Abstract
The effects of two functional domains, the membrane-proximal YXXΦ motif and the membrane-distal inhibitory sequence in the long cytoplasmic tail of the human immunodeficiency virus type 1 (HIV-1) envelope protein (Env), on immunogenicity of the envelope protein were investigated. Genes with codons optimized for mammalian expression were synthesized for the HIV 89.6 Env and a truncated Env with 50 amino acids in the cytoplasmic domain to delete the membrane distal inhibitory sequence for surface expression. Additional genes were generated in which the tyrosine residue in the YXXΦ motif was changed into a serine. Pulse-chase radioactive labeling and immunoprecipitation studies indicated that both domains can mediate endocytosis of the HIV Env, and removal of both domains is required to enhance HIV Env protein surface stability. Analysis of immune responses induced by DNA immunization of mice showed that the DNA construct for the mutant Env exhibiting enhanced surface stability induced significantly higher levels of antibody responses against the HIV Env protein. Our results suggest that the HIV Env cytoplasmic domain may play important roles in virus infection and pathogenesis by modulating its immunogenicity.
The human immunodeficiency virus (HIV) envelope glycoprotein (Env) mediates virus entry into cells and is also a major target for both cellular and antibody responses (21, 28). It is synthesized as a precursor molecule, gp160, which is subsequently processed into the surface subunit (SU) gp120 and the transmembrane subunit (TM) gp41 by a cellular protease, and exists as a trimer of gp120-gp41 heterodimers on viral or cell membranes (18, 52). gp120 interacts with receptor and coreceptor molecules for HIV and mediates virus attachment to the cell, while gp41 causes subsequent fusion between viral and cell membranes for releasing viral core components into the cell during the initial infection process (15). The TM protein consists of three distinct domains: the extracellular domain, the transmembrane domain, and the cytoplasmic domain.
The Env protein of HIV as well as other lentiviruses has a long cytoplasmic domain with over 150 amino acids, in comparison to those of other retroviruses, which are about 30 to 50 amino acids in length. Early studies have shown that the long cytoplasmic domain of HIV Env plays important roles in regulating Env protein function and virus infectivity (17, 24, 33, 51) and have identified several structural features modulating these functions, such as modulating surface expression of the Env protein (3, 4, 6, 27, 32, 45, 53), targeting Env protein to specific membrane microdomains for assembly (14, 34, 35, 44, 55), and interacting with the viral matrix protein for incorporation of the Env protein into released virions (1, 11, 16, 20, 22, 39, 57), as well as interacting with other cellular proteins (3, 29, 38, 40, 53). Of particular interest, two distinct regions have been identified in the long cytoplasmic domain of the HIV Env protein and shown to regulate its surface expression, a membrane-proximal Tyr-based (YXXΦ) endocytosis motif (amino acids 710 to 713 in the HIV 89.6 Env protein), in which Φ represents a hydrophobic amino acid with a large aliphatic side chain, and a membrane-distal dileucine-like motif (amino acids 750 to 785) (6, 45). Of these, the Tyr-based YXXΦ motif is well conserved in the cytoplasmic domains of retrovirus Env proteins (37). It has been implicated in membrane fusion activity of the HIV Env protein, Env incorporation into virions, and virus infectivity (7, 12, 31, 32, 45, 46, 50). Several studies have demonstrated that the YXXΦ motif functions as a sorting signal to mediate Env protein-directed release of HIV virions from basolateral surfaces of polarized epithelial cells and release of HIV and simian immunodeficiency virus (SIV) virions from specific membrane locations of lymphocytes (14, 34, 35). Moreover, the YXXΦ motif in the HIV Env cytoplasmic domain has been shown to serve as a potent endocytosis signal to mediate retrieval of the Env protein from the plasma membrane surface through interactions with the cellular proteins of the clathrin adaptor protein family (3, 4). Furthermore, evidence from these studies also indicates that other sequences besides the YXXΦ motif in the HIV Env cytoplasmic domain play important roles in modulating surface expression of the HIV Env protein. Recently, Bultmann et al. identified a membrane-distal surface expression inhibitory sequence (dileucine-like motif) in the cytoplasmic domain of the HIV Env which is located about 40 amino acids downstream of the YXXΦ motif (6). This segment is conserved in different clades of HIV type 1 (HIV-1) isolates and overlaps with the conserved LLP2 sequences in the HIV Env cytoplasmic domain. Fultz et al. showed that while mutation of the Tyr residue in the YXXΦ motif of the SIVmac239 Env protein did not significantly affect virus infectivity and cytopathicity in tissue cultures, replication and pathogenesis of the mutant virus were attenuated during in vivo infection of rhesus monkeys (23), indicating that the endocytosis signals may play important roles in regulating virus replication in the process of in vivo infection. However, the role of these endocytosis motifs in HIV replication and pathogenesis remains to be elucidated.
In this study, we examined the roles of the identified endocytosis motifs individually and in combination on the immunogenicity of the HIV Env protein by DNA immunization, which mimics viral protein synthesis, processing, and presentation during infection. Our results show that mutations in the HIV Env protein cytoplasmic domain to disrupt or delete these endocytosis motifs have a profound effect on induction of immune responses against the HIV Env protein, indicating another potential mechanism by which HIV evades the immune system.
MATERIALS AND METHODS
Synthesis of codon-optimized genes for the wild-type and mutant HIV Env proteins.
A synthetic gene for the HIV 89.6 Env protein with codons optimized for mammalian usage was synthesized by PCR assembly of long single-strand DNA templates (100 bases in length) as described in our previous studies (5). Mutation of the Tyr residue Y710 and truncation of the cytoplasmic domain were carried out by site-directed PCR mutagenesis. All genes were confirmed by sequencing and then cloned into a plasmid vector pCAGGS (kindly provided by Y. Kawaoka) under the control of a chicken β-actin promoter.
Protein expression analysis.
