Abstract
Poultry birds are asymptomatic reservoir of Salmonella Typhimurium (S. Typhimurium) but act as source of human infection for this bacterium. Inside the poultry, S. Typhimurium experiences several stresses, 42°C body temperature of birds is one of them. Proteins are highly susceptible to temperature mediated damage. Conversion of protein bound aspartate (Asp) residues to iso-aspartate (iso-Asp) is one of such modifications that occur at elevated temperature. Iso-Asp formation has been linked to protein inactivation and compromised cellular survival. Protein-L-isoaspartyl methyltransferase (PIMT) can repair iso-Asp back to Asp, thus enhances the cellular survival at elevated temperature. Here, we show that the pimt gene deletion strain of S. Typhimurium (Δpimt mutant strain) is hypersensitive to 42°C in vitro. The hypersusceptibility of Δpimt strain is partially reversed by plasmid based complementation (trans-complementation) of Δpimt strain. Following oral inoculation, Δpimt strain showed defective colonization in poultry caecum, and compromised dissemination to spleen and liver. Interestingly, we have observed three and half folds induction of the PIMT protein following exposure of S. Typhimurium to 42°C. Our data suggest a novel role of pimt gene in the survival of S. Typhimurium at elevated temperature and virulence.
Keywords: isoaspartate, PIMT, Salmonella, 42°C, poultry
Introduction
The enteric human pathogen Salmonella Typhimurium (S. Typhimurium) has worldwide prevalence and is a leading cause of food borne gastroenteritis (Yeung et al., 2014; Khoo et al., 2015). There are two types of Salmonella infections, including typhoidal and non-typhoidal (Darwin and Miller, 1999; Gal-Mor et al., 2014). Although typhoid fever caused by S. Typhi is more fatal, non-typhoidal Salmonella organisms are most common foodborne pathogens. The manifestations of non-typhoidal salmonellosis include mild to moderate gastroenteritis consisting of diarrhea, abdominal cramps, vomiting, and fever (Griffin and McSorley, 2011). The invasive infections can lead to septicaemia (Cohen et al., 1987). Non-typhoidal salmonellosis accounts for about 93.8 million cases with 155,000 deaths annually around the globe (Majowicz et al., 2010).
Among non-typhoidal Salmonella, Salmonella Enteritidis (S. Enteritidis) and S. Typhimurium are most frequently associated serovars with food poisoning in human (Oliveira et al., 2002). While in Europe, S. Enteritidis is more prevalent, in USA and India S. Typhimurium predominates (Besser et al., 2000; Rahman, 2002; Zhang et al., 2003). S. Typhimurium is major invasive non-typhoidal Salmonella (iNTS) in countries of Sub-Saharan Africa (Gal-Mor et al., 2014; MacLennan et al., 2014). Human acquire Salmonella infection mostly from the contaminated food, such as pork, beef, milk, milk products, poultry meat, eggs, and contaminated water. Poultry meat and eggs are the most common sources of Salmonella infections in human (Linam and Gerber, 2007). Poultry birds harbor S. Enteritidis and S. Typhimurium in their caecum without manifesting any symptoms to very mild enteritis.
To colonize and survive inside the poultry, Salmonella must need to defend against various stresses that it encounters inside the body. Along with other stresses (like oxidants, limited availability of nutrients, etc.), high body temperature of birds exert an additional threat to Salmonella. Salmonella is a mesophilic bacterium that can survive and replicate at a range of temperatures. This suggests the existence of mechanism(s) that can combat temperature stress encountered by Salmonella. By decreasing ratio of unsaturated to saturated fatty acids, S. Typhimurium modulates fatty acid composition and fluidity of the membrane, a phenomenon that has been correlated with thermotolerance of this bacterium at 45°C (Casadei et al., 2002; Sampathkumar et al., 2004; Álvarez-Ordóñez et al., 2008). Among macromolecules, proteins are the primary targets of temperature mediated inactivation. Temperature mediated modifications include unfolding, aggregation and covalent modifications in the proteins. Chaperones can refold unfolded proteins. Temperature induced expression of chaperones have been reported, suggesting their crucial roles in cellular survival under heat stress (Foster and Spector, 1995; Grimshaw et al., 2003; Waldminghaus et al., 2007). Heat shock protein htrA is shown to be helpful in survival of S. Enteritidis in egg white at the body temperature of the poultry (42°C) (Raspoet et al., 2014).
Aspartate (Asp) residues in the proteins have been shown to be prone to stress conditions. Under stress Asp converts into iso-aspartate (iso-Asp) that can introduce kink in to the polypeptide and subsequently leads in to unfolding of the proteins. Unfolded proteins are prone to make aggregates which have compromised function(s) and can affect cellular survival. For proper refolding of unfolded proteins, repair of covalently modified amino acid residues, like iso-Asp is required before chaperone function. Protein-iso-aspartyl-methyltransferase (PIMT) can repair iso-Asp back to Asp thus enhances cellular survival under stress conditions. PIMT activity has been found in various bacteria, such as S. Typhimurium, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Enterobacter aerogenes (Li and Clarke, 1992). The pimt gene knock out strains of Caenorhabditis elegans (Khare et al., 2009) and E. coli (Hicks et al., 2005) were found to be hypersensitive to various stress conditions such as oxidative stress, methanol and elevated temperature of 43°C.
