Abstract
Cutaneous leishmaniasis (CL) is one of the most common parasitic diseases and public health problems in Iran. CL is endemic in most parts of Ilam province, in the west of Iran. The distance from the center of country, the great number of divers rural areas, and lack of specialists and laboratory facilities have been the major causes of Leishmania species remaining unknown in this region. Polymerase chain reaction followed by restriction fragment length polymorphism was performed to identify the Leishmania species in 61 patients with cutaneous lesions. Eventually L. major was confirmed as the cause of cutaneous leishmaniasis in Ilam province, the west of Iran.
Keywords: PCR RFLP, L. major, Ilam province, Iran
Introduction
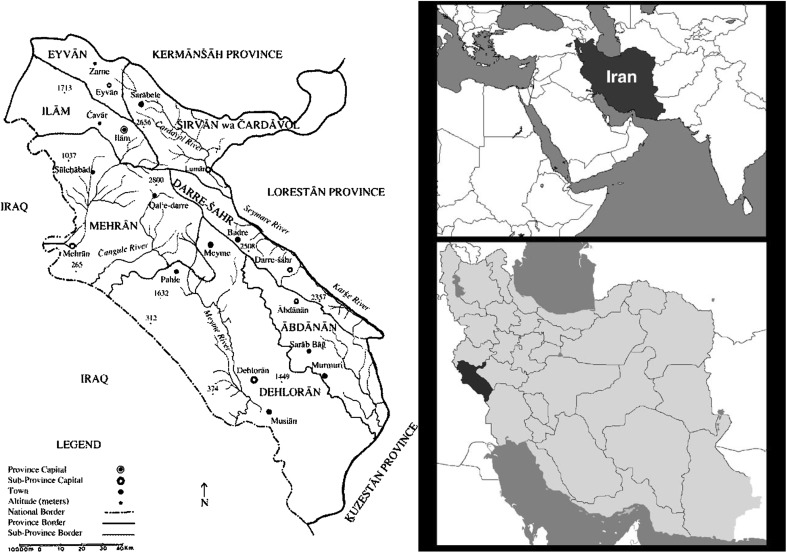
Leishmaniasis is a complex disease and a major health problem, caused by protozoan parasite of the genus Leishmania (Bailey and Lockwood 2007). Cutaneous leishmaniasis (CL) is the most common form of the disease with annual report of 1–1.5 million (World Health Organization 2002). Zoonotic cutaneous leishmaniasis (ZCL) caused by L. major and anthroponotic cutaneous leishmaniasis (ACL) caused by L. tropica are endemic in different parts of Iran. The prevalence of CL varies from 1.8 to 37.9 % in different provinces (Nadim 2000; Yaghoobi-Ershadi et al. 2002). Ilam province with 20.133 Km2 located in the west of Iran and according to 2010 census, the population has been 566,332. Ilam is a CL endemic foci with more than 1500 reported cases annually (kassiri et al. 2012; Yazdanpanah and Rostamianpur 2013). In north parts of province which is mountainous region, it has a cold climate and in the west and south which is desert lands and borders with Khuzestan province and Iraq, the weather is hot. In recent years, especially after oil and gas exploration activities in the region, the rate of CL has increased and become a public health concern (kassiri et al. 2012). Diagnosis and identification of Leishmania species are important for treatment, epidemiological studies, and control strategies (Schönian et al. 2003; Hajjaran et al. 2004; Rotureau et al. 2006). Although traditional methods of direct smear is still the golden standard technique of diagnosis of CL, but identification of Leishmania required molecular methods or animal model, because of morphological similarities between the species (Marfurt et al. 2003a). Monoclonal antibodies, Isoenzyme study, and molecular methods are used for identification of Leishmania species (Evans. 1989; Ardehali et al. 2000; Reithinger and Dujardin 2007). Currently, PCR techniques using different Leishmania genomic targets are the most useful and operational method with high sensitivity and specificity to identify Leishmania species (Singh and Sivakumar 2003; Tavares et al. 2003; Vega-López 2003). Internal transcribed spacer 1 (ITS1) region of the rRNA gene is reported as a sensitive target for this purpose (Marfurt et al. 2003a, b; Schönian et al. 2003). This study aimed to identify Leishmania species causing CL in Ilam as a not well known foci of CL in Iran (Fig. 1).
