Abstract
We performed a phylogenetic study of the E2-L2 region of human mucosal papillomaviruses (PVs) and of the proteins therein encoded. Hitherto, proteins codified in this region were known as E5 proteins. We show that many of these proteins could be spurious translations, according to phylogenetic and chemical coherence criteria between similar protein sequences. We show that there are four separate families of E5 proteins, with different characteristics of phylogeny, chemistry, and rate of evolution. For the sake of clarity, we propose a change in the present nomenclature. E5α is present in groups A5, A6, A7, A9, and A11, PVs highly associated with malignant carcinomas of the cervix and penis. E5β is present in groups A2, A3, A4, and A12, i.e., viruses associated with certain warts. E5γ is present in group A10, and E5δ is encoded in groups A1, A8, and A10, which are associated with benign transformations. The phylogenetic relationships between mucosal human PVs are the same when considering the oncoproteins E6 and E7 and the E5 proteins and differ from the phylogeny estimated for the structural proteins L1 and L2. Besides, the protein divergence rate is higher in early proteins than in late proteins, increasing in the order L1 < L2 < E6 ≈ E7 < E5. Moreover, the same proteins have diverged more rapidly in viruses associated with malignant transformations than in viruses associated with benign transformations. The E5 proteins display, therefore, evolutionary characteristics similar to those of the E6 and E7 oncoproteins. This could reflect a differential involvement of the E5 types in the transformation processes.
Papillomaviruses (PV) are a group of small DNA viruses that infect vertebrates. They are related to virtually all clinical cases of cervical cancer and are also involved in other benign and malignant proliferative disorders, such as skin warts, genital warts, laryngeal papillomas, and possibly non-melanoma skin cancer (21, 25, 26, 32, 43, 56). Cladistic relationships between PVs have been described on the basis of their phylogeny (9, 22). Briefly, the major categories are the supergroup, the group, the type, the subtype, and the variant. Supergroups are identified as clades with well-recognized differences in biology, whereas the rest of the categories are customarily described on the basis of decreasing phylogenetic distance (9, 22). Supergroup A unites PV types associated with genital lesions in humans and primates, such as human PV type 16 (HPV16), HPV18, or HPV6, although other PVs associated with cutaneous lesions, such as HPV2 or HPV7, are also encompassed within this supergroup. This work will focus on PVs belonging to supergroup A. For the sake of simplicity and attending to the tissue tropism of most of the members of this supergroup, we will name them mucosal HPVs. According to their epidemiological association with cancer, mucosal HPVs are classified as high-risk or low-risk types (11, 34). The paradigms of high-risk viruses are HPV16 and HPV18, but members of the groups they belong to, such as A9 and A7, and some other PVs from groups A5, A6, and A11 also appear to be systematically associated with malignant growth (11, 34). Mucosal HPVs bear two oncogenes, E6 and E7, which are expressed in the early stages of the infection process and are largely responsible for the changes related to the process of malignancy (33). The E5 gene, another early gene downstream of the E6 and E7 genes, is also slightly oncogenic but strongly enhances the transforming potential of E7 (4, 30, 48, 53). By contrast, the paralog bovine PV (BPV) E5 gene is the main gene responsible for the cell transformation in these viruses and is also capable of transforming human fibroblasts and keratinocytes (53).
The most studied mucosal HPV E5 protein is HPV16 E5. It is a small, highly hydrophobic protein, 83 amino acids (aa) long (6, 44), which localizes in the Golgi and in the endoplasmic reticulum (37). According to in silico predictions and to circular dichroism analysis, it has three hydrophobic domains with an alpha helix structure, which could cooperate in rendering the final spatial arrangement (2). Many disparate functions have been described for HPV16 E5, but we still lack a proper hypothesis bringing them all together into a comprehensible framework. Thus, the expression of HPV16 E5 upregulates the signal cascade initiated by the epidermal growth factor receptor upon ligand binding, through mitogen-activated protein kinases (14, 47). E5 also binds to the 16-kDa subunit of the membrane H+-ATPase, responsible of the acidification of the late endosomes (1, 12). HPV16 E5 modifies the cell response leading to initiation of apoptosis, both ligand mediated and stress induced. Thus, E5-expressing cells are less sensitive to Fas and apoptosis induced by the tumor necrosis factor alpha-related apoptosis-inducing ligand (24) and also less prone to apoptosis after UV irradiation (55). Besides, HPV16 E5 reduces gap junction-mediated intercellular communication via dephosphorylation of connexin 43 (36). This results in the cease of tissue homeostatic feedback, which has also been described as an early event in carcinogenesis progression (27). Finally, the expression of E5 blocks the traffic to the plasma membrane of major histocompatibility complex class I (MHC-I) and MHC-II molecules, thus hampering antigen presentation and T-cell recognition (7, 54). This finding correlates with the in vivo MHC-I diminished surface expression in premalignant lesions and in most carcinomas of the cervix (13, 19, 42).
Only a certain amount of working knowledge about HPV16 E5 is available thus far, and only scattered reports on the functions of other mucosal HPV E5 proteins have been published. However, it can be hypothesized that whatever the mechanisms connecting the disparate effects associated with HPV16 E5, they emerge from a central effect related to the hydrophobic character of the protein and its localization in the Golgi apparatus (15). In this sense, the only feature common to all E5 proteins is their highly hydrophobic nature and their location in the PV genetic map, and the genetic map is strictly conserved in PVs (45). E5 proteins are encoded in the E2-L2 region of the PV genome. This region is present in mucosal HPVs (supergroup A), ungulate fibropapillomaviruses (supergroup C), and animal and human cutaneous PVs (supergroup E); it is absent in EV- and melanoma-associated PVs (supergroup B) (9). The only criterion hitherto used to name a putative open reading frame (ORF) as E5 was its presence in this E2-L2 segment. This fact has led to a proliferation of putative E5 proteins even within a single genome, as is the case of E5a, E5b, and E5c proteins in HPV18 and in HPV54. Moreover, some ORFs in the E2-L2 region have been identified as E5 despite the absence of a start codon, as is the case of HPV26 E5 or HPV30 E5. Finally, some ORFs that are not encoded in the E2-L2 region but overlap E2 and/or L2 have also been termed E5, as in BPV4 E5 or HPV1 E5. The number of sequences identified as putative E5 proteins has therefore increased to 110, but their chemistry, biology, and phylogeny are largely unknown.
In the present work we have analyzed the phylogenetic and chemical relationships between the mucosal HPVs E5 proteins and have identified four different families of related E5 proteins. We describe here the evolutionary characteristics of these proteins and compare them with those of the early oncoproteins E6 and E7 and with those of the structural proteins L1 and L2. The divergence rate and overall evolutionary pattern of the E5 proteins resemble those of the oncoproteins E6 and E7 and differ from that of the late proteins L1 and L2. Furthermore, we illustrate here for the first time a correlation between the phylogenetic classification of the mucosal HPVs attending to the E5 proteins and their involvement in cervical cancer.
MATERIALS AND METHODS
DNA and protein sequences.
The E2-L2 segment sequences were retrieved either from the Los Alamos HPV Sequence Database (http://hpv-web.lanl.gov/stdgen/virus/hpv/) or from the public databases at EMBL. The E2-L2 sequences analyzed corresponded to the following viruses: HPV types 1, 2, 3, 6, 7, 10, 11, 13, 16, 18, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 39, 40, 41, 42, 44, 45, 51, 52, 54, 55, 57, 58, 59, 61, 66, 67, 68, 69, 70, 71, 72, 73, 74AE10, 77, 83, 84, 86, 87, 89, 90, and 91; common chimpanzee PV type 1; pigmy chimpanzee PV type 1; rhesus monkey PV; bovine PV types 1 and 2; reindeer PV; ovine PV (OPV) types 1 and 2; European elk PV; deer PV; Equus caballus PV; canine oral PV; cottontail rabbit PV; rabbit oral PV; Felis domesticus PV; and Phocoena spinipinnis PV.
