Abstract
Human metapneumovirus (HMPV) has emerged as an important human respiratory pathogen causing upper and lower respiratory tract infections in young children and older adults. Recent epidemiological evidence indicates that HMPV may cocirculate with respiratory syncytial virus, and HMPV infection has been associated with other respiratory diseases. In this study, we show that BALB/c mice are susceptible to HMPV infection, the virus replicates in the lungs with biphasic growth kinetics in which peak titers occur at days 7 and 14 postinfection (p.i.), and infectious HMPV can be recovered from lungs up to day 60 p.i. In addition, we show that genomic HMPV RNA can be detected in the lungs for ≥180 days p.i. by reverse transcription-PCR; however, neither HMPV RNA nor infectious virus can be detected in serum, spleen, kidneys, heart, trachea, and brain tissue. Lung histopathology revealed prevalent mononuclear cell infiltration in the interstitium beginning at day 2 p.i. and peaking at day 4 p.i. which decreased by day 14 p.i. and was associated with airway remodeling. Increased mucus production evident at day 2 p.i. was concordant with increased bronchial and bronchiolar inflammation. HMPV-specific antibodies were detected by day 14 p.i., neutralizing antibody titers reached ≥6.46 log2 end-point titers by day 28 p.i., and depletion of T cells or NK cells resulted in increased HMPV titers in the lungs, suggesting some immune control of viral persistence. This study shows that BALB/c mice are amenable for HMPV studies and indicates that HMPV persists as infectious virus in the lungs of normal mice for several weeks postinfection.
Human metapneumovirus (HMPV) is tentatively a member of the Metapneumovirus genus based on genetic sequence similarity to the type species avian metapneumovirus (46). HMPV was first identified in respiratory specimens from young children hospitalized with mild to severe lower respiratory tract illness (46), and recent studies indicate that HMPV may cause upper and lower respiratory tract illness in patients between the ages of 2 months and 87 years (5, 6, 8, 23, 49, 50). The disease burden associated with HMPV infection is not well known and may be complicated by the ability of HMPV to cocirculate with respiratory syncytial virus (RSV) in the community (28, 37, 50). A recent prospective study of young and older adults hospitalized for respiratory infections during the RSV season showed that HMPV was associated with approximately 11% of the illnesses, was detected in some control patients and adults of all ages, and was the only pathogen isolated from the respiratory tracts of several patients (8). The substantial disease pathogenesis associated with HMPV infection in young children and older adults (5, 8, 12, 47-49) emphasizes the need for a better understanding of HMPV immunity and disease pathogenesis.
Recently, several small-animal and nonhuman primate models of HMPV infection have been reported (21, 35). An examination of the growth properties of CAN98-75 and CAN97-83 HMPV strains in rodents showed that both strains replicated to high titers in the upper respiratory tract of hamsters (≥6.0 log10) and to moderate titers in the lower respiratory tract (≥3.6 log10) (35). In chimpanzees, HMPV replicated to relatively low titers (1.8 to 2.0 log10); however, infected animals developed mild colds (35). In a similar study, small-animal models, including mice, cotton rats, hamsters, and ferrets, and two primate species (rhesus macaques and African green monkeys) were evaluated for HMPV replication in the respiratory tract (21). The results showed that hamsters, ferrets, and African green monkeys supported HMPV replication and produced high levels of HMPV-neutralizing antibody titers; however, BALB/c mice were less permissive following intranasal (i.n.) challenge with 1.3 × 106 PFU of HMPV/NL/1/00 in which 2.4 log10 PFU/g of lung tissue was detected at day 4 p.i. Small-animal and nonhuman primate models, including BALB/c mice, cotton rats, ferrets, guinea pigs, and New and Old World primates, have been used to investigate clinically related human RSV(4, 11, 19, 31, 32, 40). RSV and HMPV are members of the Pneumovirinae subfamily of paramyxoviruses; however, RSV differs from HMPV by genomic organization. The RSV genome contains two nonstructural (NS2 and NS1) genes followed by nucleocapsid (N), phosphoprotein (P), matrix (M), small hydrophobic (SH), attachment (G), fusion (F), second matrix (M2), and RNA-dependent RNA polymerase (L) genes in the order 3′-NS1-NS2-N-P-M-SH-G-F-M2-L-5′ (20). In contrast, the HMPV genome lacks nonstructural genes and has a gene order of 3′-N-P-M-F-M2-SH-G-L-5′ (45). RSV is the most important cause of serious lower respiratory tract illness in infants and young children worldwide, causing repeat infections throughout life with serious complications occurring in the elderly and immune-compromised patients (7, 9, 24). The evidence that the same or different strains of RSV can cause repeat infections throughout life (25, 39) suggests that RSV does not engender durable immunity, and studies with animal models suggest that RSV infection may result in latency or persistence (41). For example, human RSV may persist in the guinea pig lung (18) and in murine macrophage cell lines and macrophage culture for weeks or months after infection (13), and recent studies have shown RSV latency (virus RNA) and persistence (mRNA) in BALB/c mice despite the presence of RSV-specific cytotoxic T lymphocytes and RSV-specific serum immunoglobulin G (IgG) (34).
In this study, we show that BALB/c mice are susceptible to HMPV infection, that infectious HMPV may persist in the lungs for up to 60 days postinfection (p.i.), and that genomic RNA can be detected for ≥180 days p.i. despite the presence of neutralizing antibodies. Lung histopathology shows predominant mononuclear cell inflammatory infiltration beginning at day 2 p.i. and peaking at day 4 p.i., with airway epithelial injury, remodeling, and mucus production concordant with increased bronchial and bronchiolar inflammation. Antibody depletion of T cells or NK cells resulted in increased HMPV titers in the lungs, suggesting some immune regulation of viral persistence.
MATERIALS AND METHODS
Animals.