Fig. 1.
Location of Ilam province
Materials and methods
-
Sampling and population During 2013–2014, 95 clinical samples were collected from microscopically confirmed CL patients living in Mehran, Dehloran, and Dasht E-abbas cities and suburban rural areas where they attended local health centers. Informed consent was obtained from every patient and the protocol was confirmed by ethics committee of Iran University of Medical Sciences.
Sample from CL lesion was aseptically inoculated to Novy–McNeal–Nicolle (NNN) media and transported to department of parasitology, Iran University of Medical Sciences, Tehran, Iran and incubated at 25 ± 1 °C. the media was examined every 3–4 days up to 30 days, and positive samples were sub passaged into RPMI 1640 supplemented with 10 % fetal bovine serum (Gibco) for mass culture.
DNA extraction L. major (MHOM/IR/75/ER) and L. tropica (MHOM/IR/99) were used as reference strain. Approximately 106 Leishmania promastigotes were washed 3× and used for DNA extraction using DNA extraction kit (Roche) according to the manufacturer’s instruction. Using spectrophotometer, the extracted DNA samples were quantified and stored at 4 °C.
PCR amplification The ribosomal internal transcribed spacer 1 (ITS1) region using the primers LITSR (5′-CTGGATCATTTTCCGATG-3′) and L5.8S (5′-TGATACCACTTATCGCAC-TT-3′) was amplified in volume of 20 µl. 5 µl of DNA samples were added to a PCR Master Mix, containing 2.0 mM Mgcl2, 200 µM dNTP’s, 20 pmol of each primers and 2U of Taq polymerase (Amplicon A190301) in the PCR reaction and amplified in thermal cycler (Gene Atlas, Astec–Japan) with initial denaturation at 95 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 48 °C for 30 s, and 72 °C for 1 min (Daliva and Momen 2002; Schönian et al. 2003). PCR products were analyzed by electrophoresis at 100 V using 1.5 % agarose gel in Tris–acetate–EDTA buffer staining with ethidium bromide, visualized by UV light.
-
RFLP analysis of the ITS1 PCR amplicon and Sequencing According to the manufacturer’s instructions, 12 µl of each PCR products was digested 2 h at 37 °C using BsuR1 (MBI Fermentas). Then, restriction fragments were analyzed by gel electrophoresis using 2.5 % agarose gels at 120 V in Tris–acetate–EDTA buffer (Daliva and Momen 2002; Schönian et al. 2003).
The fragments were visualized by UV light and the sizes of the restriction products, comparison with the references strain, determined to Leishmania species identification.
Sequencing of amplicons was performed using Sanger method (Bioneer Inc. South Korea).
Nucleotide sequence accession numbers
Some of sequences were submitted to GenBank under accession number KP773406-13.
Results
In Table 1, the patient’s characteristic and locations of the lesions are presented.
Table 1.
