Abstract
The structure of the nucleocapsid protein of bunyaviruses has not been defined. Earlier we have shown that Tula hantavirus N protein oligomerization is dependent on the C-terminal domains. Of them, the helix-loop-helix motif was found to be an essential structure. Computer modeling predicted that oligomerization occurs via helix protrusions, and the shared hydrophobic space formed by amino acids residues 380-IILLF-384 in the first helix and 413-LI-414 in the second helix is responsible for stabilizing the interaction. The model was validated by two approaches. First, analysis of the oligomerization capacity of the N protein mutants performed with the mammalian two-hybrid system showed that both preservation of the helix structure and formation of the shared hydrophobic space are crucial for the interaction. Second, oligomerization was shown to be a prerequisite for the granular pattern of transiently expressed N protein in transfected cells. N protein trimerization was supported by three-dimensional reconstruction of the N protein by electron microscopy after negative staining. Finally, we discuss how N protein trimerization could occur.
Hantaviruses (genus Hantavirus, family Bunyaviridae) are known as the most widely distributed zoonotic viruses carried by rodents. They are enveloped viruses that contain a tripartite, negative-stranded RNA genome encoding an RNA-dependent RNA polymerase (L), two integral membrane glycoproteins (G1 and G2), and a nucleocapsid (N) protein. Hantaviruses are prime examples of emerging viruses and important human pathogens (5). Many rodent species carry hantaviruses which can be transmitted to humans by inhalation of aerosolized excreta.
The N protein is the most abundant viral component in both virions and infected cells (5) and the major antigen in early serological response in humans (32). It has multiple functions in the viral replication cycle, e.g., during RNA synthesis and virus assembly. Although the three-dimensional structure of the N protein has not been solved yet, several functionally important regions have been defined, e.g., those involved in oligomerization and RNA binding. Most importantly, the N protein (amino acids 175 to 217 in Hantaan virus) (33) associates with viral RNA that is a prerequisite for ribonucleoprotein (RNP) formation (30, 31). Hantavirus N protein has also been shown to associate with membranes via electrostatic interactions, and the C-terminal 141 amino acids were found to be responsible for the Golgi localization of Black Creek Canal hantavirus N protein in the perinuclear area (25).
Recent findings suggest that, in infected cells, the N protein interacts with the L protein (13) and with the cytoplasmic tail of G1 glycoprotein (V. Koistinen, personal communication). These interactions are crucial for viral RNA transcription and replication and for virus assembly, respectively. The N protein is also able to serve some “ambassador” functions, interacting with cellular proteins. It interacts with actin filaments, which could transport newly synthesized viral RNPs to the plasma membrane (26). Recently, Daxx and SUMO-1 pathway components were found to bind to the N protein, but the significance of these interactions remains to be determined (11, 15, 17, 19).
Our study focused on mapping of the Tula hantavirus N protein domains that participate in homotypic interaction. Previously published data have shown that the N-terminal and C-terminal regions are involved in the hantavirus N protein interaction (1, 9, 10, 34). The presence of coiled-coil motifs has been demonstrated in the N-terminal region (1, 2). Our previous data suggested that the C-terminal region is more important for the interaction, at least as seen in the mammalian two-hybrid system (10). While the deletion of 43 N-terminal amino acids from the N protein in both the DNA-binding and activation domains reduced the interaction slightly, the deletion of the 37 C-terminal residues even from one partner abolished the interaction completely. These data prompted us to focus our studies on the C-terminal region.
We have now combined secondary-structure predictions and computer modeling with mammalian two-hybrid and immunofluorescent assays to define structural requirements for oligomerization of the C-terminal region of Tula virus N protein. We describe the N protein-N protein interaction involving the helix-loop-helix structure and pinpoint amino acids that are crucial for the interaction. We present a model of how N protein trimerization could occur.
MATERIALS AND METHODS
Secondary-structure predictions.
The secondary-structure predictions for the N proteins of Tula virus, Sin Nombre virus, and Hantaan virus was done with JPRED (4) and Predict Protein (27).
Computer modeling of the C-terminal interaction region.
The analysis of the probable domain architecture was done with SMART (16). Proteins having homologous sequences and partially similar secondary structures within the C-terminal region of the N protein were selected. The backbones of similar regions were used to make different configurations for the backbone of the C-terminal region of the N protein. The side chains were modeled on the backbone with the MaxSprout program (8). The C-terminal model structure was constructed with the molecular modeling system Insight II, version 2000 (Accelrys Inc.), on a Silicon Graphics workstation. The docking of the two C-terminal helices was done in Insight II, considering the different properties of the side chains on the helices and loop. The Builder module of Insight II was used to combine different parts of the C-terminal structure. For the loop region, different models were used for the hit-and-trial method together with the experimental data for finding proper docking for the C-terminal helices. The flexibility of the G389 conformations was done with the torsion module in Insight II to assign different psi and phi angles around the c-alpha atom of glycine. A change in glycine conformations produces major changes in the locations of other structures on the peptide chain. Those changes were also taken into consideration and used for docking of the C-terminal helices.
