Abstract
During the initial stages of Epstein-Barr virus (EBV) infection of peripheral resting B cells, transcription of the six genes encoding the EBV latency-associated nuclear antigens (EBNAs) is driven from Wp, a promoter that is present in multiple copies within the EBV major internal repeat. As infection progresses, transcription from Wp is downregulated following upregulation of EBNA gene transcription driven from a promoter, Cp, located ca. 3 kb upstream of the first copy of Wp. Recently published data have provided evidence that, concomitant with the switch in EBNA gene promoter usage, Wp becomes heavily methylated (R. J. Tierney et al., J. Virol. 74:10468-10479, 2000). Based on this observation, it has been argued that methylation of Wp plays a pivotal role in suppressing Wp activity in EBV-immortalized B-lymphoblastoid cell lines (LCLs). Here we present data compiled from analyses of Wp methylation in eight randomly selected low-passage-number B-LCLs. These data demonstrate that there is considerable variability in Wp methylation, both between different cell lines and within clonal LCLs. Overall, less methylation of Wp was noted in established, low-passage-number LCLs than was previously observed in bulk cultures of infected B cells at days 18 and 21 postinfection. Importantly, the majority of LCLs examined harbored both unmethylated and methylated copies of Wp. In addition, all low-passage-number LCLs examined contained both Cp- and Wp-initiated EBNA transcripts, arguing for the presence of some transcriptionally active copies of Wp. Taken together, these data argue that other factors, perhaps in conjunction with Wp methylation, play a role in suppressing Wp activity in LCLs.
Background.
Epstein-Barr virus (EBV), a DNA tumor virus and member of the herpesvirus family, efficiently transforms primary B cells and is an etiologic agent of infectious mononucleosis. In addition, numerous studies have implicated EBV in the development of various human lymphoid and epithelial malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's lymphoma, immunoblastic lymphoma, peripheral T-cell lymphoma, and gastric carcinoma (4, 13, 19).
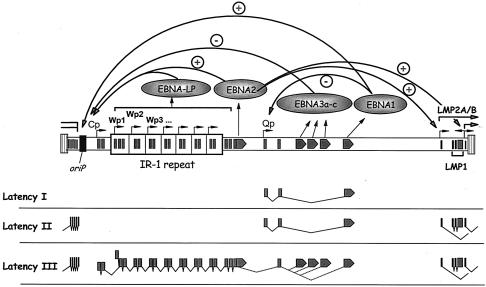
EBV primarily establishes a latent infection in which the viral genome persists as an episome (2). Several different programs of latency-associated EBV gene expression have been described. During infectious mononucleosis, during ex vivo infection of B cells, and in EBV-associated posttransplant lymphoproliferative disease, EBV enters a program of gene expression known as latency III, which results in B-cell growth transformation (13). The latency III program is characterized by the expression of a wide array of viral proteins, including six different Epstein-Barr nuclear antigens (EBNAs 1, 2, 3a, 3b, 3c, and 4) and three latency-associated membrane proteins (LMPs 1, 2a, and 2b). Transcription of the EBNAs initiates at one of two promoters, Cp or Wp (Fig. 1) (18). Notably, Wp is encoded within the major internal repeat (IR-1) of EBV and as such is present in multiple copies (Fig. 1). Alternative 3′ processing and splicing of long primary transcripts results in the production of the mature transcripts encoding the various EBNAs (Fig. 1). Upon ex vivo infection of B cells, EBNA gene transcription initially arises from the proximal EBNA gene promoter Wp, followed by induction of EBNA gene transcription from the distal EBNA gene promoter Cp (23). EBNA 1 upregulates transcription from Cp and Wp by binding to oriP, the latency-associated origin of replication located upstream of Cp (12) (Fig. 1), which also functions to maintain the viral episome in proliferating B cells (1). EBNA 2, which acts through a direct interaction with the cellular transcription factor CBF1/RBP-Jκ, also upregulates EBNA gene transcription from Cp via an enhancer located just upstream of Cp (3, 6, 20) (Fig. 1). The EBNA 3 family also interacts with CBF1/RBP-Jκ and serves to negatively regulate EBNA gene transcription initiation from Cp (27) (Fig. 1). Thus, EBNA gene transcription during type III latency is autoregulated by the EBNA gene products (Fig. 1).