Four- to six-week-old, specific-pathogen-free female BALB/c mice were purchased from Harlan Sprague-Dawley Laboratories (Indianapolis, Ind.). The mice were housed in microisolator cages and fed sterilized water and food ad libitum.
Virus preparation and cell lines.
Vero E6 cells were maintained in tissue culture medium (TCM) consisting of minimal essential medium (MEM; GIBCO Invitrogen, Carlsbad, Calif.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah). HMPV stocks were prepared in Vero E6 cells. Briefly, subconfluent Vero E6 cells in serum-free MEM were infected with plaque-purified strain HMPV/CAN98-75 (3) (GenBank accession number AY297748) at a multiplicity of infection of 0.1. The virus was allowed to adsorb for 1 h at 37°C after which TCM was added. Infected cells were incubated for 72 h at 37°C until a >90% cytopathic effect was observed by light microscopy. Infected cells were harvested by removal of the medium and replacement with a minimal volume of serum-free MEM followed by two freeze-thaw cycles at −70 and 4°C, respectively. The contents were collected and centrifuged at 4,000 × g for 20 min at 4°C to remove cell debris, and the titer was determined by plaque assay as described below.
Infection, sampling, and virus titers.
BALB/c mice were anesthetized by intraperitoneal (i.p.) administration of 2,2,2-tribromoethanol (Avertin) and infected i.n. with 106 PFU of HMPV. Prior to removal of lungs or tissues on days 0, 3, 4, 5, 7, 10, 14, 28, 60, 90, 150, or 180 p.i., anesthetized mice were exsanguinated by severing the right caudal artery, and the blood was collected for serum antibody analysis. Lung tissue was collected on ice in phosphate-buffered saline (PBS; GIBCO Invitrogen) to determine virus titers or with RNA Later (Ambion, Austin, Tex.) for reverse transcription (RT)-PCR analysis. Since HMPV does not form readily detectable plaques in Vero E6 cells, HMPV titers from homogenized lungs were determined by plaque assay using immunostaining to detect HMPV N protein as previously described (2). Briefly, HMPV-infected and uninfected lungs were collected, and 1.0 g of tissue was homogenized in 1 ml of PBS with a hand-held Tissuemiser homogenizer (Fisher Scientific, Pittsburg, Pa.). The lung homogenates were placed on ice for 15 min to allow debris to settle. Clarified lung lysates were diluted 10-fold in serum-free MEM (GIBCO Invitrogen), added to 95% confluent Vero E6 cells cultured in serum-free MEM (GIBCO Invitrogen) in 24-well plates (BD Falcon, San Jose, Calif.), and incubated for 1 h at 37°C, followed by TCM overlay. At 72 h p.i., the medium was removed from the cells on the 24-well plates, the wells were carefully washed with PBS, and the cells were fixed with acetone-methanol (60:40). After air drying, the cells were immunostained with affinity-purified hyperimmune serum reactive against a conserved metapneumovirus N protein (amino acid sequence DLSYKHAILKESQYTIKRDV) as previously described (2). The anti-N protein antibody was appropriately diluted in PBS containing blocking agents (Blotto; Bio-Rad, Hercules, Calif.) and detected by using alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma, The Woodlands, Tex.), and the plaques were enumerated with 3′,3′-diaminobenzidine (DAB; Vector Laboratories, Burlingame, Calif.). Spleen, trachea, brain, kidneys, heart, and liver were also isolated at each time point and stored at −70°C in RNA Later for RT-PCR analysis.
Histopathology, mucus staining, and immunohistochemistry.
Histopathological examination was performed for lungs, spleen, trachea, brain, kidneys, heart, and liver isolated from HMPV-infected mice. Tissues were fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin prior to light microscopy observation. To quantitate mucus staining, left lung lobe sections at 100 μm from the reference point were stained with alcian blue-periodic acid-Schiff stain to identify mucus-secreting cells as described previously (15). Briefly, the lung sections were deparaffinized in xylene and hydrated in decreasing concentrations of ethanol. The slides were then stained with alcian blue for 30 min, washed in running water for 5 min, oxidated in 1% periodic acid for 10 min, and washed in running water for 5 min. Following periodic acid-Schiff staining for 10 min, slides were rinsed three times in sodium metabisulfite, washed in running water for 10 min, and mounted following dehydration in ethanol and xylene.
For immunohistochemical staining of the airway epithelial cell marker, Clara cell secretory protein (CCSP) lung sections were prepared as described previously (16, 17). Briefly, 5-μm lung sections were deparaffinized in xylene and hydrated in graded ethanol followed by deionized water. Endogenous peroxidase activity was blocked by incubating the sections in 2% hydrogen peroxide in methanol for 1 min. Sections were stained overnight with primary rabbit antibodies against rat CCSP (a generous gift of Barry Stripp, University of Pittsburgh), rinsed in physiologic buffer, and incubated with secondary goat anti-rabbit antibodies conjugated to biotin. A streptavidin-conjugated peroxidase detection system (Vector Laboratories) was used to visualize antibody-binding complexes following incubation with diaminobenzidine. Multiple sections from each tissue block were analyzed with light microscopy.
HMPV ELISA.
Vero E6 cell lysates from uninfected and HMPV-infected cells were prepared as described above. Cell lysate supernatants were assayed for protein concentration by bicinchoninic acid protein analysis (Pierce Biotechnology, Inc., Rockford, Ill.) and diluted to 100 μg/ml in carbonate-bicarbonate buffer (pH 9.6), and 100 μl of infected or uninfected cell lysate was added to appropriate wells of an enzyme-linked immunosorbent assay (ELISA) plate (Immulon-2; Dynex Technologies, Inc., Chantilly, Va.). The plates were incubated at 4°C overnight, the supernatants were removed, and the wells were washed three times with PBS-0.05% Tween (PBS-T) and blocked with 150 μl of 5% dry milk-PBS (blocking buffer)/well. Pooled sera (n = 3 to 5 mice per time point) collected at various time points after HMPV infection were serially twofold diluted in blocking buffer, 100-μl dilutions were added in triplicate to wells of an ELISA plate, and the plate was incubated for 1 h at 37°C. The wells were washed three times with PBS-T and incubated with 100 μl of alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma) diluted in blocking buffer for 1 h at 37°C. Subsequently, the wells were decanted and washed three with PBS-T, and the reactions were developed with pNpp substrate tablets (Sigma). Absorbance was measured at 415- and 630-nm wavelengths and plotted versus days postinfection. Antibody end-point titers were determined according to the method of Reed and Muench (33) and reported as the mean reciprocal log2 ± standard deviation (SD). At least three separate ELISA assays were performed for each experiment.
