Abstract
The virion host shutoff (vhs) protein of herpes simplex virus type 1 causes the degradation of host and viral mRNA immediately upon infection of permissive cells. vhs can interact with VP16 through a 20-amino-acid binding domain, and viruses containing a deletion of this VP16-binding domain of vhs (Δ20) and a corresponding marker rescue (Δ20R) were constructed and characterized. Transient-transfection assays showed that this domain was dispensable for vhs activity. The Δ20 recombinant virus, however, was unable to induce mRNA degradation in the presence of actinomycin D, while degradation induced by Δ20R was equivalent to that for wild-type virus. Δ20, Δ20R, and KOS caused comparable RNA degradation in the absence of actinomycin D. Western blot analysis of infected cells indicated that comparable levels of vhs were expressed by Δ20, Δ20R, and KOS, and there was only a modest reduction of vhs packaging in Δ20. Immunoprecipitation of protein from cells infected with Δ20 and Δ20R showed equivalent coprecipitation of vhs and VP16. Pathogenesis studies with Δ20 showed a significant decrease in replication in the corneas, trigeminal ganglia, and brains, as well as a significant reduction in clinical disease and lethality, but no significant difference in the establishment of, or reactivation from, latency compared to results with KOS and Δ20R. These results suggest that the previously described VP16-binding domain is not required for vhs packaging or for binding to VP16. It is required, however, for RNA degradation activity of tegument-derived vhs and wild-type replication and virulence in mice.
Cells infected with herpes simplex virus (HSV) undergo a rapid shutoff of macromolecular synthesis due to the product of the UL41 gene, the virion host shutoff (vhs) protein. vhs is a 58-kDa phosphoprotein found in approximately 200 copies in the HSV tegument (8, 28). It is released directly into the cytoplasm of infected cells, where it degrades both host and viral mRNA (15, 25). vhs is capable of cleaving target RNA via an endoribonucleolytic activity at either the 5′ cap or regions of high secondary structure (3, 4, 44) and can induce degradation in a sequence-specific manner at the 3′ end of some RNA transcripts (5). vhs forms a complex with the translation initiation factor eIF4H (6), and this interaction may account for the apparent selectivity of vhs for mRNA rather than rRNA or tRNA.
vhs is conserved among the alphaherpesviruses, suggesting an important role in neurotropic herpesvirus biology. Viruses lacking the UL41 gene are viable and have only a slight in vitro growth defect (28, 37). Work with HSV has shown that vhs mutants are significantly compromised for pathogenesis (33, 36, 37). Such viruses show decreased replication in the cornea and trigeminal ganglia and decreased periocular disease following corneal inoculation (33, 36, 37). These viruses also show a significant reduction in both the establishment of and reactivation from latency (33, 36, 37). The mechanism resulting in the profoundly decreased virulence of vhs mutant viruses has not been identified.
The HSV type 1 (HSV-1) tegument contains up to 15 viral proteins (8). Assembly of the components of the viral tegument is thought to occur in both the nucleus and the cytoplasm of the infected cell (22). Several studies have shown interactions between capsid components and proteins in the viral tegument (21, 45), but little is known about how these protein-protein interactions regulate both the activity and the packaging of the individual proteins. Interestingly, VP16, a viral protein which functions as both an essential component of the tegument and a transactivator of viral immediate-early gene expression, has been shown to interact with vhs in infected cells (13, 24, 31). A 20-amino-acid domain in vhs has been identified as being sufficient for the interaction with VP16 in yeast two-hybrid and glutathione S-transferase binding assays (29). The requirement for this domain, however, has not been characterized in the context of the virus.
The apparent lack of discrimination of vhs in causing the degradation of both viral and cellular RNA means that the virus may need to regulate the activity of the de novo-synthesized vhs protein. It is likely that the interaction between vhs and VP16 is important for the regulation of this activity, and several lines of evidence support this idea. Viruses deleted in the VP16 protein show increased levels of translational arrest, and vhs activity is dampened in cells constitutively expressing VP16, suggesting that VP16 downregulates the activity of the vhs protein (16). Additionally, a virus that contains a single amino acid substitution in VP16, which precludes binding to vhs, results in a virus that is unable to mediate a productive infection, presumably due to rampant vhs activity (11). Complex formation between vhs and VP16 may thus be required to modulate the activity of de novo-synthesized vhs to a level that allows the continued synthesis of viral genes. vhs and VP16 interactions may also be important for the packaging of vhs. Indeed, a virus with a large deletion in the vhs protein that includes the VP16-binding domain fails to package vhs (28).
In this study, a virus containing a specific deletion of the 20-amino-acid VP16-binding domain of vhs (Δ20) was generated in order to examine its role in the regulation of vhs activity, packaging of vhs into the virion, and pathogenesis. Deletion of the VP16-binding domain had no effect on the RNA degradation activity of the vhs protein in transient transfections. Similarly, the activity of de novo-synthesized Δ20 vhs in the context of a recombinant virus was unaffected. In contrast, when de novo synthesis of vhs was blocked with actinomycin D, Δ20 induced no RNA degradation. Loss of the VP16-binding domain had only a modest affect on the packaging of vhs into the virion, and coprecipitation of vhs and VP16 did not require the presence of the VP16-binding domain. Interestingly, although the Δ20 virus retains nearly wild-type levels of vhs activity from the de novo-synthesized protein, this was not sufficient to mediate wild-type levels of replication in vivo or virulence, indicating the importance of the tegument-derived activity of vhs in the mouse model of pathogenesis. Moreover, Δ20 will prove useful in dissecting the relative contributions of tegument-derived and de novo-synthesized-vhs activity.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were cultured as previously described (27). The HSV-1 wild-type strain KOS was the background for all viruses. dl41, a virus in which the entire vhs open reading frame was replaced with a β-galactosidase cassette under the control of the human cytomegalovirus immediate-early promoter, was described previously (37). UL41-NHB, a virus which contains a nonsense mutation in the vhs gene, and BGS-41, a virus that contains an HCMVIE β-galactosidase cassette inserted within the vhs open reading frame, were previously described (37). 8MA is a VP16 null virus described previously (43). The vhs-null virus contains a stop codon inserted at amino acid 9, which has been shown to be deficient in vhs expression and function (S. S. Strand and D. A. Leib, unpublished data). A mock-infected lysate was generated by harvesting three confluent 175-cm2 flasks of Vero cells, using the same conditions for the generation of a high-titer virus stock as those described previously (27). Δ20, a virus containing a deletion of the VP16 binding domain, and the corresponding marker rescued virus, Δ20R, were generated as described below. Multiple- and single-step growth assays were carried out with Vero cells as described previously.
Generation of plasmids and recombinant viruses.