Patient’s characteristic and locations of the lesions
| Patient’s characteristic | |
|---|---|
| Patient’s age | Min: 5 months |
| Max: 60 years | |
| Average: 29.6 years | |
| Gender | Male: 57 % |
| Female: 43 % | |
| Number of lesions | Min: 1 |
| Max: 6 | |
| Average: 2.2 | |
| Lesion location | Face and neck: 26 % |
| Hand: 32 % | |
| Leg: 17 % | |
| Mix: 23 % | |
| Other sites: 2 % |
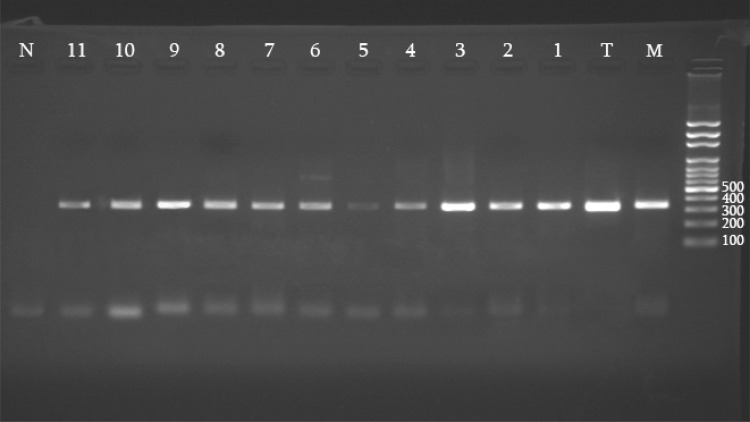
61 clinical samples were growth in NNN media and mass cultured in RPMI 1640. Sensitivity of culture method to diagnosis of Leishmania sp was 64 %. By PCR reaction, 350 bp amplicon was produced after patients DNA Amplification (Fig. 2).
Fig. 2.
Internal transcribed spacer 1 polymerase chain reaction of patient’s DNA samples. M, Leishmania major reference strain; T, Leishmania tropica reference strain. 1–11 Patient’s samples. N negative control
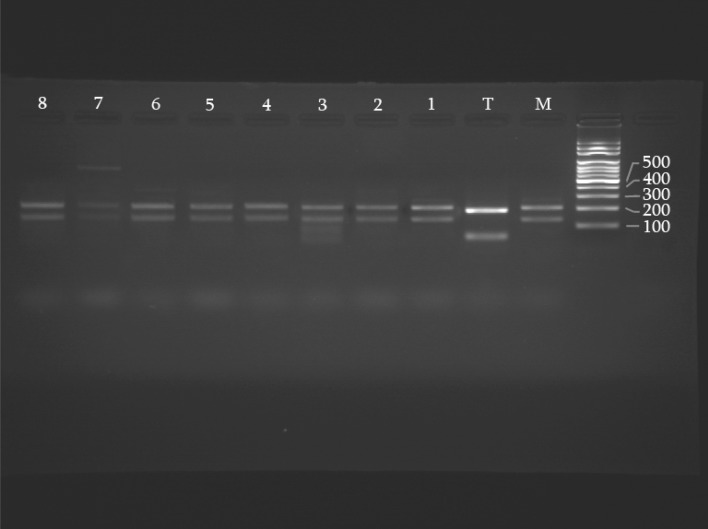
After digestion of PCR products with BsuR1 (Fermentaz) for Leishmania species identification, L. major was digested to 220 and 140 bp and L. tropica divided to 200 and 60 bp in references strain.
Based on RFLP pattern compared to reference strains, all samples were identified as L. major and no L. tropica was seen (Fig. 3).
Fig. 3.
Restriction fragment length polymorphism profiles of Leishmania species after digestion with Bsur 1. M, Leishmania major reference strain, T, Leishmania tropica reference strain. 1–8 Patient’s samples
Discussion
Iran is endemic for cutaneous leishmaniasis (Nadim 2000). Rural areas of some different provinces such as khouzestan and Isfahan are endemic for zoonotic cutaneous leishmaniasis (ZCL) and antroponic cutaneous leishmaniasis (ACL) is endemic in some cities such as Bam and Mashhad. The main goal of this study was to identifying Leishmania species in Ilam province, as a main endemic region of CL in the west of Iran. The distance from the center of country, the great number of diverse rural areas, and lack of specialists and laboratory facilities have been the major causes of Leishmania species remaining unknown in this region. Kassiri et al. (2012) during 2000–2007 was evaluated the epidemiological aspects of coutaneous leishmaniasis, this study indicated that incidence of CL was increased during recent years. In the study conducted by Tashakori and associate to characterization of Leishmania isolate in some endemic regions of Iran, Nine samples was picked up from Dehloran city, south of Ilam province and all of them was diagnosed as a L. major (2003).