HMPV neutralization assay.
HMPV-neutralizing antibody titers were determined by using an end-point dilution plaque reduction assay. Briefly, pooled serum (n = 3) collected from each time point after HMPV infection was diluted twofold in serum-free MEM and mixed 1:1 (vol/vol) with 104 PFU of HMPV. Reaction mixtures were incubated for 1 h at 37°C and added to 95% confluent Vero E6 cells in serum-free MEM in 24-well plates for 1 h at 37°C followed by TCM overlay. At 72 h p.i., the medium was removed from the cells, the wells were carefully washed with PBS, and the cells were fixed with acetone-methanol (60:40). After air drying, the cells were immunostained with anti-N protein antibody as previously described (2). The neutralizing antibody titers were calculated as the reciprocal of the highest serum dilution that showed 60% reduction (relative to the virus control) in the number of foci per well. Neutralizing antibody end-point titers were determined according to the method of Reed and Muench (33) and reported as the mean reciprocal log2 ± SD. At least three separate ELISA assays were performed for each experiment.
RT-PCR analysis of HMPV-infected lung tissue.
Total RNA was isolated from HMPV-infected BALB/c lung tissue by utilizing a commercial RNA isolation kit (Totally RNA; Ambion) according to the manufacturer's protocol. Briefly, RT-PCR primers were generated to the matrix (M) gene of HMPV/CAN98-75 (accession number AY145259). HMPV M gene forward primer (5′-AACTGTGGCACTTGATGAATAC-3′) and HMPV M gene reverse primer (5′-GCTGATGCTCTCGGCTTGAA-3′) were used for RT followed by 35 cycles of PCR. For RNA quality control, the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was amplified by using the GAPDH forward primer (5′-GGGTGGAGCCAAACGGTC-3′) and GAPDH reverse primer (5′GGAGTTGCTGTTGAAGTCGCA-3′). The expected fragment size for the HMPV M gene and GAPDH were 443 and 532 bp, respectively. RT-PCR amplimers were analyzed in a 1.0% agarose gel by gel electrophoresis at 125 V for 1 h, visualized by ethidium bromide staining, and photographed on a UV transilluminator with a Polaroid camera (Fisher Scientific).
Antibody depletion studies.
To deplete T cells, mice were treated i.p. with 150 μg of purified anti-CD3ɛ monoclonal antibody (clone 145-2C11; Accurate Chemical & Scientific Co., Westbury, N.Y.) and purified anti-αβTCR monoclonal antibody (clone H57-597) diluted in PBS. To deplete NK cells, mice were treated i.p. with 150 μg of purified anti-CD49b/Pan-NK cell monoclonal antibody (clone HMα2; BD Pharmingen, San Diego, Calif.). Control mice were treated with 150 μg of normal hamster Ig (Sigma). Mice received 150 μg of antibody treatments at day −3 and day −1 prior to HMPV infection and at day 1 p.i. Lungs, spleen, and sera (n = 3 per treatment) were collected at day 7 p.i., and HMPV lung titers were determined by plaque assay and immunostaining as described above. For day 28 and day 60 time points, mice were similarly treated; i.e., 150 μg of antibodies was administered i.p. at days 25 and 27 p.i. or at days 57 and 59 p.i., respectively.
To confirm cell depletion, spleen cells (n = 3 per treatment) were collected and appropriately stained with fluorescein isothiocyanate- or phycoerythrin-conjugated anti-CD3 (clone 17A-2) or anti-Pan-NK (clone DX5) antibody. The distribution of cell surface markers was determined from ≥10,000 events in two-color mode on a FACScan with CellQuest software (Becton Dickinson, Mountain View, Calif.), which revealed that >98% of the cells were depleted.
Statistical analysis.
Lungs were collected from three animals per time point per experiment in three to four separate experiments. For virus titer, antibody neutralization, and ELISA assays, all experiments were performed in triplicate, and the mean values ± SD determined for three separate assays. Statistical significance was determined by using a Student's t test, where a P value of <0.05 was considered statistically significant.
RESULTS
HMPV infection is associated with weight loss.
To determine if HMPV infection mediated illness, BALB/c mice were infected i.n. with 106 PFU of HMPV, and the virus titers in the lungs and weights of the mice were determined at days 0, 2, 3, 4, 5, 7, 10, 14, 28, 60, 90, and 180 p.i. Illness was also assessed by visual examination for ruffled coat, huddling, and heavy breathing. The level of illness associated with weight loss was linked to the level of virus replication (Fig. 1A). HMPV replicated in the lungs with biphasic growth kinetics reaching peak virus titers at days 7 p.i. (108 PFU/g of lung tissue) and declining to 105.8 PFU/g of lung tissue at day 10 p.i. followed by a second peak virus titer at day 14 p.i. (107 PFU/g of lung tissue). Concordantly, peak weight loss occurred at day 7 p.i. (23.5% loss of body weight), improved at day 10 p.i. (13.2% loss of body weight), and then increased again at day 14 p.i. (16.0% loss of body weight). At day 28 p.i., considerable virus titers (104.1 PFU/g of lung tissue) were detected in lung tissue; however, the weights of infected mice returned to a normal range, i.e., a 3.0% loss in body weight, and virus titers eventually decreased to 101 PFU/g of lung tissue by day 60 p.i. These results suggest that infectious HMPV persists in the lungs for at least 2 months p.i. No infectious virus was detected in lungs after day 60 p.i.; however, genomic RNA was detected at or after day 180 p.i. by RT-PCR (Fig. 1B), suggesting the possibility of latent HMPV infection. Congruent with virus titers and weight loss, peak illness assessed by visual examination for ruffled coat, huddling, and heavy breathing occurred at days 7 and 14 p.i., with no obvious signs of illness observed after day 28 p.i. Mice treated i.n. with a 106 PFU equivalent of inactivated HMPV or uninfected Vero E6 cell lysate did not exhibit weight loss or other signs of illness.