Earlier, we have shown the importance of pimt in the survival of S. Typhimurium under oxidative stress and virulence in mice (Kumawat et al., 2016). Here, we hypothesized that PIMT might play an important role in the survival of S. Typhimurium under temperature stress and aids the colonization of this bacterium in poultry. First, we deleted pimt gene from the poultry isolate of S. Typhimurium. Then we evaluated in vitro survival of pimt gene deletion strain at 42°C and in stationary phase. Subsequently, we assessed the contribution of pimt gene in the caecal colonization of S. Typhimurium.
Materials and Methods
Bacterial Strains and Culture Conditions
Five strains of S. Typhimurium, E-2375, E-4231, E-4831, E-5587, and E-5591 were procured from the repository of National Salmonella Centre (Veterinary type), Indian Veterinary Research Institute (IVRI), Izatnagar, India. S. Typhimurium was cultured in LB broth or on Hektoen enteric (HE) agar plates. The DH5α strain of E. coli was grown in LB broth or agar. Antibiotics, kanamycin (50 μg ml-1) and ampicillin (100 μg ml-1) were included in the medium for selection purposes as and when required. The cultures were grown in a 37°C incubator or in a 37°C shaker incubator at 180 rpm. In few experiments the cultures were exposed to 42°C.
PCR Confirmation of typh and Virulence Associated Genes in E-5591 Strain of S. Typhimurium
Presence of the typh (S. Typhimurium specific) and virulence associated genes in the E-5591 strain of S. Typhimurium was confirmed by polymerase chain reaction (PCR). typh (Alvarez et al., 2004), hilA (Guo et al., 2000), enterotoxin (stn) (Makino et al., 1999) and invA (Galan et al., 1992) genes were amplified using PCR.
Construction of pimt Gene Deletion Mutant in E-5591 Strain of S. Typhimurium and Its Complementation
The plasmids pKD4, pKD46, pCP20 were a kind gift from Dr. Robert J. Maier, Department of Microbiology, University of Georgia, Athens, GA, USA. Plasmid QE 60 (pQE60) was procured from Qiagen, Hilden, Germany.
The pimt gene was deleted as per the protocol described earlier (Kumawat et al., 2016). In brief, kanamycin cassette was amplified from pKD46 using pimt deletion primers given in Table 1 and PCR conditions given in Table 2. Then the pimt gene was replaced by kanamycin cassette. Following confirmation of deletion, the antibiotic cassette was removed by FLP recombinase. The plasmid based complementation was carried out using pQE60-pimt and the protocol described earlier (Kumawat et al., 2016). The mutant and complemented strains were confirmed by PCR (Kumawat et al., 2016) and designated as Δpimt and Δpimt + pQE60-pimt strains, respectively.
Table 1.
Details of the primers used in current study.
| Purpose | Primer name | Sequence (5′–3′) | Size of the product (bp) | Reference |
|---|---|---|---|---|
| Construction of pimt deletion mutant | pimt deletion | F:GATGTGGTTTCAGACTGGTTAGACAGCGTGGG | 1600 | Kumawat et al., 2016 |
| AGTTGGCACGCAGTAGGCTGGAGCTGCTTC | ||||
| R:CGTTTCGGCTTCATCAGGCGTAAGCGTGGGTG | ||||
| TTTGCAGGGCAAACATATGAATATCCTCCTTA | ||||
| pimt test | F:ATGAAGGCTACGTCTCCGTC | 872 | ||
| R:GTGACGTAAGAACCGTGCAAC |
Table 2.
Polymerase chain reaction (PCR) conditions used for amplification of different genes.
| S. No. | Gene/purpose | Initial denaturation (temp/time) | Denaturation (temp/time) | Annealing (temp/time) | Extension (temp/time) | Final extension (temp/time) |
|---|---|---|---|---|---|---|
| 1 | pimt deletion | 95°C/5 min | 95°C/30 s | 42°C/30 s | 72°C/1 min | 72°C/10 min |
| 2 | pimt test | 95°C/30 s | 95°C/30 s | 60°C/30 s | 72°C/90 s | 72°C/10 min |
Survival of Δpimt Strain at 42°C
Isolated colonies of wild type, Δpimt and Δpimt + pQE60-pimt strains of S. Typhimurium were inoculated and grown in LB broth at 37°C for overnight. The overnight grown cultures were diluted in 250 ml of fresh media at a ratio of 1: 100 (old culture: fresh media) and incubated either at 37°C or at 42°C. Aliquots were collected at different times (0, 6, 12, 24, 48, and 72 h) of incubations and serially diluted in phosphate buffered saline (PBS). The colony forming units (CFUs) were determined by plating the serial dilutions on HE agar plates.
Effect of 42°C Exposure on Expression of the PIMT Protein in S. Typhimurium
Overnight grown cultures of wild type and Δpimt strains of S. Typhimurium were diluted in fresh medium and exposed to 37 or 42°C for 12 h. Following exposure, the cells were pelleted and suspended in ice cold PBS. The cells were lysed by sonication and unbroken cells were removed by centrifugation at 7500 × g for 10 min at 4°C. Total proteins in these samples were estimated by bicinchoninic acid protein assay kit (Thermo Scientific, Rockford, IL, USA) and normalized in all samples. Fifty micrograms of proteins were resolved in a 10% SDS-gel and blotted to polyvinylidene difluoride membrane. Following blocking, the membrane was incubated in rabbit anti-PIMT antiserum. The PIMT- anti-PIMT interaction was determined by incubating the membrane in anti-rabbit IgG conjugated with alkaline phosphatase. The blot was developed in nitroblue tetrazolium and 5-Bromo-4-chloro-3-indolyl phosphate containing buffer. Replica gel loaded with same samples was stained with Coomassie Brilliant Blue (CBB) and served as a loading control.