FIG. 1.
Schematic illustration of EBNA gene transcription during different EBV latency programs. EBV encodes six EBNAs and two LMPs. In the latency I and II programs, the only EBNA gene expressed is EBNA 1, which is transcribed from the Qp promoter (16). The LMP genes are expressed during the latency II program but are absent from the latency I program. The latency III program, or growth program, involves transcription of all six EBNA genes, driven from either Cp or Wp. A combination of alternative 3′ processing and alternative splicing leads to the production of the mature transcripts encoding each EBNA gene product. In addition to the six EBNA genes, the LMP genes are also expressed during the latency III program. Transcription of the EBNA and LMP genes is autoregulated, as shown here and discussed in the text.
A significant body of data that addresses the mechanism(s) involved in downregulating EBNA gene transcription from Wp during the early stages of B-cell latency has accumulated. Many of these data are consistent with a transcriptional interference model, where upregulation of transcription from the distal Cp EBNA gene promoter leads directly to downregulation of Wp-initiated transcription (11, 12, 22, 25). The transcriptional interference model has been supported by transient transfection of reporter constructs containing a large region of the EBV genome spanning from oriP to the second W1 exon, which has shown that (i) Cp is the dominant promoter used in transfected lymphoblastoid cell lines (LCLs) to drive reporter gene transcription (11, 12, 23), (ii) mutations that impair Cp activity lead to an upregulation of Wp-initiated transcription (11, 12), and (iii) inversion of Cp leads to upregulation of Wp-initiated transcription without diminishing the level of Cp-initiated transcription (11). Analysis of recombinant EBV mutants has also provided support for the role of transcriptional interference in modulating Wp-initiated transcription: (i) mutation of the EBNA 2 enhancer upstream of Cp leads to diminished Cp activity and enhanced Wp-initiated EBNA gene transcription (25), and (ii) withdrawal of EBNA 2 function by use of the estrogen-regulated EBNA 2 conditional LCL er/eb 2.5 (7) leads to diminished Cp-initiated transcription and a concomitant upregulation of EBNA gene transcription from Wp (24).
In contrast with the data supporting the transcriptional-interference model, it has been shown that during the initial stages of EBV infection of peripheral B cells, the region around Wp becomes densely methylated by day 21 postinfection (21). The kinetics of the appearance of Wp methylation correlated well with the extinguishing of Wp-initiated transcription as detected by a reverse transcription (RT)-PCR assay (21). Consistent with this model, ex vivo methylation of Wp regulatory sequences has been shown to strongly suppress Wp-initiated transcription (5, 14, 21). Thus, those authors argue that methylation of Wp is the primary mechanism by which Wp activity is downregulated during the establishment of LCLs. This model is difficult to reconcile with the previous analyses of Cp and Wp activity from transiently transfected reporter constructs, which demonstrate a direct impact of Cp-initiated transcription on Wp-initiated transcription in the absence of any methylation of either Cp or Wp sequences (11, 12). In addition, the rapid induction of Wp-initiated transcription upon withdrawal of EBNA 2 in the er/eb 2.5 LCL (24) seems unlikely to involve alterations in the methylation status of Wp and thereby indicates that there must be unmethylated copies of Wp. Finally, previous mutagenesis (26) and methylation cassette (10) analyses have demonstrated a critical role for the enhancer region upstream of the first copy of Wp (Wp1) in Cp-initiated transcription. Based on those data, it is surprising if methylation of this enhancer region upstream of Wp1 would be tolerated in LCLs. To further address this issue, we have examined the methylation status of Wp in several low-passage-number LCLs.
Analysis of Wp methylation in low-passage-number LCLs.