FIG. 1.
HMPV infection is associated with weight loss. BALB/c mice were intranasally infected with 106 PFU of HMPV. Mice were monitored for signs of clinical illness including weight loss, huddling, ruffled fur, and decreased mobility. (A) Mice were weighed, and the lungs were harvested to determine HMPV viral titers. Each time point represents the mean titer per gram of lung tissue ± SD from three experiments using three mice per experiment. (B) Total RNA was isolated from HMPV-infected BALB/c mouse lungs and assayed by RT-PCR with HMPV M gene-specific primers (top) or GAPDH primers (bottom) as an internal control. Lane 1, 1-kb molecular size marker; lane 2, day 0 p.i.; lane 3, day 7 p.i.; lane 4, day 14 p.i.; lane 5, day 28 p.i.; lane 6, day 60 p.i.; lane 7, day 90 p.i.; lane 8, day 150 p.i.; lane 9, day 180 p.i.; lane 10, 1-kb molecular size marker.
To determine if HMPV was present in tissues other than the lung or associated with viremia, the presence of infectious virus and genomic RNA was examined in the serum, spleen, trachea, brain, kidneys, heart, and liver at days 0, 2, 3, 4, 5, 7, 10, 14, 28, 60, 90, and 180 p.i. No infectious virus or genomic RNA detected by RT-PCR was evident in any specimen examined, suggesting HMPV tropism for lung tissue.
HMPV replication in the presence of neutralizing antibody.
Since substantial titers of infectious HMPV could be recovered from the lung tissue at day 28 p.i. (Fig. 1A), we examined the antibody response to infection. Serum and neutralizing antibody titers to HMPV were determined at days 0, 3, 5, 7, 14, 28, 60, 90, 150, and 180 p.i. (Table 1). An appreciable HMPV-specific antibody response was detected at day 14 p.i. and peaked at day 28 p.i. (Table 1) and remained high throughout the time course of HMPV infection, i.e., day 180 p.i. (Fig. 2). Similar kinetics were observed for serum-neutralizing end-point titers which were detected at day 14 p.i. (≥6.40 log2), peaked at day 28 p.i. (≥7.04 log2), and remained high throughout the time course of infection (Table 1). The mean neutralizing antibody titer from individual HMPV-infected mice (n ≥ 6) was similar to the mean neutralizing antibody titer in pooled sera from HMPV-infected mice (n = 3 to 6), a finding consistent with results previously reported for HMPV-infected hamsters (35). These results indicate that infectious HMPV persists in the lungs despite the presence of a neutralizing and anti-HMPV antibody response.
TABLE 1.
HMPV persists in the lungs despite the presence of a neutralizing antibody responsea
| Day p.i. | HMPV antibody titer | Serum-neutralizing antibody titer |
|---|---|---|
| 0 | ≤1 | ≤1 |
| 3 | ≤1 | ≤1 |
| 5 | 2.99 ± 0.07 | ≤1 |
| 7 | 2.99 ± 0.40 | ≤1 |
| 14 | 6.40 ± 0.20 | 5.81 ± 0.05 |
| 28 | 7.04 ± 0.07 | 6.46 ± 0.30 |
| 60 | 7.04 ± 0.00 | 6.50 ± 0.05 |
| 90 | 7.14 ± 0.50 | 6.50 ± 0.20 |
| 150 | 7.14 ± 0.30 | 6.52 ± 0.00 |
| 180 | 7.04 ± 0.40 | 6.53 ± 0.07 |
BALB/c mice were intranasally infected with 106 PFU of HMPV. Sera were collected (n = 3) at the time points indicated, pooled, and assayed for end-point antibody titers and neutralization of HMPV infection of Vero E6 cells as described in Materials and Methods. Data are presented as the median reciprocal log2 end-point antibody titers or neutralizing antibody titers ± SD from three separate experiments using three mice per experiment.
FIG. 2.
HMPV-specific antibody response. HMPV-specific antibodies were quantitated by ELISA following intranasal infection of BALB/c mice. Each time point represents the mean optical density (OD) ± SD from three separate experiments using three mice per experiment.
HMPV infection is associated with histopathology and airway remodeling.
The magnitude of histopathology associated with HMPV infection was examined at days 0, 2, 4, 7, 10, and 14 p.i. (Fig. 3). Microscopic examination of HMPV-infected lungs showed increased inflammatory cell infiltrates, predominantly by mononuclear cells, in the lung interstitium beginning at day 2 p.i. (Fig. 3B). The intensity of inflammation peaked at day 4 p.i. (Fig. 3C) and decreased slightly between day 7 (Fig. 3D) and day 14 (Fig. 3F) p.i. No histopathological changes were observed in brain, heart, liver, spleen, and kidney (data not shown).
FIG. 3.
Histopathology of HMPV infection in mice. Interstitial inflammatory cell infiltrates were examined in the lung at (A) day 0, (B) day 2, (C) day 4, (D) day 7, (E) day 10, or (F) day 14 following hematoxylin and eosin staining. Magnification, ×25 (A to F).
