Abstract
Pathogenic bacterial contamination is a major threat to human health and safety. In this review, we summarize recent strategies for the integration of recognition elements with nanomaterials for the detection and sensing of pathogenic bacteria. Nanoprobes can provide sensitive and specific detection of bacterial cells, which can be applied across multiple applications and industries.
Table of Contents Graphic
The integration of recognition elements with nanomaterials provides a synergystic strategy for sensing of bacteria
Introduction
Rapid detection of pathogenic bacteria enables the reduction of food- and water-borne outbreaks in industrial settings, clinical and hospital diagnostics, and water and environmental quality controls as well as in resource-limited settings.1–2 Bacterial contamination of foods accounts for approximately one-third of global deaths and results in approximately 47.8 million illnesses in the United State each year, in addition to costly recalls.3 The majority of bacterial illnesses are a result of an infection or intoxication from Staphylococcus aureus (S. aureus), Salmonella typhimurium (S. typhimurium), Escherichia coli (E. coli) O157:H7, Listeria monocytogenes, Tuberculosis, Streptococcal, Clostridium perfringens, and Bacillus cereus. Although the use of antibiotics can treat most bacterial infections, several pathogenic bacteria have become resistant to one or more antibiotics, leading to a serious problem. According to World Health Organization, current antibiotics will lose effectiveness to control pathogens over the next 1–2 decades.4. In the food and hospital processing environment, the formation of bacterial biofilms on the surface of production equipment can increase fouling, promote corrosion, and contaminate product, leading to increased costs and risk.5
There is an urgent need to develop accurate and early-stage screening methods to help reduce the risk of these emerging threats in food, medical, and environmental settings. Common methods to detect and quantify bacteria include traditional plate counting and polymerase chain reaction (PCR). Plate counting allows an estimation of the number of viable bacteria in sample, while PCR enables the detection specific DNA or RNA originating from target bacterial cells. Although these methods can be sensitive and specific, they require significant sample preparation, and can increase total assay time beyond 18 hours. There is an ongoing challenge in the food and medical fields to reduce the time required for results, and improving mitigation responsiveness. Thus, a compelling and urgent need exists to improve the current methods for the rapid detection of bacteria.
Biosensor-based detection strategies are a promising tool to meet the needs described above. The key components of a biosensor include a recognition element that binds to target analytes, and a transducer that translates the binding event to a measurable signal.6 The performance of biosensors is determined by response time, dynamic range, limit of detection (LOD), single-to-noise ratio, and specificity.6 Currently, the widespread implementation of biosensors in real samples is limited by these factors. Nanomaterials can enhance the performance of biosensors, owing to their unique physicochemical properties. Nanomaterials functionalized with recognition elements have the ability to create advanced recognition and transduction processes, which can improve biosensor performance. Specifically, the large surface area of nanomaterials can allow for a more efficient capture of analytes during biosensing events.
This review will cover the synergy between recognition elements and nanomaterials in biosensors for bacterial detection. We will highlight the main biosensing recognition elements, including antibodies, aptamers, bacteriophages (phages), and electrostatic interactions. A wide range of advanced nanomaterials and biorecognition elements will be covered; however, because of the breadth of this field, the materials covered are not all inclusive. Additionally, this review will suggest future directions to build recognition element-conjugated nanoprobes to improve detection and sensing of bacterial cells.
Fabrication of nanoscale probes
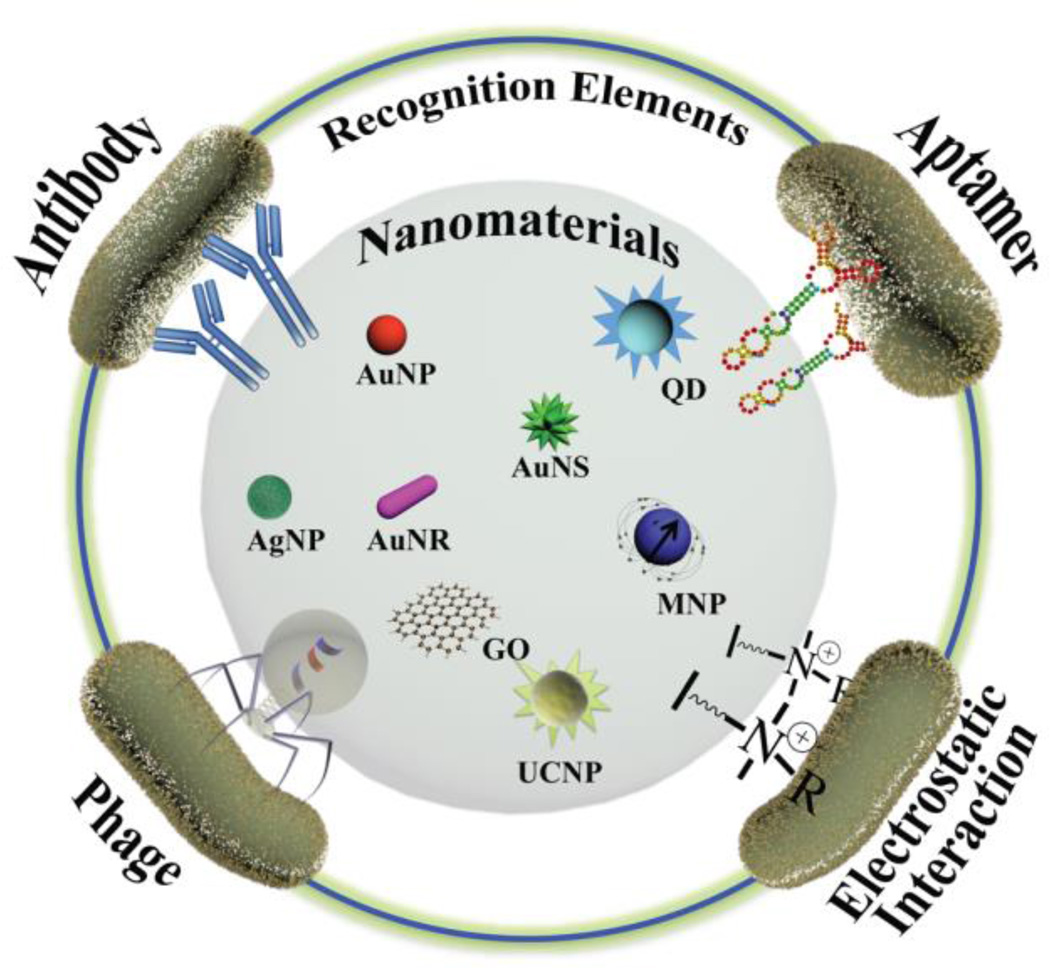
Advances in nanotechnology have allowed new nanomaterial-enabled detection strategies for the rapid and sensitive detection of pathogenic bacteria. To take advantage of the unique properties of nanomaterials and fabricate bacteria-selective components, recognition elements have been conjugated onto the surface of nanomaterials, including gold nanoparticles (AuNPs), gold nanorods (AuNRs), magnetic nanoparticles (MNPs), graphene oxide (GO), quantum dots (QDs), and upconverting nanoparticles (UCNPs). These nanomaterials have been functionalized with recognition elements, such as antibodies, aptamers, phages, and electrostatic interaction-based ligands, to specifically recognize and bind to epitopes on the surface of bacterial cells (Fig. 1).
Fig. 1.
Schematic representation showing the fabrication of recognition elements on nanomterials as bacteria-selective nanoprobes to detect and sense bacterial cells.
The immobilization of recognition elements on nanomaterials is an important step in the design of nanoscale probes to enhance biosensor performance. Several methods have been reported for the bioconjugation of nanomaterials, including physical adsorption, biotin-streptavidin binding, and covalent bonding.7 Physical adsorption is a non-specific interaction between the recognition elements and nanomaterial via hydrogen bonds, hydrophobic interactions, and van der Waals forces. Although this method is facile, the random orientation of the biomolecules can result in low target capture efficiency, and possible instability can trigger the release of target bacterial cells when the local environment is altered (e.g. variation in pH, salt concentration, and/or temperature). Since biotin-conjugated antibodies, aptamers, and engineered phages are available, the biotin-streptavidin system has become more popular due to their high binding capacity. Covalent bonding, however, provides the most robust connectivity, in particular amide bonds.
Antibody-based detection of bacteria
Antibodies (IgG) are large Y-shaped proteins that are the most commonly used biorecognition elements to capture bacterial cells due to their versatility and ease of integration into biosensing events. The three categories of IgG antibodies employed in immunology-based assays include polyclonal, monoclonal, and engineered antibody fragments.
Antibody-based Plasmonic Nanoprobes
A wide range of gold nanomaterials with varying sizes and shapes (e.g. gold nanospheres (AuNP), gold nanorods (AuNR), gold nanowires, and gold nanostars) have been fabricated.6 Differing structures and morphologies result in tunable optical/plasmonic properties in the visible to near-infrared region, providing optical signals for bacterial cell detection. The signals can be observed by monitoring the change in localized surface plasmon resonance (LSPR) within the gold nanomaterials. Although silver nanomaterials also have surface plasmon resonance, their instability to air, and potential toxicity to bacteria can limit their utilization for the detection of bacterial cells.