We randomly selected a panel of eight LCLs that we had previously established, all of which had been passaged only a limited number of times postestablishment. Among these were LCLs immortalized with two different strains of EBV (B95.8 and Akata), as well as several LCLs infected with recombinant EBVs generated by rescuing the immortalizing capacity of the P3HR-1 clone 16 virus (25, 26). Notably, our previous analyses of recombinant EBVs generated by the latter approach have demonstrated that they harbor a smaller IR-1 region, comprised of only two or three copies of the BamHI W repeat, than that of wild-type strains of EBV (25). Notably, real-time RT-PCR analysis of the abundance of EBNA gene transcription initiating from Wp or Cp in these LCLs revealed that (i) as previously shown (22), in the established and extensively passaged LCLs X50-7 and JY, EBNA gene promoter usage is strongly polarized (X50-7 cells exclusively utilize Wp and JY cells exclusively utilize Cp to drive EBNA gene transcription) and (ii) as previously shown (25), both Cp and Wp are active in low-passage-number LCLs (significant levels of Wp-initiated EBNA gene transcripts were apparent in all low-passage-number LCLs analyzed) (Table 1). The presence of Wp-initiated EBNA transcripts in these LCLs argues strongly for the presence of some hypomethylated copies of Wp in the EBV genomes harbored in these LCLs.
TABLE 1.
Both Cp and Wp are active in low-passage-number LCLsa
| LCL | % of EBNA transcripts initiated from Cp |
|---|---|
| X50-7 | <1 |
| JY | 99 |
| B95-A | 2 |
| B95-C | 37 |
| Ak-C1 | ND |
| Ak-C2 | 86 |
| 25-3 | 72 |
| 22-19 | 67 |
| 17-20 | 57 |
| 28-8 | 47 |
Relative abundances of Cp- and Wp-initiated EBNA gene transcripts were determined by using a quantitative real-time PCR assay and primers specific for splicing either from the C2 exon to the W1 exon or from the W0 exon to the W2 exon. To adjust for slight differences in the efficiencies of RT-PCR amplification of Cp- and Wp-initiated transcripts, the levels of Cp- and Wp-initiated transcripts were standardized to those of the positive controls JY (for Cp-initiated transcripts) and X50-7 (for Wp-initiated transcripts). Total RNA was prepared as previously described (12), and RT-PCR analyses were carried out with the QuantiTect SYBR green kit (QIAGEN) in accordance with the manufacturer's protocol. The primers used for PCR amplification were as follows for Cp- initiated transcripts: for the C2 exon, 5′-TTC TTA CCA TGC GGC CAT GTA G-3′, and for the W1 exon, 5′-TGG TCC AGG GCC TTC ACT TC-3′. For detection of Wp-initiated transcripts, the following primers were used: for the W0/W1 splice site, 5′-GGA GTC CAC ACA AAT CCT AGG GGA GA-3′, and for the W2 exon, 5′-TTC TTA GGA GCT GTC CGA GGG GAC-3′. ND, not determined (the Ak-C1 LCL could not be analyzed because it was lost during the course of these studies).
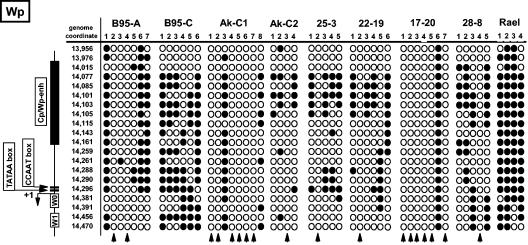
The methylation status of Wp was assessed by bisulfite PCR analysis, as previously described (9, 10). As a positive control for the presence of hypermethylation around Wp, we utilized DNA recovered from the Rael Burkitt's lymphoma cell line, which exhibits a restricted EBV latency phenotype (latency I) (8, 16). As expected, all bisulfite PCR clones recovered from the Rael cell line demonstrated that Wp is hypermethylated in this Burkitt's lymphoma cell line (Fig. 2) (17). Furthermore, in all the LCLs analyzed, there was evidence of methylation of some of the copies of Wp (Fig. 2). However, the extent of methylation was quite varied between cell lines and overall was significantly lower than that previously reported for bulk culture peripheral blood mononuclear cell infection at day 21 (21). Except with the B95-C LCL, the presence of at least one unmethylated Wp clone was detected in each cell line, indicating that unmethylated copies of Wp exist within these LCLs. Notably, the majority of bisulfite PCR products recovered from the Ak-C1 (six of eight clones analyzed) and 17-20 (six of seven clones analyzed) LCLs were devoid of any detectable methylation, suggesting that most of the IR-1 repeat region was hypomethylated. Importantly, all clones analyzed were obtained from independent bisulfite PCRs and thus do not represent the analysis of a single preferentially amplified PCR product.