In order to generate the vhs expression constructs used in the transient-transfection assays, a fragment containing the entire vhs open reading frame was generated by PCR amplification of the pUL41 plasmid that had been generated previously (37). Primers flanking the vhs open reading frame were as follows: forward primer, 5′GAACCGGAATTCCGGGTTAGGCCGATCCGCAGTTA3′; reverse primer, 5′CTTCCGGAATTCCGGGTCGTTTGTTCGGGGACAAG3′. The PCR primers were engineered such that the resulting 1.5-kb PCR product contained EcoRI-cleavable ends. PCR conditions were as follows: 1× Pfx amplification buffer, 0.3 mM deoxynucleoside triphosphate mix, 1 mM MgSO4, 0.3 μΜ forward and reverse primers, 10 pg of template DNA, 1.0 U of Pfx polymerase, and 5 μl of Pfx enhancer buffer in a total volume of 50 μl. PCR amplification conditions were as follows: 5 min, 95°C for 30 s, 95°C for 30 s, 58°C for 30 s, 72°C for 7 min, 72°C for 35 cycles. The resulting fragment was treated with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and cloned into SmaI (New England Biolabs, Beverly, Mass.)-digested, calf intestinal phosphatase (New England Biolabs)-treated pMECA (40), resulting in the construct pMECA-vhs. The 1.5-kb EcoRI fragment containing the vhs open reading frame from pMECA-vhs was ligated into EcoRI-cut pCI in both orientations to generate pCI-vhsF and pCI-vhsR. Generation of the Δ20 construct containing the deletion of the VP16-binding domain of vhs was generated as follows. An oligonucleotide complementary to 20 bp on either side of the VP16-binding domain was synthesized. Site-directed mutagenesis was performed (14), and resulting colonies were screened for the deletion of the VP16-binding domain by restriction digestion with BamHI (New England Biolabs). The resulting construct was sequenced by using Big-Dye Terminator sequencing protocol and designated pUL41-Δ20. The recombinant virus Δ20, containing the deletion of the VP16-binding domain, was generated by cotransfection of BGS-41 infectious DNA and pUL41-Δ20. Resulting white plaques were amplified in cell culture, DNA extracted, and screened by PCR. Plaques showing a deletion of the VP16-binding domain were plaque purified three times on Vero cells, and Southern blotting was performed by using the Prime-a-gene labeling system (Promega, Madison, Wis.) to generate a randomly primed pUL41 probe to confirm the genotype as described previously (27). The Δ20R virus was generated by cotransfection of Δ20 infectious DNA and pUL41. Isolation and confirmation of the genotype was carried out as for Δ20.
Functional assay for vhs activity.
Vero cells were seeded into 35-mm tissue culture dishes at a density of 2.5 × 105 cells/well. One day later, cells were transfected with 30 ng of simian virus 40 (SV40) luciferase alone or in combination with pCI-VHS (VHS WT) or pCIΔ20 (VHS Δ20) in a range of concentrations ranging from 3 to 500 ng, using lipofectamine (Gibco-BRL, Carlesbad, Calif.) according to the product literature. Forty-eight hours posttransfection, cells were harvested into 100 μl of 1× passive lysis buffer (Promega, Madison, Wis.), and luciferase readings were determined. The total protein content of the lysates was examined by using the Bradford protein assay (Bio-Rad, Hercules, Calif.). The relative light units of luciferase produced were normalized to the lowest total protein content.
RNA degradation analysis.
Confluent monolayers of Vero cells were pretreated with 10 μg of actinomycin D/ml or left untreated for 1 h prior to viral infection. Cells were infected at a multiplicity of infection (MOI) of 20 with either mock-infected lysate, KOS, UL41NHB, Δ20, or Δ20R. Cytoplasmic RNA was harvested at 4 and 8 h postinfection, using methods described previously (37). RNA was analyzed by Northern blotting, probed with either random prime-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH), using the Prime-a-Gene kit (Promega), or an oligonucleotide to the 28S rRNA end labeled with T4 polynucleotide kinase (New England Biolabs). Blots were visualized by using the Molecular Dynamics Storm Phosphorimaging system. Normalization of the loading of each lane was performed as described previously (23).
Western blot analysis.
Infected cell lysates were prepared by mock infecting or infecting Vero cells at an MOI of 20 with either KOS, dl41, Δ20, or Δ20R. Cell lysates were harvested at 16 h postinfection and placed in 1× sodium dodecyl sulfate (SDS) sample buffer, boiled, and run on an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel in 1× Tris-glycine electrophoresis buffer. Virions were gradient purified over a 10 to 30% Dextran gradient by using the following protocol. Four T175 flasks of Vero cells were infected at an MOI of 0.1 with either KOS or Δ20. The supernatant was harvested and spun at 2,500 × g for 10 min to remove cell debris. This precleared supernatant was spun at 9,000 × g for 60 min, and the resulting pellet was resuspended in 2 ml of minimal essential medium (MEM) plus 0.5% bovine serum albumin. An aliquot of this resuspended material was saved for determination of titers to measure the total amount of virus loaded on the dextran gradient. Solutions of 10 and 30% dextran were prepared in MEM plus 0.5% bovine serum albumin. A 12-ml 10 to 30% gradient was poured. A 2-ml volume containing the resuspended viral pellet was layered at the top of the gradient. The tubes were spun in an SW-41 rotor in a Beckman centrifuge at 35,000 × g for 1 h, and 0.5-ml fractions were collected. Each fraction was subjected to titer determination for the presence of infectious virus. The fractions were then spun at full speed in a microcentrifuge and resuspended in 100 μl of 1× SDS-PAGE loading dye, and 20-μl aliquots were run on an SDS-10% PAGE gel. The proteins were transferred to a polyvinylidene difluoride nitrocellulose membrane (Amersham, Piscataway, N.J.) and probed for vhs, VP5, and VP16. Western blots were visualized by using the ECL-Plus chemiluminescence detection system (Amersham) and visualized and quantified on a Molecular Dynamics Storm Phosphorimaging system. Values of individual bands were normalized to the lowest value calculated for the VP5 signal.
Antibodies.
A rabbit polyclonal antiserum (1883) to vhs protein was generated by Cocalaco Biologicals (Reamstown, Pa.), using purified vhs protein (obtained from Sullivan Read at the University of Kansas City). Briefly, rabbits were immunized with 150 μg of purified vhs protein initially and then boosted with 50 μg every 4 to 6 weeks. The vhs-specific antiserum 1883 was used at a dilution of 1:2,000. The VP5 mouse monoclonal antibody (East Coast Biologicals, New Burwick, Maine) was used at a dilution of 1:2,500. The VP16 mouse monoclonal antibody 16-25 (Santa Cruz Biologicals, Santa Cruz, Calif.) was used at a dilution of 1:2,000. Horseradish peroxidase-conjugated mouse antirabbit antibody (Bio-Rad) was used at a dilution of 1:2,000. Horseradish peroxidase-conjugated rabbit antimouse antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) was used at a dilution of 1:2,000.
Immunoprecipitations.
