Abstract
There is considerable debate about the fundamental mechanisms that underlie and restrict acquisition of human immunodeficiency virus type 1 (HIV-1) infection. In light of recent studies demonstrating the ability of C type lectins to facilitate infection with HIV-1, we explored the potential relationship between polymorphisms in the DC-SIGN promoter and risk for acquisition of HIV-1 according to route of infection. Using samples obtained from 1,611 European-American participants at risk for parenteral (n = 713) or mucosal (n = 898) infection, we identified single-nucleotide polymorphisms in the DC-SIGN promoter using single-strand conformation polymorphism. Individuals at risk for parenterally acquired infection who had −336C were more susceptible to infection than were persons with −336T (odds ratio = 1.87, P = 0.001). This association was not observed in those at risk for mucosally acquired infection. A potential role for DC-SIGN specific to systemic acquisition and dissemination of infection is suggested.
Dendritic cells (DCs) play a critical role in initiating the immune response by virtue of their ability to capture antigens and present them to T cells (1). Although the precise mechanisms of human immunodeficiency virus (HIV) acquisition are not completely understood, migration of DCs from peripheral tissues and blood to regions of draining lymphoid organs may enable their capture of HIV type 1 (HIV-1) and presentation of virus to CD4+ T cells (16). HIV-1 entry into target cells is mediated by interactions of the viral envelope glycoprotein with CD4 and the chemokine receptors CCR5 and CXCR4 on the target cell membrane, although entry may be influenced by other factors as well. DC-SIGN (DC-specific intercellular adhesion molecule 3 [ICAM-3] grabbing nonintegrin), encoded by a member of the CD209 gene family on chromosome 19p13.2-3, has recently been identified as a DC-specific adhesion receptor that mediates the interaction between DCs and resting T cells through high-affinity binding to ICAM-3 (6). DC-SIGN also binds HIV-1 gp120, facilitating capture of HIV and enhancing in vitro infection of T cells in trans. DC-SIGN-associated HIV-1 remains infectious over a prolonged period, perhaps contributing to the infectious potential of the virus during its transport by DCs from the periphery to lymphoid organs (5). This receptor may represent a common thread in many infectious diseases, as it has also been shown to facilitate direct infection of DCs by a range of infectious agents, such as Dengue virus, Ebola virus, human cytomegalovirus, Leishmania pifanoi amastigotes, and Mycobacterium tuberculosis (reviewed in reference 17). Thus it is plausible that polymorphisms in the gene might have a broad range of influence in the pathogenesis of human infectious disease in general, including HIV-1.
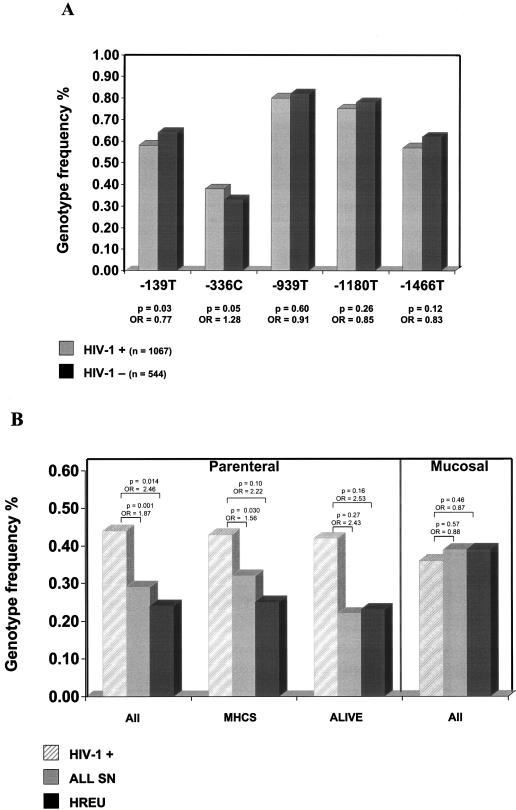
Given the functional activity of DC-SIGN, we proposed that polymorphisms in the coding region and/or promoter region of the associated gene might influence outcomes after HIV-1 exposure. Initial screening for nucleotide variation indicated conservation of DC-SIGN coding sequences. However, several single-nucleotide polymorphisms (SNPs) were identified in the region 2 kb upstream of the ATG start codon, which includes the promoter region (11). The region was amplified in four overlapping segments of 200 to 300 bp, and analysis of the products was performed using single-strand conformation polymorphism (SSCP), as previously described (3). All variants identified by SSCP were