An initial set of putative E5 sequences was obtained and analyzed. All putative ORFs carried in the E2-L2 sequences above listed, longer than 30 aa and displaying an initial methionine or leucine were identified with the ORF Finder program and included in the analysis. Additionally, other sequences named E5 in the public databases that did not fulfil these criteria were also included. Thus, E5 sequences from HPV types 5, 26, 30, 41, 66, and 69; BPV4; and PsPV identified as such by the original depositaries were included despite the absence of a starting codon. Moreover, E5 sequences from BPV4 and EcPV, also identified by the depositaries, were included despite they were not encoded in the E2-L2 segment of the corresponding viruses. The total number of putative E5 proteins in this initial data set was 119. A preliminary phylogenetic analysis was performed with these sequences, as described below. We defined two phylogenetic and chemical coherence criteria for accepting an E5-like sequence as such. We assumed first that phylogenetically close viruses should display phylogenetically close E5-like translations. Second, we assumed that phylogenetically close E5-like translations should show similar overall chemistry of the polypeptide chain. This chemical coherence was assessed as described below. Only 71 of the 119 sequences accomplished both criteria and were therefore named E5 proteins. These sequences belonged to HPV types 2, 3, 6, 7, 10, 11, 13, 16, 18, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 39, 40, 42, 44, 45, 51, 52, 54, 55, 57, 58, 59, 61, 66, 67, 68, 69, 70, 71, 72, 73, 74AE10, 77, 83, 84, 86, 87, 89, 90, and 91; CPV1; PCPV1; BPV1 and BPV2; RPV; OPV1 and OPV2; EEPV; and DPV. The corresponding E6, E7, L1, and L2 sequences form these viruses were also retrieved and analyzed in parallel to the E5 sequences.
Phylogenetic analysis.
The initial alignments were generated with TCOFFEE, which combines information for both global and local homologies (35). When E2-L2 DNA sequences were aligned, both gap opening and extension end were highly penalized to avoid high sequence alignment scores due to random similarities between long sequences. This precaution was necessary considering the differences in length among the aligned sequences. The result was the input for phylogenetic analysis with the PHYLIP program package (18). A distance matrix was generated with DNADIST or with PROTDIST with Dayhoff PAM250 as a substitution matrix. This output was analyzed with DNAPARS or with PROTPARS to generate a maximum parsimony tree, and with neighbor-joining and FITCH programs to create distance-based trees. The statistical support was assessed by 1,000 cycles of bootstrapping with the SEQBOOT and CONSENSE programs. The clusters and arrangements of individual viruses and virus groups obtained with neighbor-joining and FITCH were similar. The same procedure was performed after generating the initial alignments with CLUSTAL W (23), a progressive alignment algorithm, and with DIALIGN, a local segment alignment algorithm (31). The overall topology was the same in all cases, and only minor changes regarding distances were noticeable.
Divergence distances from the present E5, E6, E7, L1, and L2 proteins to the corresponding ancestral nodes for the group, clade, or protein ancestor were measured in the consensus phylogenetic tree. Distances for each protein were averaged, and differences were considered significant by applying the Kolmogorov-Smirnoff test, and further validated with Student's unpaired t test, when the data were consistent with a normal distribution. Additionally, individual distances for every protein in every virus were compared in pairs, with a paired Student's t test.
Protein chemistry predictions.
Hydrophobicity plots were calculated by using the Kyte-Doolitle hydropathicity scale, a main window of 13 aa, and edges of 5 aa (28). The average GRAVY values for the peptides were calculated as the sum of hydropathy values of all the amino acids, divided by the number of residues in the sequence. Topology predictions were performed at the PRED-CLASS server (http://biophysics.biol.uoa.gr/PRED-CLASS) by using cascading neural networks (41). Transmembrane segments were delineated with the TMHMM algorithm at the HUSAR server (http://genome.dkfz-heidelberg.de) by using hidden Markov model prediction (46) and confirmed at the PRED-TMR server (http://biophysics.biol.uoa.gr/PRED-TMR2) by using neural network prediction (40).
RESULTS
The E2-L2 segments of the PVs genomes show five different genetic arrangements.
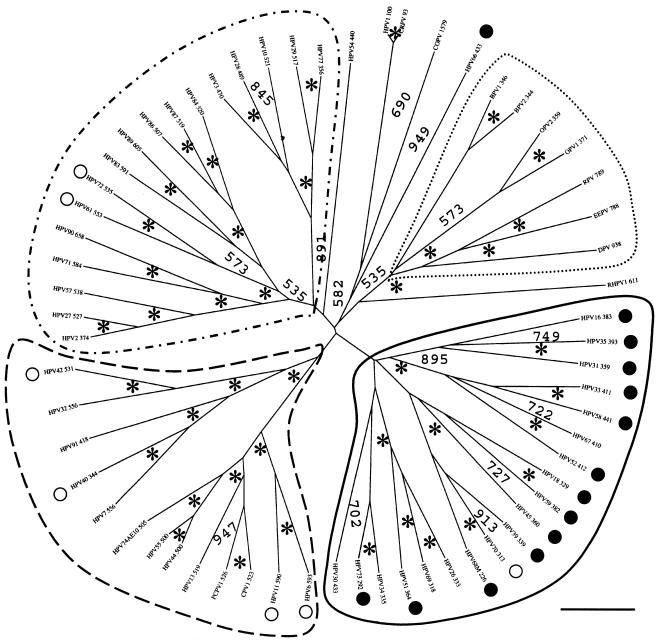
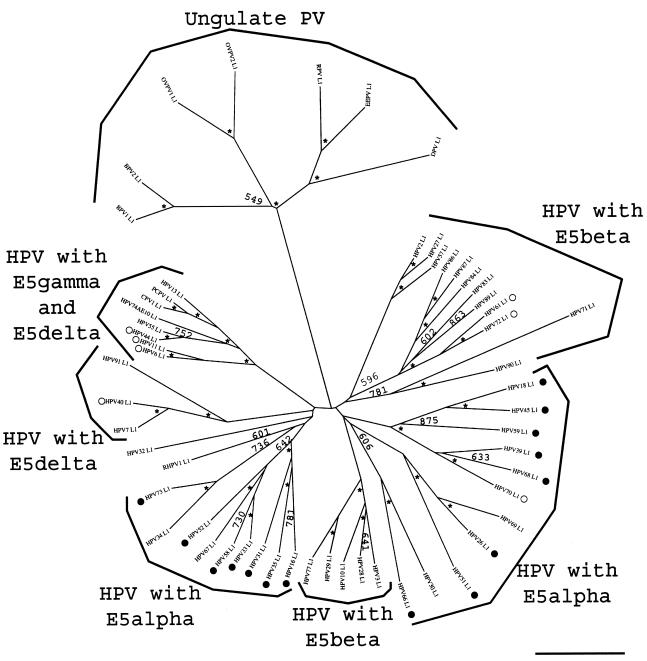
We performed a phylogenetic analysis of the E2-L2 region of 67 PVs, comprising mucosal HPVs (supergroup A), ungulate fibropapillomaviruses (supergroup C), and other phylogenetically scattered PVs (groups D and E). DNA alignments were originally generated with TCOFFEE, known to perform well in aligning sequences with high evolutionary distances and showing local homologies (29). The results were additionally confirmed with CLUSTAL W and DIALIGN. Alignments were further manually edited and phylogenies were estimated by evaluating distance matrices after 1,000 cycles of bootstrapping. Five PV branches appear clearly in the final tree, with high confidence values (Fig. 1). The first branch comprised ungulate PVs. PVs belonging to this supergroup bear an E2-L2 region between 350 and 950 bp in length and cluster together confidently 1,000 out of 1,000 times, despite the differences in length.
FIG. 1.