Sixty-millimeter plates of Vero cells were infected at an MOI of 20 with KOS, VHS-null, Δ20, or Δ20R. At 3 h postinfection, cells were incubated in cysteine and methionine-free MEM for 30 min. Medium containing 100 μCi of S35-labeled cysteine and methionine trans-label (Amersham)/mmol was added, and infected cells were harvested into 1 ml of RIPA lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) at 6 h postinfection. Lysates were spun at 67,000 × g for 30 min at 4°C to remove chromosomal DNA and aggregated proteins. Supernatant was transferred to a new tube, and lysates were precleared with 10 μl of normal rabbit serum (Sigma, St. Louis, Mo.) and 20 μl of Protein A/G Plus agarose (Santa Cruz Biologicals) for 1 h at 4°C. Immunoprecipitations were carried out with 10 μl of either VHS antibody 1883 or rabbit 1883 preimmune serum and 30 μl of Protein A/G Plus agarose (Santa Cruz Biologicals) for 1 h at 4°C. Beads were pelleted and washed twice with 1 ml of cold RIPA lysis buffer, once with 1 ml of cold IP wash buffer (0.1% [wt/vol] Triton X-100, 50 mM Tris-Cl [pH 7.4], 300 mM NaCl, 5 mM EDTA), and once with 1 ml of cold phosphate-buffered saline. Beads were resuspended in 50 μl of 1× SDS-PAGE loading dye and boiled for 5 min. Twenty-microliter samples were run on an SDS-10% PAGE gel and analyzed with autoradiography or by Western blotting for VP16, using the 16-25 antibody as described above.
Procedures with animals.
Outbred CD-1 female mice (Charles River Breeding Laboratories Inc., Kingston, N.Y.) were bilaterally scarified and infected with virus at a concentration of 2 × 106 PFU/5 μl as described previously (27, 39). Clinical disease was evaluated as previously described (32). Eye-swab material and trigeminal ganglion homogenates were subjected to titer determination as described previously (17, 37). Intracerebral inoculations were carried out as described previously (20), using 105 PFU of virus per animal. Animals were sacrificed, brains were harvested days 1 to 7 postinfection, and titers were determined as described previously (37). All statistical analyses were performed using an unpaired, two-tailed t test.
Limiting-dilution reactivation assay.
Mice were sacrificed on day 28 after infection, and the trigeminal ganglia from three to four mice were placed into 30 ml of serum-free Dulbecco's modified Eagle medium (DMEM). Ganglia were spun at 2,500 × g for 10 min, resuspended in 1 ml of dissociation medium per trigeminal ganglion (100 ml of serum-free DMEM, 1 ml of 7.5% sodium bicarbonate, 1 ml of 20 mM HEPES, 2 ml of trypsin, 2 ml of collagenase), and incubated at 37o with shaking for 1 h. Ganglia were spun at 2, 500 × g for 10 min and resuspended in 0.5 ml of DMEM/ganglion. Resuspended material was passed through a 40-μm-pore-size cell strainer, and filters were washed with 0.5 ml of DMEM/ganglion. Cells were counted by trypan blue exclusion, and the concentration was adjusted to 4 × 106 cells/ml. Twofold serial dilutions of dissociated trigeminal ganglion material were plated into collagen-coated 35-mm wells containing 2 ml of DMEM, starting at concentrations from 2 × 106 to 1.2 × 105 cells/well. Six wells were plated at each cell dilution. One day postdissociation, 5 × 104 Vero cells/ml in serum-free medium were added to each well. One hundred microliters of medium from each well was placed onto an indicator monolayer of Vero cells on days 3, 4, 5, 8, and 12 postdissociation. These monolayers were scored for the presence or absence of cytopathic effect. Data are reported as percentage of wells reactivated, per cell dilution, per virus.
Semiquantitative real-time PCR for latent genomes.
Six-week-old female CD-1 mice were infected as described above. Trigeminal ganglia were harvested on day 28 postinfection, and genomic DNA was extracted by using the RNeasy tissue preparation kit from QIAGEN (Valencia, Calif.). The number of latent genomes per trigeminal ganglion was determined using primers to the thymidine kinase (tk) gene of HSV-1 designed to amplify a 70-bp fragment. The sequence of these primers is as follows: Tk (forward), 5′CCAAAGAGGTGCGGGAGTTT3′; Tk (reverse), 5′CTTAACAGCTGTCAACAGCGTGCCG3′. PCRs were performed using the following reaction components in a total volume of 10 μl: 1× SYBR Green Super mix (Bio-Rad), 900 nM primers, 5% acetamine, and 2 μl of sample DNA. PCR conditions were as follows: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 9 min, and 40 cycles at 95°C for 15 s, 60°C for 1 min, and 72°C for 15 s. A standard curve of QIAGEN Maxi-prep purified HSV-1 bacterial artificial chromosome DNA, 17-49Bac (W. W. Gierasch and D. A. Leib, unpublished data) was diluted in the background of mouse DNA in 10-fold dilutions from 106 to 101 copies and used as a standard curve for determination of the total genome copy number in latently infected trigeminal ganglia. To control for the total amount of DNA in each trigeminal ganglion sample, real-time PCRs to the single-copy mouse adipsin gene were performed as follows. Mouse adipsin forward primer was 5′AGTGTGCGGGGATGCAGT3′; reverse primer was 5′ACGCGAGAGCCCCAGGTA3′. The PCR components, in a final volume of 10 μl, were as follows: 1× SYBR Green Super mix (Bio-Rad), 900 nm of primers, 5% acetamide, and 2 μl of sample DNA. The PCR conditions were the same as detailed above for PCRs using tk primers. Trigeminal ganglion DNA harvested from uninfected mice was used to generate mouse DNA standards from 105 to 101 copies, from which the total copy number of mouse adipsin in each of the infected trigeminal ganglion samples was determined. Each value for tk copy number was normalized to the lowest value of the mouse adipsin copy number, and the number of copies of genome per trigeminal ganglion was then expressed on a log scale. Statistical significance was calculated by using an unpaired, two-tailed t test.
RESULTS
Construction of a plasmid containing a deletion of the VP16-binding domain of vhs.
A schematic of the vhs protein indicating the four conserved domains and the VP16-binding domain is shown in Fig. 1. A deletion in the VP16-binding domain of vhs (pUL41-Δ20) was generated by site-directed mutagenesis. The 60 nucleotides encoding the 20 amino acids of the binding domain were completely deleted. The deletion of the VP16-binding domain was confirmed by a change in the size of a PCR amplification fragment. The junction resulting from deletion of the VP16-binding domain was sequenced and was found to be consistent with the predicted remaining wild-type vhs sequence (data not shown).
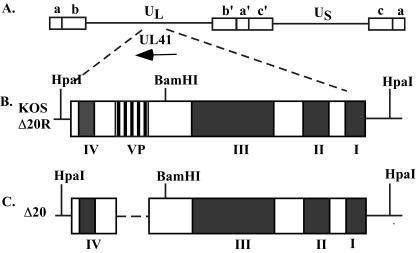
FIG. 1.
Map of the vhs (UL41) open reading frame and the recombinant viruses used in this study. (A) Schematic of the HSV-1 genome, showing the unique long and unique short regions flanked by the internal and terminal repeat sequences. The vhs open reading frame and the direction of transcription are indicated by the arrow. (B) Schematic of the wild-type vhs protein found in KOS and Δ20R. The four conserved domains important for the RNA degradation function are indicated (I to IV). The VP16-binding domain between amino acid sequences 310 and 330 is indicated (VP). BamHI and HpaI restriction enzyme sites used in Southern blotting are shown. (C) Schematic of the vhs protein containing a deletion of the VP16-binding domain found in Δ20. The nucleotides corresponding to amino acids 310 to 330 were deleted using site-directed mutagenesis.