Effect of pimt Gene Deletion on the Colonization Ability of S. Typhimurium in Poultry
All animal experimentations were carried out according to the guidelines of the institutional animal ethical committee (IAEC), IVRI, Izatnagar, India. The protocol was approved by the institutional animal ethical committee (IAEC), IVRI, Izatnagar, India. One day old chicks of the White Leghorn breed were procured from the Central Avian Research Institute (CARI), Izatnagar, India. The birds were maintained in cages as per guidelines of the CARI/IAEC and provided with ad libitum feed and water. The birds were screened for the presence of Salmonella spp. by examining the cloacal swabs, followed by PCR or serotyping. Briefly, the cloacal swabs were taken in buffered peptone water (BPW) and pre-enriched at 37°C for 6 h on a shaker incubator. The pre-enriched cultures were then diluted in the Rappaport Vassiliadis R 10 (RV-10) enrichment broth at a ratio of 1:100 and incubated at 37°C for 24 h. Following incubation, the cultures were streaked on HE agar plates and incubated at 37°C for overnight. The black centered colonies with greenish margin were selected (at least three colonies from each plate) for urease test. The urease negative colonies were tested by Salmonella specific PCR using genus specific invA primers (Galan et al., 1992) and/or by serotyping using specific anti-sera (Pavan Kumar et al., 2014). To analyze colonization abilities of different strains, Salmonella free chicks (10 per group) were orally infected with various strains of S. Typhimurium. Cloacal swabs from these birds were enriched and streaked on HE agar media. The presence of S. Typhimurium was confirmed by PCR/serotyping as described above.
Salmonella free chicks (18 per group) were orally infected with wild type or Δpimt strain of S. Typhimurium (at the dose of 109 CFUs/ bird). Actual CFUs given to the birds were determined by dilution and plating of the inoculum on HE agar plates. The colonies were enumerated following incubation of the plates at 37°C for overnight.
Caecal colonization and bacterial loads in spleen and liver were assessed at weekly intervals for up to 4 weeks. Following dissection of the birds, the caeca were collected and homogenized in 5 ml BPW. The isolation of Salmonella from such homogenates was carried out in a similar way as described in above section. The colonies were screened by S. Typhimurium specific typh primers (Alvarez et al., 2004).
Half of the spleen and 100 mg of liver were aseptically collected in 1 ml sterile PBS, triturated and serially diluted. One hundred microliters of homogenates were spread on HE agar plates and plates were incubated at 37°C for overnight. CFUs were calculated as per spleen or per gram of liver tissue.
Results
On day five post-inoculation, 60% of the birds infected with E-5591 strain of S. Typhimurium were positive for fecal shedding. While in case of E-2375, E-4231, E-4831, E-5587 strain infected birds the shedding was 0, 0, 0, and 40% respectively.
Polymerase chain reaction based analysis confirmed the presence of virulence associated genes (invA, hilA, and stn) in the E-5591 strain of S. Typhimurium (Supplementary Figure S1). The schematic used for pimt gene deletion has been shown in Figure 1A. PCR based analysis confirmed the deletion of pimt gene (Figure 1B). First recombinants showed kanamycin resistance. After removal of kanamycin cassette the mutant strain failed to grow on antibiotic containing agar plates (Figure 1C). Western blot analysis confirmed the presence of PIMT in wild type and complemented strains S. Typhimurium, while absence of this protein in Δpimt strain (not shown).
FIGURE 1.
