Abstract
The human immunodeficiency virus type 1 (HIV-1) Tat protein recruits positive transcription elongation factor b (P-TEFb) to the transactivation response (TAR) RNA structure to facilitate formation of processive transcription elongation complexes (TECs). Here we examine the role of the Tat/TAR-specified cyclin-dependent kinase 9 (CDK9) kinase activity in regulation of HIV-1 transcription elongation and histone methylation. In HIV-1 TECs, P-TEFb phosphorylates the RNA polymerase II (RNAP II) carboxyl-terminal domain (CTD) and the transcription elongation factors SPT5 and Tat-SF1 in a Tat/TAR-dependent manner. Using in vivo chromatin immunoprecipitation analysis, we demonstrate the following distinct properties of the HIV-1 transcription complexes. First, the RNAP II CTD is phosphorylated at Ser 2 and Ser 5 near the promoter and at downstream coding regions. Second, the stable association of SPT5 with the TECs is dependent upon P-TEFb kinase activity. Third, P-TEFb kinase activity is critical for the induction of methylation of histone H3 at lysine 4 and lysine 36 on HIV-1 genes. Flavopiridol, a potent P-TEFb kinase inhibitor, inhibits CTD phosphorylation, stable SPT5 binding, and histone methylation, suggesting that its potent antiviral activity is due to its ability to inhibit several critical and unique steps in HIV-1 transcription elongation.
The Tat protein encoded by human immunodeficiency virus type 1 (HIV-1) stimulates transcription elongation through its interaction with the transactivation response (TAR) RNA structure located at the 5′ end of nascent viral transcripts (2, 16, 30, 67). In view of the observations that hyperphosphorylation of the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAP II) correlates with the formation of processive elongation complexes (14) and that Tat transactivation requires RNAP II CTD (12, 48, 71), it has been proposed that a critical step in Tat transactivation is mediated through cellular kinases which are capable of phosphorylating RNAP II CTD (54, 62, 72). The RNAP II CTD is composed of multiple heptad repeats of Tyr-Ser-Pro-Thr-Ser-Pro-Ser, whose consensus is conserved among most eukaryotes. Ser 2 and Ser 5 are targets for phosphorylation and dephosphorylation during transcription (34).
Positive transcription elongation factor b (P-TEFb) (42-44) or Tat-associated kinase (23, 25, 26), composed of cyclin-dependent kinase 9 (CDK9) and cyclin T1 (23, 41, 52, 68, 70, 76, 78), is essential for Tat transactivation. Formation of the tripartite complex between Tat, cyclin T1, and TAR depends on the 5′ bulge and central loop in the TAR RNA structure (3, 18, 20, 29, 38, 68). CDK9 autophosphorylation promotes high-affinity binding of the Tat/P-TEFb complex to the TAR RNA structure (17, 19) as well as CDK9 activation (75). TAR-bound P-TEFb induces a Tat/TAR-dependent phosphorylation of RNAP II CTD to stimulate transcription elongation (33, 74). P-TEFb preferentially phosphorylates Ser 2 of the RNAP II CTD, but its substrate specificity can be modulated to target both Ser 2 and Ser 5 by Tat (74). A second CTD kinase, TFIIH (CDK7/cyclin H), phosphorylates Ser 5 of the RNAP II CTD (57, 63). Although the role of TFIIH in Tat transactivation is controversial (10, 13, 21, 50), TFIIH has been shown to regulate Tat-dependent CDK9 activation during HIV-1 transcription (75). Our group's previous results demonstrated that TFIIH and P-TEFb are both associated with HIV-1 preinitiation complexes (PICs) (74). TFIIH is released from transcription complexes between +14 and +36, while P-TEFb stays associated with transcription elongation complexes.
The RNAP II CTD also plays a role in coupling transcription with RNA processing, and this coupling appears to be regulated by CTD phosphorylation (55). Cotranscriptional mRNA capping can be induced by the presence of RNAP II CTD Ser 5 phosphorylation (56, 60). Although Ser 2 phosphorylation suffices for RNA guanylyltransferase binding, activation of the capping enzyme requires Ser 5 phosphorylation (27). Our group's previous results showed that RNAP II CTD is phosphorylated sequentially by TFIIH and that the Tat-induced P-TEFb kinase activity specifically enhances cotranscriptional capping of nascent HIV-1 transcripts (73).
The density of RNAP II within the promoter and downstream coding sequences and the CTD phosphorylation pattern appear to vary between genes and organisms. In Saccharomyces cerevisiae, chromatin immunoprecipitation (ChIP) assays have demonstrated that RNAP II is uniformly associated with transcribed genes from the promoter to the 3′ end of the gene (35, 60). However, different phosphorylated forms of RNAP II are associated with the gene at different stages of transcription. Ser 5-phosphorylated RNAP II is concentrated within the promoter-proximal region, while Ser 2-phosphorylated RNAP II is associated with the coding region (35). In higher eukaryotes, RNAP II on the dihydrofolate reductase (DHFR) and γ-actin genes is more concentrated in the promoter-proximal regions than in the interior regions (11). RNAP II phosphorylated specifically at Ser 5 is more concentrated near the promoter, while RNAP II with Ser 2 phosphorylation is distributed throughout the gene.
Coordinated recruitment of transcription stage-specific modification factors, including histone methyltransferases, correlates with active chromatin. Localized changes in chromatin structures induced by distinct histone H3 methylation play a significant role in maintaining transcriptionally active chromatin structures (22, 24). In yeast, the histone H3 lysine methyltransferases Set1 and Set2 functionally interact with phosphorylated RNAP II CTD and induce an open chromatin structure that is essential for transcription elongation (36, 39, 40, 46, 69). In particular, Set2 preferentially binds to CTD phosphorylated at Ser 2 (36, 39, 40, 69), while Set1 binds to CTD phosphorylated at Ser 5 (46).
HIV-1 genes are highly inducible and provide a particularly useful mammalian system to study regulation of processive transcription elongation and chromatin modification. Our results demonstrate that the Tat/TAR-dependent P-TEFb kinase activity phosphorylates RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription. Ser 2 and Ser 5 phosphorylation of RNAP II CTD is observed both at the promoter and in downstream coding sequences. The distribution of SPT5 on HIV-1 genes depends upon the P-TEFb kinase activity. Flavopiridol inhibition of the P-TEFb kinase activity also reduced methylation of histone H3 lysine 4 and lysine 36 on HIV-1 genes in tumor necrosis factor alpha (TNF-α)-induced cells. These observations suggest that the Tat/TAR-dependent kinase activity of P-TEFb plays a critical role in several different steps in transcription elongation and chromatin modification.
MATERIALS AND METHODS
Materials.
Recombinant baculovirus containing human P-TEFb was kindly provided by David Price (University of Iowa). The recombinant P-TEFb complex, comprised of His-tagged CDK9 and cyclin T1, was expressed in Sf9 cells by the use of recombinant baculovirus and purified as described previously (52). Flavopiridol (Aventis Inc.) was dissolved in dimethyl sulfoxide (DMSO) to 10 mM and stored at −80°C. The stock was diluted to 0.1 mM in DMSO, and a set of serial dilutions in 4% DMSO was used to give the indicated concentrations. The final concentration of DMSO in the kinase assay mixtures was less than 1%. Antibodies used here included anti-CDK9 (α-CDK9; Biodesign); α-Tat, α-Ser2P CTD (H5), and α-Ser5P CTD (H14) (Covance); α-RNAP II (N-20) and α-p62 subunit of TFIIH (Santa Cruz); α-SPT5 and α-Tat-SF1 (BD Transduction); and α-trimethyl histone H3 lysine 4 and α-dimethyl histone H3 lysine 36 (Abcam).