Due to the unique properties of LSPR, gold nanoparticles modified with antibodies have been widely used to develop bacterial biosensors. A change in the refractive index of antibody-conjugated gold nanomaterials causes a shift in the absorbance spectrum peaks, thus indicating the presence of target bacterial cells. For example, a plasmon peak shift caused by LSPR has been used to determine the presence of Salmonella cells as a result of the binding between the epitopes of target bacterial cells and antibody-conjugated AuNPs.8 However, the poor detection limits of antibody-conjugated AuNPs limit their utility for bacteria detection. Consequently, AuNRs have been used as a replacement for similar sized AuNPs due to their inherently higher sensitivity to a local dielectric environment.9
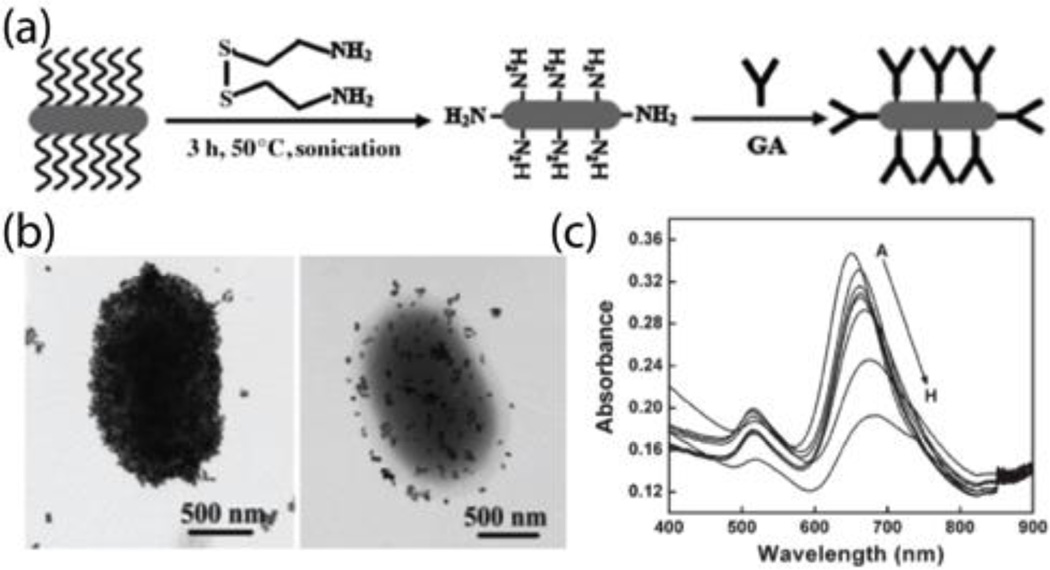
Wang et al. developed a sensor using antibody-conjugated AuNRs to detect E. coli cells. As shown in Fig. 2a, AuNR nanoprobes were fabricated by functionalizing AuNRs with anti-E. coli antibodies, which served as the recognition elements to capture target bacterial cells. The transmission electron microscopy (TEM) micrographs in Fig. 2b allowed visualization of the binding between the nanoprobes and target bacterial cells. The specific binding between the AuNR nanoprobes and bacterial cells resulted in a red shift in the AuNR plasmon band. With an increase of target E. coli cell concentration, a larger red shift and lower intensity of longitudinal peak bands were observed (Fig. 2c), with a limit of detection as low as 102 colony-forming units per mL (CFU·mL−1) was achieved in less than 30 minutes. Furthermore, multiple pathogenic bacterial strains can be detected using different types of antibody-functionalized AuNR nanoprobes. For example, anti-E. coli and anti-S. typhimurium antibodies-functionalized onto AuNRs with different aspect ratios (and hence differing optical properties) can simultaneously detect E. coli and S. typhimurium cells at the concentration of 104 CFU·mL−1.9 AuNRs bifunctionalized with magnetic nanoparticles and antibodies were also developed to detect target bacterial cells based on plasmonic resonance. Here, the magnetic properties of the binanoprobes were used to separate, purify, and concentrate the target bacterial cells.10
Fig. 2.
(a) Schematic representation of the fabrication of anti-E. coli antibodies-conjugated AuNR nanoprobes. (b) TEM images of the specific binding of anti-E. coli antibodies-conjugated AuNR nanoprobes with E. coli cells with different coverage. (c) UV-vis absorbance spectra of anti-E. coli antibodies-conjugated AuNRs with various concentrations of E. coli cells (from 102 to 106 CFU·mL−1). Reproduced with permission from ref. 9. Copyright 2008, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Although the theory behind LSPR technique is straightforward, the requirements of skilled operators and sophisticated instruments result in challenges for commercial applications in low-resource settings. Fortunately, colorimetric assays can help overcome these issues by developing portable, easy-to-use, and user-friendly devices for in-situ analysis. The aggregation and disaggregation of plasmonic nanomaterials with appropriate sizes has been reported for the analysis of a wide range of analytes.6 As a result of inter-particle crosslinking or destabilized aggregation of plasmonic gold nanomaterials in the presence of target analytes, the color of the detection solution changes from red to blue, or the reverse. This color change can be visually observed by the naked eye. The principle behind this system is that gold nanomaterials modified with antibodies reduce the distance between the individual gold nanomaterials, resulting in inter-particle plasmon coupling and color change.
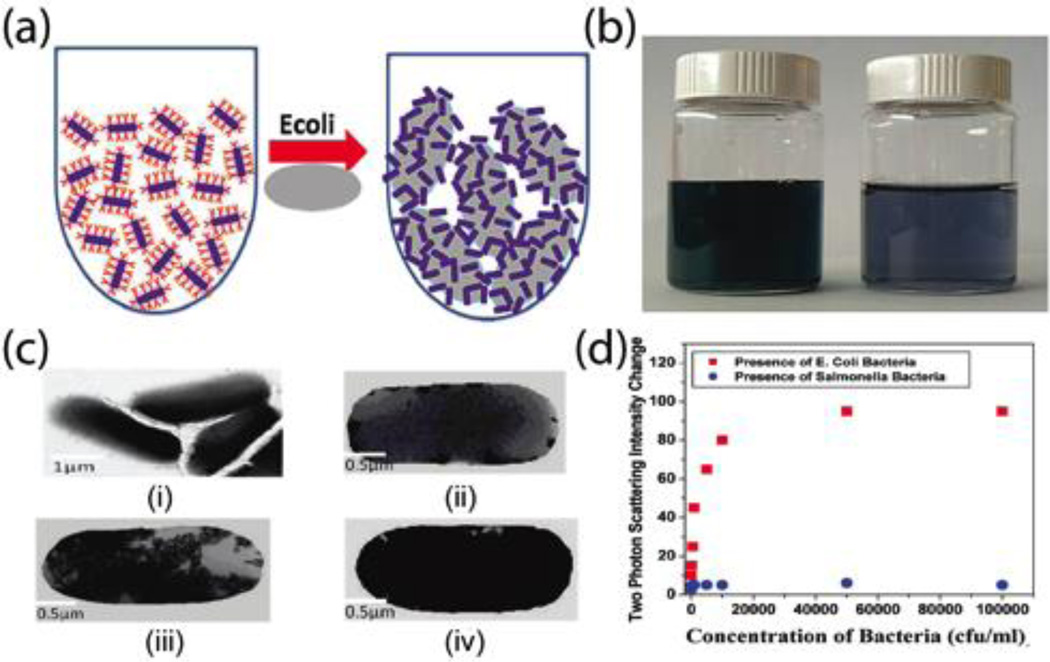
Antibodies on gold nanomaterials can specifically recognize and bind to bacterial cells through antibody-antigen interactions. Singh et al. reported anti-E. coli antibody-conjugated AuNRs to selectively detect E. coli O157:H7 in an aqueous solution at a concentration as low as 50 CFU·mL−1.11 Their results indicated the intensity of two-photon Rayleigh scattering of antibody-conjugated AuNRs increased 40-fold in the presence of various competing E. coli cell concentrations. The schematic in Fig. 3a shows the mechanism for the detection of E. coli cells using anti-E. coli antibody-conjugated AuNR nanoprobes. The size of bacterial cells (1–3 µm) is much larger than that of AuNRs, resulting in numerous antibody-conjugated AuNRs that can attach to one bacterial cell, promoting the aggregation of AuNRs. Depending on the concentration of bacterial cells, the degree of aggregation can result in different color shifts, ranging from dark green to blue (Fig. 3b). The aggregation of antibody-conjugated AuNRs on the surface of bacterial cells was imaged using TEM (Figure 3c), and the two-photon scattering intensity change of the detection solutions against various concentrations of target bacterial cells is shown in Figure 3d. The intensity of the new band appearing around 950 nm was used to indicate the aggregation of AuNRs after the addition of target bacterial cells (Figure 3d). In their report, the specificity of antibody-conjugated plasmonic nanoprobes was demonstrated against competing bacterial cells, including E. coli O157:non-H7 and E. coli O157:NM. Similarly, antibody-conjugated oval-shaped gold nanoparticles have been utilized for colorimetric detection of S. typhimurium based on the aggregation of plasmonic nanoprobes. As target bacterial cell concentrations increase, the color of the detection solutions changes from pink to blue.12
Fig. 3.
(a) Schematic representation of colorimetric detection of E. coli bacterial cells using anti-E. coli antibody-conjugated gold nanorods. (b) Photograph of the color changes before and after adding E. coli bacterial cells. (c) TEM images of aggregation of anti-E. coli antibody-conjugated AuNRs on the surface of E. coli bacterial cells with various concentrations (i. control; ii. 102; iii. 8 × 104; and iv. 107 CFU·mL−1) (d) Plot of two photon scattering intensity change against bacteria concentrations. Reproduced with permission from ref. 11. Copyright 2009, American Chemical Society.
