Abstract
The molecular biology of spuma or foamy retroviruses is different from that of the other members of the Retroviridae. Among the distinguishing features, the N-terminal domain of the foamy virus Env glycoprotein, the 16-kDa Env leader protein Elp, is a component of released, infectious virions and is required for particle budding. The transmembrane protein Elp specifically interacts with N-terminal Gag sequences during morphogenesis. In this study, we investigate the mechanism of Elp release from the Env precursor protein. By a combination of genetic, biochemical, and biophysical methods, we show that the feline foamy virus (FFV) Elp is released by a cellular furin-like protease, most likely furin itself, generating an Elp protein consisting of 127 amino acid residues. The cleavage site fully conforms to the rules for an optimal furin site. Proteolytic processing at the furin cleavage site is required for full infectivity of FFV. However, utilization of other furin proteases and/or cleavage at a suboptimal signal peptidase cleavage site can partially rescue virus viability. In addition, we show that FFV Elp carries an N-linked oligosaccharide that is not conserved among the known foamy viruses.
In retroviruses, proteolytic processing of the structural Gag and enzymatic Pol proteins is executed by the virus-encoded protease (6). In contrast, the Env glycoprotein precursors are considered to be processed by two cellular proteases (8, 13, 19). The first cleavage occurring in the endoplasmic reticulum (ER) removes the signal peptide (SP). In analogy to cellular secretory or transmembrane (TM) proteins, proteolytic removal of the SP which targets the Env precursor to the secretory pathway via the ER and Golgi apparatus is considered to be performed by cellular signal peptidases (SPases) (for a review, see references 17 and 19). The second cleavage, which separates the surface (SU) and TM domains, is catalyzed by a member of the furin convertase protease family, often the furin protease itself (8, 28). Related glycoprotein processing mechanisms also apply to a wide variety of other membrane viruses such as influenza virus and Ebola virus (4).
The processing of cellular type I membrane proteins (or even polyprotein precursors) by ER-resident SPases has been exploited by many viruses leading to SPs of about 15 to more than 50 amino acid residues independent of whether the protein is of viral or cellular origin (17). In addition, the SP is rapidly degraded by signal peptide peptidases, a process which is often directly linked to the presentation of the corresponding cleavage products by the major histocompatibility complex I for purposes of immune surveillance (17).
However, for two distinct foamy retroviruses, namely human and feline foamy virus (HFV and FFV), the 14- to 18-kDa N-terminal Env cleavage product was clearly detectable as a component of released virus particles (11, 14, 15, 35). We showed that the N terminus of FFV Env, termed the Env leader protein (Elp), is located inside particles, confirming that specific interactions between this part of Elp and Gag can occur (11, 35). Thus, the unusually large size and intrinsic stability of the N-terminal cleavage product of foamy virus (FV) Env indicate that it is significantly different from the classical SPs of cellular or viral proteins.
The uniqueness of Elp may be related to the fact that FV morphology is clearly distinct from that of the orthoretroviruses, and it is one of the criteria for their systematic placement into a separate subfamily (22). Recent studies on the genetic relatedness, mode of replication, gene expression, and, in particular, structural and functional aspects of FV morphology, morphogenesis, and Env protein function confirmed their distinct nature compared to orthoretroviruses (22). FV Env complexes have been visualized as trimeric complexes arranged in hexameric rings on the surface of negatively stained FV particles (34). Electron microscopy images of a corresponding clearness have not been obtained for any other retrovirus, supporting the view that FV Env complexes are intrinsically stable. In line with these findings, recent data even indicate that the full-length FV Env has the intrinsic capacity to direct budding of virus-like particles independent of Gag or any other viral protein (25). Finally, Elp, as a component of the released infectious virion, is absolutely required for particle budding (15, 20).
These data indicate that Elp not only has the function of a classical SP directing the viral Env proteins into the secretory pathway but also mediates Env-Gag interactions essential for particle release (35). The large size of Elp (about 16 kDa), significantly exceeding that of known SPs, and the absence of consensus motifs for cellular SPases at the appropriate position indicate that proteolytic processing of FV Elp is unique. Indeed, we recently provided evidence that a second virion-associated N-terminal FFV Env protein of 9 kDa is generated by cellular signal peptidases (11) at a site already predicted by sequence comparisons (33).