CDK9 autophosphorylation assays.
Autophosphorylation assays were performed by mixing 20 ng of P-TEFb, 100 μM ATP, and 20 μCi of [γ-32P]ATP in the absence or presence of Tat (100 ng) in buffer (50 mM Tris-HCl [pH 7.5], 5 mM dithiothreitol [DTT], 5 mM MnCl2, and 4 mM MgCl2) and incubating for 60 min at 30°C. Serial dilutions of flavopiridol in 4% DMSO were used in these assays to give the indicated concentrations. 32P-labeled CDK9 was immunoprecipitated with specific antibody and analyzed by electrophoresis on sodium dodecyl sulfate (SDS)-4 to 20% polyacrylamide gels followed by autoradiography.
CTD kinase assays.
Twenty nanograms of P-TEFb was preincubated with ATP, and CTD kinase assays were performed by adding 50 ng of glutathione S-transferase (GST)-CTD and 100 μM ATP in the absence or presence of Tat (100 ng) in buffer (50 mM Tris-HCl [pH 7.5], 5 mM DTT, 5 mM MnCl2, and 4 mM MgCl2) and incubating for 60 min at 30°C. Serial dilutions of flavopiridol in 4% DMSO were added to reaction mixtures to give the indicated concentrations. Phosphorylated GST-CTD was separated on SDS-8% polyacrylamide gels and analyzed by Western blotting with α-Ser2P CTD (H5) or α-Ser2P CTD (H14).
Purification of HIV-1 PICs.
Purification of HIV-1 PICs was carried out as described previously using biotinylated HIV-1 long terminal repeat (LTR) templates (74). HIV-1 PIC assembly reaction mixtures (30 μl) contained 15 μl of HeLa nuclear extract, 1.0 μg of biotinylated HIV-1 LTR template, and 1.0 μg of poly(dI-dC) in the absence or presence of Tat (100 ng). The in vitro transcription (IVT) buffer contained 50 mM KCl, 6.25 mM MgCl2, 20 mM HEPES (pH 7.9), 2 mM DTT, 0.5 mM EDTA (pH 8.0), 10 μM ZnSO4, 10 mM creatine phosphate, 100 μg of creatine kinase/ml, and 8.5% glycerol (1× IVT buffer). After a 30-min incubation at 30°C, streptavidin-coated magnetic beads (Dynabeads; Dynal) preequilibrated in binding buffer (20 mM HEPES [pH 7.9], 80 mM KCl, 10 mM MgCl2, 2 mM DTT, 10 μM ZnSO4, 100 μg of bovine serum albumin/ml, 0.05% Nonidet P-40 [NP-40], and 10% glycerol) were then added to the reaction mixtures, and the mixtures were further incubated for 30 min at 30°C. The immobilized PICs were then harvested by using a magnetic stand, and the PICs were washed extensively with transcription buffer. IVT and Western blot analysis could then be performed with the purified PICs assembled on the immobilized templates.
Preparation of HIV-1 TECs.
Stepwise transcription was performed as described previously (74). PICs were walked to position U14 by incubation with 50 μM dATP, CTP, GTP, and UTP for 10 min at 30°C and washed with transcription buffer. Transcription elongation complexes (TECs) stalled at U14 were incubated with RNAP II-depleted nuclear extract for 30 min at 30°C and washed to remove unbound proteins. TECs stalled at U14 were then walked stepwise along the DNA by repeated incubation with different sets of three nucleoside triphosphates (NTPs) and washed with transcription buffer to remove the unincorporated NTPs. Protein compositions of TECs stalled at different positions were analyzed by Western blotting with specific antibodies. To determine phosphorylation of proteins associated with HIV-1 TECs during transcription elongation, TECs stalled at G36 were elongated stepwise to A79 and phosphorylated proteins were labeled with [γ-32P]ATP. 32P-labeled proteins were immunoprecipitated with specific antibodies and then analyzed by electrophoresis on SDS-polyacrylamide gels followed by autoradiography.
Tat expression in OM10.1 cells by TNF-α-induction.
OM10.1 cells, a promyelocytic line containing a transcriptionally latent, single copy of wild-type HIV-1 integrated proviral DNA (7), were treated with TNF-α (10 ng/ml) for 2 h, washed, and put back into 37°C with complete medium. Cells were processed for immunoprecipitation after 0, 4, 8, 12, 16, 20, and 24 h postinduction. Approximately 1 mg of cell lysate from each treatment was cleared overnight with protein A/G at 4°C. Samples were incubated with 10 μg of α-Tat antibody at 4°C for 12 h and further incubated for 2 h after addition of protein A/G beads. Immunoprecipitates were washed with TNE 600 plus 1% NP-40 (2×) followed by a final wash of TNE 50 plus 0.1% NP-40. Tat immunoprecipitates were fractionated by electrophoresis on SDS-4 to 20% polyacrylamide gels, stained with Coomassie dye, and analyzed by Western blotting with α-Tat antibody.
ChIP assays in TNF-α-induced OM10.1 cells.
ChIP assays were performed as described previously with some modifications (49, 51). Approximately 5 × 107 OM10.1 cells were induced with 10 ng of TNF-α/ml for 2 h and subsequently treated with flavopiridol. Cells were then cross-linked (1% formaldehyde; 10 min at 37°C), and samples were sonicated to reduce DNA fragments to ∼200 to 800 bp for ChIP assays, in which DNA bound to various proteins were immunoprecipitated with antibodies as indicated in the figure legends. Specific DNA sequences in the immunoprecipitates were detected by PCR, using primers specific for HIV-1 (−92 to +180, +10 to +165, +299 to +477, and +1419 to +1763). The HIV-1 proviral DNA integrated in OM10.1 cells was derived from subtype B, LAI strain.
Transfection of HLM1 cells with Tat plasmid and ChIP assays.
HLM1cells were derived from HeLa-T4+ cells integrated with one copy of the HIV-1 genome containing a Tat-defective mutation (58). The mutation was introduced as a triple termination linker at the first AUG of the Tat gene. The HIV-1 proviral DNA was derived from pHXB2gpt. HLM1cells are negative for virus particle production but can be induced to express noninfectious HIV-1 particles after transfection with Tat cDNA or addition of Tat protein to the medium. HLM1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 100 μg of G418/ml plus 1% streptomycin and penicillin antibiotics and 1% l-glutamine (Gibco/BRL). These cells were kept at 75% confluency prior to transfection with wild-type Tat plasmid (5 μg). Cells were transfected with the Tat plasmid DNA by the calcium phosphate method. The transfected cells were washed after 4 h, and fresh complete DMEM with 10% fetal bovine serum was added for the remainder of the experiment. At day 2 (at which Tat-activated transcription, i.e., presence of singly spliced and genomic RNA, is observed), cells were processed for ChIP assays using 10 μg of α-RNAP II, α-ser2P, and α-ser5P antibodies. Specific DNA sequences in the immunoprecipitates were detected by PCR by using primers specific for the HIV-1 Env1 (8440 to 8460; 5′-AGAGAGACAGAGACAGATCCA-3′) and Env2 (8772 to 8791; 5′-AGCAAAATCCTTTCCAAGCC-3′). The final product size was 351 bp.
RESULTS
The Tat-induced kinase activity of P-TEFb is highly sensitive to flavopiridol.