Antibody-based Magnetic Nanoprobes
Immunomagnetic separation (IMS) uses antibody-conjugated magnetic beads (typically 1–3 micron in diameter) as a tool to separate, concentrate, and purify target bacterial cells from complex matrices. This method can be combined with numerous detection methods, including fluorescent, colorimetric, electrochemical, chemiluminescent, surface-enhanced Raman scattering (SERS), surface plasmon resonance (SPR), and quartz crystal microbalance (QCM). Although IMS is widely used for bacterial capture, low capture efficiencies limit its application in complex matrices. To increase capture efficiency, superparamagnetic magnetic nanoparticles (e.g. Fe3O4 and Fe2O3,) have been reported to increase capture efficiency for pathogenic bacteria separation from food samples because of their increased surface to volume ratio. 13
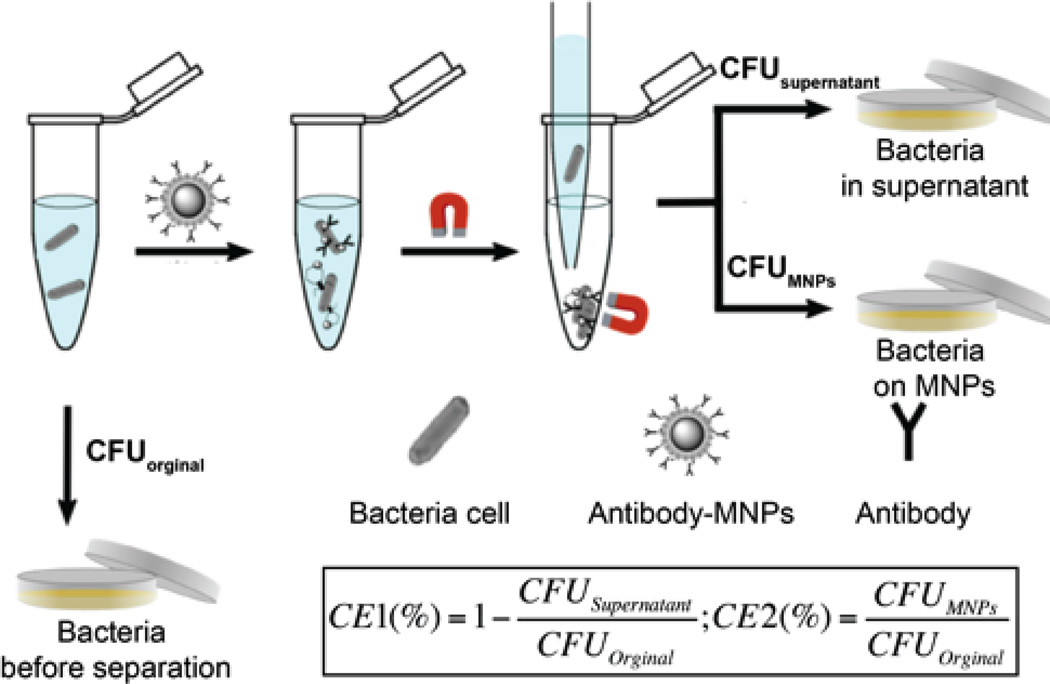
When detecting in complex matrices, magnetic separation without centrifugation can increase detection sensitivity by concentrating bacteria away from interferents. MNPs modified with antibodies can recognize and attach to the antigens on the surface of bacterial cells. As shown in Fig. 4, bound bacterial cells can be separated from sample solutions using an external magnetic field. The capture efficiency, calculated using the equation shown in Fig. 4, was introduced to measure the performance of antibody-conjugated magnetic nanoprobes.14
Fig. 4.
Magnetic separation of bound bacteria on the magnetic nanoprobes from unbound bacteria in the supernatant. Where, CFUoriginal is the total number of bacterial cells present in the initial sample, CFUsupernatant is the number of bacterial cells which remained unbound to the magnetic nanoprobes, and CFUMNPs is the number of bacterial cells bound to magnetic nanoprobes. Reproduced with permission from ref. 14. Copyright 2015, Royal Society of Chemistry.
The most commonly used magnetic nanoparticle for bacteria separation is iron oxide (Fe3O4). These nanoparticles have been reported to provide a capture efficiency of greater than 90% in real food samples.15 Unfortunately, these MNPs, having a diameter of 90 nm, required one hour for separation due to low magnetic efficiency. To decrease separation time, metal alloy MNPs, such as CoFe2O4, PtFe2O4, and MnFe2O4, were bio-functionalized for bacterial separation as well as biomedical applictions.16 Multifunctional magnetic nanoprobes have also been used to simultaneously separate and detect bacterial cells. For example, antibody-modified Fe3O4/TiO2 core/shell magnetic nanoprobes were used to separate and detect Salmonella strains.17 Wang et al. reported the use of antibody-conjugated AuNRs decorated with Fe3O4 MNPs, including Fe3O4-AuNR-Fe3O4 nanodumbbells and a Fe3O4-AuNR necklace, for multiple pathogenic bacteria detection.10
Aptamer-based detection of bacteria
Aptamers are single stranded nucleic acids (DNA or RNA) that offer several advantages over antibody-based recognition elements for the capture of bacterial cells. Aptamers are low-cost, chemically stable, and can be synthesized at a large scale. Due to their small size (typically 3–5 nm), aptamers can exhibit high binding affinity for target bacterial cells, resulting in a decrease in the overall detection limit. Aptamers can be designed for a variety of target bacteria using Systematic Evolution of Ligands by Exponential enrichment (SELEX), to bind epitopes on the surface of bacterial cells.18 During an evolution process, a large DNA library is added into a solution containing bacterial cells and incubated. The unbound DNA is then separated from bacterial cells, and bound DNA is eluted and amplified via PCR. The SELEX process is then repeated until the DNA has a high affinity for the target bacterial cells. Once an effective sequence has been identified, aptamers can be functionalized with –SH, -NH2, and -COOH groups, providing a straightforward way to immobilize aptamers to nanomaterials. Aptamer use in both fluorescence and SERS-based detection methods will be described in this section.
Aptamer-based Fluorescent Nanoprobes
Fluorescent nanomaterials feature high extinction coefficients and good photostability, relative to small molecule fluorophores. In addition to their high photostability, fluorescent nanomaterials with narrow and size tunable emission spectra allow researchers to precisely label target bacterial cells. Most importantly, fluorescent nanoparticles with differing colors, modified with diverse recognition elements can be used to detect various types of bacterial cells simultaneously.
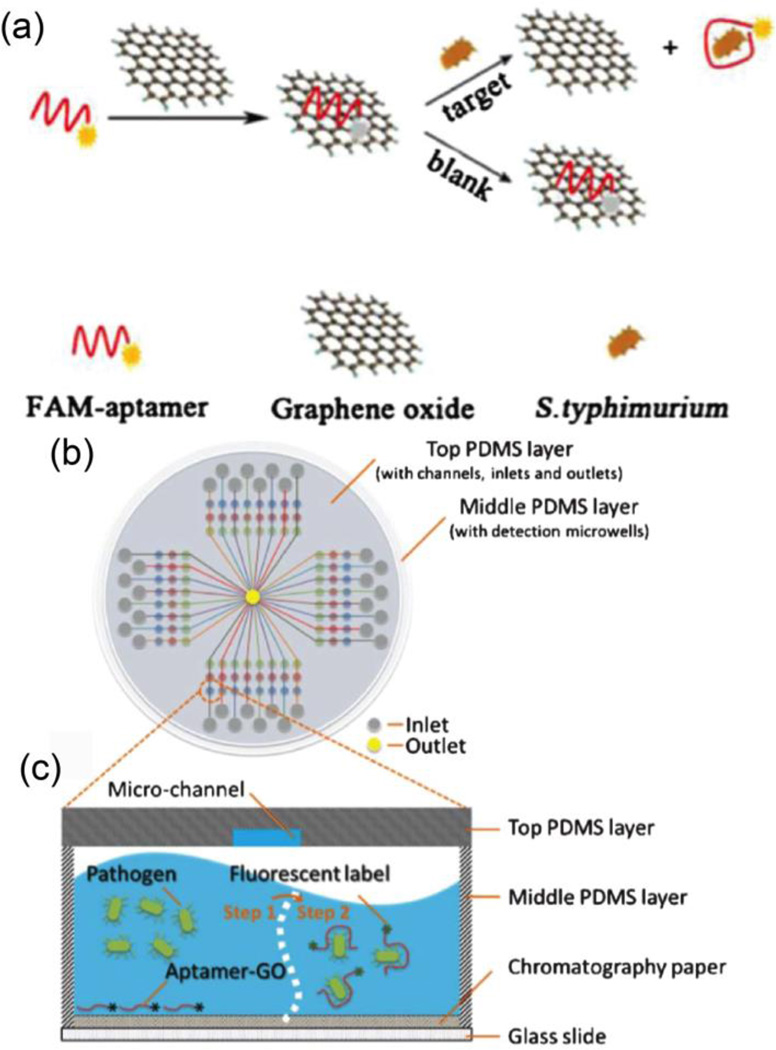
Nanomaterials, such as AuNPs and graphene oxide (GO), can serve as fluorescence quenchers to detect bacterial cells through fluorescence resonance energy transfer (FRET). Aptamers conjugated with fluorophores can interact with quencher nanomaterials, turning off the visible fluorescence via FRET. In the presence of target bacteria, the aptamers recognize the bacterial cell surface, and release from the surface of the nanomaterial quencher. The resulting increase in fluorescence is used to quantify the concentration of bacterial cells (Fig. 5a). By employing this principle, Duan et al. used an FAM-aptamer/GO complex as a nanoprobe to detect S. typhimurium with a dynamic range from 103 to 108 CFU·mL−1 and a detection limit of 100 CFU·mL−1. This system can distinguish S. typhimurium from S. paratyhhi A, S. cholera-suis, S. aureus, and E. coli K88.19 Furthermore, Zuo et al. applied aptamer-functionalized GO nanoprobes in a microfluidic biochip to detect Lactobacillus acidophilus (Fig. 5b,c).20 The limit of detection was determined to be 11 CFU·mL−1, with an assay time of 10 minutes.
Fig. 5.