Phylogenetic tree of the E2-L2 DNA sequences in PVs. DNA sequences were retrieved from the Los Alamos HPV sequence database or EMBL and aligned with the TCOFFEE algorithm. A phylogenetic tree was constructed from the multialignment by the maximum-likelihood method. Similar topologies were obtained when using DIALIGN and CLUSTAL W as alignment algorithms and neighbor-joining methods as phylogenetic methods. Numbers at nodes indicate bootstrap support values after 1,000 bootstrap cycles (only values above 500 are given). An asterisk indicates that the bootstrap support is above 950. Numbers (as in “HPV16 383”) refer to the length of the DNA sequence used for the alignment. The bar at the bottom gives the relationship between branch lengths and 0.1 matrix units. E2-L2 sequences in ungulate PVs are included as an outgroup. Mucosal HPVs can be divided into four branches according to their E2-L2 sequence. Each of these branches matches a particular type of E5 protein encoded in this segment of the viral genome. The code for the main branches corresponds to the one used in the phylogenetic tree for E5 proteins (Fig. 2). High-risk and low-risk mucosal HPVs are labeled with black and white circles, respectively. Virtually all high-risk viruses cluster together according to the E2-L2 phylogeny. The star-like appearance of the tree suggests that if there was a common ancestor to all the present E2-L2 sequences, it gave rise in a short time to the corresponding ancestors of the four branches of mucosal HPV E2-L2 sequences and also perhaps to the ungulate PV E2-L2 sequences.
The second branch (Fig. 1) enclosed all HPVs highly associated with cervical cancer and therefore identified as high-risk PV types (11, 34). PVs appearing in this branch show an E2-L2 region ranging between 290 and 440 bp (Fig. 1). These sequences clustered confidently 1,000 out of 1,000 times, and the relationships within PV groups therein are consistent with those previously described (9). Thus, all the members of groups A5, A7, A9, and A11 were encompassed in this branch, and each group formed a separate cluster, with high confidence.
The third branch of the tree (Fig. 1), embraced HPV groups A2, A3, and A4. HPV61 and HPV72 belonged to this branch and are classified as low-risk type, because of their low association with cervical cancer (11, 34). Sequences in this branch range between 370 and 605 bp, and appeared together confidently 800 out of 1,000 times. Each of the groups comprised herein clustered separately with high bootstrap values, and group A4 appears as a subtree within the A3 group (Fig. 1).
The fourth branch of the E2-L2 tree (Fig. 1), covered PV group A10. All members appeared in this branch with high bootstrap values (1,000 out of 1,000 times). This group includes not only HPV but also CPV and PCPV (Fig. 1). The closest human relative of both is HPV13, as also described for the phylogeny of L1 sequences (49). PVs in this branch are classified as low-risk types and are usually associated with nonmalignant, external lesions in the genitalia (20, 39). The E2-L2 region of PVs in this branch ranges between 500 and 600 bp.
The fifth branch of the E2-L2 tree (Fig. 1), was closely related to the fourth branch. It enclosed HPV groups A1 and A8. The low-risk HPV40 and HPV42 appeared in this branch. Sequences in this branch range between 340 and 560 bp and clustered together with high confidence (1,000 out of 1,000 times). Groups A8 and A1 were sharply discerned (Fig. 1). However, HPV54 belongs to group A1 but did not group with HPV32 and HPV42, both members of the A1 group.
Besides the sharp grouping of E2-L2 in five main branches, sequences from CRPV, COPV, and HPV1 also clustered together. These PVs belong to supergroup E PVs, and the common branching was strongly supported despite the large differences in length ranging between 100 bp and 1.6 kbp. This cobranching validated the adequacy of the approach used.
The E2-L2 region of PV codifies six different conserved E5-like proteins.
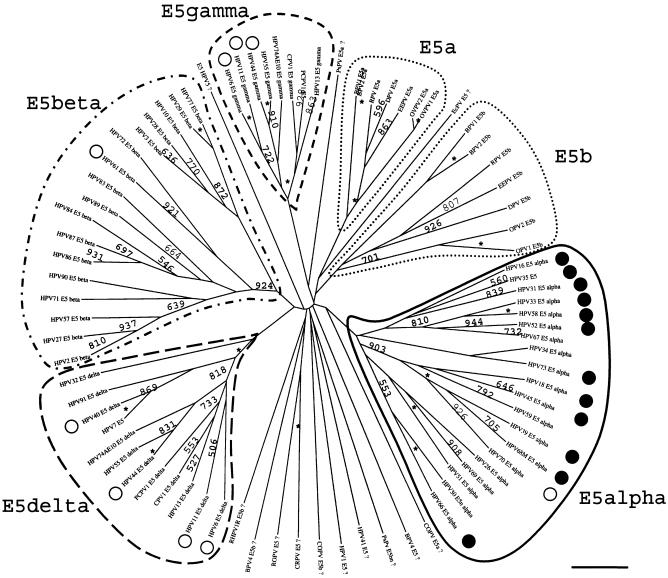
A primary set of putative E5 sequences was built as described above. A preliminary examination of these initial putative E5 sequences showed six main families of evolutionary related proteins. Many other sequences, however, showed no consistent taxonomically distribution and had no close relatives and no obvious similarities with other putative ORFs from members of the same supergroup or group; they branched alone close to a central point of the tree (data not shown). All these sequences were therefore suspected of being spurious translations. In this first stage, we first applied phylogenetic coherence criteria, assuming that phylogenetically close viruses would display phylogenetically close E5-like translations. Therefore, all the phylogenetically scattered protein sequences suspected of being spurious translations were removed, and a new analysis was performed with the remaining 84 sequences. This second sequence set showed a coherent distribution in six protein groups, fine classifications within these groups being coherent with the classification into A and C supergroups (Fig. 2) (9). In this final analysis, some of the previously discarded ORFs were included because the taxonomic diversity of the hosts prevented us to discern between true, distinct E5-like proteins and spurious translations, i.e., Phocoena spinipinnis PV, canine oral PV, cottontail rabbit PV, or rabbit oral PV. Some other translations were further included to highlight the sequence drift proposed above, such as those in rhesus monkey PV. Finally, some sequences termed E5 in the public databases but not matching any of the six groups were included in the final sequence set. This was the case of BPV4 E5a and E5b and of HPV5 E5. Neither PV bears a real E2-L2 segment, and the predicted E5 proteins overlap the corresponding E2 and/or L2 ORFs.
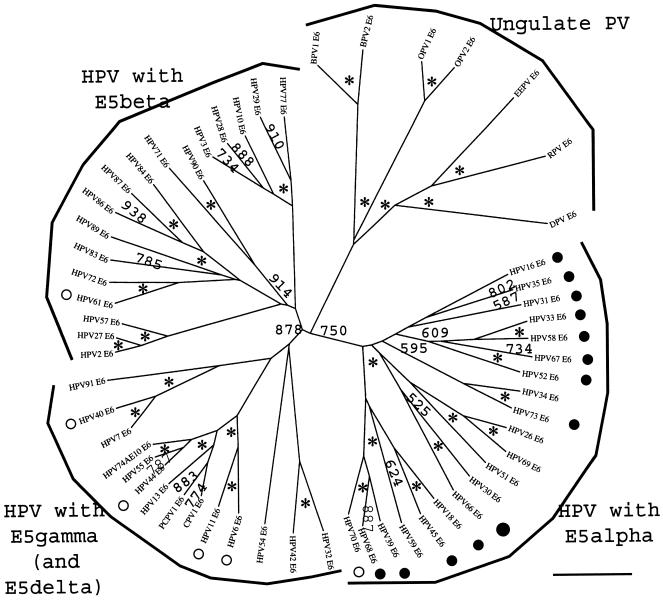
FIG. 2.