Protein expression was analyzed by radioactively labeling and immunoprecipitation followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). HeLa cells were transfected with appropriate DNA constructs by using Lipofectamine 2000 (GIBCO-BRL) following the manufacturer's protocol. At 24 h posttransfection, cells were starved in Met-Cys-deficient Dulbecco's modified Eagle's medium (DMEM) for 30 min and then labeled with [35S]Met-Cys labeling mix (Amersham) for a period of time as specified for each experiment (see the figure legends). For continuous labeling, the cells were labeled for 6 h. For pulse-chase experiments, the cells were labeled for 1 h and then chased in complete DMEM for an additional period of time as indicated in the figure legends. The cells were then lysed with lysis buffer at different time points and precipitated with antiserum against HIV-1 89.6 Env and protein A-agarose beads (Pierce). Surface expression of HIV Env protein was detected by a surface biotinylation assay as described in our previous studies (56). Briefly, after labeling and chase, cells were washed with phosphate-buffered saline (PBS) at 4°C and then incubated with 1 ml of NHS-SS-biotin dissolved in PBS (1 mg/ml) for 30 min at 4°C. The cells were then lysed, and Env protein was precipitated with antibody and protein A-agarose beads overnight at 4°C. Proteins bound to protein A were washed three times with lysis buffer, and the beads were added with 10% SDS and heated at 95°C for 15 min. Dissociated proteins were precipitated again with streptavidin agarose beads at 4°C for 3 h and then washed three times with lysis buffer. Protein samples were then prepared by addition of reducing sample buffer and heated at 95°C for 5 min prior to analysis by SDS-PAGE.
Immunization of mice.
Female BALB/c mice (H-2d) 6 to 8 weeks of age were purchased from Charles River Laboratories. Plasmids were amplified in Escherichia coli DH5α and purified with a QIAGEN Endo-Free Megaprep kit. The plasmids were then resuspended at 1 μg/μl in sterile PBS and stored at −80°C until used for immunization. Groups of mice (six per group) were immunized with a total of 100 μg of indicated DNA construct per mouse by intramuscular injection with 50 μl of DNA preparation in separate sites in both side quadriceps, followed by boosting with the same dose of DNA at week 4.
Flow cytometry analysis of immune responses.
At 14 days after the boosting DNA immunization, mice were sacrificed and spleens were removed, homogenized, and compressed through a sterile nylon membrane. Mouse splenocytes were prepared by lysis of red blood cells with ammonium chloride and washed twice with RPMI 1640. The cells were resuspended in complete culture medium (RPMI 1640 plus 10% fetal calf serum (FCS), 50 μM β-mercaptoethanol, and antibiotic mix), and the cell viability was determined by trypan blue exclusion. Cells (106) were cultured with pools of peptide (20-mers overlapping by 10 amino acids) corresponding to the HIV-1 89.6 Env protein (obtained from the NIH AIDS Research and Reference Reagent Program) for 16 h and then incubated with brefeldin A (10 μg/ml; Sigma) for an additional 5 h. For assessment of CD8 T-cell responses to a known dominant epitope in the HIV-1 89.6 Env for BALB/c mice, prepared splenocytes were incubated with the peptide IGPGRAFYAR (10 μg/ml) for 5 h in the presence of brefeldin A (10 μg/ml). Cells cultured with phorbol myristate acetate plus ionomycin were used as positive controls, whereas cells cultured with an irrelevant peptide (AMQMLKETI) corresponding to an H-2d-restricted cytotoxic T-lymphocyte epitope of the HIV Gag protein were used as negative controls (41). After stimulation, the cells were washed twice with PBS containing 3% FCS and then stained with PerCP-conjugated rat anti-mouse CD4 and phycoerythrin (PE)-conjugated rat anti-mouse CD8 antibodies (Pharmingen). Cells were then fixed and permeabilized with cytofix buffers (Caltag) and stained for intracellular gamma interferon (IFN-γ) allophycocyanin (APC)-conjugated rat anti-mouse IFN-γ antibody (Pharmingen). Flow cytometry analysis was performed on a BD FACSCalibur with CELLQuest software, and data were analyzed with Flowjo 4.2 software.
ELISA.
Mouse blood was collected by retroorbital bleeding at 14 days after final DNA immunization. Ninety-six-well plates were coated with purified HIV-1 89.6 gp120-Histag (prepared by Nicald affinity purification using a QIAGEN kit; 3 μg/ml in borate-buffered saline; pH 8.5; 100 μl per well) at 4°C overnight and blocked with PBS, 0.1% Tween 20, and 3% bovine serum album for 3 h at 37°C. Serial dilutions of mouse serum were then added to each well in triplicate and incubated at 37°C for 3 h. Horseradish peroxidase-conjugated secondary antibody (Sigma) against mouse immunoglobulin G (IgG) was added for 2 h at 37°C. After a final wash, ABTS [2,2′-azino-bis(3-ethybenz-thiazoline-6-sulfonic acid); Sigma] dissolved in citrate phosphate buffer (3 mg of ABTS in 10 ml [pH 4.2] plus 10 μl of H2O2) was added at 100 μl/well for developing color and read by an enzyme-linked immunosorbent assay (ELISA) reader at 405 nm. A standard curve for absorbance and the amount of mouse antibody absorbed to the well was obtained by coating ELISA plates with serial twofold dilutions of purified mouse antibodies followed by addition of horseradish peroxidase-conjugated secondary antibody and development of color. Data were analyzed with the Microsoft Excel program and are presented as the equivalent amount of Env-binding antibodies in mouse sera (in nanograms per milliliter), and statistical analysis was carried out using the Student t test.
Neutralizing antibody assay.
A neutralizing antibody assay was performed using a highly sensitive, single-round infectivity assay as described by Derdeyn et al. (13). The assay is based on an indicator cell line, JC53-BL (kindly provided by Tranzyme, Inc.). Briefly, JC53BL cells were seeded at 40,000 cells per well in a 96-well plate in 10% FCS-DMEM overnight at 37°C with 5% CO2. Serum samples from mice were heat inactivated at 56°C for 30 min and diluted 1:20 or 1:40 in 10% FCS-DMEM to a final volume of 25 μl and added to 25 μl of virus stock diluted in 10% FCS-DMEM containing 50 infectious particles (final serum dilution, 1:40 or 1:80). Viruses mixed with medium only were used as controls. The virus-serum mixtures were prepared in triplicate, incubated at 37°C for 1 h, and then added to the JC53-BL cells with DEAE-dextran (final concentration, 15 μg/ml). After 2 h of incubation, an additional 200 μl of 10% FCS-DMEM was added. Three days after infection, the medium was removed and the cells were fixed and stained as described in our previous studies (49). Neutralization was calculated as follows: [(average number of blue foci in control wells − average number of blue foci in virus-serum mixture sample wells)/(average number of blue foci in control wells)] × 100%. Data were analyzed by using the Microsoft Excel program and are presented as the percentage of neutralization by mouse sera, and statistical analysis was carried out using the Student t test.