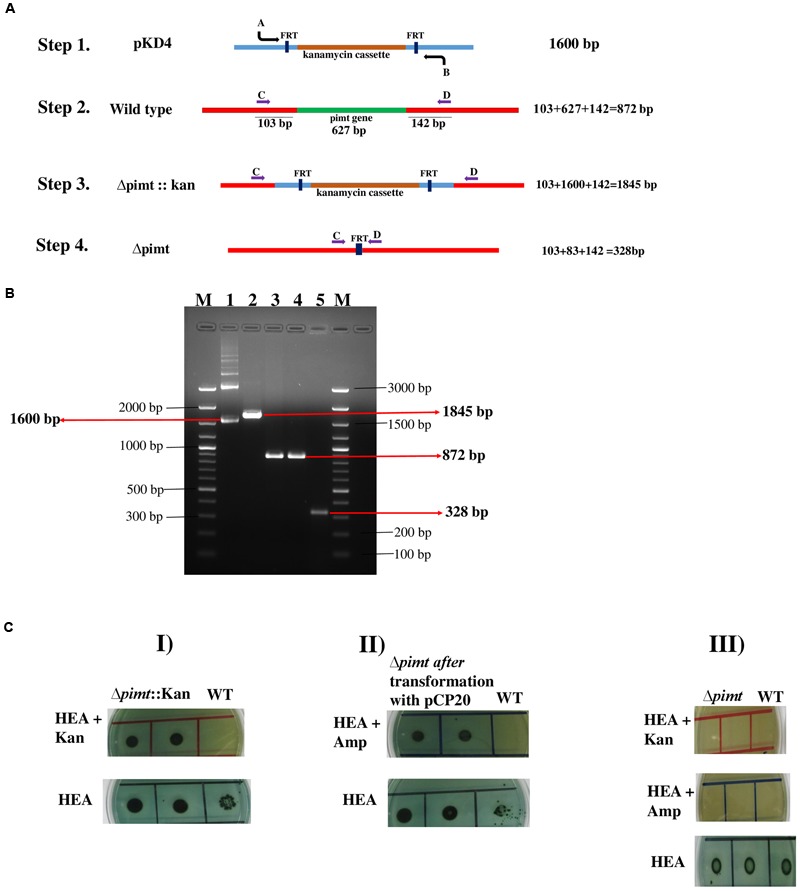
Construction and confirmation of pimt gene deletion strain of Salmonella Typhimurium. (A) Schematics. (B) Agarose gel analysis. Kanamycin cassette was amplified and fused to flanking regions of pimt gene [step 1 (A) and lane 1 (B)]. The pimt gene was replaced with kanamycin cassette; positive recombinants were selected on kanamycin plate and confirmed by PCR [step 3 (A)]. C and D primers amplified bigger and smaller PCR products in mutant and ST colonies respectively (B, lanes 2 and 3, respectively). The antibiotic cassette was removed by FLP recombinase; now C and D primers amplified similar sized PCR product in ST colony; while smaller PCR product in case of mutant colony [(A) step 4 and (B) lanes 4 and 5]. (C) Antibiotic resistance patterns observed during course of mutant construction. (I) The pimt gene was replaced by kanamycin cassette. Only Δpimt::Kan recombinants were able to grow on kanamycin containing HE agar plates. (II) Δpimt mutant after transformation with pCP20. The pCP20 transformed Δpimt mutant colonies were able to grow on HE agar-Amp plates. (III) The final Δpimt mutant after pCP20 removal. Δpimt mutant colonies were unable to grow on antibiotic containing media.
Effect of 42°C Stress on Survival of Δpimt Strain
We have evaluated the role of pimt gene in the survival of S. Typhimurium at 42°C in vitro. In comparison to wild type, the Δpimt strain was highly susceptible (p < 0.01, Figure 2A) to 42°C exposure for 48 and 72 h. The complemented strain displayed intermediate sensitivity. Following 48 h of incubation, the recoveries were (CFUs/ml as log10) 9.20 ± 0.24, 6.61 ± 0.18, and 8.08 ± 0.12 (mean ± SD) for wild type, Δpimt and Δpimt + pQE60-pimt strains of S. Typhimurium respectively. Following 72 h of incubation we did not find any viable bacteria in Δpimt samples, however, we have recovered significant numbers of live bacteria in wild type and Δpimt + pQE60-pimt cultures (Figure 2A). Interestingly, at 37°C the difference in the viability between wild type and Δpimt strains of S. Typhimurium was observed at later time point (at 72 h) and was very minor (Figure 2B). This indicates that pimt is primarily required for survival of S. Typhimurium under temperature stress and to a minor extent in late stationary phase.
FIGURE 2.
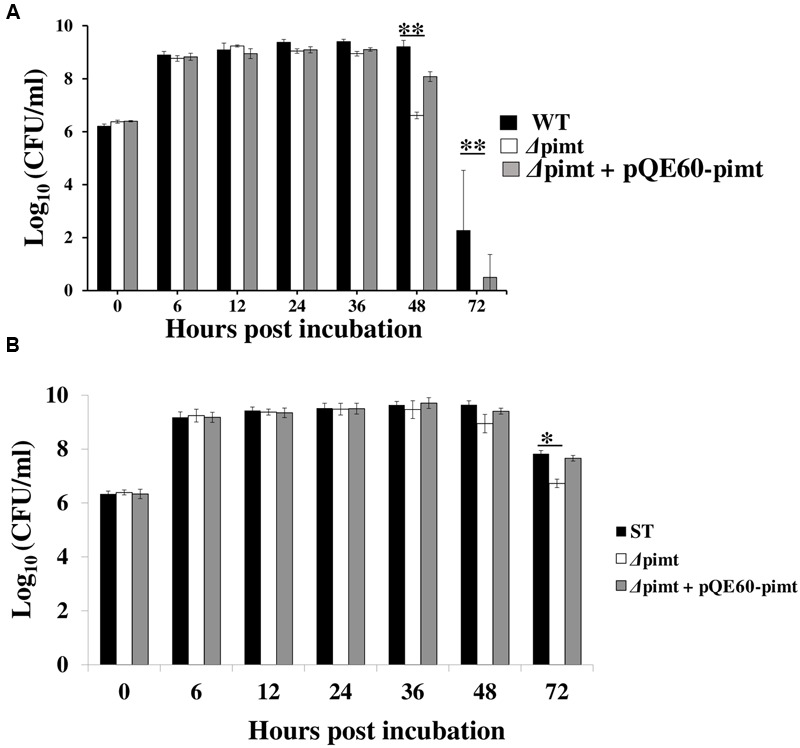
Effect of heat stress and stationary phase on in vitro survival of Δpimt strain. (A)Δpimt strain showed hypersensitivity to 42°C. Overnight grown cultures of WT, Δpimt and Δpimt + pQE60-pimt strains of S. Typhimurium were diluted and exposed to 42°C for 72 h in a shaker incubator. The aliquots were drawn at different times post-incubation, serially diluted and plated on HE agar plates. The CFUs/ml were calculated. The data was analyzed by one way ANOVA and presented as mean ± SD (n = 4). ∗∗p < 0.01. (B) Δpimt strain showed defective survival in stationary phase. Overnight grown cultures of WT and Δpimt strains of S. Typhimurium were diluted 1: 100 and incubated at 37°C for 72 h. The aliquots were drawn at different times of incubations, serially and plated on HE agar plates. The CFUs/ ml were calculated. The data was analyzed by one way ANOVA and presented as mean ± SD (n = 4). ∗p < 0.05.