Phylogenetic tree of the E5 sequences in PVs. Protein sequences were retrieved from Los Alamos HPV sequence database or TrEMBL or generated by conceptual translation of the corresponding E2-L2 sequences and aligned with the TCOFFEE algorithm. A phylogenetic tree was constructed from the multialignment by the maximum-likelihood method. Similar topologies were obtained with DIALIGN and CLUSTAL W as alignment algorithms and neighbor-joining methods as phylogenetic methods. Numbers at the nodes indicate bootstrap support values after 1,000 bootstrap cycles (only values above 500 are given). An asterisk indicates that the bootstrap support is above 950. A question mark (as in “HPV1E5?”) indicates that the sequence branch is close to the center of the tree with no clear phylogenetic relationship with other sequences and is therefore likely to be a spurious translation. The original sequence set contained 119 putative E5 sequences, and it was reduced to the 84 shown here after removal of most of the putative spurious translations. The bar at the bottom gives the relationship between branch lengths and 0.1 matrix units. High-risk and low-risk mucosal HPVs are labeled with black and white circles, respectively. E5 sequences in ungulate PVs (dotted line) are included as outgroups. E5 sequences in mucosal HPVs can be divided into four types. Four main branches can be identified, corresponding to E5α(enclosed in a solid line), E5β (enclosed in a dotted and dashed line), E5γ (enclosed in a short-dashed line), and E5δ (enclosed in a long-dashed line). There is a strong correlation between E5 tree topology and the epidemiological implications of mucosal HPVs in cervical cancer. All high-risk viruses contain the E5α protein. Viruses in the A10 group contain two E5 sequences, E5γ and E5δ. The star-like appearance of the tree suggests that if there was a common ancestor to all the present E5 sequences, it gave rise in a short time to the corresponding ancestors of the four branches of mucosal HPVs E5 sequences and also maybe to the ungulate PVs E5 sequences.
For some PVs, all of the putative ORFs carried in the E2-L2 fragment were considered spurious translations after the initial evolutionary analysis. This was the case for the E5a, E5b, and E5c proteins of HPV54 and the E5a and E5b proteins of rhesus monkey PV. None of the proteins encoded in the E2-L2 fragment of these viruses resembled any of the six main groups of E5-like proteins. This could explain the absence of phylogenetic relationship between HPV54 and group A1 and between rhesus monkey PV and group A9 when regarding E2-L2 DNA sequences. The lack of a conserved coding region would have allowed unrestricted mutations for the E2-L2 sequences of these viruses, making them drift away from the ancestral sequence. For other PVs, only one of the ORFs present in the E2-L2 region showed a consistent taxonomic distribution, while the rest appeared scattered and branched close to the center of the tree in a star-like fashion. As an example, HPV18 contains three E5-putative ORFs, but only one of them corresponded to a putative protein, according to our analysis, being spurious translations of HPV18 E5b and HPV18 E5c.
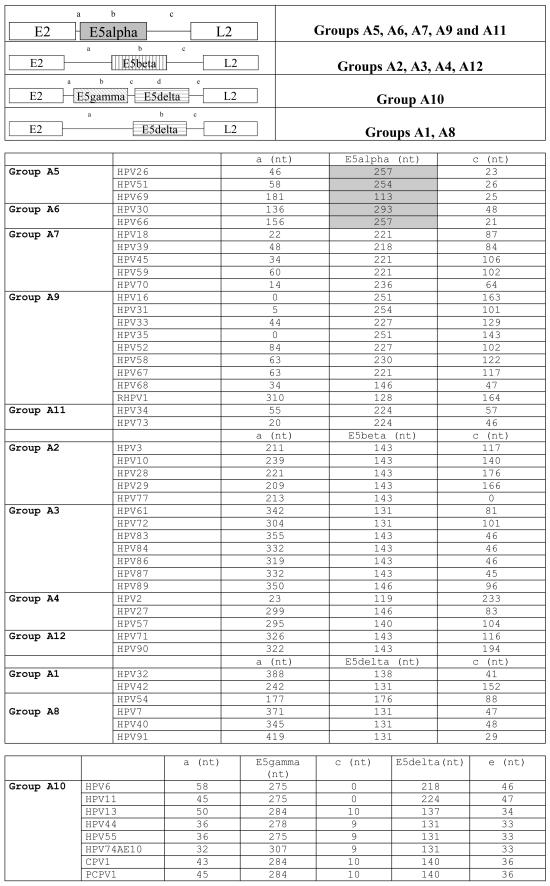
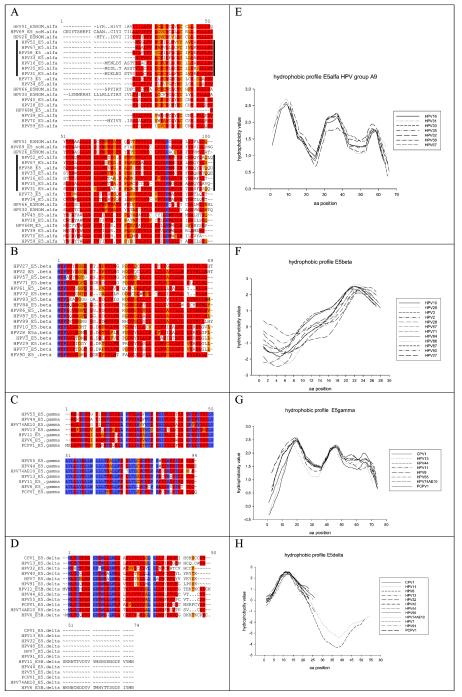
Groups A5, A6, A7, A9, and A11 showed a conserved ORF ca. 240 bp in length, starting close to the E2 stop codon but never overlapping it. This ORF encodes a protein named E5. For clarity and due to the lack of homology between the different E5 proteins, we termed it E5α. These E5α proteins are highly hydrophobic membrane proteins, with an average GRAVY index of 1.92 and average Ile+Leu+Val content of 44.2%. E5α proteins clustered together confidently, 1,000 times out of 1,000 (Fig. 2). The genetic arrangement of the E2-L2 region in these PVs is shown in Fig. 3. The best studied of these E5α is HPV16 E5α, which is 83 aa long, has a GRAVY index of 1.79 and shows up to three putative transmembrane domains at aa 11 to 29, 36 to 54, and 59 to 76. A TCOFFEE alignment of the E5α proteins is shown in Fig. 4. Amino acid identities among E5α proteins are scarce, but the global hydropathic pattern, showing three highly hydrophobic regions that could correspond to potential transmembrane regions is conserved in all of them (2). A plot of group A9 E5α proteins showing this hydrophobic profile is given in Fig. 4. The low sequence similarity between E5α proteins is also reflected in their phylogenetic distribution. Thus, they all shared an ancient common ancestor and clustered together 900 out of 1,000 times. However, an early evolutionary split made sequences in groups A9 and A11 diverge from those in groups A5, A6, and A7 (Fig. 2). The initial branching within E5α proteins and the subsequent evolutionary divergence would therefore account for the relatively low sequence homology.
FIG.3.
Genomic maps of the four types of E2-L2 segments in mucosal HPVs. The nucleotide lengths of both coding and no-coding regions are indicated. E5α proteins boxed in gray correspond to conceptual translations provided by the authors who deposited the sequence and do not display a starting methionine nor leucine. Mucosal HPVs encode four types of E5 proteins in the E2-L2 segment and show four different arrangements of the E2-L2 region. These viruses are classified here according to the E5 protein they encode.
FIG.4.
Alignments and hydrophobic profiles of the four types of E5 proteins in mucosal HPVs. Protein sequences were retrieved from the Los Alamos HPV sequence database or TrEMBL and aligned with the TCOFFEE algorithm. Color codes indicate the goodness of the alignment, decreasing in the order blue > red > orange. Hydrophobic plots were built with the Kyte-Doolittle index with a window of 10 aa and edges of 5 aa. (A) Alignment for E5α sequences from groups A5, A6, A7, and A9 displaying global similarities in highly hydrophobic segments but presenting only one conserved proline residue. Sequences for mucosal HPV group A9 are indicated with a vertical bar. They present a putative hydrophobic helix break in arginine 30, absent in the rest of the E5α proteins. (B) Alignment of E5β sequences from groups A2, A3, A4, and A12, showing a leucine-rich C terminus but lacking highly conserved particular residues. (C) Alignment of E5γ sequences present only in the A10 group, which includes human and chimpanzee PVs. A total of 40% of the residues are identical and 80% are chemically similar, accounting for a highly conserved protein. (D) Alignment of E5δ sequences present in groups A1, A8, and A10. The hydrophobic N terminus containing the motif GD(T)W(LL)LW is strictly conserved. Sequences from HPV6 and HPV11 include a long hydrophilic C terminus, absent in the rest. (E) Hydropathy plot of group A9 E5α proteins, showing three highly hydrophobic potential transmembrane domains. (F) Hydropathy plot of the E5β sequences, showing a hydrophilic N terminus and a hydrophobic C terminus, containing a potential transmembrane domain. (G) Hydropathy plot of the E5γ sequences, displaying three hydrophobic regions that could correspond to three transmembrane helices. (H) Hydropathy plot of the E5δ sequences, showing a conserved N-terminus potential transmembrane helix.