RESULTS
Surface expression of the HIV Env protein is enhanced by mutations to eliminate both endocytosis signals in the cytoplasmic domain.
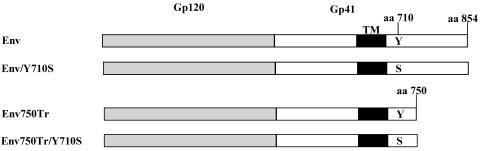
The membrane-proximal Tyr-based YXXΦ motif in the HIV Env protein cytoplasmic domain has been shown to serve as an endocytosis signal to regulate surface expression of the HIV Env protein (45). To investigate the potential effect of the YXXΦ motif on immunogenicity of the HIV Env protein, we synthesized the gene for the HIV 89.6 Env as well as a mutant HIV 89.6 Env in which the Tyr residue was changed into a serine (designated as Env/Y710S) with codons optimized for mammalian cell expression as described in our previous studies (5). The use of synthetic HIV env genes with codons optimized for mammalian expression gives high-level expression of the HIV Env proteins in a Rev-independent manner, and such constructs have been shown to induce stronger immune responses compared by DNA immunization to HIV env genes with wild-type codons (2, 26, 58). Also, we chose to mutate the conserved Tyr residue in the HIV 89.6 Env protein into a Ser residue based on the report by Deschamebault et al. (14) in which they showed that changing the Tyr residue in the YXXΦ motif into a serine exerted a more pronounced effect on surface expression of HIV Env than changing it into an alanine. Presented in Fig. 1 is a schematic diagram for the Env proteins encoded by the DNA constructs used in this study.
FIG. 1.
Schematic diagram for the HIV 89.6 Env proteins encoded by synthetic genes. Codon-optimized genes for HIV 89.6 Env, Env/Y710S, Env750Tr, and Env750Tr/Y710S were synthesized as described in Materials and Methods. The positions for the Tyr residue in the YXXΦ motif and for the Ser residue in the mutant and the lengths of each construct are shown.
For protein expression analysis and DNA immunization studies, the genes were cloned into the plasmid vector pCAGGS, driven by the chicken β-actin promoter. Expression of the HIV 89.6 Env and Env/Y710S was compared in HeLa cells by transfection with Lipofectamine and radioactive labeling followed by surface biotinylation and immunoprecipitation. As shown in Fig. 2, both Env and Env/Y710S were expressed at similar levels in the cell, and the amount of gp120 secreted into the medium was also similar. Moreover, the levels of surface expression for Env and Env/Y710S were also found to be similar, as detected by surface biotinylation. These results suggest that disrupting the YXXΦ motif alone did not significantly affect the levels of cellular expression, secretion, or surface expression of the HIV Env protein.
FIG. 2.
Mutation of the Tyr residue in the YXXΦ motif alone did not significantly affect HIV 89.6 Env protein surface expression. HeLa cells were transfected with the DNA constructs encoding HIV 89.6 Env and Env/Y710S by using Lipofectamine 2000. At 24 h posttransfection, the cells were labeled with [35S]Met-Cys labeling mix for 6 h, followed by surface biotinylation and immunoprecipitation as described in Materials and Methods. Protein samples were prepared and analyzed by SDS-PAGE. Lanes: 1, plasmid vector pCAGGS; 2, Env; 3, Env/Y710s; Pre, HIV Env precursor gp160; SU, surface subunit gp120; TM, transmembrane subunit gp41.
Previous studies have indicated that elements in the HIV Env protein cytoplasmic domain other than the YXXΦ motif may also be involved in regulating surface expression of the HIV Env protein. Bultmann et al. identified a membrane-distal dileucine-like motif in the HIV Env cytoplasmic domain that fits the profile for such an element (6). To examine the effect of this dileucine-like motif on HIV Env protein surface expression alone or in combination with the YXXΦ motif, we generated genes for truncated HIV 89.6 Env as well as Env/Y710S proteins to remove the C-terminal 100 amino acids of the cytoplasmic domain that contains the dileucine motif as a simple approach to eliminate this putative endocytosis signal, which involves a stretch of amino acids in the cytoplasmic domain. The resulting constructs were designated as Env750Tr and Env750Tr/Y710S, respectively. These genes were also cloned into the plasmid vector pCAGGS, and expression of these truncated proteins was compared with Env and Env/Y710S by pulse-chase radioactive labeling followed by surface biotinylation and immunoprecipitation to determine the effect of these putative endocytosis signals on Env protein transport to the cell surface as well as surface stability. As shown in Fig. 3, similar levels of cellular expression, secretion of gp120, and surface expression of the Env protein were detected for Env and Env/Y710S throughout the 1-h pulse and 6-h chase periods, in agreement with the continuous radioactive labeling results shown in Fig. 2. Moreover, similar levels of cellular expression, secretion of gp120, and surface expression were also observed for both Env750Tr and Env750Tr/Y710S after the 1-h pulse and up-to-4-h chase, similar to those observed for Env and Env/Y710S. However, while surface expression of Env750Tr was reduced significantly after the 6-h chase, similar to that observed for Env and Env/Y710S, surface expression for the mutant Env750Tr/Y710S stayed at high levels after the 6-h chase. Similar results were also obtained in the murine NIH 3T3 cell line, and all constructs were observed to induce extensive fusion of cells expressing receptor CD4 and coreceptor CCR5 or CXCR4 for HIV-1 (data not shown). These results showed that mutations to eliminate both motifs are necessary for obtaining enhanced surface stability of the HIV Env protein, indicating that each of the two motifs in the cytoplasmic domain alone can still mediate endocytosis of the HIV Env protein in the absence of the other.