(a) Schematic illustration of a graphene oxide sensing platform for the detection S. typhimurium (Bacterial cells concentration can be detected by turning on the fluorescence signal after the aptamer is bind to target bacterial cells). (b) Schematic representation of the PDMS/paper hybrid microfluidic chips for one-step pathogenic bacteria detection. (c) Illustration of the pathogenic bacteria detection principle inside the microfluidic channels. Reproduced with permission from ref. 19. Copyright 2014, Springer-Verlag Wien and ref. 20. Copyright 2013, Royal Society of Chemistry.
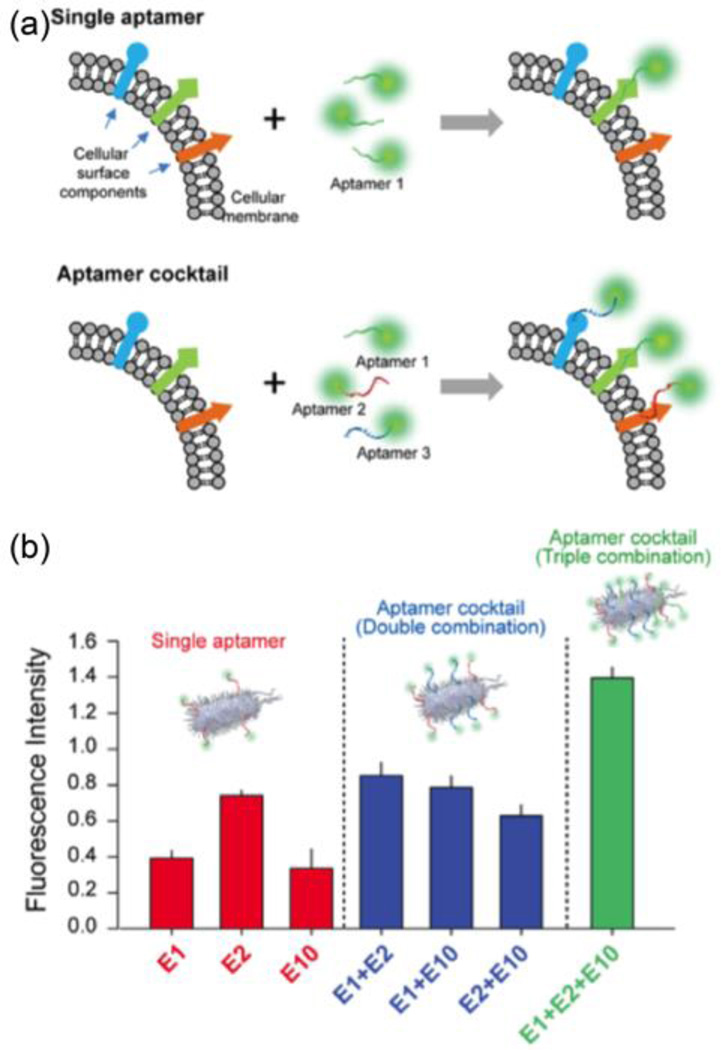
Because the bacterial surface contains numerous possible binding moieties such as polysaccharides, proteins, and flagella, Kim et al. used a cocktail of aptamers with various targets to detect E. coli cells (Fig. 6a).21 As shown in Fig. 6b, a mixture of aptamers resulted in a higher fluorescence signal than any single aptamer, indicating that a cocktail of aptamers could enhance the sensitivity. Their report indicated that an 18-fold lower limit of detection was achieved using a cocktail of aptamers to detect E. coli bacterial cells when compared with a single aptamer.
Fig. 6.
(a) Schematic illustration of single enhancement by cocktail aptamers for bacteria cell detection. (b) Fluorescence intensity of bacterial cells suspensions obtained after the fluorescence nanoprobes were labelled with single, double, and triple aptamers. Reproduced with permission from ref. 21. Copyright 2013, Elsevier B.V.
The ability to simultaneously detect multiple bacteria species is advantageous in many fields including medical diagnostics, food safety, and environmental monitoring. Researchers have demonstrated the ability to perform multiplex detection using several aptamer-nanoprobe combinations during a single assay. Duan et al. reported dual-color aptamer-conjugated fluorescent nanoprobes to simultaneously label and detect S. typhimurium and S. aureus.22 In the presence of target bacterial cells, the fluorescent nanoprobes attached to target bacterial cells, providing fluorescence readout using a 980 nm laser. This proposed method had a dynamic range of 101–105 CFU·mL−1, and a detection limit of 5 CFU·mL−1 for S. typhimurium cells and 8 CFU·mL−1 for S. aureus cells. The specificity was demonstrated against competing bacterial cells (e.g. Listeria, E. coli, Vibrio, E. sakazakii, and Streptococcus).The same researchers used aptamer-conjugated, dual-color quantum dots to simultaneously detect Vibrio parahaemolyticus and Salmonella typhimurium.23 Fluorescence-based nanoprobes containing aptamers show good promise for multiplex and sensitive detection of bacteria. However, their use in complex matrices may result in quenching of the fluorophores.
Aptamer-based SERS nanoprobes
Metallic nanoparticles have attracted increasing attention for surface-enhanced Raman scattering (SERS)-based detection of chemical and biological agents.24 The Raman signals of bacterial cells can be selectively and sensitively enhanced using plasmonic metal nanomaterials modified with recognition elements. This modification allows for the detection of bacteria by the interaction between electrons from the target bacterial cells and the metal nanomaterials.25 The vibrational or rotational transitions on the Raman spectrum correspond to specific molecular structures on the surface of bacterial cells, providing a set of chemical fingerprints. These fingerprint spectra are helpful in distinguishing different species of bacterial cells.
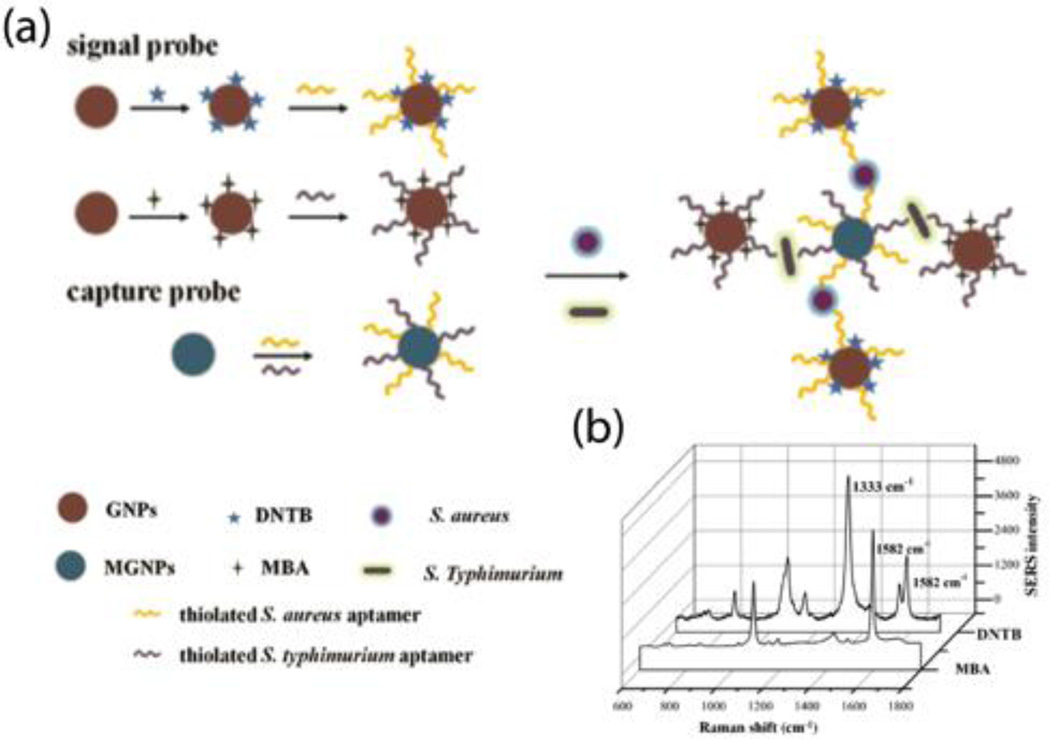
The basic principle behind aptamer-SERS detection of bacterial cells involves the utilization of aptamer-conjugated nanomaterials with numerous “hotspots”, providing a strong SERS signal. Both the aptamers and the nanoprobes are labeled with Raman reporter molecules that can specifically bind to target bacterial cells. The Raman reporter molecules are then used to quantify the concentration of bacterial cells. Ravindranath et al. demonstrated a cross-platform approach to simultaneously detect three pathogens using three aptamer-conjugated nanoprobes modified with different Raman reporter molecules.26 The total detection time was less than 45 minutes, and a detection limit of 103 CFU·mL−1 was obtained. Similarly, an AuNR-enhanced SERS aptasensor has also been developed for the simultaneous detection of S. typhimurium and S. aureus.27 As shown in Fig. 7a, AuNPs were labeled with Raman reporter molecules and two types of aptamers as nanoprobes for the detection of bacterial cells. Here, MNPs were also modified with two types of aptamers to capture and concentrate S. aureus and S. typhimurium cells. As shown in Fig. 7b, the peaks at 1333 cm−1 and 1582 cm−1 indicate the presence of S. aureus and S. typhimurium, respectively. The intensity at the specific peaks from the Raman reporter molecules contributed to the detection limit of 35 CFU·mL−1 for S. aureus and 15 CFU·mL−1 for S. typhimurium. This system has the ability to specifically detect S. aureus and S. typhimurium from E. coli, V. parahaemolyticus, B. cereus, and S. dysenteriae.
Fig. 7.
(a) Schematic illustration of the aptamer-conjugated nanoprobes for simultaneous detection of S. aureus and S. typhimurium based on SERS reporter molecules on AuNPs. (b) The Raman spectra of reporter molecules indicating the present of S. aureus and S. typhimurium in detection solution. Reproduced with permission from ref. 27. Copyright 2015, Elsevier B.V.