Mammalian two-hybrid system.
HeLa cells were cultivated in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin on 24-well culture plates to 70% confluence and transfected with 0.5 μg of pM1-TULVN and pVP16-TULVN, 0.5 μg of reporter DNA pG5luc (firefly luciferase), and 0.01 μg of control DNA pRL-SV40 (Renilla luciferase) (Promega). Plasmids encoding the full-length N protein of Tula virus strain Tula/Moravia/Ma5302V and C-terminally truncated constructs were created by PCR from S segment DNA. Transfections were performed in triplicate with 6 μl of FuGene6 reagent for each transfection according to the manufacturer's instructions (Roche Diagnostics Corporation). After 24 h luciferase activities were determined with the dual-luciferase reporter assay system (Promega). Light intensities of samples were measured with a DCR-1 luminometer (Digene Diagnostics Inc.). Due to inherent variations, Renilla luciferase values were used to normalize firefly luciferase values by the formula: normalized value of experiment A = [(Renilla luciferase value from N-N interaction/Renilla luciferase value of experiment A) × (firefly luciferase value of experiment A)]. The formula for percent interaction = (normalized value of experiment A/normalized value of N-N interaction) × 100.
Site-directed mutagenesis.
Point mutations were created in the pM1-TULVN and pVP16-TULVN plasmids with a site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All the plasmids were characterized by restriction analysis and sequenced with an ABI Prism Dye Terminator sequencing kit (Perkin-Elmer).
Immunofluorescence microscopy.
COS7 cells were cultivated on coverslips in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin in 24-well plates. Cells were transfected with 0.5 μg of pcDNA3-N constructs with different mutations in the N protein with 2 μl of FuGene6 reagent according to the manufacturer's instructions (Roche Diagnostics Corporation). Cells were fixed 24 h later with 3.2% paraformaldehyde in phosphate-buffered saline, pH 7.4 (PBS) for 15 min and permeabilized with 0.1% Triton X-100 in PBS (30 min at room temperature). Cells were stained with polyclonal N protein antibody (1:400 in PBS, 1 h at room temperature) and then with the anti-rabbit immunoglobulin G-fluorescein isothiocyanate conjugate secondary antibody (1:100 in PBS, 1 h at room temperature). The samples were examined with a Zeiss Axioplan microscope with an ×63 oil immersion lens.
RESULTS
Secondary-structure predictions for the N protein.
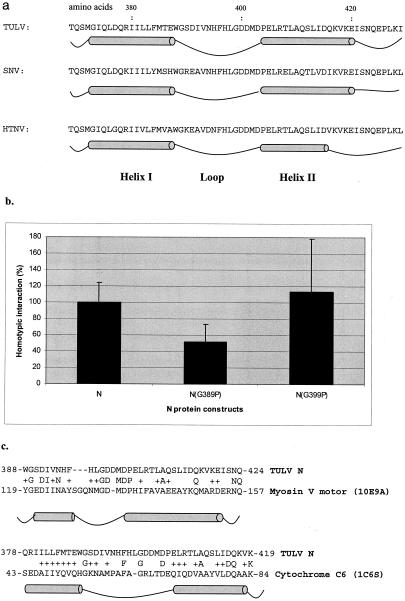
Secondary-structure predictions for the hantavirus N protein have depicted α-helical structures near the N terminus that can form a coiled-coil structure upon oligomerization (1, 2). Since the hantavirus N protein does not have a homologue of known resolved structure, we first attempted secondary-structure predictions for the C-terminal region. The JPRED and Predict Protein servers were used in the predictions. The C-terminal region was predicted to fold into a helix-loop-helix motif (Fig. 1a). For Tula virus N protein, helix I seemed to be formed by residues 373 to 387 and helix II by residues 404 to 421, separated by the loop formed by residues 388 to 403. The same overall organization was found in the N protein of the three major groups of hantaviruses (24) that are represented in Fig. 1a by Tula virus, Sin Nombre virus, and Hantaan virus. It was assumed that a similar conformation reflects a similar mode of interaction for the N proteins of different hantaviruses.
FIG. 1.
(a) Secondary-structure predictions for the C-terminal region of the N proteins of Tula, Sin Nombre, and Hantaan viruses, which represent the three major groups of hantaviruses. (b) Mammalian two-hybrid results for point mutants G389P and G399P. (c) Sequence alignment of regions corresponding to the helix-loop-helix structure in Tula virus N protein and nonhantaviral proteins. The middle line shows identical amino acid residues, and + indicates the positions of homologous amino acids residues in the two proteins.