FIG. 3.
Combination of Tyr mutation and cytoplasmic domain truncation enhances HIV Env protein stability on cell surfaces. Protein expression was carried out by transfection of HeLa cells. At 24 h posttransfection, the cells were pulse-labeled with [35S]Met-Cys labeling mix for 1 h and then chased in complete medium for 2, 4, or 6 h, followed by surface biotinylation and immunoprecipitation as described in Materials and Methods. Protein samples were prepared and analyzed by SDS-PAGE. (A) Pulse-chase radioactive labeling analysis for expression of HIV 89.6 Env and Env/Y712S. Lanes: 1, plasmid vector pCAGGS; 2, Env, pulse only; 3, Env, 2-h chase; 4, Env, 4-h chase; 5, Env, 6-h chase; 6, Env/Y710S, pulse only; 7, Env/Y710S, 2-h chase; 8, Env/Y710S, 4-h chase; 9, Env/Y710S, 6-h chase. (B) Pulse-chase radioactive labeling analysis for expression of HIV 89.6 Env750Tr and Env750Tr/Y710S. Lanes: 1, plasmid vector pCAGGS; 2, Env750Tr, pulse only; 3, Env750Tr, 2-h chase; 4, Env750Tr, 4-h chase; 5, Env750Tr, 6-h chase; 6, Env750Tr/Y710S, pulse only; 7, Env750Tr/Y710S, 2-h chase; 8, Env750Tr/Y710S, 4-h chase; 9, Env750Tr/Y710S, 6-h chase; Pre, HIV Env precursor gp160; SU, surface subunit gp120; TM, transmembrane subunit gp41.
Mutation of the Tyr residue in the YXXΦ motif and truncation of the dileucine-like endocytosis motif in the cytoplasmic domain do not affect induction of CD8 T-cell responses against the dominant epitope in the HIV 89.6 Env.
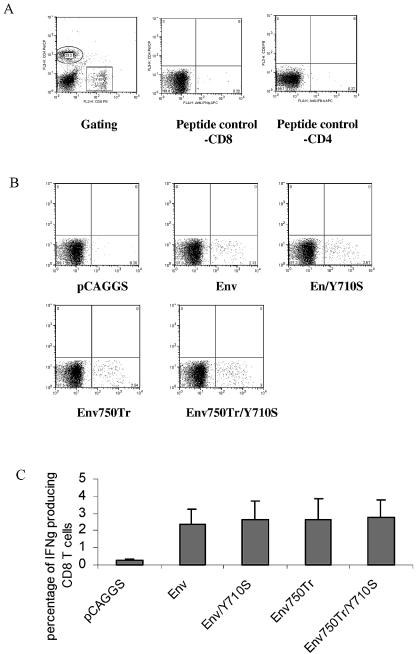
To determine the effect of membrane-proximal YXXΦ and the membrane-distal dileucine-like endocytosis motifs on immunogenicity of the HIV 89.6 Env protein, we used the DNA constructs to immunize mice and compared T-cell as well as antibody immune responses induced by these two constructs. BALB/c mice (groups of six) were immunized with Env, Env/Y710S, Env750Tr, Env750Tr/Y710S, or pCAGGS (vector control) at weeks 0 (prime) and 4 (boost) and sacrificed at week 6 for analysis of immune responses. Cellular immune responses were analyzed by intracellular cytokine staining for IFN-γ production coupled with flow cytometry (fluorescence-activated cell sorting [FACS]) of splenocytes from immunized mice after stimulation with peptide or peptide pools. The gating of CD4 and CD8 T cells from mouse splenocytes and background levels of IFN-γ production by stimulation with an irrelevant peptide (AMQMLKETI), which corresponds to an epitope in the HIV Gag protein (41), are shown in Fig. 4A. The typical results for IFN-γ-producing CD8 T cells from immunized mice after stimulation with the peptide corresponding to the dominant epitope in the HIV 89.6 Env for BALB/c mice are shown in Fig. 4B, and the average percentages of IFN-γ-producing CD8 T cells for each group are shown in Fig. 4C. These results showed that the percentages of IFN-γ-producing CD8 T cells in splenocytes from mice immunized with all four DNA constructs were similar, at 2, 1.97, 2.36, and 2.5% on average for Env, Env/Y710S, Env750Tr, and Env750Tr/Y710S, respectively. The splenocytes from the control vector pCAGGS-immunized mice showed background levels of IFN-γ-producing CD8 T cells which were similar to the levels of IFN-γ-producing CD8 T cells stimulated by the control peptide. These results demonstrated that mutation of the YXXΦ motif and truncation of the dileucine-like motif have no significant effect on induction of CD8 T-cell responses against the dominant epitope in HIV 89.6 Env protein by DNA immunization.
FIG. 4.
CD8 T-cell responses induced by DNA immunization against a dominant epitope in the HIV 89.6 Env protein were not affected by Tyr mutation and cytoplasmic domain truncation or their combination. Groups of BALB/c mice (six per group) were immunized by intramuscular injection with 100 μg of pCAGGS (vector control), Env, Env/Y710S, Env750Tr, or Env750Tr/Y710S at weeks 0 and 4. At week 6, mouse splenocytes were collected and stimulated with the peptide IGPGRAFYAR (10 μg/ml) for 6 h at 37°C. The cells were then stained for cell surface CD4 (PerCp) and CD8 (PE), as well as intracellular IFN-γ (APC), and analyzed by flow cytometry. Background IFN-γ-producing cells were determined by culturing mouse splenocytes with an irrelevant peptide (AMQMLKETI). Values in each box are the percentages of the gated population. (A) Gating of CD8 and CD4 lymphocytes and background IFN-γ production by CD8 or CD4 lymphocytes stimulated with peptide AMQMLKETI (control). (B) Representative results of FACS analysis for IFN-γ production by CD8 T cells from each immunization group stimulated with the peptide IGPGRAFYAR. Numbers in lower right boxes represent percentages of IFN-γ-positive CD8 T cells. (C) Percentages of IFN-γ-positive CD8 T cells for each immunization group after stimulation with the peptide IGPGRAFYAR. Error bars represent standard deviations for each group.