Bacteriophage-based detection of bacteria
The challenges presented by antibodies and aptamers, which include batch-to-batch variations, robustness in complex matrices, and relatively high cost, have led to research towards alternative biorecognition elements. Bacteriophages, also known as phages, are bacteria-infecting viruses consisting of nucleic acids, protein capsid, and tail fibers. The tail fibers serve as recognition elements that can specifically recognize and bind to receptors on the surface of target bacterial cells. Most phages are approximately 100 nm, making them ideal to be used as bio-nanomaterial probes for bacteria detection. Depending on the particular phage, the selectivity can be either narrow or broad. In addition to their target specificity, another key feature of phages is their ability to distinguish viable versus inactive host bacterial cells, because phages can only replicate and express enzymes within viable bacterial cells.28 The specificity of a phage-based detection system requires careful host-range screening of phage libraries. Also, it is relatively easy and inexpensive to synthesize and purify phage, providing a new platform to detect bacteria. Thus, bacteriophages offer significant advantages when considering solutions for bacterial cell detection.
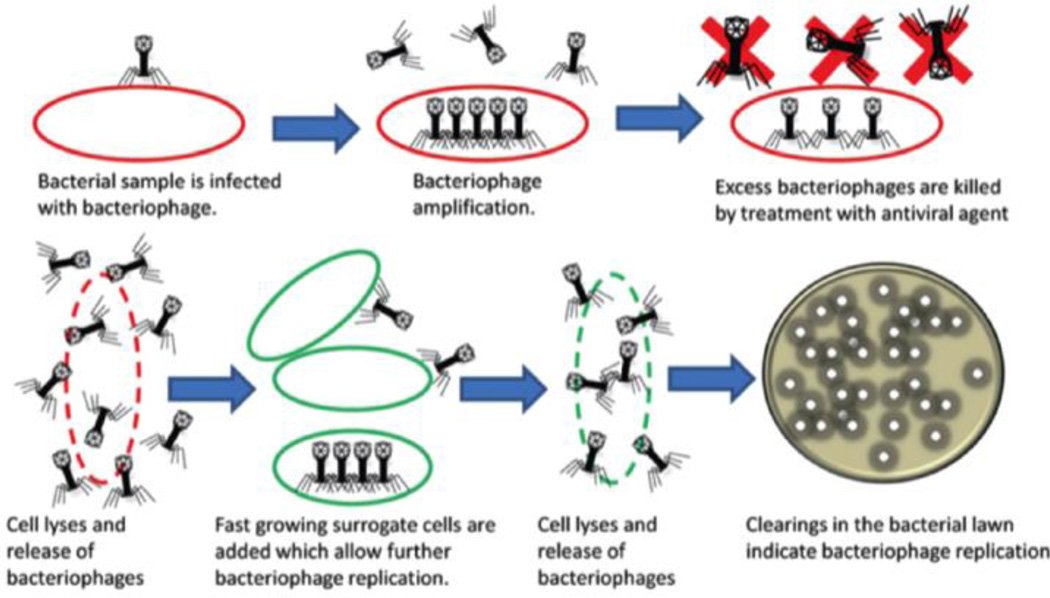
Phage lysis assay-based detection
Phages are obligate intracellular parasites, which can only replicate within target bacterial hosts.28 The phage amplification assay is illustrated in Fig. 8. Following attachment to a suitable host cell, the phage will take over the cell’s machinery to replicate DNA, synthesize proteins, and finally lyse the host cell to release the replicated virus. The released phage can then find another suitable host cell and initiate a new infection cycle. At the end of incubation, the number of phages can be quantified using conventional plating methods. The increased number of phages in solution can be used to calculate the concentration of target bacterial cells. Based on phage amplification assays, some commercial diagnostic kits are available for the detection of S. aureus, Yersinia pestis, and Mycobacterium tuberculosis. To improve the detection of pathogenic bacteria, phage amplification assays have been combined with other technologies, such as mass spectrometry, enzyme-linked immunosorbent assay (ELISA), and PCR to decrease the total detection time.29–30 For example, Martelet et al. developed a highly sensitive strategy for rapid and unambiguous detection of viable E. coli29 The amplified phages were quantified using mass spectrometry combined with liquid chromatography. Their results indicated a limit of detection as low as 1 CFU·mL−1 for viable E. coli in food matrices following an 8 hour infection.
Fig. 8.
Schematic illustrations of bacteriophage replication assays. Reproduced with permission from ref. 28. Copyright 2014, Royal Society of Chemistry.
After the phage infection cycle, the intracellular components are released from the host cell and can be used as an indicator for the target bacterial cells. Some examples of intracellular components include β-galactosidase, adenosine triphosphate (ATP), and adenylate kinase. The enzymatic activity of phage-mediated released β-galactosidase has been quantified using colorimetric, electrochemical, and chemiluminescent strategies. Derda et al. reported a colorimetric method to detect 50 CFU of E. coli cells in 1 liter of water in less than 4 hours.31 After adding luciferase and luciferin, the released ATP produced a bioluminescence signal. Their reports indicated the detection of E. coli and S. newport (selected as Gram positive and Gram negative bacteria models) with a detection limit of 103 CFU within 1 hour and 2 hours, respectively.32 Additionally, amperometric detection of Enterobacteriaceae cells in river water was performed, and reached a limit of detection at 5 × 104 CFU·mL−1 with an incubation of 2 hours, and 10 CFU·mL−1 after a pre-enrichment of 7 hours.33
Engineered Phage-based detection
Engineered phages based on genetic engineering technologies are able to transfer genes of interest into specific target bacteria, which then express and amplify the gene product. This amplification using engineered phages can improve detection of bacteria. Therefore, engineered phages provide a potential low-cost tool for specific, rapid, and sensitive detection of bacteria. To reduce the background signal and decrease the variations between different bacteria strains, phage components (e.g. capsid proteins, nucleic acid) have been labeled with fluorophores using genetic engineering. Due to the relatively large surface area of capsid proteins, phage heads labeled with a fluorescent tag can increase the sensitivity.34 A common fluorescent tag is green fluorescence protein (GFP), which is added on the small outer capsid protein on the phage head.34 Recently, engineered phages carrying an enzymatic reporter gene have attracted increasing attention for bacterial cell detection. Firefly luciferase, the most commonly used enzyme, has been reported to detect various bacterial cells.35 In addition, T7 phages were engineered to carry alkaline phosphatase or tobacco etch virus protease gene, generating phage-based platform for bacteria detection. 36–37
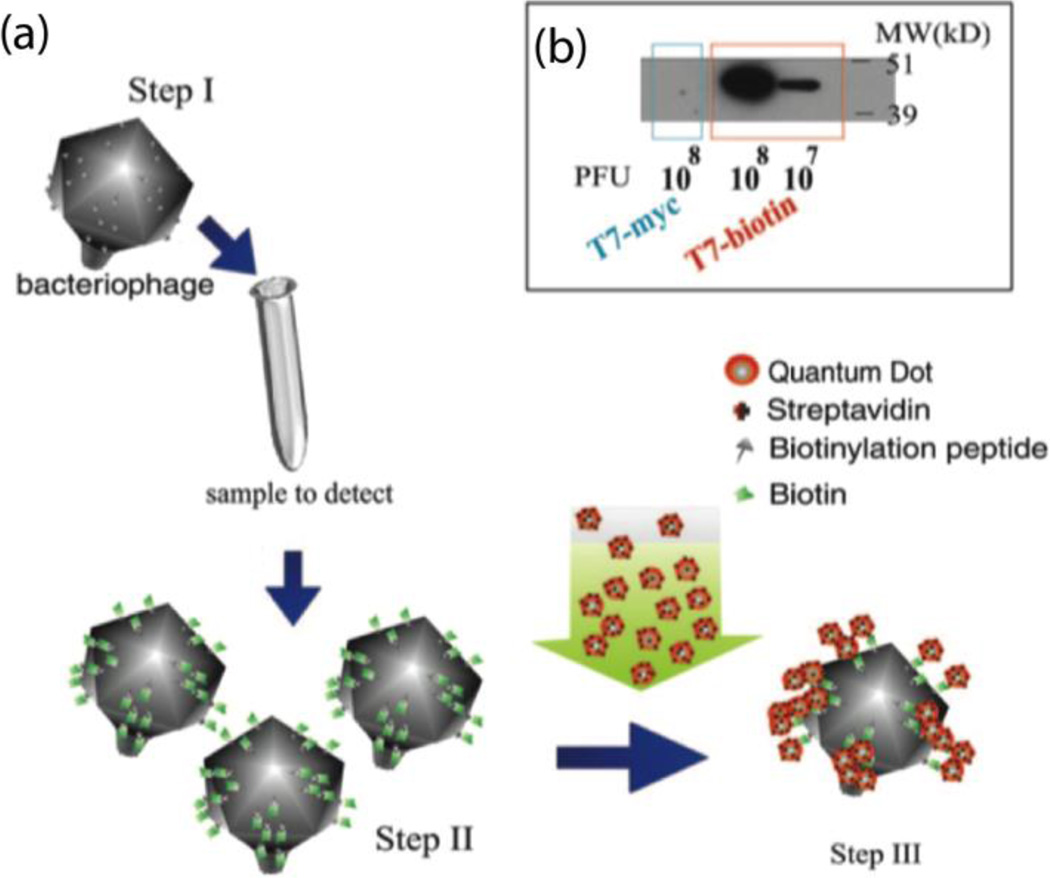
Edgar et al. reported a rapid and simple method to detect E. coli (Fig. 9).38 The T7 phage was engineered to express biotinylation peptide on the phage capsid proteins. During the phage infection cycle for bacteria cell detection, the peptide is biotinylated inside the host bacterial cells. Streptavidin-coated quantum dots were used to label biotinylated phages for fluorescent quantification. The detection limit of E. coli cells was shown to reach as low as 10 CFU·mL−1, a 100-fold amplification of readout signal over the background signal within 1 hour.