To study the C-terminal interaction regions in greater detail, we took advantage of the mammalian two-hybrid system that allows direct evaluation of the interaction capacity of the N protein mutants. First, helix II of the Tula virus N protein, which was predicted with high probability to occur between amino acids 404 and 421, was studied. When the last 25 amino acids were deleted from both interacting partners (constructs DBD-N1-404, where DBD stands for DNA-binding domain, and AD-N1-404, where AD stands for activation domain), the interaction was abolished totally (data not shown). This indicated that the last helices are crucial for the interaction.
Secondary-structure predictions gave conflicting results for a region between amino acids 388 and 394 because both servers gave low probabilities for a helix and a loop. The glycine residue G389 was hypothesized to reside in the loop, providing flexibility for the region, together with another glycine, G399. N protein mutants carrying two single-point mutations, G389P and G399P, were used to evaluate this hypothesis. The change of glycine to proline was thought to introduce inflexibility to the loop and thus hamper the interaction. It was observed in the mammalian two-hybrid assay that while the G399P mutation did not have any effect, the G389P mutation reduced the interaction by half (Fig. 1b). This could be interpreted to mean that G389 is also located in the loop because, if G389 was in the helix, the introduction of a proline should have disrupted the interaction completely. These data suggested that helix I does not extend further from the tryptophan at position 388 and that G389 contributes to the flexibility of the loop, while G399 does not.
Computer modeling and formation of a shared hydrophobic space between helices.
The SMART server (16) was used to search databases for proteins having both a homologous primary sequence and secondary structure partially similar to that of the C-terminal region of the N protein. On manual rechecking of the secondary structures, close similarity was found between residues 388 to 424 in the N protein and 119 to 157 in myosin V motor protein (accession no. 1OE9A) (amino acid similarity, 57%) (Fig. 1c), residues 378 to 419 in the N protein and 43 to 84 in Cyanobacterium synechococcus cytochrome C6 (accession no. 1C6S) (amino acid similarity, 63%) (Fig. 1c), and residues 378 to 428 in the N protein and 259 to 311 in Staphylococcus aureus tyrosyl-tRNA synthetase (accession no. AP003363) (amino acid similarity, 45%). The helix-loop-helix motif of the N protein was constructed by combining structural features of these proteins. The side chains of the N protein were added and modeled on the backbone with the MaxSprout program (8).
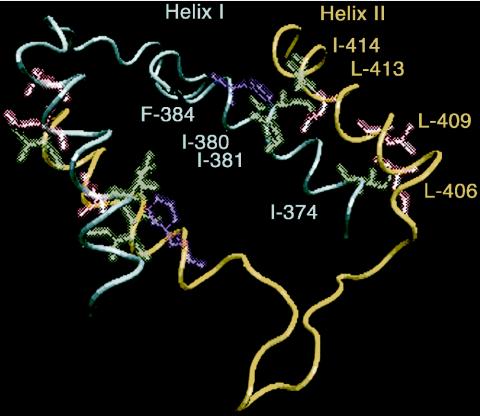
Sequence analysis revealed the stretch of highly hydrophobic amino acids 380IILLF384 in helix I (amino acids 373 to 387). The computer model showed that isoleucines 380 and 381 and also phenylalanine 384 are located on the same side of the helix in a way that the aromatic ring of F384 is bent towards the aliphatic chains of I380 and I381, forming a highly hydrophobic region. The localization of this region is optimal to attract helix II of the neighboring molecule (Fig. 2). The “docking” of the two helices suggested that the leucine at position 413 and the isoleucine at position 414 of helix II could be placed in the hydrophobic space formed by I380, I381, and F384 of helix I. In this conformation, L413 is placed between the isoleucines at positions 380 and 381, while two leucines of helix II, L406 and L409, form hydrophobic contacts with I374 of helix I. This conformation seems to give parallel docking for the two helices, in which the side chains of the hydrophobic amino acids form a shared hydrophobic space, providing stability to the N protein-N protein interaction.
FIG. 2.
Computer modeling of the C-terminal interaction regions of two N protein molecules (in blue and in yellow). Helix I and helix II of neighboring molecules interact with each other via hydrophobic contacts between amino acid side chains (depicted for one interacting interface).