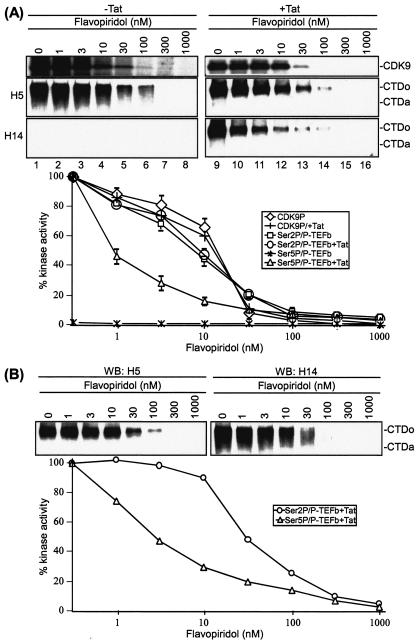
Flavopiridol, a CDK inhibitor, is one of the most potent inhibitors of HIV-1 transcription and virus replication identified to date (8, 9). A primary target of flavopiridol is the P-TEFb kinase (8). To gain insight into the inhibition of HIV-1 transcription, we first compared the inhibition curves for various substrates of P-TEFb, including CDK9 autophosphorylation and phosphorylation of Ser 2 and Ser 5 of the RNAP II CTD heptapeptide repeat. The flavopiridol sensitivity of CDK9 autophosphorylation was determined by incubating P-TEFb with [γ-32P]ATP in the absence or presence of flavopiridol. Results shown in Fig. 1A demonstrated that flavopiridol inhibited CDK9 autophosphorylation at a 50% inhibitory concentration (IC50) of 11 nM (top panel). In agreement with published reports that CDK9 autophosphorylation is not influenced by Tat in in vitro kinase reactions (19, 75), we observed little difference in flavopiridol inhibition either with or without Tat.
FIG. 1.
The Tat-induced kinase activity of P-TEFb is highly sensitive to flavopiridol. (A) The kinase activity of P-TEFb was sensitive to flavopiridol. In vitro CDK9 autophosphorylation assays were performed by incubating P-TEFb with [γ-32P]ATP in the absence (lanes 1 to 8) or presence (lanes 9 to 16) of Tat. Serial dilutions of flavopiridol were added into kinase reaction mixtures to give the indicated concentrations. (Top) 32P-labeled CDK9 was immunoprecipitated with α-CDK9 antibody and analyzed by electrophoresis on SDS-4 to 20% polyacrylamide gels followed by autoradiography. (Middle and bottom panels) In vitro CTD kinase assays were performed by incubating GST-CTD with P-TEFb in the absence (lanes 1 to 8) or presence (lanes 9 to 16) of Tat. Flavopiridol was added into reaction mixtures at a final concentration of 1 to 1,000 nM as indicated. Phosphorylated CTD was fractionated by electrophoresis on SDS-8% polyacrylamide gels and analyzed by Western blotting with α-Ser2P (H5) or α-Ser5P (H14). The hypophosphorylated (CTDa) and hyperphosphorylated (CTDo) forms of CTD are indicated. Quantitation of three independent experiments performed under similar conditions for each assay is shown at the bottom of the panels. (B) Effect of P-TEFb concentration on flavopiridol inhibition of CTD phosphorylation. In vitro CTD kinase assays were performed by incubating GST-CTD with different concentrations (20 to 100 ng) of P-TEFb in the presence of Tat. Flavopiridol was added to reaction mixtures at a final concentration of 1 to 1,000 nM as indicated. Phosphorylated CTD was fractionated by electrophoresis on SDS-8% polyacrylamide gels and analyzed by Western blotting (WB) with α-Ser2P (H5) or α-Ser5P (H14). The experiment shown is representative of three independent experiments performed with 40 ng of P-TEFb under similar conditions. The hypophosphorylated (CTDa) and hyperphosphorylated (CTDo) forms of CTD are indicated.
Next, we examined the sensitivity of the CTD kinase activity of P-TEFb. The RNAP II CTD heptad repeats of YSPTSPS may be phosphorylated at Ser 2 and Ser 5. P-TEFb normally phosphorylates the CTD at Ser 2, but in the presence of Tat it has been shown that the substrate specificity is modified to allow phosphorylation of both Ser 2 and Ser 5 (74). Phosphorylation of the CTD at Ser 2 was detected by Western blotting with antibody H5, which recognizes CTD repeats phosphorylated at Ser 2. Results shown in Fig. 1A demonstrated that Ser 2 phosphorylation was inhibited by flavopiridol at an IC50 of approximately 10 nM and that the inhibitory effect of flavopiridol on Ser 2 phosphorylation was not influenced by the addition of Tat (middle panel). Next, CTD phosphorylation at Ser 5 was detected by Western blotting with antibody H14, which recognizes CTD repeats phosphorylated at Ser 5. Remarkably, the Tat-induced CDK9 kinase activity to phosphorylate Ser 5 was highly sensitive to flavopiridol (bottom panel, lanes 9 to 16). The IC50 for this activity was calculated to be 1 nM. No CTD Ser 5 phosphorylation was detected in the absence of Tat (lanes 1 to 8).
Several control studies have been performed for flavopiridol inhibition. First, control assays demonstrated that the concentration of GST-CTD used as substrate in the kinase reactions was within the linear response of Western blotting with α-Ser2P H5 or α-Ser5P H14 (data not shown). Second, because the amount of flavopiridol required to inhibit P-TEFb depends upon the concentration of P-TEFb (9), we also determined the IC50 values of flavopiridol inhibition of P-TEFb-mediated CTD phosphorylation with a concentration range of 20 to 100 ng of P-TEFb. The concentrations of flavopiridol required for inhibition of either Ser 2 or Ser 5 phosphorylation increased with increasing concentrations of P-TEFb. Of significance, however, Tat-induced Ser 5 phosphorylation by P-TEFb was consistently 10-fold more sensitive than the Tat-independent Ser 2 phosphorylation. For example, when the concentration of P-TEFb was increased to 40 nM for the kinase assays, Ser 2 phosphorylation was inhibited by flavopiridol at an IC50 of 27 nM, while the IC50 for flavopiridol inhibition of Tat-induced Ser 5 phosphorylation was 2.8 nM (Fig. 1B).
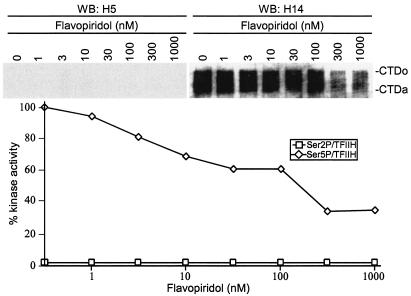
It was of interest to determine whether CTD Ser 5 phosphorylation was, in general, more sensitive to flavopiridol inhibition. To address this point, inhibition of Ser 5 phosphorylation by TFIIH (57, 63) was assayed. Using identical substrate and kinase concentrations, we found that the IC50 of Ser 5 phosphorylation by TFIIH was approximately 200 nM (Fig. 2). These results suggested that the Tat-induced CTD kinase activity of P-TEFb to phosphorylate Ser 5 of CTD is preferentially sensitive to flavopiridol.
FIG. 2.
The CTD kinase activity of TFIIH is resistant to flavopiridol. In vitro CTD kinase assays were performed by incubating GST-CTD with purified TFIIH. Flavopiridol was added to reaction mixtures at a final concentration of 1 to 1,000 nM as indicated. Phosphorylated CTD was fractionated by electrophoresis on SDS-8% polyacrylamide gels and analyzed by Western blotting (WB) with α-Ser2P (H5) or α-Ser5P (H14). The experiment shown is representative of three independent experiments performed under similar conditions. The hypophosphorylated (CTDa) and hyperphosphorylated (CTDo) forms of CTD are indicated.