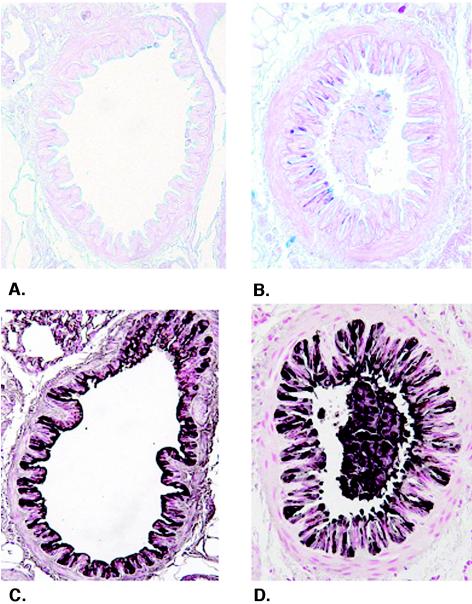
Airway epithelial cell changes during HMPV infection were assessed to determine the concordance between pathogenesis in the airways and the development of pneumonia. Histological staining of epithelial mucosubstances (Fig. 4A and B) and immunohistochemical staining of the nonciliated airway epithelial cell marker CCSP (Fig. 4C and D) were visualized by light microscopy of histological lung sections. Mucosubstances were not apparent prior to infection (day 0), consistent with the lack of observable mucus cell metaplasia in the naïve murine airway epithelium. In contrast, mucus production peaked at day 2 p.i. (Fig. 4B), with mucus production still detectable at day 4 p.i. but not apparent thereafter (data not shown). Increased mucus production was concordant with increased bronchial and bronchiolar inflammation. Immunohistochemical staining revealed that CCSP is readily apparent in airways of naïve mice (Fig. 4C). Following HMPV infection, airway epithelial injury and remodeling were observed, characterized by an altered CCSP staining pattern, increased myofibroblast thickening adjacent to the airway epithelium, and staining of cellular debris in the airways (Fig. 4D). The marked staining of CCSP in airway plugs is consistent with airway epithelial injury and sloughing following HMPV infection. Increased airway epithelial damage, as indicated by altered CCSP staining patterns in the airways and observations of cellular debris in the airway lumens, peaked at day 2 p.i., was diminished at day 4 p.i., and was not apparent thereafter (data not shown). Based on these findings, HMPV infection induces airway epithelial cell damage and promotes mucus production in the airways.
FIG. 4.
Airway epithelial changes following HMPV infection. Levels of mucus production (A and B) and the airway epithelial cell marker CCSP (C and D) were determined in lung sections from mice at day 0 (A and C) and day 2 (B and D) after HMPV infection. Mucus production (pink to purple staining) is apparent in the airways of mice. CCSP staining (brown to black staining) is associated with altered airway epithelial morphology and staining of cell debris in the airway lumen (D). Magnification, ×300.
T cells and NK cells limit HMPV replication.
The results showing that infectious HMPV persists in the lungs despite the presence of a neutralizing and anti-HMPV antibody response (Fig. 1 and Table 1) suggested that HMPV infection may be controlled in part by cytotoxic leukocytes which include T cells and NK cells. To examine this possibility, BALB/c mice were antibody depleted of T cells or NK cells prior to HMPV infection (106 PFU), and the lung virus titers were determined at days 7, 28, and 60 p.i. (Fig. 5). At day 7 p.i., moderately higher levels of infectious HMPV were detected in mice depleted of T cells (107.5 PFU/g of lung tissue) or NK cells (107.2 PFU/g of lung tissue) compared to normal Ig-treated mice (106.5 PFU/g of lung tissue), suggesting that these cell types contribute to limiting the early phase of HMPV replication. To investigate the role of these cell types in the later phases of HMPV infection, i.e., persistence, HMPV-infected mice were treated with antibody to deplete T cells or NK cells 3 days prior to lung harvest at day 28 or day 60 p.i. At day 28 p.i., significantly higher (P < 0.05) HMPV lung titers were detected in T-cell-depleted (107 PFU/g of lung tissue) and NK-cell-depleted (108 PFU/g of lung tissue) mice than in normal Ig-treated mice (104.1 PFU/g of lung tissue) (Fig. 5). Similar results occurred at day 60 p.i., during which significantly higher (P < 0.05) HMPV lung titers were detected in T-cell-depleted mice (104 PFU/g of lung tissue) and NK-cell-depleted mice (104 PFU/g of lung tissue) than in normal Ig-treated mice (101 PFU/g of lung tissue). These results suggest that both T cells and NK cells have a role in limiting early and late HMPV replication, and the 10-fold increase in HMPV lung titers in NK-cell-depleted mice (108 PFU/g of lung tissue) compared to those of T-cell-depleted mice (107 PFU/g of lung tissue) at day 28 p.i. suggests that NK cells may have a foremost role in regulating HMPV persistence.
FIG. 5.
Antibody depletion of T cells or NK cells affects HMPV replication. BALB/c mice were depleted of T cells or NK cells prior to HMPV infection (106 PFU), and the lung virus titers were determined at day 7 p.i., day 28 p.i., or day 60 p.i. Data are presented as the median virus titers ± SD from three experiments using three mice per experiment.
DISCUSSION
HMPV is a newly recognized human respiratory pathogen that causes a spectrum of respiratory illnesses ranging from asymptomatic infection to severe bronchiolitis in very young and older patients (5, 6, 8, 23, 49, 50). Genotype analysis indicates that there is little difference among HMPV isolates from patients in different areas of the world (22, 26, 28), suggesting global distribution. Little is known about HMPV pathophysiology or tropism, but HMPV infection appears to have clinical disease features similar to those of RSV. Seroprevalence studies have shown that 25% of all children aged 6 to 12 months who were tested in The Netherlands had detectable antibodies to HMPV, and by 5 years of age, 100% of patients showed evidence of past infection (28). These studies suggest that HMPV, like RSV, is an important human respiratory pathogen which requires investigational studies to better understand the biology of the virus and associated disease pathogenesis.