Fig. 9.
Schematic representation of bacteria detection using engineered T7 phages labelled with quantum dots via streptavidin-biotin interaction. (b) Western blot analysis of T7biotin and T7control phage particles. Reproduced with permission from ref. 38. Copyright 2006, National Academy of Sciences.
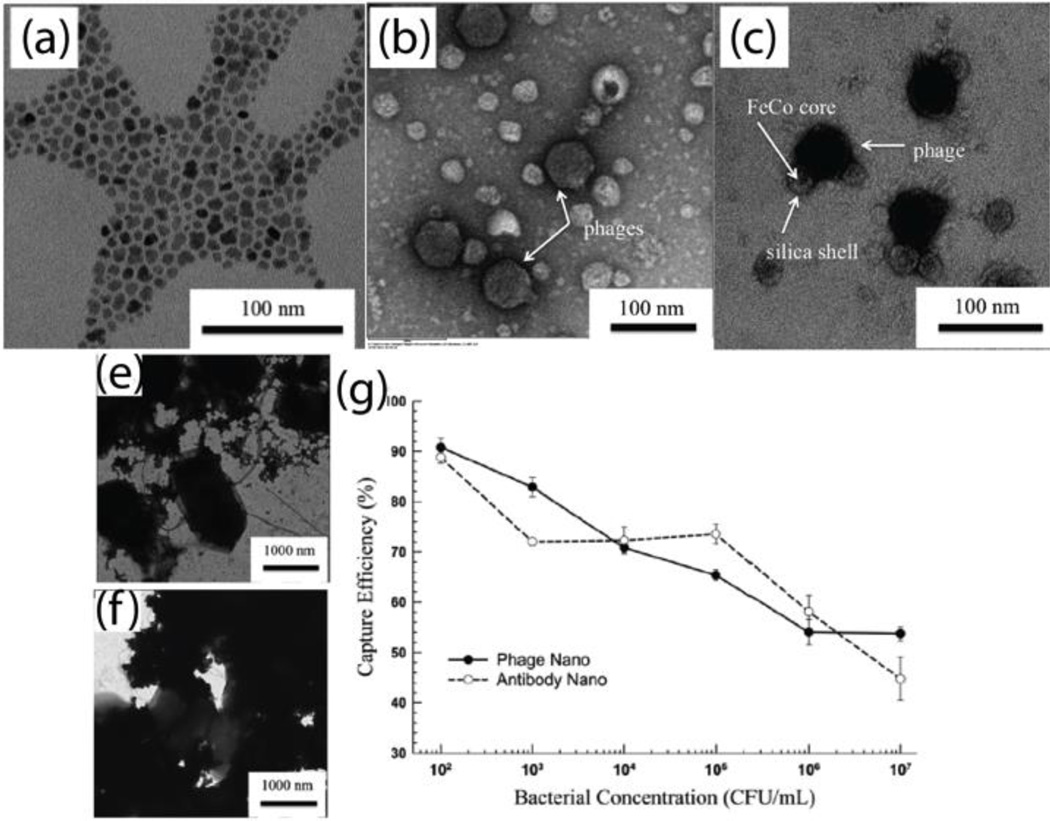
Other methods for sensing bacteria using phages include replacing antibodies with phages on magnetic beads, and phage-coating magnetic beads in combination with other technologies (e.g. SERS, SPR, and QCM). These methods have successfully detected several pathogenic bacteria.39 Taking advantage of the high surface-to-volume ratio of MNPs, Chen et al. fabricated T7 phage-based magnetic FeCo nanoprobes for bacteria separation.14 The FeCo MNPs were functionalized with streptavidin, which were bound by biotinylated T7 phages via streptavidin-biotin interaction. The TEM micrographs of streptavidin coated FeCo MNPs, biotinylated T7 phages, and T7 phage-MNPs complex are shown in Fig. 10a–c. This report indicated similar bacterial capture efficiency between antibody-and phage-conjugated nanoprobes (Fig. 10g). However, phage-decorated nanoprobes provided some advantages, including relative ease of production, ability to distinguish between viable and inactivated bacterial cells, and controllable host range. Moreover, phage-conjugated nanoprobes provide reliable specific binding, low-cost production at a large scale, as well as high tolerance to temperature and pH.14
Fig. 10.
TEM images of (a) FeCo MNPs, (b) negatively stained T7 phage particles, (c) positively stained biotinylated phage bound to streptavidin-coated FeCo MNPs, (e) antibody-conjugated MNPs attached on the surface of bacterial cells, (f) phage-modified magnetic nanoprobes attached on the surface of bacterial cells. (g) Comparison the capture efficiency between antibody- and phage-conjugated magnetic nanoprobes. Reproduced with permission from ref. 14. Copyright 2006, Royal Society of Chemistry.
Electrostatic interaction-based sensing of bacteria
Charged ligands on nanomaterials can bind to bacterial cells through electrostatic interactions, serving as the recognition element. Unlike the previous recognition elements (e.g. antibody, aptamer, and phage), electrostatic interaction-based recognition processes provide an alternative sensing strategy for bacterial cells, relying on non-specific recognition.40 These electrostatic interaction-based approaches are versatile, and can provide a sensing platform to either detect a wide range of bacterial strains, or recognize individual bacterial cell strains.
Electrostatic interaction-based detection of bacterial cells
Positively charged nanomaterials bind to negatively charged protein surfaces through electrostatic interactions. Several nanomaterials, such as gold nanoparticles, carbon nanotubes, and graphene oxide, have been reported to interact with enzymes, inhibiting enzymatic activities. Based on the reversible interaction, the recovery of enzymatic activity can be used as an indicator of bacterial cell concentration. Li et al. synthesized positively charged graphene oxide to inhibit β-galactosidase (β-gal) activity for the colorimetric detection of bacterial cells and sensing of antibiotics.41 Also, enzymatic activity has been reversibly inhibited by positively charged polyethyleneimine-coated AuNPs to detect Gram-positive or Gram-negative bacterial cells in drinking water as low as 10 CFU·mL−1 within 10 minutes using an optical reader, and within 2–3 hours via simple visual readout.42
Rotello and coworkers have reported the synthesis of cationic AuNPs for the detection of bacterial cells in multiple detection formats.43–45 Because of electrostatic interactions, the enzymatic activity of β-gal was inhibited by cationic AuNPs that featured quaternary amine head groups. In the presence of bacterial cells, AuNPs were released from the AuNPs/β-gal complex and bound to the surface of the bacterial cells, resulting in the recovery of enzymatic activity (Fig. 11a). The colorimetric results and absorbance intensities versus bacterial concentrations are shown in Fig. 11b,c, indicating an obtained detection limit of 100 CFU·mL−1 within 10 minutes.43 Furthermore, the detection system was inkjet-printed on paper for low-cost diagnostics in contaminated drinking water. As seen in Fig. 11d, the AuNPs/β-gal complex and colorimetric substrates were co-patterned on the paper strip. After dipping the test strip into a bacterial solution for 5 minutes, E. coli XL1 (102 CFU·mL−1) and B. subtilis (103 CFU·mL−1) can be detected (Fig. 11e).44 After the β-gal is released from the AuNPs/β-gal complex, the enzyme activity can also be monitored and quantified using electrochemical strategies. At the concentration of 102 CFU·mL−1, E. coli and S. aureus can be electrochemically detected within 1 hour.45
Fig. 11.
(a) Schematic illustration of electrostatic interaction detection of bacterial cells based on the switchable interaction of positively charged AuNPs and negatively charged enzyme. (b) Photograph and (c) absorbance intensity at the wavelength of 595 nm of the detection solutions of with different concentrations. (d) Inkjet printing scheme for the fabrication of test strips for bacteria detection. (e) Photograph of inkjet printed test strip for the Gram positive and Gram negative bacteria detection. Reproduced with permission from ref. 43. Copyright 2011, American Chemical Society and ref. 44. Copyright 2014, American Chemical Society.
Array-based sensing of bacteria
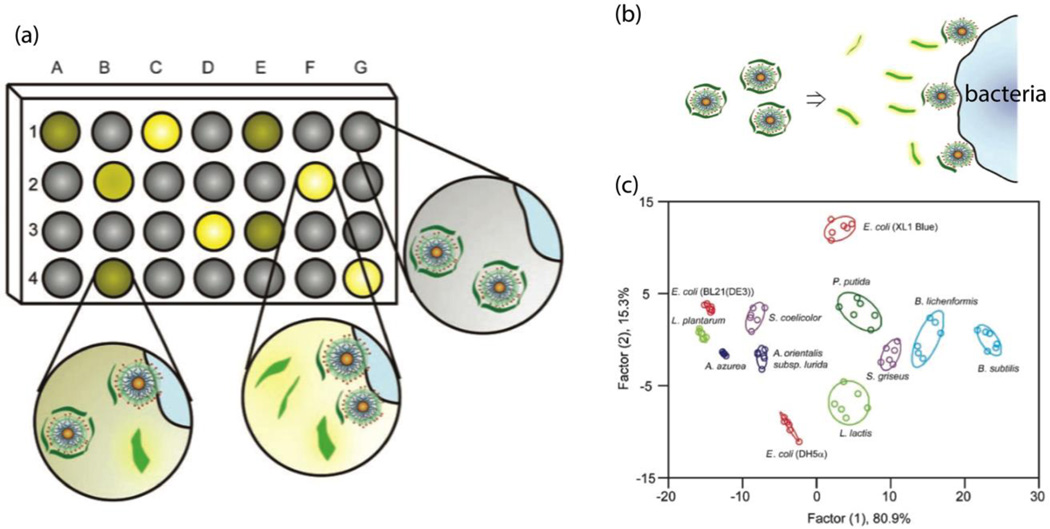
Array-based sensing with varying recognition elements provides a means of rapidly identifying bacteria species and strains. Typically, a set of nanoprobes recognizes a set of target bacterial strains, generating patterns. The patterns formed by differential interactions between nanoprobes and bacterial cells can serve as fingerprints to identify and analyze the bacterial cells.40 As shown in Fig. 12a, nanoprobes A-G were interacted with bacteria 1–4, resulting in multiple responses to form array-based patterns. The bacterial strains can be identified by analyzing the multiple response data using principle component linear discriminant analysis (LDA).40
Fig. 12.