The Tat/TAR-dependent kinase activity of P-TEFb to phosphorylate RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription elongation is highly sensitive to flavopiridol.
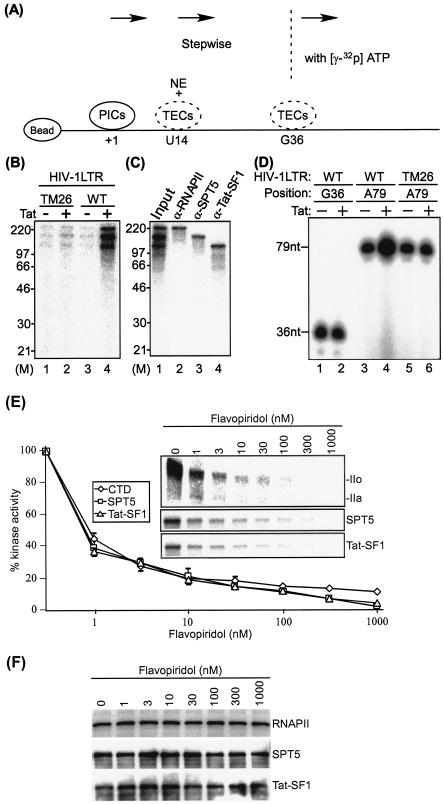
We next analyzed Tat/TAR-dependent phosphorylation of proteins associated with HIV-1 TECs, as outlined in Fig. 3A. In these assays, the wild-type HIV-1 LTR template was compared with a mutant TAR template (TM26). The mutation in TM26 includes base substitutions in the TAR RNA pyrimidine “bulge” which blocks the binding of Tat to TAR, thus interfering with Tat transactivation in vitro and in vivo (4). TECs stalled at G36 were elongated stepwise to A79, and the proteins phosphorylated during elongation reactions were labeled with [γ-32P]ATP. Results shown in Fig. 3B demonstrated that three proteins which migrated between 120 and ∼240 kDa were phosphorylated in a Tat/TAR-dependent manner (lane 4). Only background levels of the phosphorylated proteins were seen in the absence of Tat with either the TAR mutant or wild-type template (lanes 1 and 3) or in the presence of Tat with the mutant TAR template (lane 2). The 32P-labeled proteins were identified by immunoprecipitation with specific antibodies, which demonstrated that three phosphorylated proteins corresponded to the large subunit of RNAP II, SPT5, and Tat-SF1, respectively (Fig. 3C). Analysis of staged TECs demonstrated that SPT5 and Tat-SF1 entered the HIV-1 TECs between +14 and +36 (data not shown).
FIG. 3.
The Tat/TAR-dependent kinase activity of P-TEFb to phosphorylate RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription elongation is highly sensitive to flavopiridol. (A) The experimental outline was shown to detect phosphorylation of proteins associated with HIV-1 TECs. PICs assembled on biotinylated HIV-1 LTR templates were purified with streptavidin-coated magnetic beads. Purified PICs were walked to position U14 by incubation with 50 μM dATP, CTP, GTP, and UTP at 30°C for 10 min and washed with transcription buffer. TECs stalled at U14 were incubated with RNAP II-depleted nuclear extract (NE), and unbound proteins were removed by washing with transcription buffer. The TECs stalled at U14 were then walked stepwise to G36 along templates by repeated incubation with different sets of three NTPs and washed with transcription buffer to remove unincorporated NTPs. TECs stalled at G36 were elongated stepwise to A79 with [γ-32P]ATP and NTPs, and 32P-labeled proteins were analyzed by electrophoresis on SDS-4 to 20% polyacrylamide gels followed by autoradiography. (B) Tat/TAR-dependent phosphorylation of proteins associated with HIV-1 TECs during transcription elongation. TECs stalled at G36 were elongated stepwise to A79 with [γ-32P]ATP and NTPs, and 32P-labeled proteins were analyzed by electrophoresis on SDS-4 to 20% polyacrylamide gels followed by autoradiography. The wild-type (WT) HIV-1 LTR template was compared with the TAR-mutated (TM26) HIV-1 LTR template. M indicates the protein molecular weight marker. (C) Tat/TAR-dependent phosphorylation of the RNAP II CTD, SPT5, and Tat-SF1 by P-TEFb during HIV-1 transcription elongation. 32P-labeled proteins were immunoprecipitated with specific antibodies as indicated in the figure and analyzed by electrophoresis on SDS-4 to 20% polyacrylamide gels followed by autoradiography. (D) Tat/TAR-dependent phosphorylation of the RNAP II CTD, SPT5, and Tat-SF1 by P-TEFb correlated with HIV-1 transcription elongation. Nascent transcripts were labeled with [α-32P]UTP and analyzed by electrophoresis through 10% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA buffer followed by autoradiography. (E) Tat/TAR-dependent phosphorylation of the RNAP II CTD, SPT5, and Tat-SF1 by P-TEFb during HIV-1 transcription elongation was inhibited by flavopiridol. To determine flavopiridol sensitivity of the Tat/TAR-dependent kinase activity of P-TEFb during HIV-1 transcription elongation, TECs stalled at G36 were elongated stepwise to A79 in the absence or presence of flavopiridol. 32P-labeled RNAP II, SPT5, and Tat-SF1 were then immunoprecipitated with specific antibodies and analyzed by electrophoresis on SDS-4 to 20% polyacrylamide gels followed by autoradiography (inset), and three independent experiments performed under similar conditions for each assay were quantitated. The hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of RNAP II are indicated. (F) Western blot analysis of protein composition of TECs stalled at A79. TECs stalled at G36 were elongated stepwise to A79 in the absence or presence of flavopiridol. Protein composition of TECs stalled at A79 was analyzed with α-RNAP II, α-SPT5, or α-Tat-SF1 antibody.
The results shown in Fig. 3D demonstrated that coincident with phosphorylation of the RNAP II CTD, SPT-5, and Tat-SF1, there was an increase in the 79-nucleotide transcript in response to Tat with the wild-type LTR template (lanes 3 and 4). No Tat-mediated increase was observed in transcripts at G36 when the TAR RNA structure was incomplete (lanes 1 and 2) or at A79 with the mutant TAR which lacked the Tat binding site (lanes 5 and 6). The results suggested that Tat/TAR-dependent phosphorylation of RNAP II CTD, SPT5, and Tat-SF1 correlates with HIV-1 transcription elongation.
Last, we analyzed the flavopiridol sensitivity of the Tat/TAR-dependent P-TEFb kinase activity to phosphorylate RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription elongation. The stepwise transcription elongation reactions were performed in the absence or presence of flavopiridol, and 32P-labeled RNAP II, SPT5, and Tat-SF1 proteins were immunoprecipitated with specific antibodies. Results shown in Fig. 3E demonstrated that Tat/TAR-dependent phosphorylation of RNAP II CTD, SPT5, and Tat-SF1 by P-TEFb was inhibited by flavopiridol at an IC50 of approximately 1 nM. Western blot analysis showed that levels of RNAP II, SPT5, and Tat-SF1 present in the immunoprecipitates remained constant (Fig. 3F). The results suggested that the Tat/TAR-dependent P-TEFb kinase activity to phosphorylate RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription elongation is highly sensitive to flavopiridol.