In vitro vhs functional assay.
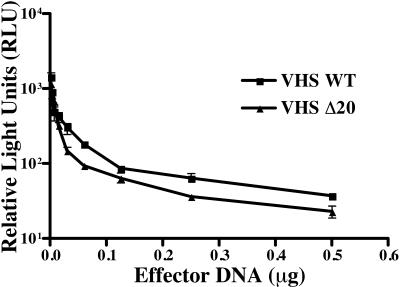
Transient-transfection assays were performed to determine whether vhs retained its RNA degradation activity in the absence of the VP16-binding domain. Vero cells were transfected with SV40 luciferase either alone or in combination with either pCI-VHS (VHS WT) or the VP16-binding domain mutant vhs (VHS Δ20) at concentrations from 3 to 500 ng. Forty-eight hours posttransfection, luciferase activities were comparable in cells transfected with either pCI-VHS or pCI-Δ20 (Fig. 2) at all concentrations used. A time point of 48 h posttransfection was chosen based on similar types of functional assays done previously (9, 26). These results therefore demonstrated at least equivalent vhs activity from both wild-type vhs (VHS WT) and the VP16-binding domain mutant (VHS Δ20), indicating that deletion of the VP16 binding domain does not significantly alter the RNA degradation activity of vhs.
FIG. 2.
Functional assay for vhs activity. Vero cells were transfected with 0.5 μg of the SV40 luciferase reporter plasmid in combination with either pCI-VHS (VHS WT), or pCI-Δ20 (VHS Δ20) in a range from 3 to 500 ng. Forty-eight hours posttransfection, cells were harvested and both a luciferase assay and a Bradford protein assay were performed. Each transfection reaction was done in triplicate, and a representative experiment is shown. All values were normalized to the total amount of protein.
Construction of a recombinant virus containing a deletion of the VP16-binding domain of vhs (Δ20) and the corresponding marker-rescued virus (Δ20R)
A recombinant virus containing a deletion of the defined VP16-binding domain of vhs (Δ20) was created by cotransfection of infectious BGS41 DNA with pUL41-Δ20. White plaques were screened by PCR and plaque purified, and the genotype was confirmed by Southern blotting (Fig. 3). A corresponding marker rescued virus was generated by cotransfection of Δ20 infectious DNA and a plasmid containing a wild-type copy of the UL41 gene (pUL41). Southern blots were probed with randomly primed pUL41 and showed fragments of 2,058 and 3,866 kb for Δ20 and fragments of 2,118 and 3,866 kb for KOS and Δ20R. The sizes of the fragments were consistent with the predicted values and indicated that the VP16-binding domain was deleted in Δ20 and restored in Δ20R.
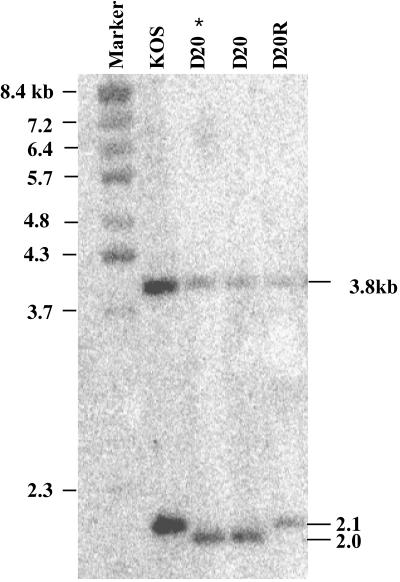
FIG. 3.
Southern blot analysis of KOS, Δ20 (D20) and Δ20R (D20R). Infectious DNA was double digested with BamHI and HpaI and probed with a randomly primed fragment of the pUL41 plasmid. Sizes of the BstEII-digested bacteriophage lambda DNA marker fragments are indicated on the left. The expected fragments for the wild-type KOS and marker rescue virus Δ20R were 3.8 and 2.1 kb. The expected fragments for the VP16-binding domain deletion virus Δ20 were 3.8 and 2.0 kb. The sizes of the expected fragments are indicated on the right. Two independent isolates of Δ20 recombinant virus were confirmed, and “D20 *” indicates the isolate that was purified and used for the experiments in this study.
Replication of Δ20 and Δ20R in vitro.
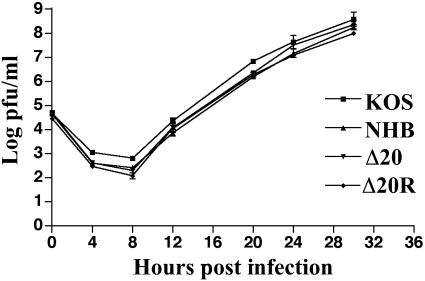
The ability of Δ20 and Δ20R to replicate in Vero cells in a multiple-step growth assay was examined relative to KOS and UL41NHB. All viruses replicated to similar levels (Fig. 4). Δ20 and Δ20R showed no difference in plaque morphology from KOS, and there was also no difference in replication in a single-step growth assay (data not shown). These results indicate that deletion of the VP16-binding domain does not affect the ability of the virus to replicate in Vero cells.
FIG. 4.
Multiple-step growth assay of KOS, UL41NHB, Δ20, and Δ20R in Vero cells at an MOI of 0.01. Time points were 4, 8, 12, 20, 24, and 30 h postinfection. The graph represents the combined data from two independent experiments.
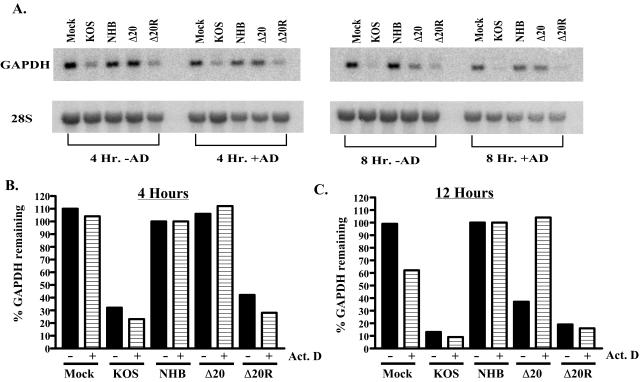
Measurement of vhs activity of Δ20 and Δ20R.
Δ20 was examined for its ability to degrade GAPDH RNA in comparison to that of KOS, UL41NHB (37), and the marker-rescued virus Δ20R. Vero cells were infected in either the presence or absence of actinomycin D, and cytoplasmic RNA was harvested and analyzed by Northern blotting. Resulting blots were probed for GAPDH and 28S RNA (Fig. 5A). At 4 h postinfection in the presence of actinomycin D, cells infected with KOS and Δ20R show significant levels of vhs activity. In contrast, there was no detectable vhs activity in cells infected with Δ20 (Fig. 5B). Indeed, even at MOIs up to 100, Δ20 failed to induce RNA degradation in Vero cells (data not shown). As would be expected, no vhs activity was shown in cells that were mock infected or infected with UL41NHB. In the absence of actinomycin D, at 4 h postinfection, KOS and Δ20R showed similar, significant levels of vhs activity, while Δ20, UL41NHB, and mock-infected cells showed no RNA degradation. At 8 h postinfection, there was significant vhs activity in cells infected with KOS and Δ20R in the presence of actinomycin D (Fig. 5C). In contrast, cells infected with Δ20 showed no vhs activity in the presence of actinomycin D, as was also true for mock-infected and UL41NHB-infected cells. In the absence of actinomycin D, however, vhs activities for Δ20, KOS, and Δ20R were comparable. This shows that by 8 h postinfection, de novo-synthesized vhs from Δ20 induces near-normal levels of RNA degradation but vhs derived from the tegument of Δ20 is inactive.