Combination of Tyr mutation and truncation of the cytoplasmic domain enhances induction of T-cell responses against subdominant epitopes in the HIV Env protein.
To determine whether mutation in the YXXΦ motif or truncation of the dileucine-like motif might affect broader T-cell responses against other potential epitopes in the HIV Env protein, we analyzed IFN-γ production by T cells in splenocytes from immunized mice stimulated with peptide pools corresponding to the HIV-1 89.6 Env protein. Four peptide pools covering the entire HIV 89.6 Env protein were used to stimulate mouse splenocytes, which were pooled for each group because of the limited supply of the peptide pools.
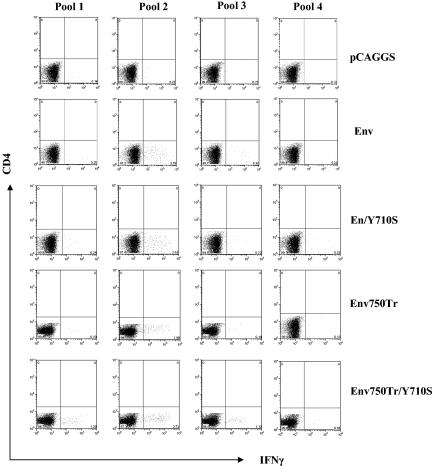
As shown in Fig. 5, by stimulation with peptide pool 2, which covers the region in the Env protein from amino acids 200 to 400 and contains the dominant epitope (amino acids 312 to 321 of the Env protein), similar percentages of CD8 T cells were induced to produce IFN-γ in splenocytes from mice immunized with all four DNA constructs (2.35% for Env, 2.76% for Env/Y710S, 1.99% for Env750Tr, and 2.73% for Env750Tr/Y710S), in agreement with the result obtained with stimulation with the dominant peptide. Also, peptide pool 2 stimulated similar percentages of IFN-γ-producing CD4 T cells from Env- and Env/Y710S-immunized mice. However, splenocytes from Env756Tr/Y710S-immunized mice gave higher levels of IFN-γ-producing CD8 T cells when stimulated with peptide pools 1 and 3, compared to splenocytes from mice immunized with other constructs. Immunization of mice with Env750Tr also induced moderately higher levels of IFN-γ-producing CD8 T cells compared to immunization with Env and Env/Y710S, which did not induce significant levels of IFN-γ-producing CD8 T cells above background when stimulated with peptide pools 1, 3, and 4. These results indicate that induction of broader CD8 T-cell responses against HIV Env by DNA immunization is not significantly affected by mutation of the YXXΦ motif, moderately enhanced by truncation of the dileucine-like motif, and further enhanced by combining mutation in the YXXΦ motif and truncation of the dileucine motif.
FIG. 5.
Combination of Tyr mutation and cytoplasmic domain truncation enhances induction of broad CD8 T-cell responses. Mouse splenocytes from each group were pooled, and the pooled splenocyte aliquots were stimulated for 16 h with each of the peptide pools covering the entire HIV 89.6 Env protein, followed by addition of brefeldin A, a further incubation of 5 h, and then stained for surface CD4 (PerCP) and CD8 (PE) and intracellular IFN-γ (APC). Numbers in lower right boxes represent percentages of IFN-γ-producing CD 8 T cells gated as shown in Fig. 4A. Peptide pools are 20-mers overlapping by 10 amino acids. Pool 1, amino acids 1 to 210; pool 2, amino acids 200 to 410; pool 3, amino acids 400 to 610; pool 4, amino acids 600 to 853.
The use of peptide pools for stimulation also allowed us to assess CD4 T-cell responses induced by immunization with different DNA constructs. As shown in Fig. 6, CD4 T cells from mice immunized with all four constructs were stimulated to produce IFN-γ by peptide pool 2 at significant levels above control vector pCAGGS-immunized mice. However, stimulation with peptide pools 1, 3, and 4 did not induce significant levels of IFN-γ-producing CD4 T cells from mice immunized with Env, Env/Y710S, and Env750Tr. In contrast, higher levels of CD4 T cells from mice immunized with Env750Tr/Y710S were stimulated to produce IFN-γ by peptide pools 1 and 3, although peptide pool 4 did not induce significant levels of IFN-γ-producing CD4 cells. Thus, the combination of a Tyr mutation in the YXXΦ motif and truncation of the dileucine-like motif also enhanced induction of CD4 T-cell responses against subdominant epitopes in the HIV 89.6 Env by DNA immunization.
FIG. 6.
Combination of Tyr mutation and cytoplasmic domain truncation enhances induction of CD4 T-cell responses. Mouse splenocytes from each group were pooled, and the pooled splenocyte aliquots (2 × 106 per well in 48-well plate) were stimulated for 16 h with each of the peptide pools covering the entire HIV 89.6 Env protein, followed by addition of brefeldin A, a further incubation of 5 h, and then staining for surface CD4 (PerCP) and CD8 (PE) and intracellular IFN-γ (APC). Numbers in lower right boxes represent percentages of IFN-γ-producing CD 4 T cells gated as shown in Fig. 4A. Peptide pools are 20-mers overlapping by 10 amino acids. Pool 1, amino acids 1 to 210; pool 2, amino acids 200 to 410; pool 3, amino acids 400 to 610; pool 4, amino acids 600 to 853.
The DNA construct for the mutant Env750Tr/Y710S is more potent in eliciting antibody responses against the HIV Env protein.