By comparative sequence analysis, we identified a furin recognition sequence RXXR↓D (the cleavage site is marked by the arrow) in all known FV Env sequences at about Env residue 130. In FFV, the sequence RRRR↓ even corresponds to a site predicted to be very efficiently processed by furin. Furin and other members of the Golgi-resident protein convertase protease family are known to cleave retroviral and nonretroviral glycoprotein precursors (28). In retroviruses, this cleavage separates the SU and TM domains of Env, an event that is absolutely required for induction of membrane fusion by TM after SU-mediated receptor recognition (3, 8, 13).
In this study we show that, in contrast to orthoretroviruses, furin itself or a furin-like protease processes the FV Env glycoprotein precursor at two distinct sites, resulting in the mature FV Elp, SU, and TM proteins.
MATERIALS AND METHODS
Virus and cell culture techniques.
293T cells, Crandell feline kidney (CRFK) cells, and FFV-FAB titration cells were grown as reported previously (36, 37). FFV isolate FUV was propagated as described previously (36). Furin-deficient human LoVo cells (kindly provided by H. Fickenscher, Heidelberg, Germany) were propagated in RPMI medium supplemented with 10% fetal calf serum (27). Transfection of recombinant DNA into 293T and LoVo cells was performed by calcium coprecipitation (32) or by using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany).
Purification of released FFV particles.
Cell culture supernatants from FFV-infected CRFK cells were harvested 5 days postinfection when the cells displayed FFV-induced cytopathic effects. Supernatants were cleared by centrifugation at 400 × g for 7 min and at 3,000 × g for 30 min, then filtered through 0.45-μm-pore-size filters, and centrifuged through 5 ml of 20% (wt/vol) sucrose in phosphate-buffered saline for 2 h at 24,000 rpm in an SW28 rotor (Beckman, Munich, Germany). The resulting pellets were gently resuspended in phosphate-buffered saline at 4°C (34).
For preparative purification of FFV Elp, supernatants from 25 150-cm2 flasks of FFV-infected CRFK cells harvested 5 days postinfection were used to enrich FFV particles by centrifugation through sucrose and banding in Optiprep as described previously (11). The pelleted virions were denatured, deglycosylated in 100 μl with 1 U of PNGase F (see below), and separated on precast 12% NuPAGE gels (Invitrogen). After nondestructive silver staining (26), the virus-specific band comigrating with the 16.5-kDa marker protein was excised, eluted, washed, digested with trypsin or chymotrypsin, and analyzed by mass spectrometry (MS) (see below).
Deglycosylation.
Before digestion of protein samples with the N-glycosidase PNGase F (New England Biolabs, Frankfurt, Germany) and O-glycosidase (Roche, Mannheim, Germany), samples were denatured in 0.5% sodium dodecyl sulfate (SDS) and 1% 2-mercaptoethanol for 10 min at 100°C (16, 30). To avoid enzyme inhibition by SDS, samples were diluted fivefold and adjusted with 1% NP-40 and 50 mM sodium phosphate, pH 7.5 (30). Reactions were performed at 37°C as indicated.
Furin digestion and inhibition of furin activity.
Proteins were digested with furin according to the manufacturer's instructions (New England Biolabs) in 100 mM HEPES-KOH (pH 7.5), 0.5% Triton X-100, 1 mM CaCl2, and 1 mM 2-mercaptoethanol. The furin inhibitor (FI) decanoyl-Arg-Val-Lys-Arg-chloromethylketone (dec-RVKR-cmk; Bachem, Heidelberg, Germany) was dissolved in dimethyl sulfoxide (DMSO) as a stock solution of 7.4 μg/μl (9, 10). For use in FFV-infected CRFK cells, the cell culture medium was supplemented with 1 mM HEPES-KOH (pH 7.5) and dec-RVKR-cmk was used at a final concentration of 14.8 μg/ml. The FI-containing medium was changed twice a day, and the used medium was pooled for purification of released particles. During in vitro protein expression, dec-RVKR-cmk was used as specified.
Molecular cloning and plasmids.