FIG. 5.
RNA degradation assay by Northern blot analysis. (A) Vero cells were mock infected or infected with KOS, UL41NHB, Δ20, and Δ20R in the presence or absence of actinomycin D (10 mg/ml). Cytoplasmic mRNA was extracted at 4 and 8 h postinfection. Panel A shows a representative Northern blot probed for GAPDH (top) and then stripped and reprobed for 28S rRNA (bottom). (B) A graphical representation of the Northern blot data in the presence and absence of actinomycin D at 4 h postinfection and (C) 12 h postinfection. All data were normalized to the lowest value of the 28S rRNA.
Western blot of infected cells.
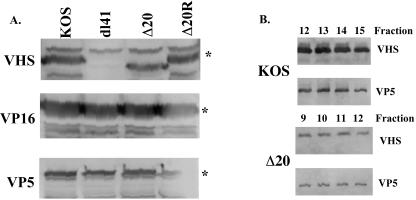
Having shown that vhs activity from the tegument of Δ20 was absent, it was critical to show that Δ20 expressed normal levels of vhs in infected cells. Western blots were performed with lysates of KOS-, Δ20-, Δ20R-, and dl41 (vhs null)-infected cells, probing for vhs, VP5, and VP16 (Fig. 6A). Western blots probed with the 1883 antiserum specific to vhs showed a reactive band at 58 kDa that was absent from dl41-infected cells. Deletion of the 20 amino acids of the VP16-binding domain in Δ20 resulted in a protein with slightly faster mobility. To normalize the amount of viral proteins produced in each cell lysate, Western blots were probed with an antiserum to the major capsid protein VP5. All lanes showed a reactive band at approximately 120 kDa, consistent with the predicted molecular mass of VP5. Normalization for VP5 in several independent experiments indicated that similar levels of vhs are expressed in cells infected with KOS, Δ20, and Δ20R. Western blots were also probed with antiserum to the VP16 protein, and a reactive species of 65 kDa, consistent with the molecular mass of VP16, was present in all lanes. Normalization to VP5 in several independent experiments indicated that similar levels of VP16 are expressed. These data indicated that deletion of the VP16-binding domain of vhs has no effect on the expression or levels of vhs synthesized in infected cells, and as would be expected, this mutation also has no effect on the expression of VP16.
FIG. 6.
Western blot of infected-cell lysates and gradient-purified virus particles. (A) Vero cells were infected with KOS, dl41, Δ20, or Δ20R at an MOI of 20 and harvested at 16 h postinfection. Western blots were probed with an antibody to vhs, VP16, or VP5. The asterisk indicates the specific protein in each blot. (B) Viral particles were purified over a 10 to 30% dextran gradient, and peak fractions of infectious virus were run on an SDS-10% polyacrylamide gel. Western blots were probed with antibodies to vhs or VP5.
Western blot of virus particles.
Deletion of the VP16-binding domain leading to disruption of the packaging of vhs into the virion would explain the lack of vhs activity in the presence of actinomycin D. To examine this, KOS and Δ20 virions were purified over a 10 to 30% dextran gradient. Fractions were run on an SDS-10% PAGE gel and subjected to Western blotting, using antisera to vhs and to the major capsid protein VP5 (Fig. 6B). The titer of each fraction was also determined with Vero cells. Infectious virus was found throughout the gradient, although fractions 12 to 15 were the peak for KOS and fractions 9 to 12 were the peak for Δ20 in the experiment shown (Fig. 6B). In all experiments, the peak of infectious particles was shifted higher in the gradient for Δ20 than for KOS. Western blots of preparations of KOS and Δ20 virions both show a vhs-reactive band at approximately 58 kDa. The presence of a VP5-reactive band was found in each of these peak fractions. These results indicate that vhs is being packaged into the Δ20 virions. An amount of 3.9 × 109 PFU/ml was found in peak fractions of KOS, and 1.4 × 109 PFU/ml was found in peak fractions of Δ20 in the experiment shown. This threefold difference in the number of infectious particles between the KOS and Δ20 fractions likely accounts for the difference in the intensities of the vhs and VP5 bands seen in the Western blot. Subsequent quantitation by phosphorimager of these blots, examining vhs in KOS and Δ20 virions compared to VP5, suggested an approximate twofold decrease in the total amount of vhs packaged in the Δ20 virus (data not shown). These results show that although the VP16-binding domain of vhs is not required for packaging, the presence of the binding domain may contribute to slightly more efficient packaging of vhs into the virion.
VP16 coprecipitates with vhs in an immunoprecipitation from infected cells.
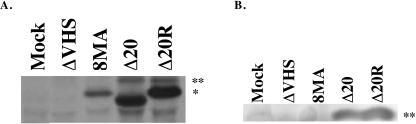
Vero cells were mock infected or infected with ΔVHS, 8MA, Δ20, or Δ20R at an MOI of 20, [S35]methionine and [S35]cysteine labeled, and lysed in RIPA buffer at 6 h postinfection. Lysates were immunoprecipitated by using the vhs antiserum 1883 and run on an SDS-PAGE gel (Fig. 7A). vhs was precipitated from lysates infected with Δ20, Δ20R, or 8MA. As was seen in Western blots (Fig. 6A), the vhs band that was precipitated from cells infected with the Δ20 virus showed a slightly faster mobility as a result of deletion of the 20-amino-acid VP16-binding domain. As expected, vhs was not precipitated from either mock- or vhs-null-infected cells. A decreased amount of vhs was precipitated from cells infected with 8MA, a virus that contains a deletion of the VP16 gene. This is most likely due to an overall decrease in viral protein production due to uncontrolled vhs activity (31). A 65-kDa protein consistent with VP16 was coprecipitated from cells infected with Δ20 and Δ20R. VP16 was not coprecipitated from cells that were either mock infected or infected with 8MA or ΔVHS, as would be expected. A nonspecific contaminating band directly below the vhs band was found in all lanes. Immunoprecipitation with preimmune sera showed no vhs- or VP16-specific bands in any of the infected-cell lysates (data not shown). To confirm that the coprecipitating band indeed was VP16, a Western blot using the VP16-specific antiserum was performed on complexes immunoprecipitated with the vhs antiserum (Fig. 7B). A VP16-specific band was present in immunoprecipitations of cells infected with Δ20 and Δ20R. As would be expected, VP16 was not present in cells that were mock infected or infected with ΔVHS or 8MA. These data show that VP16 coprecipitates with vhs in cells infected with Δ20, suggesting that the 20-amino-acid VP16-binding domain of vhs is not required for the binding of VP16.
FIG. 7.