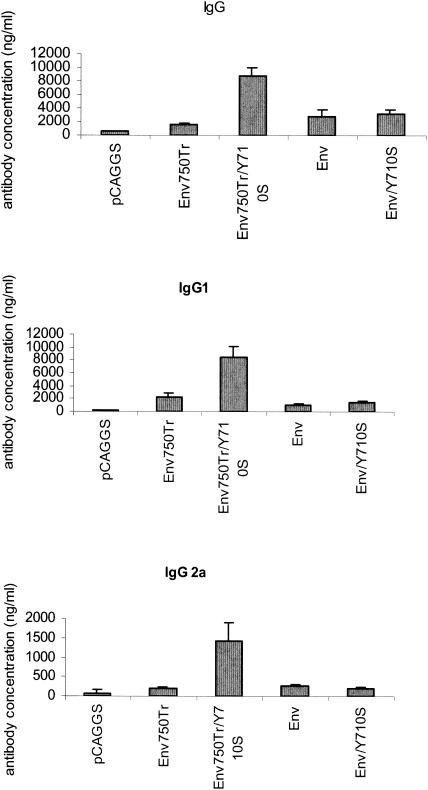
The antibody responses against the HIV 89.6 Env protein were compared by ELISA to determine the levels of binding antibody to HIV 89.6 gp120 induced by DNA immunization. As shown in Fig. 7, the DNA constructs Env, Env750Tr, and Env/Y710S induced similar levels of total IgG antibody binding to gp120, with about 1,500 ng/ml for Env750Tr and 3,000 ng/ml for Env and Env/Y710S. However, immunization of mice with Env750Tr/Y710S induced about three- to fivefold-higher levels of antibody responses against gp120, with an average of about 9,000 ng/ml. Statistical analysis showed that the anti-HIV 89.6 gp120 antibody response induced by immunization with Env750Tr/Y710S was significantly higher than the antibody responses induced by immunization with other DNA constructs (P < 0.01 versus pCAGGS; P < 0.05 versus Env750Tr, Env, and Env/Y710S), while no significant difference in antibody response levels was detected for Env-, Env/750Tr-, and Env/Y710S-immunized mice (P > 0.1). The IgG1 isotype antibody response was also increased in Env750Tr/Y710S-immunized mice about two- to fourfold compared to responses in mice immunized with the other constructs, similar to that observed for the total IgG responses. Moreover, as shown in Fig. 7C, immunization with Env750Tr/Y710S also induced a significant amount of IgG2a antibodies (about 1,300 ng/ml) against HIV 89.6 gp120, while no significant amount of IgG2a isotype gp120-binding antibody was induced by any of the other three constructs (less than 200 ng/ml). These results demonstrate that the Env750Tr/Y710S construct, which contains both a mutation in the YXXΦ motif and truncation of the dileucine motif, is more potent in inducing antibody responses than other constructs. The more pronounced increase for IgG2a antibody suggests that immune responses induced by immunization with Env750Tr/Y710S are more biased towards Th1-type immune responses, which typically exhibit an increased IgG2a/IgG1 ratio (42, 43).
FIG. 7.
The DNA construct for Env750Tr/Y710S is significantly more potent for inducing antibody responses. Sera from mice immunized with different DNA constructs were collected at 2 weeks postimmunization and analyzed for antibodies specific for HIV 89.6 Env protein by ELISA as described in Materials and Methods. The levels of antibody responses are expressed as the quantity of antibodies (total IgG, IgG1, or IgG2a) binding to HIV 89.6 gp120 in 1 ml of serum from each mouse. The results were analyzed with the Microsoft Excel program, and error bars indicate the standard deviations for each immunization group.
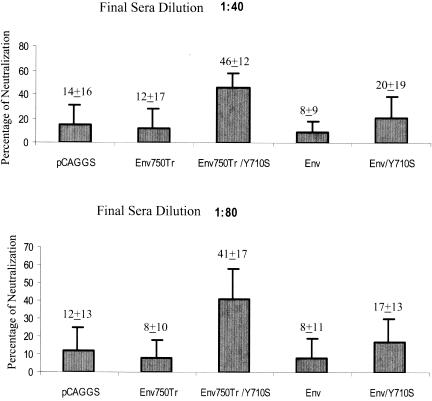
To determine whether induced antibody responses by DNA immunization are biologically active, we compared neutralizing activities of mouse sera against single-round HIV 89.6 infection of an indicator cell line, JC53-BL, which is a derivative of HeLa cells that expresses high levels of CD4, CCR5, and CXCR4 and also contains a β-galactosidase reporter cassette that is expressed from an HIV-1 long terminal repeat dependent on production of HIV-1 Tat (13). As shown in Fig. 8, at both 1:40 and 1:80 dilutions, sera from Env750Tr/Y710S-immunized mice exhibited the highest level of neutralizing activity, which was significantly higher than that in the sera from other groups of mice (P < 0.05). Although sera from Env/Y710S-immunized mice also exhibited higher levels of neutralizing activity than sera from the control vector pCAGGS-immunized mice, the differences were not statistically significant.
FIG. 8.
Neutralization of HIV 89.6 by sera from immunized mice. Sera from immunized mice were mixed with HIV 89.6 at 1:40 and 1:80 final dilutions, and neutralization of HIV 89.6 was analyzed in JC53-BL cells as described in Materials and Methods. The percentages of neutralization by each serum sample were calculated and compared. The average neutralization value and standard deviation are shown for each group.
DISCUSSION
In this study, we investigated the possible effects of the two putative endocytosis motifs in the HIV Env protein cytoplasmic domain on Env protein surface expression and immunogenicity. Codon-optimized genes were synthesized for the HIV 89.6 Env protein, and mutant HIV Env proteins with a mutation of the Tyr residue in the YXXΦ motif into a Ser or truncation of the dileucine-like motif, or their combination, were analyzed. Analysis of protein expression showed that all constructs gave similar levels of expression of the Env protein as well as secretion of gp120 into the medium. Furthermore, mutation of the Tyr residue in the YXXΦ motif or truncation of the dileucine-like motif alone did not significantly affect surface expression of the HIV Env protein. On the other hand, combination of the Tyr mutation and cytoplasmic domain truncation led to enhanced surface stability for the mutant HIV Env750Tr/Y710S, as shown by pulse-chase radioactive labeling coupled with surface biotinylation and immunoprecipitation studies. Analysis of immune responses induced by DNA immunization showed that neither mutation of the Tyr residue, truncation of the cytoplasmic domain alone, nor their combination exerted a significant effect on induction of the CD8 T-cell response against a known dominant epitope in the HIV 89.6 Env protein. However, the combination of the Tyr mutation and truncation of the cytoplasmic domain enhanced induction of CD4 T-cell responses as well as broader CD8 T-cell responses against subdominant epitopes. Moreover, the mutant Env750Tr/Y710S DNA construct was more potent in eliciting antibody responses, while mutation of the Tyr residue or truncation of the dileucine-like motif individually did not significantly affect antibody induction compared to that with the wild-type HIV 89.6 Env protein. Furthermore, immunization with Env750Tr/Y710S elicited significant levels of IgG2a antibody responses, indicating the induced immune response is more biased towards a Th1-type response in comparison to other constructs. Sera from Env750Tr/Y710S-immunized mice also showed significantly higher levels of neutralizing activity against HIV 89.6 compared with sera from the other groups. These results show that mutation to disrupt the YXXΦ motif together with truncation of the dileucine-like motif significantly enhanced immunogenicity of the HIV Env protein, indicating that the HIV Env protein may be selected to preserve both endocytosis signals to down-modulate its immunogenicity as a mechanism to evade the immune system.