RNAP II CTD phosphorylation at the HIV-1 promoter and downstream coding regions.
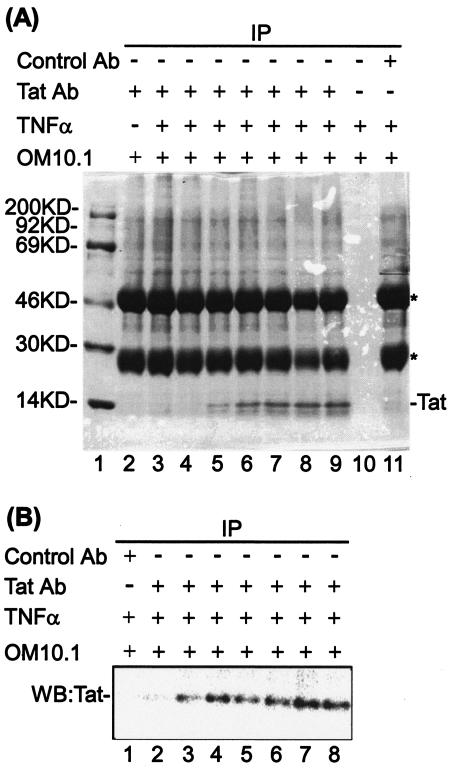
To gain insight into the action of the P-TEFb kinase activity in HIV-1 transcription, we performed ChIP assays to analyze the distribution of RNAP II and CTD phosphorylation within the promoter and downstream coding sequences. For these studies, we used OM10.1 cells, a promyelocytic line containing a single copy of a transcriptionally latent, integrated HIV-1 proviral DNA (7). DNA sequence analysis of the integrated proviral genome demonstrated that the viral LTR and Tat coding sequences were wild type (data not shown). The cells were induced with TNF-α, and Tat was immunoprecipitated with specific α-Tat antibody (Fig. 4A). Immunoblot analysis of Tat immunoprecipitates demonstrated that TNF-α treatment of OM10.1 cells induced Tat protein expression as early as 4 h postinduction (Fig. 4B, lanes 2 to 8). Thus, this cell line provides an excellent inducible system to analyze LTR activation and Tat transactivation.
FIG. 4.
TNF-α-induced Tat expression in OM10.1 cells. (A) Coomassie staining of the Tat immunoprecipitates from TNF-α-induced OM10.1 cells. OM10.1 cells, which contain one copy of latent HIV-1 infectious clone, were induced with 10 ng of TNF-α/ml for 2 h, washed, and put back into 37°C with complete medium. The cells were subjected to immunoprecipitation (IP) with specific α-Tat antibody after 0, 4, 8, 12, 16, 20, or 24 h postinduction, and Tat immunoprecipitates were analyzed by SDS-4 to 20% polyacrylamide gel electrophoresis and Coomassie staining (lanes 3 to 9). The immunoprecipitates with no antibody (lane 10) or preimmune antibody (lane 11) from the TNF-α-induced OM10.1 cells, which were incubated for 24 h postinduction, were used for controls. The immunoprecipitate from noninduced cells is shown in lane 2, and lane 1 indicates the protein molecular mass markers. The immunoglobulin G heavy and light chains are indicated by asterisks. (B) Western blot analysis of Tat from TNF-α-induced OM10.1 cells. The Tat immunoprecipitates from the TNF-α-induced OM10.1 cells, which were incubated for 0, 4, 8, 12, 16, 20, or 24 h postinduction, were analyzed by Western blotting with α-Tat antibody (lanes 2 to 8). The immunoprecipitate with preimmune antibody from the TNF-α-induced OM10.1 cells, which were incubated for 24 h postinduction, was used as a control (lane 1).
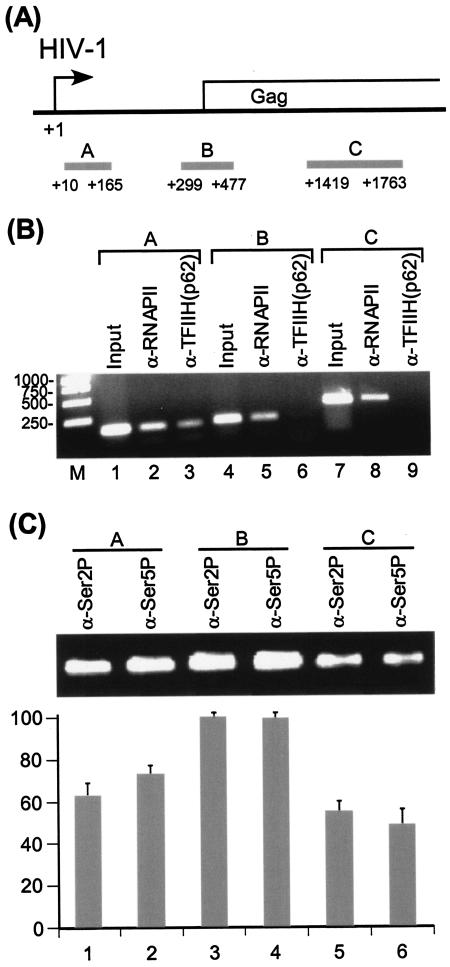
For ChIP assays, OM10.1 cells were induced with TNF-α and cross-linked with formaldehyde. The cross-linked chromatin was sonicated to 200- to ∼800-bp DNA fragments, which were immunoprecipitated with specific antibodies to obtain the protein-DNA complexes of interest. The amount of DNA immunoprecipitated was analyzed by PCR amplification with specific primers. Three sites in the HIV-1 promoter-proximal and downstream coding regions were used for the ChIP analysis (Fig. 5A). Results shown in Fig. 5B demonstrated that the distribution of total RNAP II across the HIV-1 promoter and downstream coding regions was constant (lanes 2, 5, and 8). Consistent with in vitro transcription analysis of HIV-1 transcription complexes, TFIIH was released from the transcription complex during the elongation phase of transcription (Fig. 5B, lanes 3, 6, and 9). Interestingly, results shown in Fig. 5C demonstrated that high levels of Ser 2 and Ser 5 phosphorylation were observed at both the promoter-proximal and downstream coding regions. The pattern of Ser 2 and Ser 5 phosphorylation of RNAP II CTD on HIV-1 genes appears distinct. ChIP analyses of mammalian DHFR and γ-actin genes and yeast genes have demonstrated that Ser 5 phosphorylation of the RNAP II CTD is concentrated near the promoter, while Ser 2 phosphorylation is found throughout the gene (11, 35).
FIG. 5.
Distribution of different phosphorylation forms of RNAP II on HIV-1 genes. (A) Diagram of HIV-1 genes, shown with PCR amplification fragments for the promoter-proximal (region A, TAR region +10 to +165), 5′ Gag (region B, +299 to +477), and mid-Gag (region C, +1419 to +1763) underneath the genes (gray histogram). The sequences used for PCR amplification were from subtype B, LAI strain. (B) Distribution of RNAP II and TFIIH on HIV-1 genes. OM10.1 cells were induced with 10 ng of TNF-α/ml for 2 h and washed three times with phosphate-buffered saline. The cells were subsequently incubated in fresh medium for 18 h. Under these conditions, HIV-1 RNA could be detected by standard Northern analysis (data not shown). ChIP assays were then performed with α-RNAP II (N-20) or α-p62 subunit of TFIIH antibody. (C) Distribution of different phosphorylation forms of RNAP II on HIV-1 genes. OM10.1 cells were induced with 10 ng of TNF-α/ml for 2 h and subsequently incubated in fresh medium for 18 h. ChIP assays were then performed with α-RNAP II CTD Ser2P (H5) or α-RNAP II CTD Ser5P (H14) antibody. Each ChIP result shown is an average of four experiments with the standard error of mean indicated.