The computer modeling data were first tested with the mammalian two-hybrid assay. The amino acid residues of the hydrophobic stretch (IILL) in helix I were subjected to standard alanine scanning, and the capacity of the mutant N protein for the homotypic interaction was analyzed (Fig. 3a). When I380 was replaced by alanine, resulting in the sequence AILL, the N protein-N protein interaction was reduced to approximately 60% of that with the nonmutated protein. Changing I381 (IALL) did not have any effect. However, when both isoleucines (I380 and I381) were replaced (AALL), the interaction was abolished totally and was not restored by introducing isoleucines instead of leucines at positions 382 and 383 (AAII). In contrast, the replacement of leucines L382 and L383 with alanines (IIAA) did not inhibit the N protein interaction at all. Finally, mutants with four leucines (LLLL) maintained the interaction as well as the nonmutated protein. Taken together, these data indicate that amino acids with long hydrophobic side chains at positions 380 and 381 are crucial for the interaction. The residue F384 was also found to be crucial for the interaction.
FIG. 3.
Study of the interaction capacity of N protein mutants in the mammalian two-hybrid system. The indicated mutations were introduced to both DNA-binding domain and activation domain constructs. (A and B) Two-hybrid results in which hydrophobic residues (underlined in the sequence above) were mutated in helix I (A) and in helix II (B). (C) Two-hybrid results of mutated residues (bold in the sequence above) participating in the putative cation-π interaction.
Next, the residues forming the hydrophobic core of helix II (L413 and I414) were changed to glutamic acids (the introduced charge was supposed to disrupt the hydrophobic space). The mutation L413E reduced the interaction to approximately 60% of that of the nonmutated protein, while the mutation I414E did not have any effect. The double mutation L413E/I414E nearly abolished the interaction, confirming the importance of the hydrophobic contacts between the helices (Fig. 3b). The mammalian two-hybrid data support the computer model showing that the hydrophobic residues of helices I and II can form a shared hydrophobic space.
Searching for cation-π interactions.
While it seemed logical to assume that hydrophobic interactions could provide stability to protein interaction and folding locally, electrostatic interactions could be responsible for initiating the interaction, i.e., bringing the interaction regions into close proximity. This task could be performed, e.g., by the cation-π interactions between the aromatic side chains of tryptophan or phenylalanine with neighboring positively charged arginines and/or lysines (7). Computer modeling suggested tryptophan-388 and phenylalanine-396 as potential residues that might form cation-π interactions. This hypothesis was first tested with the mutations W388A and F396A. It was observed that the second mutation did not affect the interaction at all, but mutation W388A abolished the N protein-N protein interaction (Fig. 3c). In fact, a mutation in either of the partners (DNA-binding domain or activation domain) abolished the interaction.
Next, we searched for a potential partner for W388. Since R367, R368, R379, and R407 were the most probable candidates, these residues were mutated to alanine, and the interaction capacity of the mutant N proteins was tested. Surprisingly, none of these N protein mutants showed decreased interaction (Fig. 3c), suggesting either that these residues are not involved at all or that the cation-π interaction between them and W388 or F396 is not crucial for the N protein interactions observed in our two-hybrid system.
Localization of the N protein mutants in transfected cells.
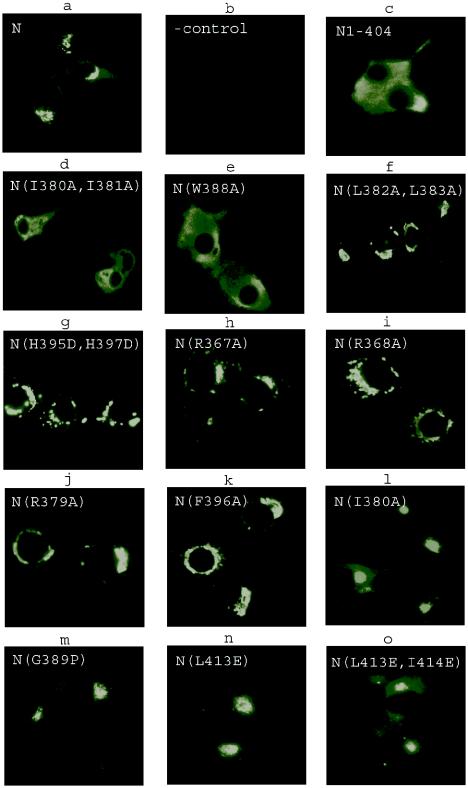
In infected cells, the hantavirus N protein has been shown to localize to the perinuclear region in the cytoplasm that may be the site of viral RNA replication (13, 25). To study the intracellular localization of the N protein mutants, the constructs were expressed in COS7 cells and the N protein was visualized by immunofluorescence microscopy (Fig. 4). Intact N protein localized to distinct structures in the perinuclear region (Fig. 4a), similar to those seen in virus-infected cells (not shown). The mutant with the 25 C-terminal residues truncated, N1-404, was dispersed throughout the cytoplasm (Fig. 4c). Similarly, mutants with longer truncations (N1-379, N1-392, and N1-398) localized diffusely (data not shown). These data showed that the C-terminal region of the hantavirus N proteins is crucial for perinuclear targeting.