(a) Schematic illustration of the signal pattern generation using array-based sensing. (b) Schematic representation of the detection of bacteria strains using turning-on fluorescence array sensors. (c) Canonical score plot for the fluorescence array patterns calculated using LDA. Reproduced with permission from ref. 46. Copyright 2008, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Philips et al. demonstrated an array-based sensing of bacteria within minutes, using AuNPs and poly (para-phenyleneethynylenen) (PPE), a fluorescent polymer.46 The AuNPs were modified with three different quaternary amine functional head groups, which can quench the fluorescent polymers through FRET. In the presence of bacterial cells, the fluorescent polymers are released from the AuNPs/PPE complex (Fig. 11b). Due to the differential binding affinities between different AuNPs and bacterial cells, multiple fluorescence readouts were recorded and analyzed using LDA. As shown in Fig. 12c, each cluster represents one strain of bacteria. Twelve bacterial strains, including Gram-negative and Gram-positive bacteria, can be detected and discriminated within 30 minutes. Interestingly, the same strains with different substrains (e.g. E. coli BL21, E. coli XL1, and E. coli DH5) can also be differentiated. Due to the complex compounds in biofilms, it is very challenging to discriminate biofilm bacteria strains. Using array-based sensing, Li et al. have developed a multichannel sensor to discriminate bacteria strains in biofilms within 20 minutes.47 The results demonstrated that the multichannel sensor could also identify nonpathogenic and pathogenic bacteria.
Other recognition element-based detection of bacteria
Some small molecules, including carbohydrates, lectin, and vancomycin, have emerged as important recognition elements on nanomaterials for the detection of bacteria. Due to their enhanced stability to temperature and pH variations, these small molecules have attracted attention for mediating interactions between nanomaterials and bacterial cells. These small recognition elements have a strong affinity to bind a broad range of bacterial cells, which are suitable for the detection of unanticipated bacteria. Compared with antibodies or aptamers, these small molecules have much higher recognition element densities on the surface of nanomaterials, providing strong affinity for the capture of bacterial cells.
Carbohydrates (polysaccharides or oligosaccharides) have been modified on nanoparticles as recognition elements to capture bacterial cells. EI-Boubbou et al. have used mannose- and galactose-functionalized MNPs to discriminate the E. coli strains.48 After carbohydrates are conjugated with proteins or lipids, the formed glycoproteins or glycolipids can also recognize and bind to bacterial cells. Lectin, the most typical glycoprotein, can bind to N-acetyl glucosamine of peptidoglycan on the surface of bacteria. Lectins have previously been bound within columns to capture bacteria in a liquid sample. Additionally, they have been bound to magnetic beads to continuously remove microorganisms from blood.49 Furthermore, vancomycin, a glycopeptide antibiotic, has been functionalized on MNPs to trap Gram positive or negative bacterial cells.50
Conclusions and prospects
There is a critical need for rapid detection and identification of pathogenic bacteria. Recognizing the importance of the interaction between recognition element and bacterial cells is helpful to design specific detection strategies. The proper choice of nanomaterials provides access to sensitive signal transduction methods to detect bacterial cells. Selection of the proper combination of recognition elements and nanomaterial transducers enables the creation of ‘next generation’ biosensors that can address societal needs. There has been considerable progress made in this area, however there remains a need to generate more sensitive systems (ability to detect bacteria with 1 CFU·mL−1) that are able to rapidly identify bacteria species and strain. An additional challenge is the ability to sense bacteria in challenging media, ranging from serum (for septicaemia) to food and environmental samples.
Because many cases of food- and water-borne infections arise in the developing world, additional challenges for the widespread implementation of bacterial biosensors exist. The economic situation in developing countries makes issues including manufacturability, and cost barriers to implementation. The advancement of nanoprobes for bacterial cell detection has the ability to address both efficiency and cost issues, impacting multiple industries, such as food safety, clinic diagnostics, and environmental monitoring.
Supplementary Material
The importance of recognition elements in the detection and sensing of pathogenic bacterial cells.
The principles for the design of recognition elements conjugated on nanomaterial
The broad diversity of recognition element-nanomaterial complexes used for the detection and sensing of pathogenic bacterial cells.
Detection and sensing mechanisms using recognition element-nanomaterial complex as nanoprobes.
The challenges and opportunities of recognition element-nanomaterial complexes for future bacterial detection.
Acknowledgments
The authors acknowledge the support of USDA NIFA 2013-02037, NIH GM077173, NSF CHE-1506725 and NSF Center for Hierarchical Manufacturing CMMI-1025020.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/c000000x/
References
References
- 1.Patel P. (Bio) Sensors for Measurement of Analytes Implicated in Food Safety: A Review. TrAC, Trends Anal. Chem. 2002;21(2):96–115. [Google Scholar]
- 2.Whitesides GM. The Origins and the Future of Microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 3.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global Trends in Emerging Infectious Diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. Nanomaterials for Targeted Detection and Photothermal Killing of Bacteria. Chem. Soc. Rev. 2012;41(8):3193–3209. doi: 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]
- 5.Poulsen LV. Microbial Biofilm in Food Processing. LWT - Food Science and Technology. 1999;32(6):321–326. [Google Scholar]
- 6.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Gold Nanoparticles in Chemical and Biological Sensing. Chem. Rev. 2012;112(5):2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S, Worobo RW, Durst RA. Escherichia Coli O157: H7 as an Emerging Foodborne Pathogen: A Literature Review. Crit. Rev. Biotechnol. 2001;21(1):27–48. doi: 10.1080/20013891081674. [DOI] [PubMed] [Google Scholar]
- 8.Fu J, Park B, Zhao Y. Limitation of a Localized Surface Plasmon Resonance Sensor for Salmonella Detection. Sensors Actuators B: Chem. 2009;141(1):276–283. [Google Scholar]
- 9.Wang C, Irudayaraj J. Gold Nanorod Probes for the Detection of Multiple Pathogens. Small. 2008;4(12):2204–2208. doi: 10.1002/smll.200800309. [DOI] [PubMed] [Google Scholar]
- 10.Wang CG, Irudayaraj J. Multifunctional Magnetic-Optical Nanoparticle Probes for Simultaneous Detection, Separation, and Thermal Ablation of Multiple Pathogens. Small. 2010;6(2):283–289. doi: 10.1002/smll.200901596. [DOI] [PubMed] [Google Scholar]
- 11.Singh AK, Senapati D, Wang S, Griffin J, Neely A, Candice P, Naylor KM, Varisli B, Kalluri JR, Ray PC. Gold Nanorod Based Selective Identification of Escherichia Coli Bacteria Using Two-Photon Rayleigh Scattering Spectroscopy. Acs Nano. 2009;3(7):1906–1912. doi: 10.1021/nn9005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Singh AK, Senapati D, Neely A, Yu H, Ray PC. Rapid Colorimetric Identification and Targeted Photothermal Lysis of Salmonella Bacteria by Using Bioconjugated Oval - Shaped Gold Nanoparticles. Chemistry-A European Journal. 2010;16(19):5600–5606. doi: 10.1002/chem.201000176. [DOI] [PubMed] [Google Scholar]
- 13.Gao J, Gu H, Xu B. Multifunctional Magnetic Nanoparticles: Design, Synthesis, and Biomedical Applications. Acc. Chem. Res. 2009;42(8):1097–1107. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Duncan B, Wang Z, Wang LS, Rotello VM, Nugen SR. Bacteriophage-Based Nanoprobes for Rapid Bacteria Separation. Nanoscale. 2015;7(39):16230–16236. doi: 10.1039/c5nr03779d. [DOI] [PubMed] [Google Scholar]
- 15.Varshney M, Yang LJ, Su XL, Li YB. Magnetic Nanoparticle-Antibody Conjugates for the Separation of Escherichia Coli O157 : H7 in Ground Beef. J. Food Prot. 2005;68(9):1804–1811. doi: 10.4315/0362-028x-68.9.1804. [DOI] [PubMed] [Google Scholar]
- 16.Frey NA, Peng S, Cheng K, Sun SH. Magnetic Nanoparticles: Synthesis, Functionalization, and Applications in Bioimaging and Magnetic Energy Storage. Chem. Soc. Rev. 2009;38(9):2532–2542. doi: 10.1039/b815548h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W-J, Tsai P-J, Chen Y-C. Functional Fe3o4/Tio2 Core/Shell Magnetic Nanoparticles as Photokilling Agents for Pathogenic Bacteria. Small. 2008;4(4):485–491. doi: 10.1002/smll.200701164. [DOI] [PubMed] [Google Scholar]