Effect of 42°C Exposure on Induction of PIMT Protein
After observing the importance of pimt gene in the survival of S. Typhimurium under temperature stress in vitro, we hypothesized that exposure of S. Typhimurium to 42°C might induce PIMT protein. Western blot analysis showed a thin band of PIMT in S. Typhimurium grown at 37°C, the intensity of which increased about 3.5-fold following exposure of S. Typhimurium to 42°C. This band was absent in the Δpimt strain lysate loaded lanes, grown either at 37 or 42°C (Figure 3). Replica gel stained with CBB showed similar amounts of protein loading in different lanes (data not shown).
FIGURE 3.
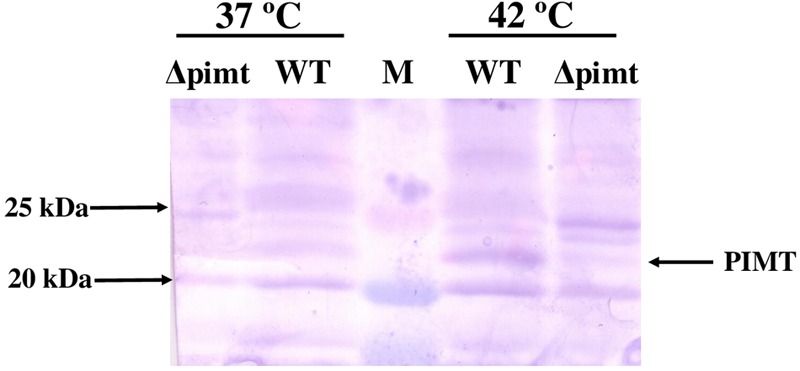
Effect of heat stress on expression of PIMT protein in S. Typhimurium. Wild type and Δpimt strains of S. Typhimurium were grown at 37 or 42°C for 12 h. Cells were then pelleted and induction of PIMT protein was analyzed by SDS-PAGE followed by Western blotting against anti-PIMT antibodies. The incubation temperatures and samples loaded in the lanes are depicted in figure itself. Lane M is protein molecular weight markers. PIMT band is marked by arrow.
Role of pimt in Caecal Colonization of S. Typhimurium
The normal body temperature of poultry is around 42°C (Donkoh, 1989; Cooper and Washburn, 1998) and we have seen hypersusceptibility of Δpimt strain to 42°C in vitro. Next we hypothesized that pimt might contribute to colonization of S. Typhimurium in the poultry. The birds were orally infected with either wild type or Δpimt strain of S. Typhimurium and colonization was assessed in the caecum. We recovered bacteria from the 100% caeca of wild type infected birds during entire course (28 days) of experiment. While the caeca of Δpimt strain infected birds showed reduced colonization during first 2 weeks (75 and 60% on days 7 and 14, respectively) and eventually cleared the bacteria on 21 days post-infection (Table 3).
Table 3.
The number of positive/infected birds on different days post-infection.
|
Salmonella positive birds |
||
|---|---|---|
| Days (post-infection) | WT inoculated birds | Δpimt inoculated birds |
| 7 | 4/4 (100%) | 3/4 (75%) |
| 14 | 5/5 (100%) | 3/5 (60%) |
| 21 | 4/4 (100%) | 0/4 (0%) |
| 28 | 5/5 (100%) | 0/5 (0%) |
Contribution of pimt in Dissemination of S. Typhimurium to Poultry Spleen and Liver
The bacterial loads in spleen and liver were determined at different times post-infection. Three days post-infection, we recovered bacteria from the spleens of wild type as well as Δpimt mutant infected birds. However, after 7, 14, and 21 days post-infection, only spleens of wild type infected birds showed bacteria (Figure 4A). In liver we observed dissemination of wild type strain of S. Typhimurium only. However, we did not recover any bacteria from the liver of Δpimt mutant infected birds at any time post-infection (Figure 4B).
FIGURE 4.