(A) Coimmunoprecipitation, using a vhs antibody, of 35S-labeled complexes of vhs and VP16 from Vero cells infected with Δvhs, 8MA, Δ20, or Δ20R at an MOI of 20. The band corresponding to the vhs protein, indicated by an asterisk, is absent from a virus with a deletion of the vhs open reading frame (ΔVHS) and from mock-infected cells. The band corresponding to the coprecipitated VP16 protein, indicated by two asterisks, is absent from cells infected with a virus that contains a deletion of the VP16 gene (8MA), from vhs-null-infected cells, and from mock-infected cells. (B) Western blot of coimmunoprecipitated proteins from Vero cells infected with ΔVHS, 8MA, Δ20, or Δ20R at an MOI of 20. Vero cells were immunoprecipitated as described in the legend to panel A, and blots were probed with an antibody to VP16. A 65-kDa band is present in cells infected with Δ20 and Δ20R and absent from cells that were mock infected or infected with Δvhs or 8MA.
Effect of deletion of the VP16-binding domain of vhs on pathogenesis.
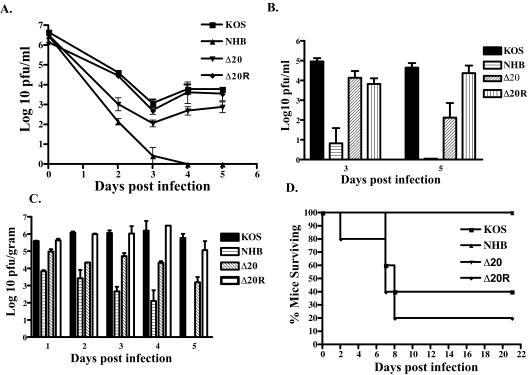
Although the VP16-binding domain of vhs is not necessary for VP16 binding, it is required for the activity of tegument-derived vhs, as was shown in the RNA degradation assays. Deletion of the binding domain thus results in a virus in which only the de novo-synthesized pool of vhs is active. This virus therefore provides an interesting tool with which to examine the contribution of tegument-derived vhs to pathogenesis. Six-week-old female CD-1 mice were bilaterally scarified and inoculated with 106 PFU of KOS, UL41NHB, Δ20, or Δ20R per eye. Acute replication in the cornea was analyzed on days 2 to 5 postinfection (Fig. 8A). As expected, relative to KOS, replication of the vhs-null virus UL41NHB was significantly reduced by 2 to 3 logs on days 2 and 3 postinfection (P < 0.001) and was cleared from the eye by day 4. In contrast, the vhs-sufficient viruses KOS and Δ20R showed indistinguishable high levels of replication on days 2 to 5 postinfection, and the virus was cleared from the eye by day 7. In contrast, Δ20 replicated approximately 1 to 2 logs less than KOS and Δ20R on days 2 to 5 postinfection (P < 0.005) and was also cleared from the eye by day 7. Clinical signs were evaluated on day 11 postinfection and showed KOS and Δ20R with an average score of 1.9 out of a possible score of 4 and NHB and Δ20 with an average score of 0.15 out of a possible score of 4, demonstrating a compromise in the ability of the Δ20 virus to induce clinical disease at the periphery.
FIG. 8.
Acute replication in murine corneas and trigeminal ganglia following corneal inoculation with 2 × 106 PFU of virus per eye. (A) Mice were infected with KOS, UL41NHB, Δ20, or Δ20R, and eyeswabs were taken on days 2, 3, 4, and 5 postinfection. (B) Trigeminal ganglia were harvested on days 3 and 5 postinfection. Data represent the logarithmic mean for four to six eyes or trigeminal ganglia per virus per day. (C) Mice were inoculated with 2 × 105 PFU of virus intracerebrally (KOS, UL41NHB, Δ20, or Δ20R). Brains were harvested on days 1, 2, 3, 4, and 5 postinfection. All samples were normalized per gram weight of tissue. Data represent the logarithmic mean for two to four brains per virus per day. (D) Survival of mice following intracerebral injection of 105 PFU of KOS, UL41NHB, Δ20, or Δ20R.
Acute replication in trigeminal ganglia was analyzed on days 3 and 5 postinfection (Fig. 8B). Both KOS and Δ20R replicated similarly and to high levels on days 3 and 5 postinfection. As expected, the vhs-deficient virus UL41NHB showed barely detectable levels of replication on day 3 and was completely cleared from the trigeminal ganglion by day 5 postinfection. Interestingly, Δ20 showed replication levels equivalent to those of the wild type on day 3, but by day 5 there was an approximately 3-log decrease in viral titers in comparison to results for KOS and Δ20R (P < 0.05), suggesting an impaired ability of the Δ20 virus to sustain replication in the nervous system.
To further investigate the replication of Δ20 in the nervous system, 6-week-old CD-1 female mice were inoculated with 105 PFU virus intracerebrally. Titers of virus in the brain were examined on days 1 to 5 postinfection (Fig. 8C). At all days postinfection, KOS and Δ20R showed equivalent high levels of replication. Replication of a vhs-deficient virus, UL41NHB, showed a 2- to 4-log decrease in replication on all days postinfection. Δ20, though it did not replicate as poorly as UL41NHB, showed a 1- to 2-log difference in replication in the brain (P < 0.003) on all days postinfection. Lethality studies showed 60 to 80% of KOS and Δ20R dying before day 21 after infection with 105 PFU (Fig. 8D). In contrast, a dose of 105 PFU of NHB or Δ20 did not kill any mice up to at least 21 days postinfection (P < 0.05), consistent with a defect in the ability of Δ20 to replicate in the nervous system.
Establishment of and reactivation from latency.
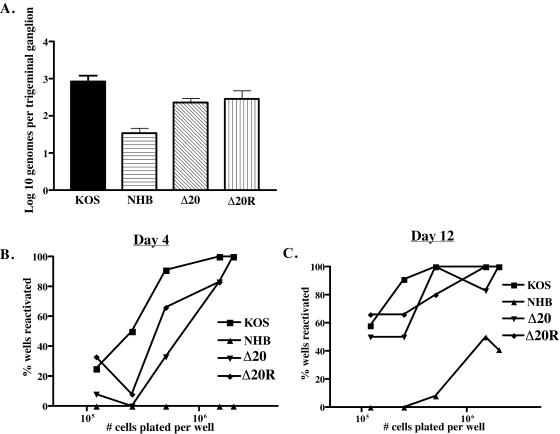
The ability of the Δ20 virus to establish and reactivate from latency was analyzed on day 28 postinfection. The number of latent genomes per trigeminal ganglion was analyzed using real-time PCR for the thymidine kinase gene of HSV-1 (Fig. 9A). All samples were normalized to the single-copy mouse adipsin gene. KOS, Δ20, and Δ20R all show approximately 103 genomes per trigeminal ganglion, indicating no significant difference in the abilities of Δ20 and Δ20R to establish latency. This finding suggests that deletion of the VP16-binding domain of vhs has no impact on the ability of HSV-1 to establish latency. Approximately 5 × 101 HSV-1 genomes per trigeminal ganglion were found in mice infected with UL41NHB, a significant difference (P value, <0.001) in comparison to the above-mentioned viruses. This suggests a reduced ability of vhs deletion viruses to establish latency, a finding that had been documented previously (37).