To functionally inactivate the furin recognition site in the infectious FFV provirus, a fusion PCR-mediated mutagenesis was performed as described previously (1). FFV Env sequences were amplified by PCRs with primers F6550s (5′-TCAATAGTTACCTTGAGTACA-3′) plus ΔRRa (5′-GGTATTGTATATCAGATCTCCGCGAGGAGGAAGAGGAAGTATTTCC-3′; altered residues are underlined, and numbers refer to the 5′ end of the primers in the FFV genome) (36) and ΔRs (5′-CGGAGATCTGATATACAATACCACAAAC-3′) plus F7325a (5′-GCAAAGGATTGTTGTACCGA-3′) with cloned pFeFV-7 DNA as the template (37) and the proofreading polymerase Pfu (Stratagene, Amsterdam, The Netherlands). The gel-purified PCR products were combined and used as a template for a final PCR with primers F6550s and F7325a. The amplicon of about 800 nucleotides (nt) was digested with AflII and PmlI and inserted into the correspondingly digested FFV genome pFeFV-7 (37). Independent correct clones were identified and designated pFeFV-ARRA.
For in vitro expression of FFV Env, env and bel sequences were excised from pFeFV-7 with BsrGI (nt position 6169) and XbaI (nt position 9958) and inserted into the Acc65I- and XbaI-digested vector pBluescriptSK+ (Stratagene), generating plasmid pBl-FFV-Env. To truncate the FFV env sequences in pBl-FFV-Env after residue 146, the N-terminal 146 residues of Elp were amplified by PCR with primers Env-s (5′-GCTCATGATGGAACAAGAACATGTG-3′; the introduced BspHI site is underlined) and Elp-as (5′-GGAATTCTCAACCTTGTGGGATCCCTGA-3′; the introduced EcoRI site is underlined). The amplicon was digested with SalI (nt position 6393) and inserted into the SalI- and Ecl136II-digested vector pBl-FFV-Env. The truncated clone was designated pBl-FFV-EnvN.
The eukaryotic FFV Env expression plasmid pBC-FFV-Env was constructed by PCR with pFeFV-7 DNA as a template, Pfu polymerase, and primers 6319s (5′-GCCACCATGGAACAAGAACATGTGATGAC-3′) and 9267as (5′-GGCCCCCGGGTTATTGGTCCTTCTTCCGGGTA-3′; the XmaI site is underlined). The amplicon was digested with XmaI and inserted between the blunt-ended HindIII and XmaI sites of vector pBC12CMV (7).
Plasmid pSG-furin, driving the expression of bovine furin, and the empty vector backbone pSG5, together with a rabbit serum directed against bovine furin, were obtained from Wolfgang Garten, Marburg, Germany (23).
In vitro expression and processing of radioactively labeled Elp.
The expression of pEnvN from the recombinant plasmid pBl-FFV-EnvN was conducted in the TNT coupled reticulocyte lysate (Promega, Mannheim, Germany) by using the prokaryotic T7 promoter located directly upstream of the FFV env sequences. TNT reactions were run with and without canine microsomes, according to the manufacturer's protocol (Promega). Proteins were labeled by incorporation of [35S]methionine (Amersham Biosciences, Freiburg, Germany) during translation. Samples were separated by polyacrylamide gel electrophoresis (PAGE), and the gel was treated with Amplify (Amersham Biosciences), dried, and exposed to X-ray films.
Immunoblotting.
Sampling of cellular and viral antigens and immunoblotting of proteins separated on denaturing gels with antisera against FFV proteins and cat serum 8014 by enhanced chemiluminescence (ECL) were as described previously (1, 2, 35).
MS analysis of proteolytically digested proteins.
Silver-stained protein bands from SDS gels were repeatedly washed with water, 40 mM ammonium bicarbonate-ethanol (1:1, vol/vol), reduced with 10 mM dithiothreitol at 56°C for 1 h, and alkylated with 55 mM iodoacetamide at 25°C for 30 min in the dark. After alkylation, gel bands were repeatedly washed with 40 mM ammonium bicarbonate and ethanol, dehydrated with 100% acetonitrile, and air dried for 15 min. Digestion with trypsin (10 ng/μl) was performed in 40 mM ammonium bicarbonate at 37°C overnight. When chymotrypsin was used, the bicarbonate buffer was replaced with 100 mM Tris-HCl (pH 7.8) for reduction, alkylation, and washing of protein bands. Digestion with chymotrypsin (17 ng/μl) was carried out in 100 mM Tris-HCl (pH 7.8) and 10 mM CaCl2 at 25°C for 3 h.