confirmed by sequencing analysis. We genotyped samples from 1,611 European-American (EA) participants in five well-characterized HIV-1 cohorts at risk for parenteral (Multicenter Hemophilia Cohort Study [7], AIDS Linked to Intravenous Experience [18], and Hemophilia Growth and Development Study [9]) or mucosal acquisition (Multicenter AIDS Cohort Study [13] and San Francisco City Clinic Cohort [2]) of HIV-1 infection for the DC-SIGN promoter polymorphisms. Six common and 10 rare variants that defined eight haplotypes with frequencies >1% were identified (Table 1). The frequency distributions of the most common DC-SIGN promoter SNPs (−139, −336, −939, −1180, and −1466) were compared among 544 uninfected EA participants and 1,067 HIV-1-infected EA participants. The −139T SNP (odds ratio [OR] = 0.77, P = 0.03) was slightly overrepresented among HIV-1-uninfected participants, whereas the opposite was found for the −336C SNP (Fig. 1A). In addition, 244 uninfected and 520 infected African-Americans within these cohorts were genotyped, but no significant differences in genotype frequencies were observed in this group (data not shown).
TABLE 1.
DC-SIGN promoter region haplotypes
| Haplotypea | fb | Nucleotide at position:
|
||||||
|---|---|---|---|---|---|---|---|---|
| −139 | −336 | −939 | −1089 | −1180 | −1466 | −1530 | ||
| 1 | 0.28 | C | T | T | C | T | C | A |
| 2 | 0.26 | T | T | C | C | A | T | A |
| 3 | 0.15 | C | T | T | C | T | T | A |
| 4 | 0.07 | C | C | C | C | A | C | A |
| 5 | 0.07 | T | T | C | A | A | T | A |
| 6 | 0.05 | C | C | C | C | A | C | C |
| 7 | 0.04 | T | T | C | C | A | C | A |
| 8 | 0.03 | C | C | C | C | T | C | A |
Only haplotypes with a frequency of >1% are shown. Haplotypes were estimated in compound heterozygotes with the expectation maximization algorithm.
f, frequency.
FIG. 1.
DC-SIGN promoter polymorphisms and susceptibility to parenterally and mucosally acquired infection. (A) Frequency of DC-SIGN promoter polymorphisms in HIV-1-infected individuals (HIV-1 +) and HIV-seronegative individuals (HIV-1 −). P values and ORs are listed below each SNP. (B) Frequency of −336C in HIV-1-infected individuals and HIV-negative individuals (HREU, high risk exposed uninfected; all SN, all HIV-seronegative individuals) from all cohorts combined (all) and in individual cohorts (MHCS [Multicenter Hemophilia Cohort Study] and ALIVE [AIDS Linked to Intravenous Experience]).
It has been suggested that DC-SIGN might not be involved in sexual transmission of HIV because it is not expressed by Langerhans cells, mucosal DCs in the genital tract (14). Therefore, further analyses were performed, with participants stratified according to route of infection. No significant difference in risk was seen for any of the SNPs in the mucosal group (Table 2). However, among all parenterally exposed participants there was a significant association with increased likelihood of infection for those with the −336C SNP (OR = 1.87, P = 0.001). This association remained statistically significant after correction for multiple tests (P = 0.005). The strength of the association was increased when the analysis was restricted to HIV-1-negative individuals at highest risk for infection (those with documented receipt of HIV-1-contaminated clotting factor; OR = 2.46), though significance was diminished to some extent (P = 0.014), probably as a consequence of the reduced numbers (50 high-risk individuals versus a total of 365 uninfected individuals) (Fig. 1B). Importantly, similar trends were seen in individual cohorts (Fig. 1B).