FIG. 9.
Analysis of establishment and reactivation from latency. (A) Real-time PCR for the HSV-1 tk gene was performed on individual trigeminal ganglia from latently infected mice. All samples were normalized to the single-copy mouse adipsin gene. A minimum of six trigeminal ganglia were analyzed per virus. The data are reported as the logarithmic mean of the number of genome copies per trigeminal ganglion. Trigeminal ganglia from latently infected mice were dissociated from a single-cell suspension and then plated in twofold dilutions. (B) Supernatant samples from each cell dilution were taken on day 4 and (C) day 12 postdissociation and plated on an indicator monolayer of Vero cells. This indicator monolayer was then scored for the presence or absence of cytopathic effect, and the results were recorded as the percentage of wells positive for reactivation at each cell dilution. A minimum of six trigeminal ganglion per virus were pooled and used for the reactivation assay. Six wells were plated at each cell dilution per virus infection.
The trigeminal ganglia from each virus group were pooled, and a limiting-dilution reactivation assay was performed by culturing twofold dilutions of dissociated latently infected trigeminal ganglion cells, starting with a concentration of 2 × 106 cells/well and ending at a concentration of 1.2 × 105 cells/well (Fig. 9B and C). Each cell dilution contained six replicates. Supernatant samples from each well of each dilution were plated onto Vero cells days 4, 5, 8, and 12 postdissociation and scored for cytopathic effect. The data are reported as the number of wells positive for reactivation per virus at each cell dilution. As shown in Fig. 9B, Δ20 showed reduced reactivation at all cell dilutions in comparison to KOS on day 4 postdissociation. Δ20R shows slightly reduced reactivation in comparison to KOS but does not appear to be as compromised as Δ20. NHB shows no reactivation at any cell dilution on day 4 postdissociation. By day 12 postdissocation KOS, Δ20, and Δ20R show similar levels of reactivation at all cell dilutions (Fig. 9C). In contrast, NHB shows a significant attenuation in the ability to reactivate from latency at all cell dilutions. In summary, Δ20 shows near-wild-type reactivation in these assays.
DISCUSSION
The mRNA degradation function of the vhs protein has been studied in detail both in cell-free systems and in the context of viral infection. Although the RNA degradation activity is dependent upon four well-studied conserved domains of HSV-1 (2, 9, 26), the functions of the nonconserved regions of the protein are less well defined. The small modular domain between residues 310 and 330 of the vhs protein has been proposed to be important for the interaction between vhs and VP16, and this interaction, in turn, has been proposed to regulate vhs activity (30, 31). This is the first study to examine the function of this domain directly in the context of virus infection. In this study, recombinant viruses were generated in which the 20 amino acids of the VP16-binding domain of vhs were deleted and restored.
Immunoprecipitation of infected cells showed unaltered coprecipitation of vhs and VP16 in Δ20, indicating that the VP16-binding domain of vhs is not necessary for vhs-VP16 interactions in the context of the virus. We propose two possible explanations for this finding. First, the avidity of the complex between vhs and VP16 is changed, but not enough to preclude binding. No quantitative biochemical analyses of the strength of the complex in Δ20 compared to that in KOS have been attempted, but they would be of interest in order to address the effect of deletion of the binding domain on the strength of the vhs-VP16 interaction. One line of evidence to suggest a potential difference in the strength of the complex comes from the analysis of vhs packaging. Western blots of viral particles indicated about a twofold decrease in the amount of vhs packaged into the virion. The details of the assembly of the viral tegument are still largely obscure, and little is known about the regulation of packaging of individual proteins and how specific protein-protein interactions facilitate this process. One example of the importance of protein-protein interactions in the packaging of the proteins has been shown in pseudorabies virus, where the packaging of UL36 is dependent on the presence of UL37 (10). If vhs is indeed packaged as a result of its interactions with VP16, a partial disruption in the strength of the complex may be reflected in the total amount of vhs packaged into the virion. The data in this paper show that the VP16-binding domain is not necessary for the interaction between vhs and VP16 in the context of the virus. Although the domain is sufficient for the interaction in vitro (29), these in vitro assays do not replicate the complex milieu of protein-protein interactions that can occur in the context of other viral proteins. Second, it is possible that additional, yet-uncharacterized protein-protein interactions between vhs, VP16, and other tegument proteins may contribute to stabilizing a suboptimal interaction between vhs and VP16.
The RNA degradation phenotype of Δ20 is one of the most interesting effects of the loss of the VP16-binding domain. Transient-transfection assays using vhs expression constructs and RNA degradation experiments with the recombinant virus Δ20 showed that the VP16-binding domain is dispensable for vhs activity from de novo-synthesized vhs protein. This result is consistent with the VP16-binding domain being outside of the conserved domains identified to be critical for vhs activity (2, 9, 26). This study, however, has clearly shown a requirement for this 20-amino-acid domain for the RNA degradation activity mediated by tegument-derived vhs. Even in the absence of tegument-derived vhs activity, the activity of the de novo-synthesized protein from Δ20 is nearly equivalent to the vhs activity in KOS- and Δ20R-infected cells. Δ20 thus allows for the differentiation of tegument-derived and de novo-synthesized vhs activity. It has been previously observed that a virus that contains a deletion of the VP16 gene shows an increased level of translational arrest, which is reversed upon simultaneous deletion of vhs (16). This suggests that de novo-synthesized vhs is active but that its activity is either dampened or precluded by the binding of VP16. In this study it was shown that Δ20 vhs retains its ability to interact with VP16 and yet still mediates a significant level of RNA degradation. This observation was unexpected for a virus which has lost tegument-derived vhs activity yet has retained its ability to interact with VP16. This observation could be explained in two ways. While loss of the VP16-binding domain does not destroy the ability of vhs and VP16 to interact, it may change the avidity or structure of the complex such that de novo-synthesized vhs retains some activity. Alternatively, in wild-type infection, it is possible that the synthesis and localization of de novo-synthesized vhs allow a modest level of vhs activity, which is inhibited upon subsequent synthesis and binding to VP16. In either case, this suggests that de novo-synthesized vhs may contribute to more of the RNA degradation activity in infected cells than was previously thought.
The absence of tegument-derived vhs activity from Δ20 is most intriguing, and a number of explanations exist. First, vhs is a phosphoprotein (28), and it is unknown whether its phosphorylation is required for function. An alteration of the phosphorylation state of Δ20 vhs could result in a change in activity. A limited analysis of the phosphorylation state of Δ20 vhs in comparison to that of wild-type vhs revealed no change (data not shown). This analysis does not, however, distinguish between similar levels of phosphorylation at different locations on the protein. A change in the structure of the vhs protein or the vhs-VP16 complex due to loss of the binding domain may hide or expose residues in which phosphorylation and dephosphorylation are critical for function. Further analyses of the posttranslational modifications required for vhs activity, using phosphopeptide mapping, for example, need to be performed. Second, it has been shown that disruption of the tegument, and therefore correct localization of tegument proteins upon entry to a susceptible cell, requires phosphorylation (23). It is possible that vhs from the Δ20 virus does not get appropriately dissociated or associated from or with the other tegument or cellular proteins upon entry and is therefore, unable to mediate RNA degradation. The events required for the disruption of the tegument and protein-protein interactions in the tegument remain poorly understood. Finally, changes in phosphorylation or association and dissociation from tegument or cellular proteins may in turn disrupt the ability of vhs to interact with the translation initiation factor eIF4H, a protein that seems to be critical for vhs activity (6, 19).