FIG. 2.
Analysis of Wp methylation in low-passage-number LCLs. Genomic DNA was isolated from the indicated LCLs and from the EBV-positive Burkitt's lymphoma cell line Rael. As a negative control, DNA was also isolated from the EBV-negative Burkitt's lymphoma cell line DG75. DNA was bisulfite treated, amplified with primers designed to amplify bisulfite-modified DNA, cloned, and sequenced as previously described (9, 10). The primers used were those previously described (9, 10). The genome coordinates of the CpGs in the amplified region are shown on the left side of the panel. Also shown are the positions of known regulatory regions. ○, unmethylated CpGs; •, methylated CpGs. Each numbered column shows the data obtained from an independent bisulfite PCR amplification. The vertical arrows denote those bisulfite PCR clones for which no CpG methylation was observed. The B95-A and B95-C LCLs were established by infection of peripheral blood B cells with B95.8 virus. The Ak-C1 and Ak-C2 LCLs were established by infection of peripheral blood B cells with Akata virus. The 25-3, 22-19, 17-20, and 28-8 LCLs were established by infection of peripheral blood B cells with immortalizing virus recovered from P2HR-1 clone 16 cells transfected with a plasmid containing the EBV BamHI W, Y, and H fragments, the 3′ EBNA-leader protein-encoding exons, the entire EBNA 2 coding exon, and flanking sequences, as previously described (26, 27).
Conclusions.
Analysis of eight low-passage-number LCLs demonstrated that the IR-1 region of the EBV genome was indeed methylated, as previously reported (21). However, the extent of methylation within this region was quite varied. Furthermore, we detected in all but one of the LCLs examined the presence of unmethylated copies of Wp. Thus, the importance of DNA methylation in extinguishing Wp activity during the establishment of LCLs is unclear. As discussed above, previous data have provided strong evidence that transcriptional interference is the primary mechanism of quenching downstream Wp-initiated transcription (11, 12, 24-26).
Analysis of the methylation status of the EBV genome during restricted viral latency in the memory B-cell compartment has provided a strong correlation between methylation of Wp and Cp and the absence of detectable Cp- or Wp-initiated transcripts (10, 15). Based on those data, we have argued that targeted methylation of the IR-1 region of the viral genome is important for long-term maintenance of virus infection in vivo (10). Perhaps the hypermethylation of Wp observed at day 21 postinfection (21) reflects (i) viral genome methylation in B cells that then fail to progress to LCLs and (ii) the establishment of a restricted form of EBV latency in tissue culture. Thus, only those EBV-infected B cells in which there was low-to-modest methylation of the IR-1 repeat region would become immortalized and grow out in culture. Because the previously reported RT-PCR analyses of Cp- and Wp-initiated EBNA gene transcription (21) were not carried out with a limiting dilution analysis, this group could not assess whether the EBNA gene transcripts detected arise from B cells harboring viral genomes in which Wp is hypermethylated. The analyses of EBV genome methylation and viral gene transcription in peripheral B cells during the acute phase of infectious mononucleosis (21) also suffer from the same experimental limitation. Comparison of the levels of Wp methylation observed at day 21 postinfection (21) to the levels found in our analysis of eight low-passage-number LCLs argues that there is selection against EBV genomes in which Wp is hypermethylated. Further careful analysis of the relationship between IR-1 methylation and the transition to Cp utilization will be required before it can be resolved whether methylation plays a role in EBNA gene promoter utilization.
Acknowledgments
This research was supported by NIH research grants R01-CA58524 and R01-CA43143 to S.H.S.
We thank members of the Speck lab for helpful advice on this research.