TABLE 2.
DC-SIGN promoter polymorphisms and susceptibility to parenteral and mucosal infection
| Variant | Parenteral infection
|
Mucosal infection
|
||||||
|---|---|---|---|---|---|---|---|---|
| Phenotypic frequency
|
OR | P | Phenotypic frequency
|
OR | P | |||
| HIV-1+ (n = 348) | HIV-1− (n = 365) | HIV-1+ (n = 719) | HIV-1− (n = 179) | |||||
| −139T | 0.53 | 0.63 | 0.66 | 0.03 | 0.60 | 0.66 | 0.76 | 0.17 |
| −336C | 0.44 | 0.29 | 1.87 | 0.001 | 0.36 | 0.39 | 0.87 | 0.46 |
| −939T | 0.52 | 0.61 | 0.69 | 0.05 | 0.60 | 0.64 | 0.84 | 0.41 |
| −1180T | 0.75 | 0.81 | 0.73 | 0.15 | 0.82 | 0.83 | 0.94 | 0.81 |
| −1466T | 0.69 | 0.77 | 0.69 | 0.08 | 0.77 | 0.80 | 0.85 | 0.51 |
Haplotype analysis revealed that haplotype 4, which contains all of the high-risk SNPs, was also associated with parenterally acquired infection (OR = 2.62, P = 0.018). Indeed, the effect is somewhat stronger, although less significant, and significance was lost after correction for multiple tests. None of the other haplotypes showed a significant association with infection. The haplotype containing all protective SNPs (−139T, −336T, −939T, −1180T, and −1466T) was quite rare (frequency ≤ 0.001), so it was not possible to test the influence of this haplotype on the risk of HIV-1 infection. There was no significant association seen with HIV-1 disease progression for any of the individual SNPs or haplotypes.
DCs are believed to play a central role in HIV-1 infection as they may be among the first cells encountered by virus (5). DC-SIGN, a C-type lectin receptor that is exclusively expressed by DCs, can enhance virus infection by efficiently binding to the envelope glycoprotein of both R5 and X4 viruses (5). Levels of DC-SIGN expression on subsets of DCs are variable, however, and Langerhans cells, a subset of DCs that are present in the genital epithelium, do not express DC-SIGN at all (6). Furthermore, infection of these cells is blocked by inhibitors of CCR5 expression and not by C type lectin inhibitors (10). Thus, it is plausible that CCR5-dependent infection of Langerhans cells, and not DC-SIGN-mediated capture of HIV-1, is the major pathway involved in sexual transmission of HIV-1 (14), accounting for the failure to demonstrate a significant association of DC-SIGN polymorphism and mucosal infection. On the other hand, DC-SIGN expressed in lymph nodes and circulating DCs may play a role in HIV-1 acquisition via the parenteral route. In this regard, heterogeneity in expression of DC-SIGN among healthy subjects (4) may determine the ability of these cells to potentiate infection. Polymorphism in the promoter region of CCR5 has been shown to have an effect on HIV-1 disease progression (12), CCR5 surface density (15), and susceptibility of cells to HIV-1 infection (10, 15). Here we show that a polymorphism in the putative promoter region of DC-SIGN is associated with risk of infection via the parenteral route but not the mucosal route. A potential mechanism for this may involve differential constitutive or inducible expression of DC-SIGN on blood DCs as a result of this polymorphism, but this remains to be demonstrated. −336C is located 214 bp upstream of the major transcription start site (11) and is therefore likely to be in a region containing potential binding sites for transcriptional factors. Indeed, a database search (TRANSFAC database) (8) identified a potential Sp1 binding site in the presence of −336C, which is lost when −336T is present. Functional analyses should determine whether or not this is biologically significant.
Although the −336C SNP was quite common in African-Americans, there was no significant association with risk of infection seen for this group. This raises the possibility that the effect seen in Caucasians is not due directly to −336C, but rather to another as yet unidentified SNP on chromosome 19p13.2-3 that is in linkage disequilibrium with −336C in Caucasians but not in African Americans. Alternatively, −336C may exert its protective effect only in the presence of another variant that is common in Caucasians but missing in African-Americans.