In this study, we investigated BALB/c mice as a model for HMPV infection and show that BALB/c mice are susceptible to infection, HMPV replicates in the lungs with biphasic growth kinetics in which peak titers occur at days 7 and 14 p.i., and infectious HMPV can be recovered from lungs up to day 60 p.i. In addition, we show that genomic HMPV RNA can be detected in the lungs for ≥180 days p.i. by RT-PCR; however, neither HMPV RNA nor infectious virus can be detected in serum, spleen, kidneys, heart, trachea, or brain tissue. To some extent, the kinetics of early HMPV infection are similar to what has been shown for RSV infection in BALB/c mice. For example, intranasal administration of a high-titered RSV inoculum results in replication in the lungs and nasal passages, with peak lung virus titers occurring between days 5 and 7 p.i. (11, 40). RSV titers up to 106.9 PFU/g of lung tissue can be attained in older mice (11); however, RSV titers recovered in 6- to 12-week-old mice generally range between 104.5 to 106 PFU/g of lung tissue, and the virus is cleared between days 7 and 10 p.i. (11). HMPV lung replication differs from that of RSV in that HMPV has a biphasic replication pattern. BALB/c mice intranasally inoculated with 106 PFU of HMPV have an initial peak virus titer at day 7 p.i. (108 PFU/g of lung tissue) which subsequently declines to 105.8 PFU/g of lung tissue at day 10 p.i. but is followed by a second peak virus titer at day 14 p.i. (107 PFU/g of lung tissue). Since neither infectious HMPV nor HMPV RNA was detected in the serum or the other tissues examined in this study, HMPV, like RSV, replicates in the lungs but is not cleared with the same kinetics as RSV, i.e., between days 7 and 10 p.i. In addition, HMPV persists in the lungs of infected mice up to day 60 p.i., exhibiting titers of 104 PFU/g of lung tissue at day 28 p.i. and 101 PFU/g of lung tissue at day 60 p.i.
In a recent study, four small-animal models (BALB/c mice, hamsters, cotton rats, and ferrets) were tested for permissiveness to HMPV infection; however, only hamsters and ferrets supported high-titer replication (4 to 5 log10 PFU/g of lung tissue) of HMPV/NL/17/00, with neutralizing antibody titers ranging from 3 to 8 log2 per animal, and no animal model examined exhibited signs of illness (21). In contrast, we show that BALB/c mice are readily infected with HMPV/CAN98-75; infection is associated with early lung histopathology, weight loss, and the development of a neutralizing antibody response; and infectious virus persists in the lung up to day 60 p.i. These results suggest that HMPV strain differences may affect host replication permissibility and disease pathogenesis. An examination of HMPV permissiveness in rhesus macaques and African green monkeys has shown that African green monkeys are most permissive for HMPV/NL/17/00, HMPV/CAN98-75, and HMPV/CAN97-83, although infections in nonhuman primates do not appear to mimic human disease signs (21, 35). Interestingly, an examination of the level of replication and immunogenicity of HMPV/CAN98-75 and HMPV/CAN97-83 in African green monkeys revealed that HMPV replicated to high titers in the lower respiratory tract of infected African green monkeys, the infected monkeys developed high titers of serum-neutralizing antibodies, and the two HMPV genetic lineages were found to be highly antigenically related, i.e., 64 to 99%, in heterologous challenge studies (21, 35).
In this study, HMPV infection in BALB/c mice was associated with extensive histological changes in the lungs and airway remodeling, features that are also associated with RSV infection in BALB/c mice (10, 27, 40). In addition, HMPV infection was associated with some pathophysiologic parameters associated with exacerbated lung disease, i.e., airway remodeling and mucus production, and, like RSV, induced increased CCSP expression (51). These results suggest that HMPV may infect the small airways and interact with Clara cells in the bronchioles. Consistent with this possibility, HMPV infection was associated with airway epithelial injury and remodeling that was characterized by an altered CCSP staining pattern, increased myofibroblast thickening adjacent to the airway epithelium, and staining of cellular debris in the airways.
Several lines of evidence suggest that RSV infection may result in latency or persistence, including detection of RSV proteins and genomic RNA in the lungs of RSV-infected guinea pigs for ≥60 days p.i. (18), in murine macrophage-like cell lines and macrophage cultures (13), and in B lymphocytes following infection with bovine RSV (44). Recent RT-PCR studies examining RSV persistence in BALB/c mice have shown the presence of genomic RNA and mRNA encoding the G and F glycoproteins, the matrix protein, and NS1 and NS2 proteins in lungs of BALB/c mice up to 100 days p.i. despite the presence of RSV-specific cytotoxic T lymphocytes and RSV-specific serum IgG (34). Interestingly, low levels of infectious RSV were recovered from the lungs of some mice depleted of T cells 150 days postinfection, suggesting that RSV persists by means of low-grade replication in the lungs. These findings are similar to what is reported in this study for HMPV infection of BALB/c mice; i.e., genomic RNA detected ≥180 days p.i. in the lungs of infected mice, HMPV replication in the presence of serum and neutralizing antibodies, and antibody depletion of T cells or NK cells at late time points postinfection, i.e., day 28 or day 60 p.i., result in increased HMPV titers in the lung. These studies suggest that BALB/c mice are a useful small-animal model to study the pathophysiology associated with acute or persistent HMPV.