The Δ20 virus also has interesting implications for HSV-1 pathogenesis. Although the de novo-synthesized form of vhs retains a high level of RNA degradation, this is clearly insufficient for wild-type pathogenesis, since Δ20 is significantly attenuated, although not as compromised as a vhs-null virus (35-37). This shows that tegument-derived vhs contributes to pathogenesis. The attenuation of Δ20 could be explained by consideration of the early events in pathogenesis that may be modulated by the preformed vhs protein found in the tegument. vhs-mediated RNA degradation has been implicated in regulating the innate immune response (38). Infection of susceptible cells results in synthesis of alpha and beta interferon (IFN), which then stimulate a cascade of events leading to the induction of an antiviral state (7, 34). It has been demonstrated that in some cell lines, vhs mutants are more sensitive to the effects of IFN (38), and alpha and beta IFN receptor knockout mice show partial restoration of virulence to a vhs-deficient virus (18). Incoming vhs may be necessary, therefore, for modulating the interferon response, and lack of the preformed protein from the tegument is only partially compensated for by the de novo-synthesized protein. vhs activity has also been implicated in regulation of the adaptive immune response by decreasing the amount of major histocompatibility complex classes 1 and 2 on the surface of infected cells (1, 12, 41, 42). The immediate down-regulation of these molecules upon entry of the virus particle may be more advantageous than the down-regulation afforded at a later time upon de novo synthesis of vhs protein. In general, it is interesting to consider that the events mediated by the incoming tegument protein may be different from those mediated by the de novo-synthesized protein, each fraction being critical for pathogenesis in different ways. Additional studies to examine the specific differences in the contribution of tegument-derived vhs to pathogenesis versus that of de novo-synthesized vhs are under way.
Acknowledgments
We thank Sullivan Read for providing the vhs protein used in the generation of the vhs-specific antibody (1883). We thank Martha Kramer for helpful advice and primer sequences required for the development of the real-time PCR assay for viral genomes. We thank Diane Alexander for technical assistance in development of the real-time PCR assay for both the tk gene and the mouse adipsin gene. We also thank the members of our Friday morning virology group meeting for helpful discussions.
This work was supported by NIH grants to David A. Leib (EY10707) and the Department of Ophthalmology and Visual Sciences (P30EY02687). Support for the department from Research to Prevent Blindness and a Lew Wasserman Scholarship to David A. Leib are gratefully acknowledged.
REFERENCES
- 1.Ambagala, A. P., S. Hinkley, and S. Srikumaran. 2000. An early pseudorabies virus protein down-regulates porcine MHC class I expression by inhibition of transporter associated with antigen processing (TAP). J. Immunol. 164:93-99. [DOI] [PubMed] [Google Scholar]
- 2.Berthomme, H., B. Jacquemont, and A. Epstein. 1993. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology 193:1028-1032. [DOI] [PubMed] [Google Scholar]
- 3.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgadi, M. M., and J. R. Smiley. 1999. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J. Virol. 73:9222-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA 101:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 8.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, F. E., C. A. Smibert, and J. R. Smiley. 1995. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J. Virol. 69:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knez, J., P. T. Bilan, and J. P. Capone. 2003. A single amino acid substitution in herpes simplex virus type 1 VP16 inhibits binding to the virion host shutoff protein and is incompatible with virus growth. J. Virol. 77:2892-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppers-Lalic, D., F. A. Rijsewijk, S. B. Verschuren, J. A. van Gaans-Van den Brink, A. Neisig, M. E. Ressing, J. Neefjes, and E. J. Wiertz. 2001. The UL41-encoded virion host shutoff (vhs) protein and vhs-independent mechanisms are responsible for down-regulation of MHC class I molecules by bovine herpesvirus 1. J. Gen. Virol. 82:2071-2081. [DOI] [PubMed] [Google Scholar]
- 13.Kristie, T. M., and B. Roizman. 1986. Alpha 4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of alpha genes and of selected other viral genes. Proc. Natl. Acad. Sci. USA 83:3218-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 17.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, P., F. E. Jones, H. A. Saffran, and J. R. Smiley. 2001. Herpes simplex virus virion host shutoff protein requires a mammalian factor for efficient in vitro endoribonuclease activity. J. Virol. 75:1172-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynas, C., T. J. Hill, N. J. Maitland, and S. Love. 1993. Latent infection with the MS strain of herpes simplex virus type 2 in the mouse following intracerebral inoculation. J. Neurol. Sci. 120:107-114. [DOI] [PubMed] [Google Scholar]
- 21.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 22.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pak, A. S., D. N. Everly, K. Knight, and G. S. Read. 1995. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology 211:491-506. [DOI] [PubMed] [Google Scholar]
- 27.Rader, K. A., C. E. Ackland-Berglund, J. K. Miller, J. S. Pepose, and D. A. Leib. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J. Gen. Virol. 74:1859-1869. [DOI] [PubMed] [Google Scholar]
- 28.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmelter, J., J. Knez, J. R. Smiley, and J. P. Capone. 1996. Identification and characterization of a small modular domain in the herpes simplex virus host shutoff protein sufficient for interaction with VP16. J. Virol. 70:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smibert, C. A., D. C. Johnson, and J. R. Smiley. 1992. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J. Gen. Virol. 73:467-470. [DOI] [PubMed] [Google Scholar]
- 31.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, T. J., C. E. Ackland-Berglund, and D. A. Leib. 2000. Herpes simplex virus virion host shutoff (vhs) activity alters periocular disease in mice. J. Virol. 74:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. J., L. A. Morrison, and D. A. Leib. 2002. Pathogenesis of herpes simplex virus type 2 virion host shutoff (vhs) mutants. J. Virol. 76:2054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 35.Strelow, L., T. Smith, and D. Leib. 1997. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology 231:28-34. [DOI] [PubMed] [Google Scholar]
- 36.Strelow, L. I., and D. A. Leib. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 70:5665-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 39.Tenser, R. B., R. L. Miller, and F. Rapp. 1979. Trigeminal ganglion infection by thymidine kinase-negative mutants of herpes simplex virus. Science 205:915-917. [DOI] [PubMed] [Google Scholar]
- 40.Thomson, J. M., and W. A. Parrott. 1998. pMECA: a cloning plasmid with 44 unique restriction sites that allows selection of recombinants based on colony size. BioTechniques 24:922-924 [DOI] [PubMed] [Google Scholar]
- 41.Tigges, M. A., S. Leng, D. C. Johnson, and R. L. Burke. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901-3910. [PubMed] [Google Scholar]
- 42.Trgovcich, J., D. Johnson, and B. Roizman. 2002. Cell surface major histocompatibility complex class II proteins are regulated by the products of the gamma(1)34.5 and U(L)41 genes of herpes simplex virus 1. J. Virol. 76:6974-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]