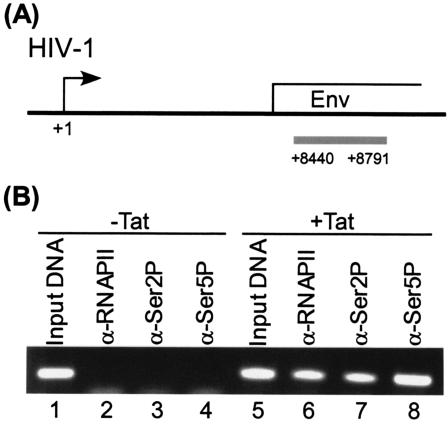
To demonstrate that Ser 2 and Ser 5 phosphorylation of the RNAP II CTD on HIV-1 genes was due to Tat transactivation, HLM-1 cells (58), which contain an integrated copy of a Tat-defective proviral clone, were transfected with a Tat expression plasmid. ChIP analysis demonstrated that Tat induced high levels of RNAP II and Ser 2 and Ser 5 phosphorylation within the far downstream Env coding region (Fig. 6).
FIG. 6.
Distribution of different phosphorylation forms of RNAP II on HIV-1 genes is Tat dependent. (A) Diagram of HIV-1 genes, with the PCR amplification fragment for the Env region shown underneath (gray histogram). The HIV-1 proviral DNA integrated in HLM1 cells was derived from pHXB2gpt. (B) Tat-dependent distribution of different phosphorylation forms of RNAP II on HIV-1 genes. HLM1 cells, which were derived from HeLa-T4+ cells integrated with one copy of HIV-1 genome containing a Tat-defective mutation, were grown in DMEM containing 100 μg of G418/ml plus 1% streptomycin, penicillin antibiotics, and 1% l-glutamine (Gibco/BRL). Cells were transfected with the Tat plasmid DNA (5 μg) by the calcium phosphate method when cells reached 75% confluence. The transfected cells were washed after 4 h posttransfection, and fresh complete DMEM with 10% fetal bovine serum was added for the remainder of the experiment. At day 2 cells were processed for ChIP assay by using α-RNAP II, α-Ser2P, and α-Ser5P antibodies. Specific DNA sequences in the immunoprecipitates were detected by PCR with primers specific for the HIV-1 env gene.
We next analyzed the flavopiridol sensitivity of Ser 2 and Ser 5 phosphorylation at different regions of HIV-1 genes. TNF-α-induced OM10.1 cells were treated with flavopiridol, and then the levels of Ser 2 and Ser 5 phosphorylation at the promoter and downstream coding regions were determined by ChIP assays. The results of these studies demonstrated that flavopiridol inhibition of the P-TEFb kinase activity caused a significant decrease of RNAP II CTD Ser 2 phosphorylation on HIV-1 genes at both the promoter and downstream coding regions (data not shown). At the promoter region, Ser 5 phosphorylation was resistant to flavopiridol inhibition, consistent with the CTD kinase activity of TFIIH being responsible for Ser 5 phosphorylation at this region. However, flavopiridol treatment resulted in a dramatic decrease of Ser 5 phosphorylation in downstream coding regions, with the greatest decrease at the mid-Gag region (10-fold), which is consistent with the Tat/TAR-dependent kinase activity of P-TEFb being responsible for Ser 5 phosphorylation in downstream coding regions. While the amount and distribution of hyperphosphorylated RNAP II epitopes changed dramatically, there was no significant change in total RNAP II in untreated versus flavopiridol-treated cells (data not shown).
Association of SPT5, but not Tat-SF1, with HIV-1 transcription complexes is dependent on P-TEFb kinase activity.
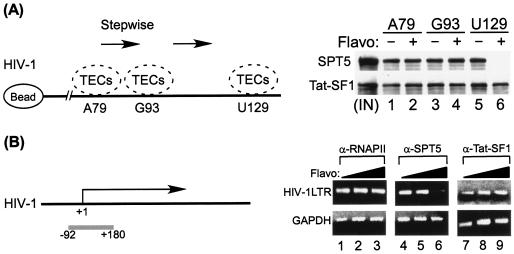
To assess the significance of phosphorylation of SPT5 and Tat-SF1 during HIV-1 transcription elongation, TECs stalled at A79 were elongated stepwise to U129 in the absence or presence of flavopiridol. Protein components of TECs stalled at different positions were analyzed by Western blotting with α-SPT5 or α-Tat-SF1, respectively. Results shown in Fig. 7A demonstrated that flavopiridol inhibition of the P-TEFb kinase activity prompted the dissociation of SPT5 from the HIV-1 TECs between G93 and U129. In contrast, flavopiridol inhibition of the P-TEFb kinase activity had little effect on the stable association of Tat-SF1 with the HIV-1 TECs. These results suggested that the association of STP5 with HIV-1 TECs is dependent on the kinase activity of P-TEFb during transcription elongation.
FIG. 7.
(A) Association of SPT5, but not Tat-SF1, with HIV-1 TECs depends upon the kinase activity of P-TEFb. TECs stalled at A79 were elongated stepwise to U129 in the absence or presence of flavopiridol (50 nM). Protein components of TECs stalled at different positions were analyzed by Western blotting with either α-SPT5 or α-Tat-SF1 antibody. A diagram of the TECs' stepwise walk is shown on the left. (B) ChIP analysis of the distribution of SPT5 and Tat-SF1 on the HIV-1 promoter-proximal region. HIV-1 latently infected OM10.1 cells in the mid-log phase of growth were induced with 10 ng of TNF-α/ml for 2 h and washed three times with phosphate-buffered saline. Cells were then incubated for 48 h with 0 to 75 nM flavopiridol (Flavo). ChIP assays were performed with α-RNAP II (N-20), α-SPT5, or α-Tat-SF1 antibody. Immunoprecipitated DNAs were detected by PCR utilizing primers specific for the HIV-1 LTR (upper panel) from −92 to +180 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (lower panel) from −85 to +81. The diagram of HIV-1 genes is shown at the left of the panel with the PCR fragment shown underneath the genes in a gray histogram. The sequence in the promoter region (from −92 to +180) used for PCR amplification was from subtype B, LAI strain.
To gain insight into the effect of P-TEFb kinase activity on the association of SPT5 and Tat-SF1 with HIV-1 transcription complexes in vivo, we performed ChIP assays to analyze the distribution of SPT5 and Tat-SF1 on HIV-1 genes at the promoter-proximal region. TNF-α-induced OM10.1 cells were treated with flavopiridol and then subjected to ChIP assays. Results shown in Fig. 7B demonstrated that flavopiridol inhibition of the P-TEFb kinase activity caused a significant decrease in the binding of SPT5 to the HIV-1 transcription complex. In contrast, the distribution of Tat-SF1 in flavopiridol-treated cells was not significantly changed.
Loss of P-TEFb kinase activity leads to decreased histone H3 lysine methylation on HIV-1 genes.