The HIV Env protein contains a long cytoplasmic domain in which several functional domains or motifs, such as the conserved Cys residue for palmitoylation, two lentivirus lytic peptide, calmodulin binding site, the membrane-proximal Tyr-based YXXΦ motif, and the membrane-distal dileucine-like motif, have been identified (3, 6, 29, 38, 40, 44, 45, 53, 55). It is interesting that in addition to the membrane-proximal YXXΦ motif, which is conserved among retrovirus glycoproteins, the HIV Env protein also retained a dileucine-like motif to regulate its surface expression (6). While the two motifs seem to be redundant with respect to their functions to mediate HIV Env protein endocytosis, preserving both motifs may be advantageous for HIV to ensure strict regulation of its Env protein surface expression and minimize induction of immune responses. Our results show that the presence of either motif alone can still effectively mediate Env protein endocytosis, as observed for Env750Tr and Env/Y710S. Furthermore, disrupting both endocytosis motifs in the HIV Env protein cytoplasmic domain is required to augment its ability to elicit antibody responses. Moreover, cellular expression of the HIV Env protein and secretion of gp120 were not significantly affected by these mutations, indicating that the enhanced antibody responses may result from the increased surface stability of the mutant Env750Tr/Y710S protein. In our previous studies with SIV, we found that immunization of mice with DNA constructs encoding SIV Env proteins with a full-length or truncated cytoplasmic domain induced similar levels of antibody responses, although increased Env protein expression was found for the DNA construct encoding the truncated SIV Env protein, which has 18 amino acids in its cytoplasmic domain and contains the intact YXXΦ motif (48). Taken together, these results suggest that surface stability, rather than the level of surface expression, plays an important role in modulating HIV Env protein immunogenicity.
Although the mechanism by which endocytosis signals regulate HIV Env protein immunogenicity remains to be determined, it is possible that rapid endocytosis of HIV Env proteins from the surface of antigen-presenting cells leads to reduced exposure to the B-cell receptors and therefore reduced stimulation of antibody production. The regulation of immunogenicity by surface stability also provides a possible explanation for the results reported by Fultz et al. (23), in which they showed that a mutant SIV with mutation of the Tyr residue in the YXXΦ motif was attenuated during in vivo infection. It is possible that the mutant virus was more susceptible to recognition and attack by the immune system with increased surface stability of the Env protein. However, we cannot rule out other possible factors that may also contribute to regulate the immunogenicity of the HIV Env protein. For example, truncation of the cytoplasmic domain also deleted the two LLP segments as well as the calmodulin-binding domain, which mediate cytopathicity of the HIV Env protein and thus prolong the surviving time of the Env protein-expressing cells for activation of the immune response. Furthermore, mutation of the Tyr residue may prevent the Env protein from clustering at special membrane locations, whereas truncation of the cytoplasmic domain removes a palmitoylation signal that is critical for its localization in lipid rafts, as shown in other studies (44). Moreover, Egan et al. reported that in HIV-infected cells, the activity of the YXXΦ endocytosis motif is suppressed by coexpressed Gag protein (19). It is also interesting that the relative amount of uncleaved gp160 on the cell surface seems to be higher for the truncated Env proteins, Env750Tr and Env750Tr/Y710S, than for the full-length Env proteins, Env and Env/Y710S (compare Fig. 3A and B surface panels). Earlier studies have shown that uncleaved HIV Env proteins are more potent in eliciting antibody responses (8, 30). This raises the possibility that the long cytoplasmic domain of the HIV Env protein may also function to delay Env protein transport to the cell surface to ensure more complete processing for quality control. If this were true, identification of such potential functional domains in the HIV Env protein cytoplasmic tail might aid in the design of mutant HIV Env proteins with enhanced surface stability and modified processing efficiency for vaccine development. Therefore, it is also possible that these as well as other yet-to-be-identified factors may act together to regulate immunogenicity of the HIV Env protein. Analysis of T-cell responses showed that although induction of CD8 T cells specific for the dominant epitope in the HIV 89.6 Env protein was not affected, mutations of both endocytosis signals moderately enhanced induction of CD8 T cells against potential subdominant epitopes as well as IFN-γ-producing CD4 T cells, as determined by stimulation using peptide pools. Although the observed enhancement for T-cell responses is modest, it may be translated into a more drastic effect on antibody induction through the action of T helper cells. This is evidenced by the induction of relatively higher levels of IgG2a antibodies by immunization with Env750Tr/Y710S, which indicates the presence of Th1 helper cells that typically produce IFN-γ upon stimulation (42, 43). Thus, it is also possible that mutations of the endocytosis signals affect induction of T-cell responses in DNA immunization, which in turn modulate antibody induction.
Previous studies have shown that in HIV-infected cells Fas ligand expression is increased due to expression of the HIV Nef protein, which leads to killing of adjacent T cells through Fas-mediated apoptosis (25, 54). Furthermore, it has been shown that Nef protein also down-regulates surface expression of major histocompatibility complex class I molecules in infected cells and thus enables these cells to evade recognition by cytotoxic T cells (9, 10, 36, 47). Our results indicate that modulation of Env protein surface expression by the endocytosis signals in the Env protein cytoplasmic domain may serve as another strategy for HIV evasion of the immune system. Day et al. reported that both Nef and the YXXL endocytosis motif act independently to modulate HIV infectivity (12). It will be interesting to determine whether their roles in conferring HIV evasion of the immune system are connected or independent of each other. Future studies to delineate the possible mechanism will provide more insight into HIV pathogenesis and its interplay with the host immune system, as well as the development of AIDS vaccines that can induce more potent antibody responses against the HIV Env protein.