FIG. 4.
Localization of transiently expressed N protein mutants in COS7 cells. Immunofluorescence was used to visualize N proteins in cells transfected with a construct expressing full-length N protein (a) or mock transfected (b). Mutant N proteins localizing diffusely (c to e), mutant N proteins localizing in the perinuclear region (f to k) and mutant N proteins localizing both diffusely and in perinuclear region (l to o) are shown.
Furthermore, since the full-length N protein interacted in the mammalian two-hybrid assay and truncated N proteins did not, there was a good correlation between the capacity of mutants to interact in the mammalian two-hybrid system and their perinuclear localization. This pattern held up when the N protein point mutants were studied. The mutants that were unable to interact in the two-hybrid system (I380A/I381A and W388A) localized diffusely (Fig. 4d and e), while mutants capable of homotypic interaction localized to the perinuclear region (Fig. 4f to k). Four mutants (I380A, G389P, L413E and L413E/I414E) showed a mixed pattern and localized both diffusely and in the perinuclear region (Fig. 4l to o). These mutants showed medium values in the two-hybrid assay (Fig. 3a and b), confirming that undisturbed interaction is required for perinuclear localization.
Model for the N protein trimer.
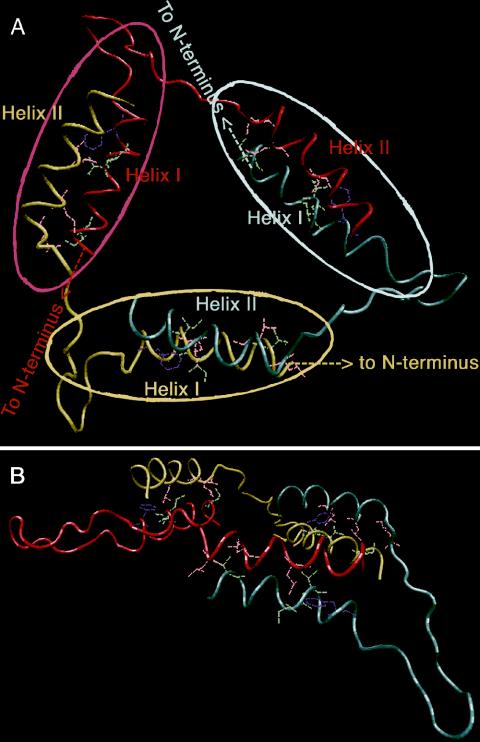
Taken together, in silico and in vitro data led to a model for trimerization of the N protein's C-terminal region. The model shows that when the C termini of the three monomers are brought in close proximity to each other (most probably via interaction of the N-terminal coiled coils; see the Discussion), the steric hindrance in the C-terminal region is relieved when helix II protrudes from the N protein and makes contact with helix I of a neighboring monomer. Particularly, G389 could form a hinge that allows the loop to orient helix II properly. The reduced interaction of mutant G389P in the mammalian two-hybrid system suggests that the proline introduces stiffness into the loop and thereby inhibits the efficient protrusion of helix II from the N protein core that is needed for the interaction. The two helices are placed in such a way that a shared hydrophobic space is formed. The basic feature of the model is that the trimer structure is stabilized by helix exchange in a cyclic manner from one monomer to another (Fig. 5A, top view). When viewed from the side, the helices form a planar structure (Fig. 5B).
FIG. 5.
Computer modeling of the C-terminal region of the N protein trimer. The C-terminal helix-loop-helix structure of three N protein molecules (yellow, red, and blue) is represented. The core of each monomer is illustrated with colored ovals. The arrow points towards the N terminus of each monomer. The helix-loop-helix structure is shown as a top view (A) and as a side view (B).
DISCUSSION
Mechanisms of hantavirus N protein oligomerization.
Several studies have investigated the oligomerization capacity of viral nucleocapsid proteins. Nucleocapsid proteins of nonsegmented, negative-strand RNA viruses (such as viruses belonging to the Paramyxoviridae and Rhabdoviridae families) seem to have a similar structural organization: an N-terminal globular domain is responsible for self-assembly and RNA binding, while an exposed C-terminal domain interacts with other viral proteins such as the polymerase, P protein, or M protein (3, 12, 21, 22). Structural studies have been hampered by the ability of N proteins to oligomerize, both in self-assembly and probably also association with cellular RNAs. This has been probably the main obstacle to crystallization of the N protein. Our study presents a model for hantavirus N protein trimerization that could facilitate further structural studies.