The E2-L2 region of PV groups A2, A3, A4, and A12 also codifies for only one conserved ORF ca. 140 bp long (Fig. 4). This ORF starts ca. 300 bp downstream of the E2 stop codon. The putative protein encoded here has also been named E5, despite the absence of obvious sequence similarity with the former E5α described. We therefore designate it E5β. Sequence homology between E5β proteins is relatively high, as shown in the alignment in Fig. 4. They present one hydrophobic, putative transmembrane region and show an average GRAVY index of 1.24; the average Ile+Leu+Val content is 46.0%. As an example, HPV2 E5β is 48 aa long, has a GRAVY value of 1.03, and shows one putative transmembrane domain (aa 25 to 42). A simultaneous hydrophobic plot for the E5β sequences is given in Fig. 4. The global similarities between E5β proteins can be seen, as they display a hydrophilic N terminus and a putative transmembrane region close to the C terminus.
HPV groups A1, A8, and A10 possess a long E2-L2 region, with ca. 600 bp (Fig. 4). In group A10, the first half of this segment encodes an extremely well-conserved putative protein ca. 90 aa long that we have named E5gamma. Like all E5-like proteins, E5γ are highly hydrophobic membrane proteins, with an average GRAVY index of 1.60 and average Ile+Leu+Val content of 46.0%. As an example, HPV11 E5γ is 91 aa long, has a GRAVY value of 1.83, and contains up to three putative transmembrane domains (aa 13 to 37, 42 to 61, and 68 to 87). Almost half of the E5γ amino acid sequences are identical, and more than 80% residues are similar. The corresponding TCOFFEE alignments and simultaneous hydrophobic plots of the E5γ proteins are given in Fig. 4. E5γ proteins are therefore highly conserved and are present exclusively in the A10 group, which encompasses PVs infecting humans, chimpanzees, and pigmy chimpanzees. These two facts combined suggest a conserved role for this putative protein in the biology of these viruses.
In the second half of the E2-L2 segment, groups A1, A8, and A10 share a conserved short ORF ca. 150 bp long. We named the putative protein expressed here E5δ. HPV6 and HPV11 E5δ proteins additionally present an extended C terminus, ca. 30 aa in length. All E5δ proteins show a highly hydrophobic, potential transmembrane region of conserved amino acids. The average GRAVY index of E5δ proteins is 1.02, and the average Ile+Leu+Val content is 36.1%. As an example, HPV13 E5δ is 45 aa long, has a GRAVY value of 0.98, and shows a putative transmembrane domain (aa 11 to 33). Certain stretches of the sequence are extremely conserved, such as the pattern GDXW(L, M)XLW or the hydrophobic box downstream (Fig. 4).
The phylogenetic relationships of these E5α, -β, -γ, and -δ proteins; E5a and E5b from ungulates; and some other conceptual translations from PV sequences from the E2-L2 region are depicted in Fig. 2. Each of these proteins clustered separately and confidently, and no closer evolutionary relationship between them could be inferred. This means that if there was a unique ancestor for all of them, it predated the split ungulate-primate group, and it gave rise to six evolutionary pathways leading to six different proteins in a very short time, yielding this star-like pattern of the phylogenetic tree.
The evolutionary pattern of E5-like proteins is different from that of the late proteins L1 and L2, coincides with that of early proteins E6 and E7, and correlates with the clinical manifestations of the viral infection.
The HPV E5-like ORFs identified here have been classified into four different groups according to their chemical characteristics and their phylogenetic relationships, and all ORFs carried in the E2-L2 region and suspected of being spurious translations were removed and not analyzed. However, to rule out the possibility that our study dealt only with conceptual translations and had no biological significance, we analyzed the phylogenetic relationships within the early proteins E6 and E7 and the late proteins L1 and L2 in PVs with an E5 gene-like ORF and compared both results. The corresponding protein sequences were aligned by TCOFFEE, and phylogeny was estimated by evaluating the distance matrices after 1,000 cycles of bootstrapping. The corresponding trees for L1 and E6 are depicted in Fig. 5 and 6, respectively. The topology of the trees for L2 and E7 was similar to that of L1 and E6, respectively (data not shown).
FIG. 5.
Phylogenetic tree of the L1 sequences in mucosal HPVs. Protein sequences were retrieved from the Los Alamos HPV sequence database or TrEMBL and aligned with the TCOFFEE algorithm. A phylogenetic tree was constructed from the multiple alignments by the maximum-likelihood method. Similar topologies were obtained when using DIALIGN and CLUSTAL W as alignment algorithms and neighbor-joining methods as phylogenetic methods. Numbers at the nodes indicate bootstrap support values after 1,000 bootstrap cycles (only values above 500 are given). An asterisk indicates that the bootstrap support is above 950. The bar at the bottom gives the relationship between branch lengths and 0.1 matrix units. High-risk and low-risk mucosal HPVs are labeled with black and white circles, respectively. L1 sequences in ungulate PVs are included as outgroups. An early split event separated mucosal HPV L1 proteins in two main branches, close to the divergence point with the ancestor from ungulate PV L1 proteins. The phylogenetic relationships, according to L1 proteins, do not match the epidemiological classification of these viruses. The phylogenetic relationships among viruses were the same as those for L1 and L2 but differed for E5, E6, and E7 proteins.
FIG. 6.
Phylogenetic tree of the E6 sequences in mucosal HPVs. Protein sequences were retrieved from the Los Alamos HPV sequence database or TrEMBL and aligned with the TCOFFEE algorithm. A phylogenetic tree was constructed from the multiple alignments by the maximum-likelihood method. Similar topologies were obtained when using DIALIGN and CLUSTAL W as alignment algorithms and neighbor-joining methods as phylogenetic methods. Numbers at nodes indicate bootstrap support values after 1,000 bootstrap cycles (only values above 500 are given). An asterisk indicates that the bootstrap support is above 950. The bar at the bottom gives the relationship between branch lengths and 0.1 matrix units. High-risk and low-risk mucosal HPVs are labeled with black and white circles, respectively. E6 sequences in ungulate PVs are included as outgroups. The tree topology coincides with the corresponding topology for E5 (Fig. 2) and E7 proteins: viruses encoding E5α, E5β, and E5δ proteins cluster in different branches. The phylogeny of E6 and E7 proteins showed a clear parallel with the epidemiological classification of these viruses.
The clustering of mucosal PVs at the group level is the same, with some exceptions, notably RHPV1, independent of the protein analyzed. However, the relationships between groups are not. Thus, according to the late proteins L1 and L2, there was an ancient event separating groups A2, A3, A4, A5, A6, A7, and A12 from groups A1, A8, A9, A10, and A11. This early splitting event appeared with good bootstrap support (500 times out of 1,000) in the phylogenies of both proteins L1 and L2. The two sets of PV groups segregated in this event do not show homogeneous biological characteristics. Thus, high-risk PVs (groups A5, A6, A7, A9, and A11) appeared separately in both group sets. Consequently, the biological roles of L1 and L2 can be disconnected from the malignancy of the infection of the corresponding PV.
The topologies of the phylogenetic trees for the early proteins E6 and E7 are different from those of L1 and L2 and match the description provided above for E5-like proteins. In these early genes studied, there was an ancient splitting event separating three main branches of mucosal HPVs (Fig. 5). The first one comprised groups A5, A6, A7, A9, and A11. These groups enclose all the PVs identified as high-risk PVs and correspond to those encoding an E5α protein. The second branch included groups A2, A3, A4, and A12 and matches those described as encoding an E5β protein. Finally, the third branch encompassed groups A1, A8, and A10—those groups containing an E5δ ORF and also an E5γ ORF in the case of the A10 group. E5-like proteins therefore show the same evolutionary topology as the early proteins E6 and E7. This fact reinforces the validity of our identification of four putative types of mucosal HPV E5-like ORFs (genotypes) as real ORFs whose translations could correlate with the malignancy and clinical manifestations of the infection phenotypes.