Acknowledgments
This work was supported by Public Health Service grant AI47018 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Akari, H., A. Yoshida, T. Fukumori, and A. Adachi. 2000. Host cell-dependent replication of HIV-1 mutants with deletions in gp41 cytoplasmic tail region is independent of the function of Vif. Microbes Infect. 2:1019-1023. [DOI] [PubMed] [Google Scholar]
- 2.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, L. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 5.Bu, Z., L. Ye, M. J. Skeen, H. K. Ziegler, R. W. Compans, and C. Yang. 2003. Enhancement of immune responses to an HIV env DNA vaccine by a C-terminal segment of listeriolysin O. AIDS Res. Hum. Retrovir. 19:409-420. [DOI] [PubMed] [Google Scholar]
- 6.Bultmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 75:5263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cervantes-Acosta, G., R. Lodge, G. Lemay, and E. A. Cohen. 2001. Influence of human immunodeficiency virus type 1 envelope glycoprotein YXXL endocytosis/polarization signal on viral accessory protein functions. J. Hum. Virol. 4:249-259. [PubMed] [Google Scholar]
- 8.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, G. B., R. T. Gandhi, D. M. Davis, O. Mandelboim, B. K. Chen, J. L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 10.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 11.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 12.Day, J. R., C. Munk, and J. C. Guatelli. 2004. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J. Virol. 78:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W.A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deschambeault, J., J. P. Lalonde, G. Cervantes-Acosta, R. Lodge, E. A. Cohen, and G. Lemay. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J. Virol. 73:5010-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68:1689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egan, M. A., L. M. Carruth, J. F. Rowell, X. Yu, and R. F. Siliciano. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Facke, M., A. Janetzko, R. L. Shoeman, and H. G. Krausslich. 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 67:4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed, E. O., and M. A. Martin. 1995. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J. Biol. Chem. 270:23883-23886. [DOI] [PubMed] [Google Scholar]
- 22.Freed E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fultz, P. N., P. J. Vance, M. J. Endres, B. Tao, J. D. Dvorin, I. C. Davis, J. D. Lifson, D. C. Montefiori, M. Marsh, M. H. Malim, and J. A. Hoxie. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J. Virol. 75:278-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geleziunas, R., W. Xu, K. Takeda, H. Ichijo, and W. C. Greene. 2001. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature 410:834-838. [DOI] [PubMed] [Google Scholar]
- 26.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 27.Haffar, O. K., G. R. Nakamura, and P. W. Berman. 1990. The carboxy terminus of human immunodeficiency virus type 1 gp160 limits its proteolytic processing and transport in transfected cell lines. J. Virol. 64:3100-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa, H., M. Sasaki, S. Noda, and Y. Koga. 1998. Apoptosis induction by the binding of the carboxyl terminus of human immunodeficiency virus type 1 gp160 to calmodulin. J. Virol. 72:6574-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieny, M. P., R. Lathe, Y. Riviere, K. Dott, D. Schmitt, M. Girard, L. Montagnier, and J. Lecocq. 1988. Improved antigenicity of the HIV env protein by cleavage site removal. Protein Eng. 2:219-225. [DOI] [PubMed] [Google Scholar]
- 31.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, and J. A. Hoxie. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68:5509-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. J., W. Hu, A. G. Fisher, D. J. Looney, V. F. Kao, H. Mitsuya, L. Ratner, and F. Wong-Staal. 1989. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res. Hum. Retrovir. 5:441-449. [DOI] [PubMed] [Google Scholar]
- 34.Lodge, R., H. Gottlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodge, R., J. P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangasarian, A., V. Piguet, J. K. Wang, Y. L. Chen, and D. Trono. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J. Virol. 73:1964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsh, M., and A. Pelchen-Matthews. 2000. Endocytosis in viral replication. Traffic 1:525-532. [DOI] [PubMed] [Google Scholar]
- 38.Miller, M. A., T. A. Mietzner, M. W. Cloyd, W. G. Robey, and R. C. Montelaro. 1993. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retrovir. 11:1057-1066. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 41.Qiu, J. T., B. Liu, C. Tian, G. N. Pavlakis, and X. F. Yu. 2000. Enhancement of primary and secondary cellular immune responses against human immunodeficiency virus type 1 Gag by using DNA expression vectors that target Gag antigen to the secretory pathway. J. Virol. 74:5997-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raz, E., H. Tighe, Y. Sato, M. Corr, J. A. Dudler, M. Roman, S. L. Swain, H. L. Spiegelberg, and D. A. Carson. 1996. Preferential induction of a Th1 immune response and inhibition of specific IgE antibody formation by plasmid DNA immunization. Proc. Natl. Acad. Sci. USA 93:5141-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romagnani, S. 1997. The Th1/Th2 paradigm. Immunol. Today 18:263-266. [DOI] [PubMed] [Google Scholar]
- 44.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowell, J. F., P. E. Stanhope, and R. F. Siliciano. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J. Immunol. 155:473-488. [PubMed] [Google Scholar]
- 46.Sauter, M. M., A. Pelchen-Matthews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J. Cell Biol. 132:795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 48.Vzorov, A. N., D. Lea-Fox, and R. W. Compans. 1999. Immunogenicity of full length and truncated SIV envelope proteins. Viral Immunol. 12:205-215. [DOI] [PubMed] [Google Scholar]
- 49.Vzorov, A. N., and R. W. Compans. 2000. Effect of the cytoplasmic domain of the simian immunodeficiency virus envelope protein on incorporation of heterologous envelope proteins and sensitivity to neutralization. J. Virol. 74:8219-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 53.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, X. N., B. Laffert, G. R. Screaton, M. Kraft, D. Wolf, W. Kolanus, J. Mongkolsapay, A. J. McMichael, and A. S. Baur. 1999. Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor zeta chain. J. Exp. Med. 189:1489-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, C., and R. W. Compans. 1996. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J. Virol. 70:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, X., X. Yuan, M. F. McLane, T. H. Lee, and M. Essex. 1993. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 67:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]