The results from this study are similar to those of a recent study showing that RSV may persist in the lungs of BALB/c mice despite an established immune response (34). Viruses that persist despite an ostensibly intact immune response have been shown to use a variety of strategies including immune evasion, mimicking host proteins, and replicating at immune-privileged sites (1). Nothing is known about the mechanisms that HMPV may use to persist; however, a recent study has shown that RSV may resist host antiviral mechanisms by modulating the type I interferon pathway by mechanisms associated with the expression of nonstructural NS1 and NS2 proteins (36), through G glycoprotein CX3C chemokine mimicry (43), and by displaying a conformationally altered mature envelope protein that is less susceptible to an anti-F glycoprotein neutralizing antibody response (29). In addition, G glycoprotein expression is associated with abherrent cytokine and altered chemokine mRNA expression in pulmonary leukocytes responding to infection as well as altered pulmonary leukocyte trafficking (42). In this study, depletion of T cells or NK cells was associated with significantly increased HMPV titers in the lungs of depleted mice, suggesting that these cell types contribute to HMPV immune surveillance and control. One possible explanation for the ability of HMPV to replicate in the presence of a neutralizing antibody response may be antigenic variation of neutralizing epitopes. It is known that the RSV G glycoprotein exhibits extensive antigenic and genetic diversity, and variations exist not only in gene coding sequences but also in the signals that control gene expression (14, 38). It is possible that the known genetic diversity in the HMPV G glycoprotein (30) may contribute to antigenic variation of neutralizing epitopes. It is also possible that HMPV may persist in immune-privileged sites such as neurons or macrophages. Since the HMPV genome does not contain NS1 or NS2 genes (45), it is unlikely that HMPV modulates the type I interferon pathway in a fashion similar to that of RSV (36); however, it is possible that other HMPV proteins may alter the antiviral cytokine or chemokine response.
In summary, the development of a highly reproducible small-animal model for HMPV infection, i.e., the BALB/c mouse model, will likely be beneficial to advance our understanding of the pathophysiology associated with HMPV infection as well as the mechanisms that contribute to immunity and disease pathogenesis.
REFERENCES
- 1.Ahmed, R., L. A. Morrison, and D. M. Knipe. 1996. Persistence of viruses, p. 219-249. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 2.Alvarez, R., L. P. Jones, B. S. Seal, D. R. Kapczynski, and R. A. Tripp. 2004. Serological cross-reactivity of members of the Metapneumovirus genus. Virus Res. 105:67-73. [DOI] [PubMed] [Google Scholar]
- 3.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshe, R. B., L. S. Richardson, W. T. London, D. L. Sly, J. H. Lorfeld, E. Camargo, D. A. Prevar, and R. M. Chanock. 1977. Experimental respiratory syncytial virus infection of four species of primates. J. Med. Virol. 1:157-162. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Côté, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. [DOI] [PubMed] [Google Scholar]
- 6.Boivin, G., G. De Serres, S. Côté, R. Gilca, Y. Abed, L. Rochette, M. G. Bergeron, and P. Dery. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanock, R. M., R. H. Parrott, M. Connors, P. L. Collins, and B. R. Murphy. 1992. Serious respiratory tract disease caused by respiratory syncytial virus: prospects for improved therapy and effective immunization. Pediatrics 90:137-143. [PubMed] [Google Scholar]
- 8.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 9.Fixler, D. E. 1996. Respiratory syncytial virus infection in children with congenital heart disease: a review. Pediatr. Cardiol. 17:163-168. [DOI] [PubMed] [Google Scholar]
- 10.Graham, B. S., L. A. Bunton, P. F. Wright, and D. T. Karzon. 1991. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J. Clin. Investig. 88:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, B. S., M. D. Perkins, P. F. Wright, and D. T. Karzon. 1988. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 26:153-162. [DOI] [PubMed] [Google Scholar]
- 12.Greensill, J., P. S. McNamara, W. Dove, B. Flanagan, R. L. Smyth, and C. A. Hart. 2003. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg. Infect. Dis. 9:372-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrero-Plata, A., E. Ortega, V. Ortiz-Navarrete, and B. Gomez. 2004. Antigen presentation by a macrophage-like cell line persistently infected with respiratory syncytial virus. Virus Res. 99:95-100. [DOI] [PubMed] [Google Scholar]
- 14.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harkema, J. R., and J. A. Hotchkiss. 1992. In vivo effects of endotoxin on intraepithelial mucosubstances in rat pulmonary airways. Quantitative histochemistry. Am. J. Pathol. 141:307-317. [PMC free article] [PubMed] [Google Scholar]
- 16.Harrod, K. S., R. J. Jaramillo, C. L. Rosenberger, S. Z. Wang, J. A. Berger, J. D. McDonald, and M. D. Reed. 2003. Increased susceptibility to RSV infection by exposure to inhaled diesel engine emissions. Am. J. Respir. Cell Mol. Biol. 28:451-463. [DOI] [PubMed] [Google Scholar]
- 17.Hayashida, S., K. S. Harrod, and J. A. Whitsett. 2000. Regulation and function of CCSP during pulmonary Pseudomonas aeruginosa infection in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:L452-L459. [DOI] [PubMed] [Google Scholar]
- 18.Hegele, R. G., S. Hayashi, A. M. Bramley, and J. C. Hogg. 1994. Persistence of respiratory syncytial virus genome and protein after acute bronchiolitis in guinea pigs. Chest 105:1848-1854. [DOI] [PubMed] [Google Scholar]
- 19.Hegele, R. G., P. J. Robinson, S. Gonzalez, and J. C. Hogg. 1993. Production of acute bronchiolitis in guinea-pigs by human respiratory syncytial virus. Eur. Respir. J. 6:1324-1331. [PubMed] [Google Scholar]