The overall topology of the evolutionary trees of early and late genes in mucosal HPVs is not superimposable. It can then be inferred that the selection pressures driving the evolution of L1 and L2 proteins and those driving the evolution of the early genes E6 and E7, the E2-L2 segment, and the E5-like proteins are different and have led to different evolutionary paths that can be accurately tracked. There is therefore a different evolutionary pattern, which parallels the different functions of L1 and L2 (involved in the first contact between virus host and in virus assembly and release) and those of E6, E7, and E5 (involved in the early steps of viral infection).
Early proteins E5, E6, and E7 diverged more than the late proteins L1 and L2, and those in high-risk viruses evolved more rapidly than those in low-risk viruses.
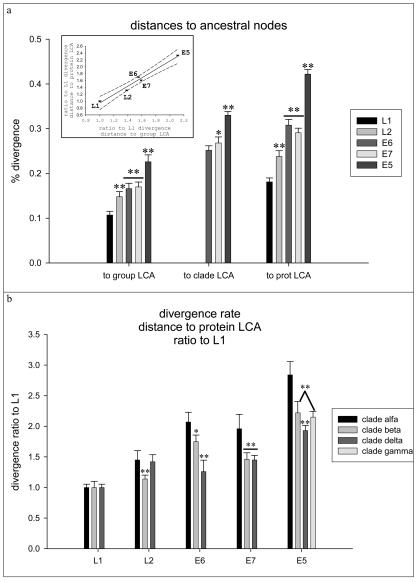
Having proven that the evolutionary pattern of HPV early and late genes was different, we addressed the question whether there were also differences in the evolutionary rate between both protein types. We measured and compared the corresponding distances from the present viral proteins to the last common ancestor (LCA) of the group to the LCAs of the clade and the protein. Here we will define the LCA for every clade α, β, and δ as the last common node having given rise to all PVs encoding E5α, E5β, and E5δ proteins, respectively. The position of the putative protein LCA was estimated considering the branching point of the trees giving rise to the corresponding ungulate PV protein. Results are depicted in Fig. 7. When comparing distances from present proteins to group and protein LCAs, the divergence percentage increased in the order L1 < L2 < E6 ≈ E7 < E5. When comparing distances from present proteins to clade LCAs, the divergence percentage increased in the order E6 ≤ E7 < E5 (Fig. 7). Thus, while present L1 proteins diverged ca. 18% from the putative LCA, L2 proteins diverged ca. 24%, E6 and E7 diverged ca. 30%, and E5 diverged ca. 42%. These differences reflect again that the selection pressures pushing the evolution of early and late genes in mucosal HPVs are different. The dissimilarities in rate evolution between early and late proteins are proportionally the same when looking at the group and protein LCAs (Fig. 7, inset). Thus, assuming that there was a single ancestor for every of the present L1, L2, E5, E6, and E7 proteins and that these common ancestors were contemporary and assuming a constant mutation rate for the HPVs, the early genes have sustainedly evolved more rapidly than the late genes.
FIG. 7.
Divergence rates of L1, L2, E5, E6, and E7 proteins in mucosal HPVs. Protein sequences were retrieved from the Los Alamos HPV sequence database or TrEMBL and aligned with the TCOFFEE algorithm, and their phylogeny was estimated by the maximum-likelihood method. An asterisk indicates a significant difference with P values of <0.05. Two asterisks indicate a significant difference with P values of <0.01. Bars enclose 95% confidence intervals of the corresponding mean. (a) Percentage of divergence between the present proteins and the putative LCA of the HPV group protein and the protein LCA. Early proteins have diverged significantly more quickly than late proteins, and the divergence percentage increases in the order L1 < L2 < E6 ≈ E7 < E5. The phylogeny according to E5, E6, and E7 is overimposable and allows the definition of an additional category, namely the clade, regarding the type of E5 protein encoded in the genome. The divergence rate between present proteins and clade LCA proteins also matches the sequence E6 < E7 < E5. (Inset) Divergence percentages are expressed as ratios with respect to the corresponding L1 divergence percentage. There is a strong correlation between the divergence ratios regarding the distance to protein LCA and the distance to protein group LCA. Early proteins therefore evolved sustainedly faster than structural proteins in mucosal HPVs. Error bars enclose 95% confidence intervals for the mean. The dashed lines enclose the 95% confidence interval for the lineal regression of the data. (b) Divergence percentages normalized with respect to the L1 divergence percentage for each virus. Viruses are classified as belonging to clade α, β, or δ, according to the E5 protein type they encode. Viruses in clade α are highly associated with malignant transformations. Viruses in clades β and δ are associated with benign transformations. Viruses in group A10 contain an additional E5γ protein, which is referred to here as clade γ. Proteins E5, E6, and E7 diverged more in high-risk viruses than in their low-risk counterparts.
Early proteins in mucosal HPVs have diverged more than late proteins. Since the early genes E6 and E7, and likely also E5, are involved in the processes of malignancy leading to the establishment of neoplasias (17), we compared the correlation of the evolutionary rates of early and late genes with the epidemiologic classification of viruses into high- and low-risk types (11, 34). Distances to L1, L2, E6, E7, and E5 LCAs were measured for every virus and normalized with respect to the corresponding L1 distances. Paired comparison of these normalized divergences confirmed the gradient in evolutionary rate described above, with late proteins evolving more slowly than early proteins (Fig. 7). The individual results were then combined for viruses containing E5α, E5β, E5γ, and E5δ proteins, and the corresponding values were compared. As shown in Fig. 7b, E6, E7, and E5-like early proteins diverged significantly more in high-risk viruses than their counterparts in low-risk viruses, while there are only marginal differences in the evolutionary rate of late proteins. Thus, while E5α has diverged ca. three times faster than the corresponding L1, E5β, E5γ, and E5δ evolved only approximately two times faster than the corresponding L1. Similarly, E6 and E7 in high-risk viruses have diverged approximately two times faster than L1, while in low-risk viruses the ratio reaches only ca. 1.5 times the divergence of L1. Thus, both the evolutionary pattern and the evolutionary rate differ between early genes and late genes in mucosal HPVs and also between high-risk and low-risk PVs.
DISCUSSION
We performed a phylogenetic analysis of the E2-L2 region in mucosal HPVs at both DNA and protein levels. The global topology of both phylogenetic trees is comparable. The overall view of the phylogeny according to the E2-L2 segment and to the proteins encoded therein is that there is a sharp correlation between the evolutionary history (the genotype) and the clinical manifestations of the infection (the phenotype).
The E2-L2 segment usually encodes short, hydrophobic proteins, named E5 or E5a, E5b, and E5c in PVs containing more than one of these putative ORFs. We propose a classification of the ORFs carried in the E2-L2 region of the mucosal HPVs as a result of having applied two coherence criteria for the E5-like proteins: phylogenetic coherence (phylogenetically close E5 proteins are expected to appear in phylogenetically close viruses) and chemical coherence (phylogenetically close proteins are expected to display similar basic chemical characteristics). Underlying these assumptions is the basic hypothesis that chemistry is the main restriction for protein evolution (3). First, we identified many of these putative proteins as spurious translations, on the basis of their incongruent phylogenetic distribution. We propose therefore that all these ORFs so far designated E5 should not be named as such. The list of ORFs that meet our criteria of chemical and phylogenetic congruence and therefore should be named E5 is provided in Fig. 3. Our results predict that the average divergence between present E5-like proteins and the LCA is more than 40%. On this basis, we propose a change in the nomenclature to sharply designate with different names what, in reality, could be different polypeptides. We therefore suggest that E5-like proteins be named E5α, E5β, E5γ, and E5δ. This nomenclature reflects simultaneously the homogeneity of the different proteins regarding their chemistry and their evolutionary patterns and matches the epidemiological characteristics of the different viruses bearing these ORFs.