Accurate diagnosis of cutaneous Leishmania species is critical for effective treatment and planning control program in endemic regions. Parasitological methods have remained a golden standard of diagnosis of CL patients, but these conventional methods are not capable of identifying the species of Leishmania parasites (Marfurt et al. 2003a, b). In previous study in hyper endemic cities of Isfahan and Bam cities, the center of Iran, ITS1-PCR followed by RFLP and sequencing, is capable to identifying the Leishmania species and demonstrated a polymorphism between species in that regions (Doudi et al. 2010). This study has been carried out based on the previous studies for its higher sensitivity and reliability of ITS1-PCR followed by RFLP to identifying Leishmania species (Al-Jawabreh et al. 2006; Doudi et al. 2010).
In this study, similar to the same studies, culture method is not very sensitive to the diagnosis of cutaneous leishmaniasi and cannot be used for species identification (Andresen et al. 1996; Aviles et al. 1999). Based on clinical features of patient’s lesions, permanent present of wild rodent in rural areas, geographical location of endemic regions, and also rare previous studies (Tashakori et al. 2003) it was conjectured that L. major is the main cause of cutaneous leishmaniasis throughout the endemic areas of Ilam province, the west of Iran. This hypothesis was proved by the results of present study.
Domination of L. major in this region is maybe to a weather and climate condition and the great number of wild rodent (gerbils) in dried and semi-dried areas (kassiri et al. 2012; Yazdanpanah and Rostamianpur 2013). Also recent activities for oil and gas exploration and making changes in rodent housing may have played a role in the increase of prevalence of disease and should be regarded. To control and eradicate the CL, we have proposed more public education and further studies with focus on reservoir hosts and vectors (sandflies) in these regions.
Acknowledgments
This project was supported by Iran University of Medical Sciences project No. 91-01-30-16956. We are grateful to the staff of local health centers of Ilam University of Medical Sciences.
References
- Al-Jawabreh A, Schönian G, Hamarsheh O, Presber W. Clinical diagnosis of cutaneous leishmaniasis: a comparison study between standardized graded direct microscopy and ITS1-PCR of Giemsa-stained smears. Acta Trop. 2006;99(1):55–61. doi: 10.1016/j.actatropica.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Andresen K, Gaafar A, El-Hasan A, Ismail A, Dafaila M, Theander TG, Kharazmi A. Evaluation of the polymerase chain reaction in the diagnosis of cutaneous leishmaniasis due to Leishmania major: a comparison with direct microscopy of smears and sections from lesions. Trans R Soc Trop Med Hyg. 1996;90:133–135. doi: 10.1016/S0035-9203(96)90112-1. [DOI] [PubMed] [Google Scholar]
- Ardehali S, Moattari A, Hatam GR, Hosseini SMH, Sharifi I. Characterization of Leishmania isolated in Iran: 1. Serotyping with species specific monoclonal antibodies. Acta Trop. 2000;75(3):301–307. doi: 10.1016/S0001-706X(00)00064-4. [DOI] [PubMed] [Google Scholar]
- Aviles H, Belli A, Armijos R, Monroy F, Harris E. PCR detection and identification of Leishmania parasites in clinical specimens in Ecuador: a comparison with classical diagnostic methods. J Parasitol. 1999;85:181–187. doi: 10.2307/3285616. [DOI] [PubMed] [Google Scholar]
- Bailey MS, Lockwood DNJ. Cutaneous leishmaniasis. Clin Dermatol. 2007;25:203–211. doi: 10.1016/j.clindermatol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Daliva AM, Momen H. Internal-trascribed-spacer (ITS) sequences used to explore polygenetic relationships within Leishmania. Ann Trop Med Parasitol. 2002;94(6):651–654. doi: 10.1080/00034980050152085. [DOI] [PubMed] [Google Scholar]