Previously, we have shown that the hantavirus N protein forms trimers and to a lesser extent dimers in infected cells (9). These trimers were thought to serve as intermediates in viral RNP formation. It was noticed that the N protein molecules interacted noncovalently and that divalent cations enhanced the interaction. In the study that followed (10), we used the mammalian two-hybrid assay, peptide scanning, and alanine scanning techniques to study oligomerization in greater detail. Two regions involved in the N protein-N protein interaction were found. The first is located in the N-terminal part of the molecule, within the first 43 amino acid residues. The second region was found within the C terminus of the molecule; the construct containing amino acid residues 1 to 398 was the longest one that still maintained the interaction with the full-length N protein in the mammalian two-hybrid system. Longer truncations, e.g., removal of amino acids 393 to 398 (VNHFHL), totally abolished the interaction. The construct N(1-398) was not able to interact with the C-terminally truncated construct N(1-404) (10).
These data, together with the data presented in this paper, support a model, in which the interaction between the C-terminal regions of two N protein molecules is based on the helix I-loop-helix II structure that is formed by residues 373 to 387, 388 to 403, and 404 to 421, respectively. The important feature of the model is that the interacting helices I and II belong to two different N protein molecules. The very C-terminal residues of the N protein, 422 to 429, apparently do not contribute to the interaction (or at least to the activity observed in the mammalian two-hybrid system). Ultimately, this model can only be verified by crystallization of the N protein or the domain in question. A recent publication on the crystal structure of Borna disease virus nucleoprotein provides a cheerful example. Notably, the structural data suggest that the multimer (tetramer) is the “biologically active entity” of Borna disease virus N protein (28).
Computer modeling suggests that the interaction occurs between helices I and II in a way that helix II protrudes from one N protein molecule and interacts with helix I of a neighboring molecule. The hydrophobic residues in both helices form a shared hydrophobic space that stabilizes the interaction. The loop region that includes the above-mentioned amino acid residues, 393-VNHFHL-398, does not seem to be directly involved in establishing the connection between the C-terminal regions. Instead, this region can present a steric hindrance to the approaching N protein monomer and thus stimulate the exchange of helices II. However, we were not able to confirm this hypothesis experimentally by mutating histidines 395 and 397 or phenylalanine 396.
In addition to hydrophobic contacts, the possibility that the tryptophan at position 388 participates in the cation-π interaction was studied. Although no supportive evidence for this type of interaction was obtained, the W388 mutant was shown to be absolutely crucial for the N protein-N protein interaction in the mammalian two-hybrid system. One can conclude either that the residue alone is crucial or that the mutation disrupts the folding of the C-terminal region and destroys the interaction. Interestingly, W119 of Seoul virus N protein was shown to have a similar effect on N protein interaction in the yeast two-hybrid system (34). These data suggest that the tryptophan helps to maintain the overall structure of the N protein.
The inability of the N protein mutants to interact in the mammalian two-hybrid system was nicely reflected in their cellular localization; the interacting mutants localized to the perinuclear region, while noninteracting mutants were uniformly distributed throughout the cytoplasm (Fig. 4). The mutants showing intermediate values in the interaction assay localized both diffusely and perinuclearly. In the case of the L413E/I414E, which showed a more punctate than diffuse staining pattern (Fig. 4o), it seems that, in this case, the relatively low interaction value (15%) is still sufficient to allow N proteins to accumulate in the perinuclear region. Perinuclear localization of hantavirus N protein has been reported for Black Creek Canal hantavirus, and it was shown that the last 141 residues are sufficient for targeting (25). It has been suggested that perinuclear membranes (Golgi complex) could be the site of viral RNA replication (13, 25). Our result showing that the N protein-N protein interaction is required for perinuclear localization is not contrary to this speculation.
Overall model of the hantavirus N protein trimer.
Since hydrophobic contacts in the C-terminal region may not necessarily initiate N protein trimerization and since the cation-π interaction was not observed, other regions may be involved. Previously published data suggest the N-terminal region as one of the candidates: (i) the N-terminal regions of Tula virus N protein (residues 1 to 43) and Sin Nombre virus N proteins (residues 1 to 39) were shown to participate in the homotypic interaction in the two-hybrid assays (1, 10), (ii) the N-terminal residues of hantavirus N proteins were predicted to form coiled coils (1), and (iii) oligomerization studies of N protein peptides corresponding to residues 3 to 75 assembled into trimeric coiled coils (2). Other regions, e.g., the one that includes amino acid residues 100 to 125, may also be essential for the interaction (34). As soon as the C-terminal regions of three N protein monomers are brought closer to each other, this will allow helix II-helix I contact (as described in this paper), which occurs in a cyclic manner between neighboring N protein molecules, completing trimer formation. The cryoelectron microscopic analysis of recombinant N protein after class averaging confirmed the existence of the trimer (14). From the side view, contacts were seen at both ends of the molecule.