The phylogenetic trees of the E2-L2 (Fig. 1) and of the E5 (Fig. 2) DNA segments show a star-like pattern. In both trees, the main branches emerge close to a putative central point, and the relative distances between clades are comparable. It could be claimed therefore that we have compared sequences which do not share any common ancestor and that this fact is responsible for the star-like appearance of the final trees. Evidence, however, suggests that all the present E2-L2 mucosal HPV sequences and the true E5 proteins could have shared a common ancestor. The E2-L2 segment could be a hypervariable region in the mucosal HPVs and is therefore likely to have undergone rapid evolution, as well as insertions, deletions, or recombinations (22). The star-like appearance of the phylogenetic tree of the E2-L2 region DNA sequences would therefore reflect such hypervariability. We have further provided additional evidence regarding the relative evolutionary distances between the present E5-like proteins and the respective LCAs, compared with the corresponding distances for other four genes in the PV genome. Concerning the four groups of HPV E5 sequences, we have shown that there is no evident sequence similarity between them and that the evolutionary divergence between present proteins in different groups rises to 80%. The highly hydrophobicity, the high Ile+Leu+Val content, and the presence of transmembrane regions are the only common characters for all E5α, -β, -γ, and -δ proteins. Of all E5-like proteins, only the biology of HPV16 E5α is partially known. It localizes mainly in the Golgi apparatus and has been associated with several disconnected effects related to differential response to growth factors and stress, apoptosis initiation, and MHC surface expression (5, 16). These multiple effects could arise from local changes in the membrane chemistry, related to the highly hydrophobic nature of the protein and its transmembrane potential (14). This is the only characteristic common to all E5-like proteins that could account for the multiple effects hitherto associated with them. Experimental data related to other E5 types also point in this direction. Thus, HPV2 E5β is also a Golgi protein and blocks the surface expression of MHC-II molecules (8). In addition, both HPV6 E5γ and HPV11 E5γ localize in the Golgi and associate with the 16-kDa pore-forming protein component of the vacuolar ATPase (10, 12), also known to be an interaction partner of HPV16 E5α (1, 12). The overall data suggest, therefore, that mucosal HPV E5 proteins share a common ancient ancestor and that they underwent a rapid early divergence process that gave rise to the present four E5 families. The particular composition of the E5 proteins, where the three amino acids Ile, Leu, and Val (representing 13 possible codons) account for more than 45% of the sequence, could have eased the sequence drift here proposed.
The E5-like proteins display the same evolutionary characteristics as E6 and E7. The phylogeny of human mucosal PV, according to L1 and L2, is the same and matches previous reports (9, 22, 38, 50, 51) but does not coincide with that of the early genes. The strong correlation between phylogeny and epidemiology in all the early proteins studied is absent in the corresponding analysis for the late proteins L1 and L2. This fact shows that the structural proteins L1 and L2 have a secondary role, if any, in the malignant transformations associated with viral infection.
The divergence rate at the protein level increases in the progression L1 < L2 < E6 ≈ E7 < E5. There is, therefore, a clear gradient in the rate of divergence from late genes, which evolve more slowly, to early genes, which evolve more quickly. In the same direction, the divergence rate of the different E5-like proteins followed the progression E5α < E5β ≈ E5δ < E5γ. This reinforces again our proposal that the E5-like proteins here identified are real proteins and that there is a correlation between the E5 version encoded in a given PV genome and its higher or lower association with the development of neoplasia.
The findings that early proteins have diverged more than late proteins and that early proteins in high-risk viruses have evolved more than early proteins in low-risk viruses match with the involvement of early proteins in the initial transformation processes of the viral infection (17). The expression of E6 and E7 modifies the normal cell cycle and alters the differentiation program of the keratinocyte, thus allowing viral DNA replication. E6 and E7 initially bind p53 and retinoblastoma protein p105RB, respectively, although both are known to have other cellular targets (17, 52). The expression of E5, on the other hand, raises a multitude of apparently disconnected effects that enhance those of E6 and E7 (5, 16) and which could arise from a modification of cell membrane chemistry (15). The cellular binding partners of L1 and L2 are still unknown, but it can be inferred from our results that they will not be involved in cellular homeostasis to the same extent as those of E6, E7, and E5. The increased divergence rate in early genes, especially in high-risk PVs, could have arisen as a result of a coevolutionary arms race between virus and host. In the case of E5, the high hydrophobic content would have potentiated the divergence. A complementary view of the increased divergence rate of early genes compared to late genes could explain this fact as a reflection of a high number of interaction partners of these early proteins. Thus, the higher the number of interaction partners of a protein, the broader its effects are and the higher its divergence rate will be. This view would match a scenario where the number of interaction partners and the multiplicity of biological effects on the infected cell also increase in the sequence L1 < L2 < E6 ≈ E7 < E5.
E5-like proteins can be classified into four groups according to their chemical characteristics and evolutionary relationships. This classification matches the epidemiological characteristics of the mucosal HPVs and their differential association with cancer development (11, 34). Moreover, the evolutionary pattern and divergence rate of the E5 proteins agree with those of the early genes E6 and E7, but not with those of the late genes L1 and L2. To date, most of the data available refer to the E5α protein, and few reports are available about the biological effects of E5β, E5γ, and E5δ. The different evolutionary history of the early and the late genes raises the question of which gene (if any) reflects the true evolutionary history of the PV; it does not exclude the presence of an initial period where recombination and horizontal exchange of genetic material between viruses could have been possible. Finally, the properties here analyzed and predicted for these proteins suggest that their characterization could provide us with new insights into the biology and the diversity of clinical manifestations of the PV infection in humans.
Acknowledgments
We thank Santiago García-Vallvé for his useful comments and improvements to an initial draft of the manuscript.
I.G.B. is the recipient of a Fundación Ramón Areces postdoctoral fellowship.
REFERENCES
- 1.Adam, J. L., M. W. Briggs, and D. J. McCance. 2000. A mutagenic analysis of the E5 protein of human papillomavirus type 16 reveals that E5 binding to the vacuolar H+-ATPase is not sufficient for biological activity, using mammalian and yeast expression systems. Virology 272:315-325. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, A., and J. Reed. 2002. Modelling of the human papillomavirus type 16 E5 protein. Biochim. Biophys. Acta 1601:9-18. [DOI] [PubMed] [Google Scholar]
- 3.Babbit, P. C., and J. A. Gerlt. 1997. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J. Biol. Chem. 272:30591-30594. [DOI] [PubMed] [Google Scholar]
- 4.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, I. G., A. Alonso, and E. Auvinen. 2004. Human papillomavirus type 16 E5 protein. Papillomavirus Rep. 15:1-6. [Google Scholar]
- 6.Bubb, V., D. J. McCance, and R. Schlegel. 1988. DNA sequence of the HPV-16 E5 ORF and the structural conservation of its encoded protein. Virology 163:243-246. [DOI] [PubMed] [Google Scholar]
- 7.Campo, M. S. 2002. Animal models of papillomavirus pathogenesis. Virus Res. 89:249-261. [DOI] [PubMed] [Google Scholar]
- 8.Cartin, W., and A. Alonso. 2003. The human papillomavirus HPV2a E5 protein localizes to the Golgi apparatus and modulates signal transduction. Virology 314:572-579. [DOI] [PubMed] [Google Scholar]
- 9.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, S. L., T. Z. Tsai, C. P. Han, and Y. P. Tsao. 1996. Mutational analysis of human papillomavirus type 11 E5a oncoprotein. J. Virol. 70:3502-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad, M., V. J. Bubb, and R. Schlegel. 1993. The human papillomavirus type 6 and 16 E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. J. Virol. 67:6170-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cromme, F. V., C. J. Meijer, P. J. Snijders, A. Uyterlinde, P. Kenemans, T. Helmerhorst, P. L. Stern, A. J. van den Brule, and J. M. Walboomers. 1993. Analysis of MHC class I and II expression in relation to presence of HPV genotypes in premalignant and malignant cervical lesions. Br. J. Cancer 67:1372-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crusius, K., E. Auvinen, B. Steuer, H. Gaissert, and A. Alonso. 1998. The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp. Cell Res. 241:76-83. [DOI] [PubMed] [Google Scholar]
- 15.Crusius, K., M. Kaszkin, V. Kinzel, and A. Alonso. 1999. The human papillomavirus type 16 E5 protein modulates phospholipase C- gamma-1 activity and phosphatidyl inositol turnover in mouse fibroblasts. Oncogene 18:6714-6718. [DOI] [PubMed] [Google Scholar]
- 16.DiMaio, D., and D. Mattoon. 2001. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene 20:7866-7873. [DOI] [PubMed] [Google Scholar]
- 17.Fehrmann, F., and L. A. Laimins. 2003. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene 22:5201-5207. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package),version 3.5c. Department of Genetics, University of Washington, Seattle. http://evolution.genetics.washington.edu/phylip.html.