REFERENCES
- 1.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker, L. L., L. D. Klaman, and D. A. Thorley-Lawson. 1996. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J. Virol. 70:3286-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 4.Imai, S., S. Koizumi, M. Sugiura, M. Tokunaga, Y. Uemura, N. Yamamoto, S. Tanaka, E. Sato, and T. Osato. 1994. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc. Natl. Acad. Sci. USA 91:9131-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansson, A., M. Masucci, and L. Rymo. 1992. Methylation of discrete sites within the enhancer region regulates the activity of the Epstein-Barr virus BamHI W promoter in Burkitt lymphoma lines. J. Virol. 66:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin, X. W., and S. H. Speck. 1992. Identification of critical cis elements involved in mediating Epstein-Barr virus nuclear antigen 2-dependent activity of an enhancer located upstream of the viral BamHI C promoter. J. Virol. 66:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempkes, B., U. Zimber-Strobl, G. Eissner, M. Pawlita, M. Falk, W. Hammerschmidt, and G. W. Bornkamm. 1996. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J. Gen. Virol. 77:227-237. [DOI] [PubMed] [Google Scholar]
- 8.Masucci, M. G., B. Contreras-Salazar, E. Ragnar, K. Falk, J. Minarovits, I. Ernberg, and G. Klein. 1989. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt's lymphoma line Rael. J. Virol. 63:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulson, E. J., J. D. Fingeroth, J. L. Yates, and S. H. Speck. 2002. Methylation of the EBV genome and establishment of restricted latency in low-passage EBV-infected 293 epithelial cells. Virology 299:109-121. [DOI] [PubMed] [Google Scholar]
- 10.Paulson, E. J., and S. H. Speck. 1999. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J. Virol. 73:9959-9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puglielli, M. T., N. Desai, and S. H. Speck. 1997. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp). J. Virol. 71:120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puglielli, M. T., M. Woisetschlaeger, and S. H. Speck. 1996. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 70:5758-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 14.Robertson, K. D., and R. F. Ambinder. 1997. Mapping promoter regions that are hypersensitive to methylation-mediated inhibition of transcription: application of the methylation cassette assay to the Epstein-Barr virus major latency promoter. J. Virol. 71:6445-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson, K. D., and R. F. Ambinder. 1997. Methylation of the Epstein-Barr virus genome in normal lymphocytes. Blood 90:4480-4484. [PubMed] [Google Scholar]
- 16.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1995. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc. Natl. Acad. Sci. USA 92:10565-10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1997. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol. Cell. Biol. 17:364-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Speck, S. H., and J. L. Strominger. 1989. Transcription of Epstein-Barr virus in latently infected, growth-transformed lymphocytes, p. 133-150. In G. Klein (ed.), Advances in viral oncology, vol. 8. Raven Press, New York, N.Y. [Google Scholar]
- 19.Sugiura, M., S. Imai, M. Tokunaga, S. Koizumi, M. Uchizawa, K. Okamoto, and T. Osato. 1996. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br. J. Cancer 74:625-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung, N. S., S. Kenney, D. Gutsch, and J. S. Pagano. 1991. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J. Virol. 65:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tierney, R. J., H. E. Kirby, J. K. Nagra, J. Desmond, A. I. Bell, and A. B. Rickinson. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 74:10468-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woisetschlaeger, M., J. L. Strominger, and S. H. Speck. 1989. Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc. Natl. Acad. Sci. USA 86:6498-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woisetschlaeger, M., C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 87:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo, L., and S. H. Speck. 2000. Determining the role of the Epstein-Barr virus Cp EBNA2-dependent enhancer during the establishment of latency by using mutant and wild-type viruses recovered from cottontop marmoset lymphoblastoid cell lines. J. Virol. 74:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo, L. I., M. Mooney, M. T. Puglielli, and S. H. Speck. 1997. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J. Virol. 71:9134-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo, L. I., J. Woloszynek, S. Templeton, and S. H. Speck. 2002. Deletion of Epstein-Barr virus regulatory sequences upstream of the EBNA gene promoter Wp1 is unfavorable for B-cell immortalization. J. Virol. 76:11763-11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]