- 18.Hamula CLA, Zhang H, Li F, Wang Z, Chris Le X, Li X-F. Selection and Analytical Applications of Aptamers Binding Microbial Pathogens. TrAC, Trends Anal. Chem. 2011;30(10):1587–1597. doi: 10.1016/j.trac.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan YF, Ning Y, Song Y, Deng L. Fluorescent Aptasensor for the Determination of Salmonella Typhimurium Based on a Graphene Oxide Platform. Microchimica Acta. 2014;181(5–6):647–653. [Google Scholar]
- 20.Zuo P, Li X, Dominguez DC, Ye B-C. A Pdms/Paper/Glass Hybrid Microfluidic Biochip Integrated with Aptamer-Functionalized Graphene Oxide Nano-Biosensors for One-Step Multiplexed Pathogen Detection. Lab on a Chip. 2013;13(19):3921–3928. doi: 10.1039/c3lc50654a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YS, Chung J, Song MY, Jurng J, Kim BC. Aptamer Cocktails: Enhancement of Sensing Signals Compared to Single Use of Aptamers for Detection of Bacteria. Biosens. Bioelectron. 2014;54:195–198. doi: 10.1016/j.bios.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Duan N, Wu SJ, Zhu CQ, Ma XY, Wang ZP, Yu Y, Jiang Y. Dual-Color Upconversion Fluorescence and Aptamer-Functionalized Magnetic Nanoparticles-Based Bioassay for the Simultaneous Detection of Salmonella Typhimurium and Staphylococcus Aureus. Anal. Chim. Acta. 2012;723:1–6. doi: 10.1016/j.aca.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Duan N, Wu S, Yu Y, Ma X, Xia Y, Chen X, Huang Y, Wang Z. A Dual-Color Flow Cytometry Protocol for the Simultaneous Detection of Vibrio Parahaemolyticus and Salmonella Typhimurium Using Aptamer Conjugated Quantum Dots as Labels. Anal. Chim. Acta. 2013;804:151–158. doi: 10.1016/j.aca.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 24.Guerrini L, Graham D. Molecularly-Mediated Assemblies of Plasmonic Nanoparticles for Surface-Enhanced Raman Spectroscopy Applications. Chem. Soc. Rev. 2012;41(21):7085–7107. doi: 10.1039/c2cs35118h. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez-Puebla RA, Liz-Marzan LM. Traps and Cages for Universal Sers Detection. Chem. Soc. Rev. 2012;41(1):43–51. doi: 10.1039/c1cs15155j. [DOI] [PubMed] [Google Scholar]
- 26.Ravindranath SP, Wang Y, Irudayaraj J. Sers Driven Cross-Platform Based Multiplex Pathogen Detection. Sensors Actuators B: Chem. 2011;152(2):183–190. [Google Scholar]
- 27.Zhang H, Ma X, Liu Y, Duan N, Wu S, Wang Z, Xu B. Gold Nanoparticles Enhanced Sers Aptasensor for the Simultaneous Detection of Salmonella Typhimurium and Staphylococcus Aureus. Biosens. Bioelectron. 2015;74:872–877. doi: 10.1016/j.bios.2015.07.033. [DOI] [PubMed] [Google Scholar]
- 28.van der Merwe RG, van Helden PD, Warren RM, Sampson SL, Gey van Pittius NC. Phage-Based Detection of Bacterial Pathogens. Analyst. 2014;139(11):2617–2626. doi: 10.1039/c4an00208c. [DOI] [PubMed] [Google Scholar]
- 29.Martelet A, L’Hostis G, Nevers M-C, Volland H, Junot C, Becher F, Muller BH. Phage Amplification and Immunomagnetic Separation Combined with Targeted Mass Spectrometry for Sensitive Detection of Viable Bacteria in Complex Food Matrices. Anal. Chem. 2015 doi: 10.1021/ac504508a. [DOI] [PubMed] [Google Scholar]
- 30.Pierce CL, Rees JC, Fernández FM, Barr JR. Detection of Staphylococcus Aureus Using 15n–Labeled Bacteriophage Amplification Coupled with Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry. Anal. Chem. 2011;83(6):2286–2293. doi: 10.1021/ac103024m. [DOI] [PubMed] [Google Scholar]
- 31.Derda R, Lockett MR, Tang SKY, Fuller RC, Maxwell EJ, Breiten B, Cuddemi CA, Ozdogan A, Whitesides GM. Filter-Based Assay for Escherichia Coli in Aqueous Samples Using Bacteriophage-Based Amplification. Anal. Chem. 2013;85(15):7213–7220. doi: 10.1021/ac400961b. [DOI] [PubMed] [Google Scholar]
- 32.Blasco R, Murphy M, Sanders M, Squirrell D. Specific Assays for Bacteria Using Phage Mediated Release of Adenylate Kinase. J. Appl. Microbiol. 1998;84(4):661–666. doi: 10.1046/j.1365-2672.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 33.Laczka O, Garcia-Aljaro C, del Campo FJ, Pascual FXM, MasGordi J, Baldrich E. Amperometric Detection of Enterobacteriaceae in River Water by Measuring Beta-Galactosidase Activity at Interdigitated Microelectrode Arrays. Anal. Chim. Acta. 2010;677(2):156–161. doi: 10.1016/j.aca.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Oda M, Morita M, Unno H, Tanji Y. Rapid Detection of Escherichia Coli O157: H7 by Using Green Fluorescent Protein-Labeled Pp01 Bacteriophage. Appl. Environ. Microbiol. 2004;70(1):527–534. doi: 10.1128/AEM.70.1.527-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schofield DA, Bull CT, Rubio I, Wechter WP, Westwater C, Molineux IJ. Development of an Engineered Bioluminescent Reporter Phage for Detection of Bacterial Blight of Crucifers. Appl. Environ. Microbiol. 2012;78(10):3592–3598. doi: 10.1128/AEM.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcaine SD, Tilton L, Serrano MAC, Wang M, Vachet RW, Nugen SR. Phage-Protease-Peptide: A Novel Trifecta Enabling Multiplex Detection of Viable Bacterial Pathogens. Appl. Microbiol. Biotechnol. 2015;99(19):8177–8185. doi: 10.1007/s00253-015-6867-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcaine SD, Pacitto D, Sela DA, Nugen SR. Phage & Phosphatase: A Novel Phage-Based Probe for Rapid, Multi-Platform Detection of Bacteria. Analyst. 2015;140(22):7629–7636. doi: 10.1039/c5an01181g. [DOI] [PubMed] [Google Scholar]
- 38.Edgar R, McKinstry M, Hwang J, Oppenheim AB, Fekete RA, Giulian G, Merril C, Nagashima K, Adhya S. High-Sensitivity Bacterial Detection Using Biotin-Tagged Phage and Quantum-Dot Nanocomplexes. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Alcaine SD, Jiang Z, Rotello VM, Nugen SR. Detection of Escherichia Coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015;87(17):8977–8984. doi: 10.1021/acs.analchem.5b02175. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Z, Le ND, Gupta A, Rotello VM. Cell Surface-Based Sensing with Metallic Nanoparticles. Chem. Soc. Rev. 2015:4264–4274. doi: 10.1039/c4cs00387j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Wu LJ, Guo SS, Fu HE, Chen GN, Yang HH. Simple Colorimetric Bacterial Detection and High-Throughput Drug Screening Based on a Graphene-Enzyme Complex. Nanoscale. 2013;5(2):619–623. doi: 10.1039/c2nr32704j. [DOI] [PubMed] [Google Scholar]
- 42.Thiramanas R, Laocharoensuk R. Competitive Binding of Polyethyleneimine-Coated Gold Nanoparticles to Enzymes and Bacteria: A Key Mechanism for Low-Level Colorimetric Detection of Gram-Positive and Gram-Negative Bacteria. Microchimica Acta. 2015:1–8. [Google Scholar]
- 43.Miranda OR, Li X, Garcia-Gonzalez L, Zhu Z-J, Yan B, Bunz UH, Rotello VM. Colorimetric Bacteria Sensing Using a Supramolecular Enzyme-Nanoparticle Biosensor. J. Am. Chem. Soc. 2011;133(25):9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creran B, Li X, Duncan B, Kim CS, Moyano DF, Rotello VM. Detection of Bacteria Using Inkjet-Printed Enzymatic Test Strips. ACS Appl. Mater. Interfaces. 2014;6(22):19525–19530. doi: 10.1021/am505689g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Jiang Z, Ackerman JD, Yazdani M, Hou S, Nugen SR, Rotello VM. Electrochemical Nanoparticle-Enzyme Sensors for Screening Bacterial Contamination in Drinking Water. Analyst. 2015;140(15):4991–4996. doi: 10.1039/c5an00637f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips RL, Miranda OR, You C-C, Rotello VM, Bunz UHF. Rapid and Efficient Identification of Bacteria Using Gold-Nanoparticle–Poly(Para-Phenyleneethynylene) Constructs. Angew. Chem. Int. Ed. 2008;47(14):2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Kong H, Mout R, Saha K, Moyano DF, Robinson SM, Rana S, Zhang X, Riley MA, Rotello VM. Rapid Identification of Bacterial Biofilms and Biofilm Wound Models Using a Multichannel Nanosensor. ACS Nano. 2014;8(12):12014–12019. doi: 10.1021/nn505753s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Boubbou K, Gruden C, Huang X. Magnetic Glyco-Nanoparticles: A Unique Tool for Rapid Pathogen Detection, Decontamination, and Strain Differentiation. J. Am. Chem. Soc. 2007;129(44):13392–13393. doi: 10.1021/ja076086e. [DOI] [PubMed] [Google Scholar]
- 49.Cooper RM, Leslie DC, Domansky K, Jain A, Yung C, Cho M, Workman S, Super M, Ingber DE. A Microdevice for Rapid Optical Detection of Magnetically Captured Rare Blood Pathogens. Lab on a Chip. 2014;14(1):182–188. doi: 10.1039/c3lc50935d. [DOI] [PubMed] [Google Scholar]
- 50.Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, Bergeron MG, Simard B. Vancomycin-Modified Nanoparticles for Efficient Targeting and Preconcentration of Gram-Positive and Gram-Negative Bacteria. Acs Nano. 2008;2(9):1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.