Matrix-assisted laser desorption ionization (MALDI) mass spectra were recorded in the positive-ion mode with delayed extraction on a Reflex II (Bruker-Daltonik) time of flight (TOF) instrument equipped with a SCOUT-26 inlet and a 337-nm nitrogen laser. Mass spectra were obtained by accumulation of 50 to 200 individual laser shots. Calibration of the spectra was performed internally by a two-point linear fit by using the autolysis products of trypsin at an m/z of 842.50 and an m/z of 2,211.10 or chymotryptic autolysis products at an m/z of 705.47 and an m/z of 1,783.88.
For the MS analysis of tryptic and chymotryptic digests, MALDI samples were prepared by reverse-phase extraction by using ZipTip C18 according to the manufacturer's instructions (Millipore). Elution and spotting onto individual spots on the target were obtained with a 50% acetonitrile-water solution saturated with α-cyano-4-hydroxycinnamic acid (21).
Postsource decay analysis was performed in the positive-ion reflector mode with delayed extraction by setting an ion gate width of 40 Da around the ion of interest. Data were acquired in 14 segments by decreasing the reflector voltage in a stepwise fashion. For each segment, 100 to 200 individual laser shots were accumulated. The fragment ion spectrum was obtained by pasting together all segments to a single spectrum by using the FAST software provided by Bruker. Fragment ion calibration was performed externally with the fragment masses of the adrenocorticotropic hormone 18-39 clip (12).
RESULTS
Identification of a furin consensus site in the N terminus of FFV Env.
A recognition site for a cellular furin protease, RRRR↓X (Fig. 1A), is located between FFV Env residues 124 and 128, with the cleavage predicted to occur between residues 127 and 128. To study whether this sequence can be cleaved by the prototypic furin protease, the synthetic peptide NH2-TSSSSRRRRDIQYHK-COOH (the furin recognition motif is in boldface type; Peptide Specialty Laboratories GmbH, Heidelberg, Germany) encompassing FFV Env residues 119 to 133 (Fig. 1A) was subjected to furin cleavage. Reaction products were analyzed by MALDI-MS. Whereas the substrate peptide at an m/z of 1,876.99 was no longer detectable upon furin digestion, an ion signal at an m/z of 1,092.64, corresponding to the N-terminal sequence TSSSSRRRR was clearly detectable, confirming the presence of the predicted furin cleavage site (data not shown). The C-terminal cleavage product (m/z of 803.41) was not detectable due to the presence of strong ion signals in this mass range originating from detergent in the cleavage buffer (5). No other cleavage product could be observed in the corresponding MALDI spectrum.
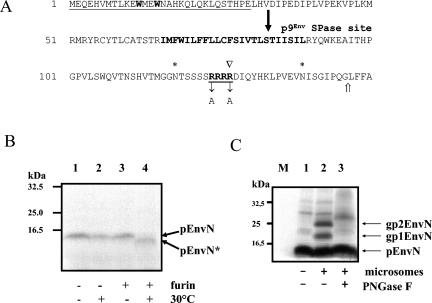
FIG. 1.