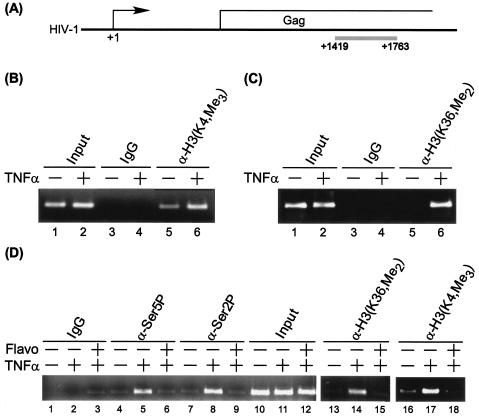
Transcriptionally active chromatin can be methylated at H3-K4, H3-K36, and H3-K79, whereas transcriptionally repressed chromatin can be methylated at H3-K27 and H4-K20 (22, 24). Furthermore, it has been reported that the yeast Set1 and Set2 enzyme complexes, which mediate histone lysine methylation, interact with phosphorylated RNAP II (36, 39, 40, 46, 69). To gain insight into the role of methylation in HIV transcription, methylation of histone H3 on HIV-1 genes at a downstream coding region was analyzed by ChIP assays (Fig. 8A). Results shown in Fig. 8B demonstrated that trimethylation of H3-K4 on HIV-1 genes increased in Tat-expressing OM10.1 cells (lanes 5 and 6). There was also a marked induction of H3-K36 dimethylation on HIV-1 genes (Fig. 8C, lanes 5 and 6). In contrast, histone H3-K27 methylation showed little or no change in Tat-expressing HIV-1 cells (data not shown). Interestingly, histone H3 lysine methylation on HIV-1 genes decreased significantly in flavopiridol-treated cells in parallel with RNAP II CTD Ser 2 and Ser 5 phosphorylation (Fig. 8D, lanes 4 to 9 and 13 to 18). These results suggest that the P-TEFb kinase activity plays an important role in regulating downstream chromatin modifications, such as histone methylation during HIV-1 transcription elongation. We propose that Ser 2 and Ser 5 phosphorylation of RNAP II CTD by the P-TEFb kinase activity is critical for efficiently recruiting histone lysine methyltransferases during HIV-1 gene expression. Consistent with the increase in methylation of lysine 4 of histone H3, in preliminary experiments we observed binding of Set9 to the HIV template during transcription elongation (data not shown). These results are of interest since Set9 has been reported to bind specifically to the H3 tail and, through methylation of K4, displaces the NuRD complex, which negatively regulates transcription (47).
FIG. 8.
Loss of P-TEFb kinase activity reduces methylation of histone H3 at lysine 4 and lysine 36 on HIV-1 genes. (A) Diagram of HIV-1 genes with the PCR fragment shown underneath the genes in a gray histogram. The sequence used for amplification was from subtype B, LAI strain. (B and C) Methylation of histone H3 at lysine 4 and lysine 36 on HIV-1 genes was induced by TNF-α in HIV-1 latently infected cells. HIV-1 latently infected OM10.1 cells, in log phase, were treated with 10 ng of TNF-α/ml for 2 h. The cells were washed and incubated in complete medium at 37°C for 24 h. After 24 h, the cells were cross-linked with 1% formaldehyde, and ChIP assays were performed with antibody against trimethyl-H3-lysine 4 [α-H3(K4,Me3)], dimethyl-H3-lysine 36 [α-H3(K36,Me2)], or control immunoglobulin G (IgG). Specific DNA sequence of the Gag region in the immunoprecipitates was detected by PCR under conditions in which the yield was dependent on the amount of input DNA. (D) Flavopiridol (Flavo) treatment significantly decreased methylation of histone H3 at lysine 4 and lysine 36 on HIV-1 genes in TNF-α-induced HIV-1 latent cells. The TNF-α-induced OM10.1 cells were treated with 75 nM flavopiridol at 37°C for 24 h. After 24 h, the cells were cross-linked with 1% formaldehyde, and ChIP assays were performed with antibody against trimethyl-H3-lysine 4, dimethyl-H3-lysine 36, RNAP II CTD Ser 2P (H5), and Ser 5P (H14), or control IgG, respectively. Specific DNA sequence of the Gag region in the immunoprecipitates was detected by PCR under conditions in which the yield was dependent on the amount of input DNA.
DISCUSSION
Tat recruits P-TEFb to the TAR RNA structure to facilitate the formation of processive transcription elongation complexes. Here we demonstrate that P-TEFb functionally contributes to HIV-1 gene expression at multiple levels. P-TEFb associated with the HIV-1 TECs phosphorylates RNAP II CTD, SPT5, and Tat-SF1 in a Tat/TAR-dependent manner. Flavopiridol inhibition of the P-TEFb kinase activity leads to a prominent decrease in phosphorylation of RNAP II CTD, SPT5, and Tat-SF1 during HIV-1 transcription elongation. A unique phosphorylation pattern of the RNAP II CTD at Ser 2 and Ser 5 on HIV-1 genes is found at the promoter and downstream coding regions. High levels of Ser 2 and Ser 5 phosphorylation are distributed throughout HIV-1 genes. The stable association of SPT5 with HIV-1 transcription complexes is dependent upon the P-TEFb kinase activity. Interestingly, the P-TEFb kinase activity also correlates with histone H3 methylation on HIV-1 genes in the downstream coding region. Loss of the P-TEFb kinase activity results in a significant decrease of H3-K4 trimethylation and H3-K36 dimethylation in flavopiridol-treated HIV-1.
It has been shown that, in yeast, CTD Ser 5 phosphorylation occurs primarily at promoter regions and is dependent upon TFIIH (35). Ser 2 phosphorylation, in contrast, is seen primarily in coding regions. In the human DHFR and γ-actin genes, like yeast, Ser 5 phosphorylation is concentrated near the promoter (11). Ser 2 phosphorylation, in contrast to the phosphorylation pattern in yeast, is observed at the promoter and downstream coding sequences. In this study, ChIP experiments demonstrated that the CTD phosphorylation pattern on HIV-1 genes was unique. Most notably, Ser 5 CTD phosphorylation was observed not only at the promoter but also at equally high levels in the downstream coding sequences. The appearance of Ser 2 and Ser 5 phosphorylation is consistent with our previous findings that Tat modifies the kinase activity of P-TEFb to phosphorylate Ser 2 and Ser 5 of the RNAP II CTD (74). Hyperphosphorylation of the RNAP II CTD by the Tat/TAR-dependent P-TEFb kinase activity leads to a transition from nonprocessive to processive transcription, which overcomes promoter-proximal pausing of RNAP II and results in the uniform distribution of total RNAP II along HIV-1 genes.
Our results further demonstrate that P-TEFb kinase activity is required for the stable association of SPT5 (64) with the HIV-1 TECs. In contrast, the stable association of Tat-SF1 (77) with the HIV-1 TECs does not require the P-TEFb kinase activity. On basal promoters, the SPT4-SPT5 heterodimer is associated with the hypophosphorylated form of RNAP II. Under active transcription elongation, SPT5 is associated with the hyperphosphorylated form of RNAP II in conjunction with P-TEFb and SPT6 (1, 5, 31, 65). Phosphorylation of SPT5 is thought to play a critical role in this transition, converting SPT5 from an inhibitor of transcriptional elongation to a stimulator of elongation (5, 28, 32, 53, 61, 65). It has been recently shown that SPT5 is methylated at its arginine residues (37). Interestingly, mutation of SPT5 methylation sites increases the ability to stimulate transcription elongation.