- 19.Gill, D. K., J. M. Bible, C. Biswas, B. Kell, J. M. Best, N. A. Punchard, and J. Cason. 1998. Proliferative T-cell responses to human papillomavirus type 16 E5 are decreased amongst women with high-grade neoplasia. J. Gen. Virol. 79:1971-1976. [DOI] [PubMed] [Google Scholar]
- 20.Gross, G. 1987. Lesions of the male and female external genitalia associated with human papillomaviruses, p. 197-234. In K. Syrjänen, L. Gissmann, and L. G. Koss (ed.), Papillomaviruses and human diseases. Springer-Verlag, Heidelberg, Germany.
- 21.Grussendorf-Conen, E. I. 1987. Papillomavirus-induced tumors of the skin: cutaneous warts and epidermodysplasia verruciformis, p. 159-181. In K. Syrjänen, L. Gissmann, and L. G. Koss (ed.), Papillomaviruses and human diseases. Springer-Verlag, Heidelberg, Germany.
- 22.Halpern, A. L. 2000. Comparison of papillomavirus and immunodeficiency virus evolutionary patterns in the context of a papillomavirus vacine. J. Clin. Virol. 19:43-56. [DOI] [PubMed] [Google Scholar]
- 23.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch, K., and A. Alonso. 2002. The human papillomavirus type 16 E5 protein impairs TRAIL- and FasL-mediated apoptosis in HaCaT cells by different mechanisms. J. Virol. 76:12162-12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaya, H., E. Kotiloglu, S. Inanli, G. Ekicioglu, S.-U. Bozkurt, A. Tutkum, and S. Kullu. 2001. Prevalence of human papillomavirus (HPV) DNA in larynx and lung carcinomas. Pathologica 93:531-534. [PubMed] [Google Scholar]
- 26.Keefe, M., A. al-Ghamdi, N. J. Maitland, P. Egger, C. J. Keefe, A. Carey, and C. M. Sanders. 1994. Cutaneous warts in butchers. Br. J. Dermatol. 130:9-14. [DOI] [PubMed] [Google Scholar]
- 27.King, T., L. Fukushima, A. Hieber, K. Shimabukuro, W. Sakr, and J. Bertram. 2000. Reduced levels of connexin43 in cervical dysplasia: inducible expression in a cervical carcinoma cell line decreases neoplastic potential with implications for tumor progression. Carcinogenesis 21:1097-1109. [PubMed] [Google Scholar]
- 28.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydrophatic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 29.Lassmann, T., and E. L. L. Sonnhammer. 2002. Quality assessment of multiple alignment programs. FEBS Lett. 529:126-130. [DOI] [PubMed] [Google Scholar]
- 30.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 31.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 32.Multhaupt, H. A., J. N. Fessler, and M. J. Warhol. 1994. Detection of human papillomavirus in laryngeal lesions by in situ hybridization. Hum. Pathol. 25:1302-1305. [DOI] [PubMed] [Google Scholar]
- 33.Munger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 34.Munoz, N., F. X. Bosch, S. de Sanjosé, R. Herrero, X. Castellsagué, K. V. Shah, P. J. F. Snijders, and C. J. L. M. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 35.Notredame, C., D. Higgins, and J. Heringa. 2000. T-Coffee: A novel method for multiple sequence alignments. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 36.Oelze, I., J. Kartenbeck, K. Crusius, and A. Alonso. 1995. Human papillomavirus type 16 E5 protein affects cell-cell communication in an epithelial cell line. J. Virol. 69:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oetke, C., E. Auvinen, M. Pawlita, and A. Alonso. 2000. Human papillomavirus type 16 E5 protein localizes to the Golgi apparatus but does not grossly affect cellular glycosylation. Arch. Virol. 145:2183-2191. [DOI] [PubMed] [Google Scholar]
- 38.Ong, C. K., S. Nee, A. Rambaut, H. U. Bernard, and P. H. Harvey. 1997. Elucidating the population histories and transmission dynamics of papillomaviruses using phylogenetic trees. J. Mol. Evol. 44:199-206. [DOI] [PubMed] [Google Scholar]
- 39.Oriel, J. D. 1987. Genital and anal papillomavirus infections in human males, p. 183-196. In K. Syrjänen, L. Gissmann, and L. G. Koss (ed.), Papillomaviruses and human diseases. Springer-Verlag, Heidelberg, Germany.
- 40.Pasquier, C., and S. J. Hamodrakas. 1999. An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng. 12:631-634. [DOI] [PubMed] [Google Scholar]
- 41.Pasquier, C., V. J. Promponas, and S. J. Hamodrakas. 2001. PRED-CLASS: cascading neural networks for generalized protein classification and genome-wide applications. Proteins 44:361-369. [DOI] [PubMed] [Google Scholar]
- 42.Ritz, U., F. Momburg, H. Pilch, C. Huber, M. J. Maeurer, and B. Seliger. 2001. Deficient expression of components of the MHC class I antigen processing machinery in human cervical carcinoma. Int. J. Oncol. 19:1211-1220. [DOI] [PubMed] [Google Scholar]
- 43.Rudlinger, R., M. H. Bunney, R. HuGrob, and J. A. Hunter. 1989. Warts in fish handlers. Br. J. Dermatol. 120:375-381. [DOI] [PubMed] [Google Scholar]
- 44.Seedorf, K., T. Oltersdorf, G. Krammer, and W. Rowekamp. 1987. Identification of early proteins of the human papilloma viruses type 16 (HPV 16) and type 18 (HPV 18) in cervical carcinoma cells. EMBO J. 6:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shadan, F., and L. Villarreal. 1996. The evolution of small DNA viruses of eukaryotes: past and present considerations. Virus Genes 11:239-257. [DOI] [PubMed] [Google Scholar]
- 46.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 47.Straight, S. W., P. M. Hinkle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle, G. F., and L. Banks. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239-1245. [DOI] [PubMed] [Google Scholar]
- 49.Van Ranst, M., A. Fuse, P. Fiten, E. Beuken, H. Pfister, R. D. Burk, and G. Opnedakker. 1992. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology 190:587-596. [DOI] [PubMed] [Google Scholar]
- 50.Van Ranst, M., J. B. Kaplan, and R. D. Burk. 1992. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J. Gen. Virol. 73:2653-2660. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Q., L. A. Salter, and D. K. Pearl. 2002. Estimation of evolutionary parameters with phylogenetic trees. J. Mol. Evol. 55:684-695. [DOI] [PubMed] [Google Scholar]
- 52.Woodworth, C. 2002. HPV innate immunity. Front. Biosci. 7:2058-2071. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Y. C., B. A. Spalholz, M. S. Rabson, and P. M. Howley. 1985. Dissociation of transforming and trans-activation functions for bovine papillomavirus type 1. Nature 318:575-577. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, B., P. Li, E. Wang, Z. Brahmi, K. W. Dunn, J. S. Blum, and A. Roman. 2003. The E5 protein of human papillomavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-gamma. Virology 310:100-108. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, B., D. F. Spandau, and A. Roman. 2002. E5 protein of human papillomavirus type 16 protects human foreskin keratinocytes from UV B-irradiation-induced apoptosis. J. Virol. 76:220-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]