Identification of a furin cleavage site between FFV Env residues 127 and 128 by using in vitro-synthesized pEnvN protein. (A) Sequence of the N-terminal 150 amino acid residues (in single-letter code) of FFV Env with the following features marked: residues W12 and W15 required for Gag-Env interaction in boldface type; the region against which the Elp serum is directed is underlined; the hydrophobic TM anchor is marked in boldface type; potential N-glycosylation sites are marked by asterisks above the sequence; the RRRR motif specifying a furin cleavage site (open arrowhead) is underlined and in boldface type, with the Arg-Ala exchanges of the mutant pFeFV-ARRA indicated below the sequence; the end of the in vitro-synthesized N-terminal Env protein pEnvN is marked by an open arrow. The solid arrow marks a potential SPase cleavage site (11, 18, 33). (B) To characterize in vitro-synthesized pEnvN, TNT reactions were run with pBl-FFV-EnvN template DNA, supplemented with furin buffer, and incubated at 30°C for 120 min in the absence (−) and presence (+) of furin, as indicated below the autoradiogram. Samples in lanes 1 and 3 were taken before the reactions were started. The reaction products were analyzed by gel electrophoresis and autoradiography. The in vitro-synthesized pEnvN protein with an apparent molecular mass of 14 kDa is marked by an arrow. Incubation with furin (lane 4) resulted in the disappearance of the 14-kDa band and the appearance of the 12-kDa pEnvN* furin cleavage product (arrow). (C) In vitro glycosylation of pEnvN. As above, pEnvN was in vitro synthesized in the absence (lane 1) or presence of microsomes, as indicated below the lanes. The reaction products were separated by PAGE and visualized by autoradiography. In all reactions, the primary translation product pEnvN is clearly present. The addition of microsomes induced two specific bands of about 19 (gp1EnvN) and 23 (gp2EnvN) kDa (lane 2) that were both susceptible to digestion with the N-glycosidase PNGaseF (lanes 3). The positions of marker proteins separated in parallel are shown to the left.
Characterization of in vitro-synthesized Elp.
To substantiate the furin-mediated processing of the FFV Env peptide, we used a eukaryotic in vitro transcription and translation system (TNT; Promega) to generate a radioactively labeled N-terminal Env protein for biochemical analyses. For this purpose, FFV Env residues 1 to 146 covering the furin consensus site were cloned 3′ of the T7 RNA polymerase promoter in the pBluescriptSK+ vector and used for in vitro Env expression. The encoded pEnvN protein encompassing Env residues 1 to 146 corresponds in size to that predicted for HFV Elp (Fig. 1A) (15).
Initially, TNT reactions were run with and without pBl-FFV-EnvN template DNA. Reactions with the Env template followed by gel electrophoresis and autoradiography yielded a clear band with an apparent molecular mass of 14 kDa and no reaction products in the absence of the Env template DNA (data not shown). To investigate whether the in vitro-synthesized 14-kDa pEnvN can be processed by furin, pEnvN TNT reactions were supplemented with furin, incubated at 30°C for 2 h, and analyzed (Fig. 1B, lanes 1 to 4). During furin incubation, pEnvN was processed to pEnvN* of about 12 kDa (Fig. 1B, lane 4; Fig. 2B [schematic presentation of TNT-derived Elp proteins]). This processing was specifically abolished by the addition of the furin inhibitor (FI) dec-RVKR-cmk (10) (data not shown), confirming the presence of a cleavable furin site in FFV pEnvN.
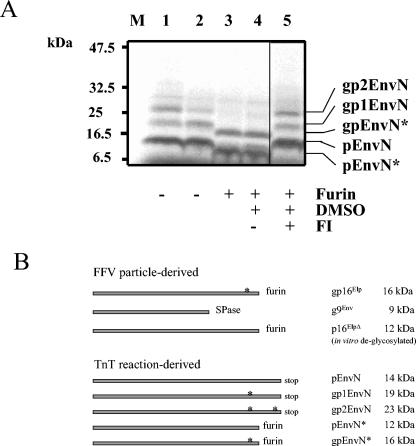
FIG. 2.