An active chromatin structure stays open during transcription so that RNAP II can traverse through nucleosomes. The maintenance of an active chromatin structure on a given gene correlates with histone methylation. Transcriptionally active euchromatin can be methylated at H3-K4, H3-K36, and H3-K79 (22, 24). Recent studies have demonstrated that the yeast histone methyltransferases Set1 and Set2 are recruited to coding regions by the PAF transcription elongation factor complex in a manner dependent upon the phosphorylated state of the RNAP II CTD (36, 46). Accordingly, Ctk1-mediated Ser 2 phosphorylation and Kin28-mediated Ser 5 phosphorylation recruit Set2 and Set1, respectively, and in both cases the PAF complex is required. Our results show that specific inhibition of the P-TEFb kinase activity causes a reduction in phosphorylation of RNAP II CTD at either Ser 2 or Ser 5 and a defect in methylation of histone H3 lysine 4 and lysine 36 on HIV-1 genes. We propose that during mammalian gene expression, phosphorylation of RNAP II by P-TEFb is critical for efficiently recruiting histone methylation machinery.
A significant number of antiviral drugs are designed to target viral proteins to attain specificity and avoid toxicity. Unfortunately, these drugs commonly select for drug-resistant viral mutants and exhibit activity against only a few closely related viruses. During the past few years, pharmacological CDK inhibitors have been used effectively to inhibit the replication of human cytomegalovirus, herpes simplex virus, and HIV (6, 8, 45, 59, 66). Of the pharmacological CDK inhibitors developed or tested to date, flavopiridol is by far the most potent and effective inhibitor of HIV-1 transcription and virus replication identified. Flavopiridol binds to the ATP pocket of CDK9 and by virtue of salt bridges between Lys 20/29 and Glu 92 is able to form a stable, noncompetitive interaction with the enzyme (8, 9, 15). Flavopiridol inhibits HIV-1 transcription at 1 to 10 nM, while it inhibits cellular transcription at much higher concentrations (8, 9). We propose that the results presented in this study explain, at least in part, the specificity toward HIV-1 transcription. P-TEFb phosphorylates not one, but three critical cofactors in a Tat/TAR-dependent manner. The exact molecular mechanism of how RNAP II CTD, SPT5, and Tat-SF1 phosphorylation enhances HIV-1 transcription remains to be elucidated. Given the effectiveness of flavopiridol as an antiviral drug, it will be of interest to explore derivatives which are more effective but exhibit less toxicity in patients.
Acknowledgments
We thank Shanese Baylor and Rebecca Linton for their editorial assistance.
This work was supported by the King Fahd endowment (A.K.), grants from the George Washington University REF funds (F.K.), National Institutes of Health grants AI054222 (A.K.), AI44357, AI43894, and A13969 (F.K.) and a grant from the National Institutes of Health Intramural AIDS Targeted Antiviral Program (IATAP).
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boris-Lawrie, K. A., J. N. Brady, and A. Kumar. 1992. Sequences within the R region of the long terminal repeat activate basal transcription from the HIV-1 promoter. Gene Expr. 2:215-230. [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., T. Albrecht, and E. A. Thompson. 1998. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J. Biol. Chem. 273:22075-22082. [DOI] [PubMed] [Google Scholar]
- 7.Butera, S. T., B. D. Roberts, L. Lam, T. Hodge, and T. M. Folks. 1994. Human immunodeficiency virus type 1 RNA expression by four chronically infected cell lines indicates multiple mechanisms of latency. J. Virol. 68:2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 9.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D., and Q. Zhou. 1999. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol. Cell. Biol. 19:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, R. F., and K. T. Jeang. 1996. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J. Biol. Chem. 271:27888-27894. [DOI] [PubMed] [Google Scholar]
- 13.Cujec, T. P., H. Okamoto, K. Fujinaga, J. Meyer, H. Chamberlin, D. O. Morgan, and B. M. Peterlin. 1997. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 15.de, Azevedo, W. F., Jr., F. Canduri, and N. J. da Silveira. 2002. Structural basis for inhibition of cyclin-dependent kinase 9 by flavopiridol. Biochem. Biophys. Res. Commun. 293:566-571. [DOI] [PubMed] [Google Scholar]
- 16.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Martinez, L. F., G. Mavankal, J. M. Neveu, W. S. Lane, D. Ivanov, and R. B. Gaynor. 1997. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 16:2836-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 23.Gold, M. O., X. Yang, C. H. Herrmann, and A. P. Rice. 1998. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol. 72:4448-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann, C. H., and A. P. Rice. 1993. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology 197:601-608. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for Tat activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 30.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 33.Kim, Y. K., C. F. Bourgeois, C. Isel, M. J. Churcher, and J. Karn. 2002. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 22:4622-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 35.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak, Y. T., J. Guo, S. Prajapati, K. J. Park, R. M. Surabhi, B. Miller, P. Gehrig, and R. B. Gaynor. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11:1055-1066. [DOI] [PubMed] [Google Scholar]
- 38.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 39.Li, B., L. Howe, S. Anderson, J. R. Yates, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 40.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 41.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 43.Marshall, N. F., and D. H. Price. 1992. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 12:2078-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, P. J., I. H. Gelman, and P. E. Klotman. 2001. Suppression of HIV-1 expression by inhibitors of cyclin-dependent kinases promotes differentiation of infected podocytes. J. Am. Soc. Nephrol. 12:2827-2831. [DOI] [PubMed] [Google Scholar]
- 46.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 47.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto, H., C. T. Sheline, J. L. Corden, K. A. Jones, and B. M. Peterlin. 1996. trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA 93:11575-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 50.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 51.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 52.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 54.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermeulen, J. P. Tassan, L. Schaeffer, E. A. Nigg, J. H. Hoeijmakers, and J. M. Egly. 1994. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 79:1093-1101. [DOI] [PubMed] [Google Scholar]
- 58.Sadaie, M. R., and G. L. Hager. 1994. Induction of developmentally programmed cell death and activation of HIV by sodium butyrate. Virology 202:513-518. [DOI] [PubMed] [Google Scholar]
- 59.Schang, L. M., A. Rosenberg, and P. A. Schaffer. 2000. Roscovitine, a specific inhibitor of cellular cyclin-dependent kinases, inhibits herpes simplex virus DNA synthesis in the presence of viral early proteins. J. Virol. 74:2107-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 63.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 273:6769-6775. [DOI] [PubMed] [Google Scholar]
- 64.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, D., C. de la Fuente, L. Deng, L. Wang, I. Zilberman, C. Eadie, M. Healey, D. Stein, T. Denny, L. E. Harrison, L. Meijer, and F. Kashanchi. 2001. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J. Virol. 75:7266-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weeks, K. M., C. Ampe, S. C. Schultz, T. A. Steitz, and D. M. Crothers. 1990. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 249:1281-1285. [DOI] [PubMed] [Google Scholar]
- 68.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 69.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, X., M. O. Gold, D. N. Tang, D. E. Lewis, E. Aguilar-Cordova, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 94:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, X., C. H. Herrmann, and A. P. Rice. 1996. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J. Virol. 70:4576-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yankulov, K., and D. Bentley. 1998. Transcriptional control: Tat cofactors and transcriptional elongation. Curr. Biol. 8:R447-R449. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, M., L. Deng, F. Kashanchi, J. N. Brady, A. J. Shatkin, and A. Kumar. 2003. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. USA 100:12666-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, M., S. Nekhai, D. C. Bharucha, A. Kumar, H. Ge, D. H. Price, J. M. Egly, and J. N. Brady. 2001. TFIIH inhibits CDK9 phosphorylation during human immunodeficiency virus type 1 transcription. J. Biol. Chem. 276:44633-44640. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 78.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]