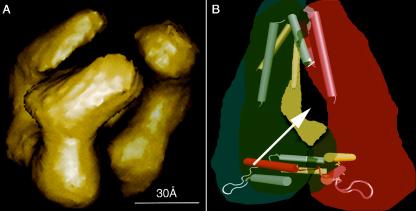
Our first attempt at three-dimensional reconstruction of recombinant N protein by electron microscopy after negative staining shows a trimer with monomers attached to each other at both ends (Fig. 6A). The dimensions of each monomer are approximately 8 by 3 nm. The N protein forms a curved structure that resembles influenza and rabies virus N protein monomers in the reconstructed three-dimensional models (20, 29). As the C-terminal interactions of the hantavirus N protein are more dominant than the N-terminal ones, we assume that the bottom end on the figure is the C-terminal end of the protein. Thus, N protein trimerization seems to depend on contacts made by both the N and C termini of the protein (Fig. 6B).
FIG. 6.
Three-dimensional model of an N protein trimer reconstructed from electron micrographs of a negatively stained sample. The recombinant N protein (9) was applied to carbon film-coated 300- or 400-mesh Au grids (Quantifoil) diluted, i.e., floated in sequence in 1% uranyl acetate and negatively stained. Electron microscopic pictures were collected at ×50,000 at 80 kV with a Jeol 1200EX microscope, and negatives were scanned at 4,000 dots per inch (Nikon LS-8000 ED). For picking projections (≈38,000) of proteins from electron micrographs, the random extensive sampling method was used (6, 14) when distinct particles were difficult or impossible to identify. For making the three-dimensional reconstruction, EMAN was used (18; EMAN web-site, http://ncmi.bcm.tmc.edu/≈s̈tevel/EMAN/doc/index.htm) (A). The N protein trimer model showing three monomers (in red, in yellow, and in blue) and their interacting helices (cylinders): coiled-coil motifs in the N-terminal region (above) and the helix-loop-helix in the C-terminal region (bottom). Helices of the same color belong to the same monomer (B). The arrow depicts a possible orientation of the viral RNA.
Hantavirus RNP formation is dependent not only on interactions between the N protein trimers previously bound to the RNA but also on interactions of the trimers with viral RNA. According to the scheme depicted in Fig. 6B, the monomers, interacting via the N-terminal domains, assemble around the viral RNA molecule in such a way that it traverses the cavity in the middle of the trimer. The RNA molecule is not seen in the electron microscopic reconstruction, but it is obviously able to pass horizontally through the cavity (the diameter of the linear, single-stranded RNA is approximately 10 Å). The middle part of the Hantaan virus N protein has been shown to carry an RNA-binding region (33). Electron microscopic analysis has been used to visualize the N proteins of influenza A virus and rabies virus (20, 29). In these studies, small N protein-RNA rings, in which the N proteins are bound to short RNAs, were examined. These structures were more regular and therefore more suitable for three-dimensional structural reconstruction than full-length viral RNPs. Three-dimensional reconstructions of the influenza A virus and rabies virus mini-RNPs showed, for the N protein, a curved structure similar to that of the hantavirus N protein. This suggests a similar mode of assembly for the hantavirus RNPs. Consequently, the hantavirus N protein monomers may show some flexibility in the presence of RNA, allowing the formation of a helical structure, for which the N trimers could serve as intermediates. The pathway for hantavirus RNP assembly in vivo remains largely unknown.
The hantavirus N protein is not the only one that can form trimers in vitro and in vivo; the N proteins of other bunyaviruses, Bunyamwera virus (genus Orthobunyavirus) (23) and Uukuniemi virus (genus Phlebovirus) (R. Petterson, personal communication), have this capacity as well. Although the assembly of helical nucleocapsids of many negative-stranded RNA viruses occurs without defined intermediate structures, in bunyaviruses trimers can act as assembly intermediates during RNP formation. It seems safe to assume that both the overall folding of the N protein and the mode of its trimerization in other bunyaviruses may resemble, at least in some aspects, those of the hantavirus N protein.
Now that we better understand the structural requirements for the homotypic N protein interaction, a structural basis for designing drugs that interfere with the very basic functions of the hantavirus N protein is set. This can also create a moderate optimism about other members of the Bunyaviridae family, which includes more than 300 different viruses, some of which are important pathogens of humans, livestock, and plants.
Acknowledgments
We thank Olli Vapalahti for helpful discussion and Sarah Butcher for critical review of the manuscript. Leena Kostamovaara and Pirjo Sarjakivi are acknowledged for expert technical assistance.
This work was supported by EU grants QLRT-1999-01119 and QLK2-CT-2002-01358 and the Sigrid Jusélius Foundation, Helsinki.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 3.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 4.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, R. M., M. Bouloy, C. H. Calisher, R. Goldbach, J. T. Moyer, S. T. Nichol, R. Petterson, A. Plyusnin, and C. Schmaljohn. 2000. Bunyaviridae, p. 599-621. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: the classification and nomenclature of viruses. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 6.Engelhardt, P. 2002. Electron tomography—a many splendored thing, p. 16-17. In Proceedings of the 53rd Annual Meeting of the Scandinavian Society for Electron Microscopy, Tampere, Finland.