In vitro glycosylation and furin processing of pEnvN and schematic presentation of reaction products. (A) pEnvN was in vitro synthesized in the presence of microsomes. After completion of the TNT reaction, an aliquot was directly mixed with gel loading buffer (lane 1), the rest was supplemented with furin reaction buffer, and aliquots were incubated for 4 h at 30°C with (+) 0.1 U of furin (lanes 3 to 5) or without (−) furin (lane 2). As a control for solvent effects, the sample in lane 4 was supplemented with DMSO, the sample in lane 5 contained 620-μg/ml FI in DMSO, and lane 3 did not contain DMSO or FI. Reaction products were separated by PAGE and visualized by autoradiography. In the reaction mixtures in lanes 1 and 2, the primary translation product pEnvN and the glycosylated forms gp1EnvN and gp2EnvN were clearly present. The addition of furin (lanes 3 and 4) resulted in the disappearance of these bands and the appearance of two new bands of 16 (gpEnvN*) and 12 (pEnvN*) kDa that were both repressed by FI (lane 5). The positions of marker proteins separated in parallel are shown on the left. (B) Comparative schematic presentation of FFV virion-derived Elp and p9Env proteins (top) and the pEnvN-derived proteins generated in TNT reactions (bottom). The sizes of the different proteins starting at Env residue 1 are schematically given by the size of the bars. The C termini were generated either by furin, SPase, or a stop codon in the TNT template DNA, as indicated. Protein modifications by glycosylation are marked by asterisks. For each protein, the name and its apparent molecular mass are given. Note that gp16Elp and p16ElpΔ are identical to gpEnvN* and pEnvN*, respectively.
Modification of in vitro-synthesized Elp.
To study the modification and processing of Elp, TNT synthesis of pEnvN was performed in the presence of microsomes. Microsomes are membrane vesicles derived from the ER and the Golgi apparatus known to contain enzymes required for the processing and modification of retroviral Env glycoproteins (13).
The major TNT translation product generated in the absence and presence of microsomes (Fig. 1C, lanes 1 and 2) was the 14-kDa pEnvN protein. The reaction with microsomes clearly resulted in two additional bands of 19 and 23 kDa (Fig. 1C, lane 2, gp1EnvN and gp2EnvN; Fig. 2B). Both bands were completely sensitive to digestion with the N-glycosidase PNGaseF (lane 3). The detection of two distinct N-glycosylation products of pEnvN directly corresponds to the presence of two potential N-glycosylation sites at Env residues N118 and N139 in the recombinant pEnvN (Fig. 1A). The glycosylation site at N139 is C-terminal to the furin cleavage site and should thus not be present in the mature Elp that is expected to extend only to R127. Further processing was not detectable in TNT reactions supplemented with microsomes.
In vitro-derived Elp is similar to that from FFV particles.
TNT reactions supplemented with microsomes were performed in the presence and absence of furin to mimic the situation in FFV-infected cells. Without furin, the primary translation product pEnvN and the two different N-glycosylated forms gp1EnvN and gp2EnvN were present (Fig. 2A, lanes 1 and 2, and Fig. 2B). All three bands almost completely disappeared upon addition of furin (lanes 3 and 4). Instead, two strong bands corresponding in size to the virion-derived glycosylated Elp (designated gpEnvN*) as well as the 12-kDa furin cleavage product pEnvN* derived from the unglycosylated pEnvN appeared (Fig. 1B). Reaction products marked by asterisks (Fig. 2B) had been generated by furin, since the FI dec-RVKR-cmk (but not the solvent DMSO) (lane 4) completely suppressed these bands (Fig. 2A, lane 5).
To directly compare the in vitro-processed Elp protein with the authentic, virion-derived Elp, we performed pEnvN TNT reactions with microsomes and furin and analyzed the reaction products in parallel with Elp from FFV particles subjected to in vitro furin digestion (Fig. 3). The in vitro-generated pEnvN proteins were visualized by autoradiography, whereas the virion-derived Elp and p9Env were detected by immunoblotting with the Elp serum. Alignment of the images and comparisons with the marker proteins separated in parallel show that the in vitro-generated gpEnvN* perfectly comigrates with the virion-derived mature Elp protein (Fig. 3, lanes 2 and 3), consistent with their identical molecular structures, depicted in Fig. 2B.
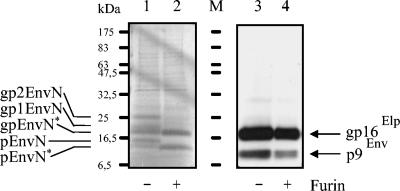
FIG. 3.
