. Gettins, and L. Rong. 2003. Kinetic analysis of binding interaction between the subgroup A Rous sarcoma virus glycoprotein SU and its cognate receptor Tva: calcium is not required for ligand binding. J. Virol. 77:7517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, X., L. Li, J. Jung, S. Xiang, C. Hollmann, and Y. S. Choi. 2001. The distinct roles of T cell-derived cytokines and a novel follicular dendritic cell-signaling molecule 8D6 in germinal center-B cell differentiation. J. Immunol. 167:49-56. [DOI] [PubMed] [Google Scholar]
- 55.Zingler, K., C. Belanger, R. Peters, D. Agard, and J. A. T. Young. 1995. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J. Virol. 69:4261-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zingler, K., and J. A. T. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]