- 7.Gallivan, J. P., and D. A. Dougherty. 1999. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA 96:9459-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm, L., and C. Sander. 1991. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J. Mol. Biol. 218:183-194. [DOI] [PubMed] [Google Scholar]
- 9.Kaukinen, P., V. Koistinen, O. Vapalahti, A. Vaheri, and A. Plyusnin. 2001. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 82:1845-1853. [DOI] [PubMed] [Google Scholar]
- 10.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J. Virol. 77:10910-10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 92:37-45. [DOI] [PubMed] [Google Scholar]
- 12.Kouznetzoff, A., M. Buckle, and N. Tordo. 1998. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J. Gen. Virol. 79:1005-1013. [DOI] [PubMed] [Google Scholar]
- 13.Kukkonen, S. K. J., A. Vaheri, and A. Plyusnin. Tula hantavirus L protein is a 250 kDa perinuclear membrane-associated protein. J. Gen. Virol. 85:1181-1189. [DOI] [PubMed]
- 14.Kumar, V., J. Heikkonen, P. Engelhardt, and K. Kaski. 2004. Robust filtering and particle picking in micrograph images towards 3D reconstruction of purified proteins with cryo-electron microscopy. J. Struct. Biol. 145:41-51. [DOI] [PubMed] [Google Scholar]
- 15.Lee, B. H., K. Yoshimatsu, A. Maeda, K. Ochiai, M. Morimatsu, K. Araki, M. Ogino, S. Morikawa, and J. Arikawa. 2003. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 98:83-91. [DOI] [PubMed] [Google Scholar]
- 16.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32(database issue):D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, X. D., T. P. Makela, D. Guo, R. Soliymani, V. Koistinen, O. Vapalahti, A. Vaheri, and H. Lankinen. 2002. Hantavirus nucleocapsid protein interacts with the Fas-mediated apoptosis enhancer Daxx. J. Gen. Virol. 83:759-766. [DOI] [PubMed] [Google Scholar]
- 18.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, A., B. H. Lee, K. Yoshimatsu, M. Saijo, I. Kurane, J. Arikawa, and S. Morikawa. 2003. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (HTNV) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology 305:288-297. [DOI] [PubMed] [Google Scholar]
- 20.Martin-Benito, J., E. Area, J. Ortega, O. Llorca, J. M. Valpuesta, J. L. Carrascosa, and J. Ortin. 2001. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers, T. M., A. Pieters, and S. A. Moyer. 1997. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology 229:322-335. [DOI] [PubMed] [Google Scholar]
- 22.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, S. Kusagawa, H. Komada, and Y. Ito. 1999. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 80:2017-2022. [DOI] [PubMed] [Google Scholar]
- 23.Osborne, J. C., and R. M. Elliott. 2000. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946-9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plyusnin, A. 2002. Genetics of hantaviruses: implications to taxonomy. Arch. Virol. 147:665-682. [DOI] [PubMed] [Google Scholar]
- 25.Ravkov, E. V., and R. W. Compans. 2001. Hantavirus nucleocapsid protein is expressed as a membrane-associated protein in the perinuclear region. J. Virol. 75:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravkov, E. V., S. T. Nichol, C. J. Peters, and R. W. Compans. 1998. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J. Virol. 72:2865-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 28.Rudolph, M. G., I. Kraus, A. Dickmanns, M. Eickmann, W. Garten, and R. Ficner. 2003. Crystal structure of the borna disease virus nucleoprotein. Structure (Cambridge) 11:1219-1226. [DOI] [PubMed] [Google Scholar]
- 29.Schoehn, G., F. Iseni, M. Mavrakis, D. Blondel, and R. W. Ruigrok. 2001. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J. Virol. 75:490-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 31.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vapalahti, O., H. Kallio-Kokko, A. Narvanen, I. Julkunen, A. Lundkvist, A. Plyusnin, H. Lehvaslaiho, M. Brummer-Korvenkontio, A. Vaheri, and H. Lankinen. 1995. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 46:293-303. [DOI] [PubMed] [Google Scholar]
- 33.Xu, X., W. Severson, N. Villegas, C. S. Schmaljohn, and C. B. Jonsson. 2002. The RNA binding domain of the Hantaan virus N protein maps to a central, conserved region. J. Virol. 76:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimatsu, K., B. H. Lee, K. Araki, M. Morimatsu, M. Ogino, H. Ebihara, and J. Arikawa. 2003. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 77:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]