Comigration of virion-derived and in vitro-synthesized Elp. pEnvN was in vitro synthesized in the TNT system in the presence of microsomes, and an aliquot of the reaction product was subsequently digested with furin (lanes 1 and 2, indicated below the lanes). In parallel, released FFV particles were enriched from supernatants of FFV-infected cells by sedimentation through a sucrose cushion. An aliquot of the FFV particles was digested with furin in parallel with the in vitro-synthesized pEnvN proteins (lanes 3 and 4, indicated below the lanes). The reaction products were analyzed on a single gel separated by a lane loaded with prestained molecular mass markers (lane M). Lanes 1 and 2 together with a part of the marker lane were dried, and the labeled reaction products were visualized by autoradiography (marked on the left). The remainder of the gel (part of lane M and lanes 3 and 4) was subjected to immunoblotting with the Elp serum and detected by ECL (marked on the right). The alignment of the images and comparisons with the marker proteins show that the in vitro-generated gpEnvN* perfectly comigrates with the mature virion-derived gp16Elp. The positions of marker proteins are given on the left. +, present; −, absent.
For HFV, Elp was suggested to extend about 20 residues beyond the furin site described here (15). FFV Elp is unlikely to exceed the furin site, since furin treatment of FFV particles (under conditions suited for the complete digestion of the engineered pEnvN) did not result in detectable digestion of Elp (Fig. 3, lane 4).
Inactivation of the N-terminal furin consensus site in FFV Env results in reduced Elp amounts.
The function of the furin consensus site RRRR↓D (Fig. 1A), located between FFV Env residues 127 and 128, was analyzed in the proviral context. To this end, the two arginine residues (Fig. 1A) essential for furin cleavage (28) were converted to alanines, as indicated. The R-A substitution ARRA was introduced into the full-length infectious FFV DNA clone pFeFV-7 to study the phenotype of these exchanges on the replication of the resulting pFeFV-ARRA mutant.
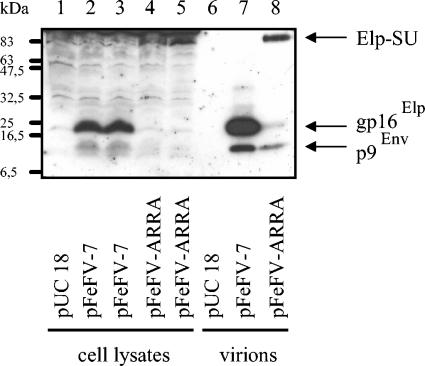
FFV mutant pFeFV-ARRA, wild-type (wt) clone pFeFV-7, and pUC18 control DNA were transiently transfected in duplicate into 293T cells. Three days after transfection, syncytium formation of clone pFeFV-ARRA was clearly reduced relative to the wt genome. At this time, released FFV particles were enriched by sedimentation through 20% sucrose. Equal volumes of the particles were analyzed by immunoblotting. As determined with cat serum 8014 (2), the processing of Gag and Pol proteins was not affected by the mutation. However, the overall yield of particles was reduced (data not shown). Using an Elp-specific antiserum, the authentic 16-kDa Elp (also designated gp16Elp) and the 9-kDa N-terminal Env SP p9Env (11) were detectable in pFeFV-7-transfected cell culture supernatants (Fig. 4, lane 7). In supernatants from pFeFV-ARRA-transfected cells (Fig. 4, lane 8), the concentration of the authentic Elp was reduced to trace amounts, whereas the intensity of the p9Env SP was only moderately decreased. However, an Elp-reactive protein of about 83 kDa was now detectable. Provided that the R-A mutations had inhibited the proteolytic release of Elp, this novel 83-kDa protein should represent the Elp-SU fusion protein (as indicated). In line with this assumption, the 83-kDa Elp-SU fusion protein was also detected with an FFV Elp-SU antiserum (data not shown). Correspondingly, the mature Elp was not detectable in lysates of pFeFV-ARRA-transfected cells, whereas it was visible in wt-transfected cells (Fig. 4, lanes 2 to 5). Again, the 83-kDa Elp-SU was exclusively present in pFeFV-ARRA-transfected cells. The unprocessed Env precursor of about 130 kDa was not detectable due to unspecific background reactivity in the lysates.
FIG. 4.