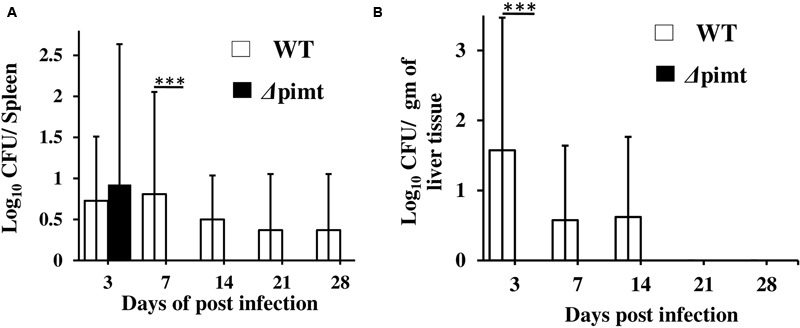
(A) Quantification of bacterial burdens in the spleen of the chicks following oral infection with WT or Δpimt strains of S. Typhimurium. The spleens were collected, triturated in sterile PBS and 100 μl of such homogenates were plated on HE agar plates. The colonies were counted following incubations of the plates and CFUs/spleen were calculated. The data are presented as mean ± SD, n = 5. (B) Quantification of bacterial burdens in the liver of the chicks following oral infection with wild type or Δpimt strains of S. Typhimurium. The liver tissues were collected, 100 mg of tissue was triturated in sterile PBS and 100 μl of such homogenates were plated on HE agar plates. The colonies were counted following incubations of the plates. CFUs/gram of liver were calculated. The data are presented as mean ± SD, n = 5. ∗∗∗p < 0.001.
Discussion
Many covalent modifications to amino acid have been described. Conversion of Asp residues to iso-Asp is one of these several covalent modifications that occur in proteins. Iso-Asp formation has been linked with modulation of protein function(s) (David et al., 1999; Lee et al., 2012; Dimitrijevic et al., 2014). PIMT can repair iso-Asp back to normal Asp. Role of pimt gene in the survival of various organisms under various stress conditions has been reported (Khare et al., 2009). Recently, we have reported the contribution of pimt in the survival of S. Typhimurium under oxidative stress and virulence in mice model (Kumawat et al., 2016). In current study, we have evaluated the contribution of pimt gene in the survival of S. Typhimurium under temperature stress and in stationary phase. Subsequently, we have assessed the contribution of PIMT in the virulence of S. Typhimurium in poultry.
First, we procured five strains of S. Typhimurium and tested their colonization abilities in the poultry. Based on fecal shedding analysis, E-5591 strain of S. Typhimurium showed efficient colonization abilities than other tested strains. After selecting the strain, we constructed pimt gene deletion and complemented strains (Figure 1).
In comparison to the wild type, the Δpimt strain of S. Typhimurium was about 438-fold (p < 0.01) more susceptible to 42°C exposure. Complemented strain showed intermediate sensitivity which may be due to expression of PIMT from plasmid pQE-60 in complemented strain (Figure 2A). At 37°C the growth of Δpimt strain was similar as of S. Typhimurium for up to 36 h. However, the mutant strain (as compared to wild type) showed significant growth difference after 72 h (i.e., in very late stationary phase) but the susceptibility was only 12-fold more than wild type (Figure 2B).
Next, we wondered if PIMT protein gets induced following exposure of S. Typhimurium to elevated temperature. Interestingly, temperature stress (42°C) has more effect on PIMT induction (Figure 3, about 3.5-fold) as compare to oxidative stress as observed in our earlier study (Kumawat et al., 2016, 1.5-fold). Taken together, our current and previous data (Kumawat et al., 2016) suggest that pimt is primarily required for S. Typhimurium survival against temperature stress and secondarily aids to the survival of this bacterium against oxidants and in stationary phase.
pimt gene contributes to the survival of C. elegans (Khare et al., 2009), E. coli and Salmonella Typhimurium (Kumawat et al., 2016) under oxidative stress conditions. Few studies have suggested the importance of pimt gene in survival of E. coli under heat stress and in stationary phase (Visick et al., 1998). Further, in E. coli over expression of PIMT inhibited the aggregation of β-galactosidase at 43°C (a protein that aggregates at 43°C), decreases the level of iso-aspartates in this protein, and increases its thermal stability (Kern et al., 2005), suggesting a direct role of PIMT in protection of proteins at elevated temperatures. Interestingly, significantly higher PIMT specific activities were observed following exposure of HeLa cells and Arabidopsis at elevated temperatures (Ladino and O’Connor, 1990; Villa et al., 2006).
Oxidative stress and high body temperature (42°C) of the birds are two important stresses that Salmonella encounters inside the poultry. We evaluated the colonization abilities of Δpimt strain in poultry. Wild type S. Typhimurium colonized in caecum of 100% infected birds. However, Δpimt strain showed some colonization initially but was failed produce a chronic infection (Table 3). Similarly, Δpimt showed compromised dissemination to spleen and not at all able to invade liver (Figure 4). Taken together, our data suggest that pimt contributes to the S. Typhimurium virulence in poultry.
Bacteria encode an array of factors which help them to alleviate various environmental and host generated stress that they encounter. A combination of these virulence factors ensures successful bacterial colonization and survival in the host. Protein repair enzymes can serve as one category of such virulence factors. Protein repair enzymes, including PIMT may not act directly as virulence factors. However, by repairing key iso-aspartate residues in proteins (iso-Asp containing proteins), PIMT might be help bacterial survival in the host (heat stress in this case of S. Typhimurium). The observed phenotype of Δpimt mutant might be a combination effect of functions of these target genes which required during colonization of S. Typhimurium. Therefore, it would be interesting to identify these iso-Asp containing proteins and assess their contribution in the survival of S. Typhimurium under heat stress and virulence.