- Doudi M, Hejazi H, Razavi M, Narimani M, Khanjani S, Eslami G. Comparative molecular epidemiology of Leishmania major and Leishmania tropica by PCR-RFLP technique in hyper endemic cities of Isfahan and Bam, Iran. Med Sci Monit. 2010;16(11):CR530–CR535. [PubMed] [Google Scholar]
- Evans D. Handbook on isolation, characterization and cryopreservation of Leishmania. Geneva: World Health Organization; 1989. [Google Scholar]
- Hajjaran H, Mohebali M, Razavi MR, et al. Identification of Leishmania species isolated from human cutaneous leishmaniasis, using random amplified polymorphic DNA (RAPD-PCR) Iran J Publ Health. 2004;33:8–15. [Google Scholar]
- Kassiri H, Sharifinia N, Jalilian M, Shemshad K. Epidemiological aspects of cutaneous leishmaniasis in Ilam province, west of Iran (2000–2007) Asian Pac J Trop Dis. 2012;2(Suppl 1):382–386. doi: 10.1016/S2222-1808(12)60186-8. [DOI] [Google Scholar]
- Marfurt J, Nasereddin A, Niederwieser I, et al. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J Clin Microbiol. 2003;41:3147–3153. doi: 10.1128/JCM.41.7.3147-3153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfurt J, Niederwieser I, Makia ND, Beck HP, Felger I. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn Microbiol Infect Dis. 2003;46:115–124. doi: 10.1016/S0732-8893(03)00040-3. [DOI] [PubMed] [Google Scholar]
- Nadim A. Leishmaniasis. In: Azizi F, Janghorbani M, Hatam H, editors. Epidemiology and control of common disorders in Iran. 2. Tehran: Metabolism Research Center, Shaheed Beheshti University of Medical Sciences; 2000. pp. 524–534. [Google Scholar]
- Reithinger R, Dujardin JC. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol. 2007;45(1):21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotureau B, Ravel C, Couppié P, et al. Use of PCR-restriction fragment length polymorphism analysis to identify the main new world Leishmania species and analyze their taxonomic properties and polymorphism by application of the assay to clinical samples. J Clin Microbiol. 2006;44:459–467. doi: 10.1128/JCM.44.2.459-467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönian G, Nasereddin A, Dinse N, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–358. doi: 10.1016/S0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- Singh S, Sivakumar R. Recent advances in the diagnosis of leishmaniasis. J Postgrad Med. 2003;49:55–60. doi: 10.4103/0022-3859.927. [DOI] [PubMed] [Google Scholar]
- Tashakori M, Ajdary S, Kariminia A, et al. Characterization of Leishmania Species and L. major strains in different endemic areas of leishmaniasis in Iran. Iran Biomed J. 2003;7:43–50. [Google Scholar]
- Tavares CA, Fernandes AP, Melo MN. Molecular diagnosis of leishmaniasis. Expert Rev Mol Diagn. 2003;3:657–667. doi: 10.1586/14737159.3.5.657. [DOI] [PubMed] [Google Scholar]
- Vega-López F. Diagnosis of cutaneous leishmaniasis. Curr Opin Infect Dis. 2003;16:97–101. doi: 10.1097/00001432-200304000-00006. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2002) TDR strategic direction: leishmaniasis. http://www.who.int/tdr/diseases/leish
- Yaghoobi-Ershadi MR, Hanafi-Bojd AA, Javadian E, Jafari R, Zahraei-Ramazani AR, Mohebali M. A new focus of cutaneous leishmaniasis caused by Leishmania tropica. Saudi Med J. 2002;23:291–294. [PubMed] [Google Scholar]
- Yazdanpanah H, Rostamianpur M. Analysis of spatial distribution of leishmaniasis and its relationship with climatic parameters (case study: Ilam Province) Bull Environ Pharmacol Life Sci. 2013;2(12):80–86. [Google Scholar]