Further studies are necessary to demonstrate the role of DC-SIGN promoter polymorphisms identified in this study in the function of DC-SIGN, and consequently their effect on HIV-1 infection. It will also be of interest to determine whether the −336C SNP is associated with protection from or susceptibility to other pathogens known to bind DC-SIGN.
Acknowledgments
This work was supported with federal funds from the U.S. National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 2.Buchbinder, S. P., M. H. Katz, N. A. Hessol, P. M. O'Malley, and S. D. Holmberg. 1994. Long-term HIV-1 infection without immunologic progression. AIDS 8:1123-1128. [DOI] [PubMed] [Google Scholar]
- 3.Cullen, M., J. Noble, H. Erlich, K. Thorpe, S. Beck, W. Klitz, J. Trowsdale, and M. Carrington. 1997. Characterization of recombination in the HLA class II region. Am. J. Hum. Genet. 60:397-407. [PMC free article] [PubMed] [Google Scholar]
- 4.Engering, A., S. J. Van Vliet, T. B. Geijtenbeek, and Y. Van Kooyk. 2002. Subset of DC-SIGN+ dendritic cells in human blood transmits HIV-1 to T lymphocytes. Blood 100:1780-1786. [DOI] [PubMed] [Google Scholar]
- 5.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 6.Geijtenbeek, T. B., R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, G. J. Adema, Y. van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 7.Goedert, J. J., C. M. Kessler, L. M. Aledort, R. J. Biggar, W. A. Andes, G. C. White II, J. E. Drummond, K. Vaidya, D. L. Mann, M. E. Eyster, et al. 1989. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321:1141-1148. [DOI] [PubMed] [Google Scholar]
- 8.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilgartner, M. W., S. M. Donfield, A. Willoughby, C. F. Contant, Jr., B. L. Evatt, E. D. Gomperts, W. K. Hoots, J. Jason, K. A. Loveland, S. M. McKinlay, et al. 1993. Hemophilia growth and development study. Design, methods, and entry data. Am. J. Pediatr. Hematol. Oncol. 15:208-218. [DOI] [PubMed] [Google Scholar]
- 10.Kawamura, T., F. O. Gulden, M. Sugaya, D. T. McNamara, D. L. Borris, M. M. Lederman, J. M. Orenstein, P. A. Zimmerman, and A. Blauvelt. 2003. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. USA 100:8401-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, H., W. Yu, L. Y. Liou, and A. P. Rice. 2003. Isolation and characterization of the human DC-SIGN and DC-SIGNR promoters. Gene 313:149-159. [DOI] [PubMed] [Google Scholar]
- 12.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 13.Phair, J., L. Jacobson, R. Detels, C. Rinaldo, A. Saah, L. Schrager, and A. Munoz. 1992. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J. Acquir. Immune Defic. Syndr. 5:490-496. [PubMed] [Google Scholar]
- 14.Piguet, V., and A. Blauvelt. 2002. Essential roles for dendritic cells in the pathogenesis and potential treatment of HIV disease. J. Investig. Dermatol. 119:365-369. [DOI] [PubMed] [Google Scholar]
- 15.Salkowitz, J. R., S. E. Bruse, H. Meyerson, H. Valdez, D. E. Mosier, C. V. Harding, P. A. Zimmerman, and M. M. Lederman. 2003. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin. Immunol. 108:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinman, R. M., A. Granelli-Piperno, M. Pope, C. Trumpfheller, R. Ignatius, G. Arrode, P. Racz, and K. Tenner-Racz. 2003. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 276:1-30. [DOI] [PubMed] [Google Scholar]
- 17.van Kooyk, Y., and T. B. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 18.Vlahov, D., J. C. Anthony, A. Munoz, J. Margolick, K. E. Nelson, D. D. Celentano, L. Solomon, and B. F. Polk. 1991. The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res. Monogr. 109:75-100. [PubMed] [Google Scholar]