Author Contributions
PP, TG, RA, and MM designed the experiments. PP, MK, PB, and SD performed all the experiments. PP and MK analyzed the data. PP and MM wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LS and handling Editor declared their shared affiliation and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The funds for current study were provided by NASF, ICAR, India. We acknowledge the support and facilities provided by the Director, IVRI.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00361/full#supplementary-material
Polymerase chain reaction confirmation of presence of virulence associated (stn, hilA, and invA) and Typhimurium specific (typh) genes in ST E-5591. Above mentioned genes were amplified from genomic DNA of S. Typhimurium E-5591 and analyzed on 1% agarose gel. PCR products are marked by arrows.
References
- Alvarez J., Sota M., Vivanco A. B., Perales I., Cisterna R., Rementeria A., et al. (2004). Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42 1734–1738. 10.1128/JCM.42.4.1734-1738.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Ordóñez A., Fernández A., López M., Arenas R., Bernardo A. (2008). Modifications in membrane fatty acid composition of Salmonella typhimurium in response to growth conditions and their effect on heat resistance. Int. J. Food Microbiol. 123 212–219. 10.1016/j.ijfoodmicro.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Besser T. E., Goldoft M., Pritchett L. C., Khakhria R., Hancock D. D., Rice D. H., et al. (2000). Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol. Infect. 124 193–200. 10.1017/S0950268899003283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadei M. A., Manas P., Niven G., Needs E., Mackey B. M. (2002). Role of membrane fluidity in pressure resistance of Escherichia coli NCTC 8164. Appl. Environ. Microbiol. 68 5965–5972. 10.1128/AEM.68.12.5965-5972.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Bartlett J. A., Corey G. R. (1987). Extra-intestinal manifestations of Salmonella infections. Medicine 66 349–388. 10.1097/00005792-198709000-00003 [DOI] [PubMed] [Google Scholar]
- Cooper M. A., Washburn K. W. (1998). The relationships of body temperature to weight gain, feed consumption, and feed utilization in broilers under heat stress. Poult. Sci. 77 237–242. 10.1093/ps/77.2.237 [DOI] [PubMed] [Google Scholar]
- Darwin K. H., Miller V. L. (1999). Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12 405–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. L., Keener J., Aswad D. W. (1999). Isoaspartate in ribosomal protein S11 of Escherichia coli. J. Bacteriol. 181 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrijevic A., Qin Z., Aswad D. W. (2014). Isoaspartyl formation in creatine kinase B is associated with loss of enzymatic activity; implications for the linkage of isoaspartate accumulation and neurological dysfunction in the PIMT knockout mouse. PLoS ONE 9:e100622 10.1371/journal.pone.0100622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkoh A. (1989). Ambient temperature: a factor affecting performance and physiological response of broiler chickens. Int. J. Biometeorol. 33 259–265. 10.1007/BF01051087 [DOI] [PubMed] [Google Scholar]
- Foster J. W., Spector M. P. (1995). How Salmonella survive against the odds. Annu. Rev. Microbiol. 49 145–174. 10.1146/annurev.mi.49.100195.001045 [DOI] [PubMed] [Google Scholar]
- Galan J. E., Ginocchio C., Costeas P. (1992). Molecular and functional characterization of the Salmonella invasion gene invA: homology of invA to members of a new protein family. J. Bacteriol. 174 4338–4349. 10.1128/jb.174.13.4338-4349.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O., Boyle E. C., Grassl G. A. (2014). Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5:391 10.3389/fmicb.2014.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin A. J., McSorley S. J. (2011). Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 4 371–382. 10.1038/mi.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw J. P., Jelesarov I., Siegenthaler R. K., Christen P. (2003). Thermosensor action of GrpE. The DnaK chaperone system at heat shock temperatures. J. Biol. Chem. 278 19048–19053. 10.1074/jbc.M300924200 [DOI] [PubMed] [Google Scholar]
- Guo X., Chen J., Beuchat L. R., Brackett R. E. (2000). PCR Detection of Salmonella enterica serotype Montevideo in and on raw tomatoes using primers derived from hilA. Appl. Environ. Microbiol. 66 5248–5252. 10.1128/AEM.66.12.5248-5252.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks W. M., Kotlajich M. V., Visick J. E. (2005). Recovery from long-term stationary phase and stress survival in Escherichia coli require the L-isoaspartyl protein carboxyl methyltransferase at alkaline pH. Microbiology 151 2151–2158. 10.1099/mic.0.27835-0 [DOI] [PubMed] [Google Scholar]