- 20.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacPhail, M., J. H. Schickli, R. S. Tang, J. Kaur, C. Robinson, R. A. Fouchier, A. D. Osterhaus, R. R. Spaete, and A. A. Haller. 2004. Identification of small-animal and primate models for evaluation of vaccine candidates for human metapneumovirus (hMPV) and implications for hMPV vaccine design. J. Gen. Virol. 85:1655-1663. [DOI] [PubMed] [Google Scholar]
- 22.Madhi, S. A., H. Ludewick, Y. Abed, K. P. Klugman, and G. Boivin. 2003. Human metapneumovirus-associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (HIV-1)-infected and HIV-1-uninfected African infants. Clin. Infect. Dis. 37:1705-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maggi, F., M. Pifferi, M. Vatteroni, C. Fornai, E. Tempestini, S. Anzilotti, L. Lanini, E. Andreoli, V. Ragazzo, M. Pistello, S. Specter, and M. Bendinelli. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIntosh, K., and J. M. Fishaut. 1980. Immunopathologic mechanisms in lower respiratory tract disease of infants due to respiratory syncytial virus. Prog. Med. Virol. 26:94-118. [PubMed] [Google Scholar]
- 25.Mufson, M. A., R. B. Belshe, C. Orvell, and E. Norrby. 1987. Subgroup characteristics of respiratory syncytial virus strains recovered from children with two consecutive infections. J. Clin. Microbiol. 25:1535-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen, M. D., D. J. Siebert, I. M. Mackay, T. P. Sloots, and S. J. Withers. 2002. Evidence of human metapneumovirus in Australian children. Med. J. Aust. 176:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Openshaw, P. J. 1995. Immunity and immunopathology to respiratory syncytial virus. The mouse model. Am. J. Respir. Crit. Care Med. 152:S59-S62. [DOI] [PubMed] [Google Scholar]
- 28.Osterhaus, A., and R. Fouchier. 2003. Human metapneumovirus in the community. Lancet 361:890-891. [DOI] [PubMed] [Google Scholar]
- 29.Parren, P. W., P. Poignard, H. J. Ditzel, R. A. Williamson, and D. R. Burton. 2000. Antibodies in human infectious disease. Immunol. Res. 21:265-278. [DOI] [PubMed] [Google Scholar]
- 30.Peret, T. C., Y. Abed, L. J. Anderson, D. D. Erdman, and G. Boivin. 2004. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J. Gen. Virol. 85:679-686. [DOI] [PubMed] [Google Scholar]
- 31.Prince, G. A., A. B. Jenson, R. L. Horswood, E. Camargo, and R. M. Chanock. 1978. The pathogenesis of respiratory syncytial virus infection in cotton rats. Am. J. Pathol. 93:771-791. [PMC free article] [PubMed] [Google Scholar]
- 32.Prince, G. A., and D. D. Porter. 1976. The pathogenesis of respiratory syncytial virus infection in infant ferrets. Am. J. Pathol. 82:339-352. [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50 percent end-points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 34.Schwarze, J., D. R. O'Donnell, A. Rohwedder, and P. J. Openshaw. 2004. Latency and persistence of respiratory syncytial virus despite T cell immunity. Am. J. Respir. Crit. Care Med. 169:801-805. [DOI] [PubMed] [Google Scholar]
- 35.Skiadopoulos, M. H., S. Biacchesi, U. J. Buchholz, J. M. Riggs, S. R. Surman, E. Amaro-Carambot, J. M. McAuliffe, W. R. Elkins, M. St. Claire, P. L. Collins, and B. R. Murphy. 2004. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J. Virol. 78:6927-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and gamma interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockton, J., I. Stephenson, D. Fleming, and M. Zambon. 2002. Human metapneumovirus as a cause of community-acquired respiratory illness. Emerg. Infect. Dis. 8:897-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullender, W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullender, W. M., M. A. Mufson, G. A. Prince, L. J. Anderson, and G. W. Wertz. 1998. Antigenic and genetic diversity among the attachment proteins of group A respiratory syncytial viruses that have caused repeat infections in children. J. Infect. Dis. 178:925-932. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, G., E. J. Stott, M. Hughes, and A. P. Collins. 1984. Respiratory syncytial virus infection in mice. Infect. Immun. 43:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tripp, R. A. 2004. The brume surrounding respiratory syncytial virus persistence. Am. J. Respir. Crit. Care Med. 169:778-779. [DOI] [PubMed] [Google Scholar]
- 42.Tripp, R. A., L. Jones, and L. J. Anderson. 2000. Respiratory syncytial virus G and/or SH glycoproteins modify CC and CXC chemokine mRNA expression in the BALB/c mouse. J. Virol. 74:6227-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripp, R. A., L. P. Jones, L. M. Haynes, H. Zheng, P. M. Murphy, and L. J. Anderson. 2001. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2:732-738. [DOI] [PubMed] [Google Scholar]
- 44.Valarcher, J. F., H. Bourhy, A. Lavenu, N. Bourges-Abella, M. Roth, O. Andreoletti, P. Ave, and F. Schelcher. 2001. Persistent infection of B lymphocytes by bovine respiratory syncytial virus. Virology 291:55-67. [DOI] [PubMed] [Google Scholar]
- 45.van den Hoogen, B. G., T. M. Bestebroer, A. D. Osterhaus, and R. A. Fouchier. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119-132. [DOI] [PubMed] [Google Scholar]
- 46.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Hoogen, B. G., D. M. Osterhaus, and R. A. Fouchier. 2004. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr. Infect. Dis. J. 23:S25-S32. [DOI] [PubMed] [Google Scholar]
- 48.van den Hoogen, B. G., G. J. van Doornum, J. C. Fockens, J. J. Cornelissen, W. E. Beyer, R. de Groot, A. D. Osterhaus, and R. A. Fouchier. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571-1577. [DOI] [PubMed] [Google Scholar]
- 49.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 41:3043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vicente, D., G. Cilla, M. Montes, and E. Perez-Trallero. 2003. Human metapneumovirus and community-acquired respiratory illness in children. Emerg. Infect. Dis. 9:602-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, S. Z., C. L. Rosenberger, Y. X. Bao, J. M. Stark, and K. S. Harrod. 2003. Clara cell secretory protein modulates lung inflammatory and immune responses to respiratory syncytial virus infection. J. Immunol. 171:1051-1060. [DOI] [PubMed] [Google Scholar]