- Kern R., Malki A., Abdallah J., Liebart J. C., Dubucs C., Yu M. H., et al. (2005). Protein isoaspartate methyltransferase is a multicopy suppressor of protein aggregation in Escherichia coli. J. Bacteriol. 187 1377–1383. 10.1128/JB.187.4.1377-1383.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Gomez T., Linster C. L., Clarke S. G. (2009). Defective responses to oxidative stress in protein L-isoaspartyl repair-deficient Caenorhabditis elegans. Mech. Ageing Dev. 130 670–680. 10.1016/j.mad.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo C. H., Sim J. H., Salleh N. A., Cheah Y. K. (2015). Pathogenicity and phenotypic analysis of sopB, sopD and pipD virulence factors in Salmonella enterica serovar typhimurium and Salmonella enterica serovar Agona. Antonie Van Leeuwenhoek 107 23–37. 10.1007/s10482-014-0300-7 [DOI] [PubMed] [Google Scholar]
- Kumawat M., Pesingi P. K., Agarwal R. K., Goswami T. K., Mahawar M. (2016). Contribution of protein isoaspartate methyl transferase (PIMT) in the survival of Salmonella Typhimurium under oxidative stress and virulence. Int. J. Med. Microbiol. 306 222–230. 10.1016/j.ijmm.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Ladino C. A., O’Connor C. M. (1990). Protein carboxyl methylation and methyl ester turnover in density-fractionated human erythrocytes. Mech. Ageing Dev. 55 123–137. 10.1016/0047-6374(90)90020-G [DOI] [PubMed] [Google Scholar]
- Lee J. C., Kang S. U., Jeon Y., Park J. W., You J. S., Ha S. W., et al. (2012). Protein L-isoaspartyl methyltransferase regulates p53 activity. Nat. Commun. 3 927 10.1038/ncomms1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Clarke S. (1992). Distribution of an L-isoaspartyl protein methyltransferase in Eubacteria. J. Bacteriol. 174 355–361. 10.1128/jb.174.2.355-361.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linam W. M., Gerber M. A. (2007). Changing epidemiology and prevention of Salmonella infections. Pediatr. Infect. Dis. J. 26 747–748. 10.1097/INF.0b013e3181376abc [DOI] [PubMed] [Google Scholar]
- MacLennan C. A., Martin L. B., Micoli F. (2014). Vaccines against invasive Salmonella disease: current status and future directions. Hum. Vaccin. Immunother. 10 1478–1493. 10.4161/hv.29054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz S. E., Musto J., Scallan E., Angulo F. J., Kirk M., O’Brien S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50 882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- Makino S. I., Kurazono H., Chongsanguam M., Hayashi H., Cheun H. I., Suzuki S., et al. (1999). Establishment of the PCR system specific to Salmonella spp. and its application for the inspection of food and fecal samples. J. Vet. Med. Sci. 61 1245–1247. 10.1292/jvms.61.1245 [DOI] [PubMed] [Google Scholar]
- Oliveira S. D., Santos L. R., Schuch D. M. T., Silva A. B., Salle C. T. P., Canal C. W. (2002). Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 87 25–35. 10.1016/S0378-1135(02)00028-7 [DOI] [PubMed] [Google Scholar]
- Pavan Kumar P., Agarwal R. K., Thomas P., Sailo B., Prasannavadhana A., Kumar A., et al. (2014). Rapid detection of Salmonella enterica subspecies enterica serovar Typhimurium by loop mediated isothermal amplification (LAMP) test from field chicken meat samples. Food Biotechnol. 28 50–62. 10.1080/08905436.2013.870911 [DOI] [Google Scholar]
- Rahman H. (2002). Some aspects of molecular epidemiology & charcterisation of Salmonella typhimurium isolated from man & animals. Indian J. Med. Res. 115 108–112. [PubMed] [Google Scholar]
- Raspoet R., Appia-Ayme C., Shearer N., Martel A., Pasmans F., Haesebrouck F., et al. (2014). Microarray-based detection of Salmonella enterica serovar Enteritidis genes involved in chicken reproductive tract colonization. Appl. Environ. Microbiol. 80 7710–7716. 10.1128/AEM.02867-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar B., Khachatourians G. G., Korber D. R. (2004). Treatment of Salmonella enterica serovar Enteritidis with a sublethal concentration of trisodium phosphate or alkaline pH induces thermotolerance. Appl. Environ. Microbiol. 70 4613–4620. 10.1128/AEM.70.8.4613-4620.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa S. T., Xu Q., Downie A. B., Clarke S. G. (2006). Arabidopsis protein repair L-isoaspartyl methyltransferases: predominant activities at lethal temperatures. Physiol. Plant. 128 581–592. 10.1111/j.1399-3054.2006.00772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick J. E., Cai H., Clarke S. (1998). The L-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J. Bacteriol. 180 2623–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T., Heidrich N., Brantl S., Narberhaus F. (2007). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65 413–424. 10.1111/j.1365-2958.2007.05794.x [DOI] [PubMed] [Google Scholar]
- Yeung C. Y., Liu C. C., Tseng Y. T., Tsai K. C., Hsieh M. A., Chan W. T., et al. (2014). Rapid identification of Salmonella using Hektoen enteric agar and 16s ribosomal DNA probe-gold nanoparticle immunochromatography assay in clinical faecal specimens. Lett. Appl. Microbiol. 58 311–317. 10.1111/lam.12191 [DOI] [PubMed] [Google Scholar]
- Zhang S., Kingsley R. A., Santos R. L., Andrews-Polymenis H., Raffatellu M., Figueiredo J., et al. (2003). Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71 1–12. 10.1128/IAI.71.1.1-12.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Polymerase chain reaction confirmation of presence of virulence associated (stn, hilA, and invA) and Typhimurium specific (typh) genes in ST E-5591. Above mentioned genes were amplified from genomic DNA of S. Typhimurium E-5591 and analyzed on 1% agarose gel. PCR products are marked by arrows.