Analysis of Env proteins expressed from mutant pFeFV-ARRA. 293T cells were transfected in duplicate with plasmids pFeFV-7 (lanes 2 and 3) and pFeFV-ARRA (lanes 4 and 5). Cellular extracts were harvested 3 days posttransfection, and equal amounts of proteins were subjected to immunoblotting with the Elp-specific antiserum. In parallel, particles were concentrated from the pooled supernatants of the cells and analyzed on the same gel (lanes 7 and 8). pUC18-transfected cells served as controls (lanes 1 and 6). The Elp-related proteins p9Env and gp16Elp and the Elp-SU fusion protein were detected by ECL. Lanes 1 to 5 were intentionally overexposed to detect FFV Elp, p9Env, and the Elp-SU fusion protein.
The FFV infectivity released from the cells was determined. The titer of the pFeFV-ARRA mutant was reduced 4- to 20-fold compared to the parental wt clone pFeFV-7.
Inhibition of Elp processing by an FI.
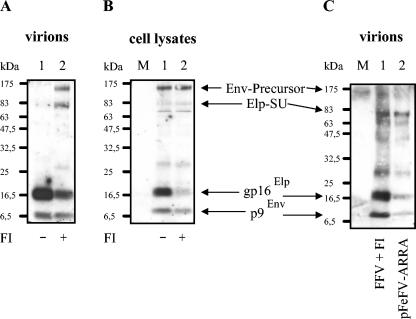
We next utilized the FI dec-RVKR-cmk to inhibit FFV Env processing (10). FI dec-RVKR-cmk is also active against other members of the furin convertase protease family (9, 10). Since furin or a furin-related protease was recently shown to mediate cleavage of HFV Env to yield the SU and TM proteins (3), we used inhibition of the FFV SU-TM cleavage as an independent measure for the efficiency of the inhibitor. CRFK cells were infected with FFV and incubated from 4 to 72 h postinfection in the presence of 7.4 μg of dec-RVKR-cmk/ml. FFV-infected CRFK cells not treated with FI served as controls. Released particles were enriched from the cell culture supernatants by sedimentation through sucrose, and equal volumes of the particles were analyzed by immunoblotting. In supernatants from untreated cells, only the mature Elp and the SP p9Env were detectable with the Elp antiserum (Fig. 5A, lane 1). In contrast, the addition of FI led to decreased Elp concentrations, whereas the unprocessed 130-kDa Env precursor and the 83-kDa Elp-SU became visible (lane 2). Both fusion proteins were absent in the sample not treated with FI (lane 1).
FIG. 5.
Inhibition of Elp processing by the FI dec-RVKR-cmk. FFV-infected CRFK cells were incubated in the presence (+) or absence (−) of 7.4 μg of the FI dec-RVKR-cmk/ml as described in Materials and Methods between 4 and 72 h postinfection. Three days postinfection, the cells and supernatants were harvested. (A) Released particles were enriched from the cell culture supernatants and analyzed by immunoblotting with the Elp antiserum. Without FI, the mature Elp and the 9-kDa SP were detectable. With FI, the unprocessed Env precursor and the 83-kDa Elp-SU protein became visible, whereas only small amounts of the mature Elp were detectable. (B) In the cell-associated antigen, the amount of Elp was significantly reduced by FI. (C) The FFV virion-specific 83-kDa Elp-specific protein generated in the presence of FI (lane 1) and the 83-kDa Elp-specific protein of pFeFV-ARRA-derived particles (lane 2, compare to Fig. 4) comigrate when analyzed by immunoblotting on adjacent gel lanes.
When the cell-associated viral proteins were detected with the Elp antiserum, a reduced processing at the Elp-SU cleavage site leading to a decreased Elp concentration was again detectable in the FI-treated sample (Fig. 5B). The data show that the FI led only to a partial inhibition of furin-mediated FFV Env processing in infected cells.
We next confirmed that the virion-associated Elp-specific protein of 83 kDa detectable upon mutagenesis of the furin consensus site and the protein induced by the FI inhibitor dec-RVKR-cmk have identical molecular masses, arguing that they represent the Elp-SU fusion protein. For this purpose, pFeFV-ARRA-derived particles and FFV virions obtained after partial furin inhibition were separated on the same gel and detected with the Elp antiserum (Fig. 5C). The 83-kDa Elp-reactive protein generated upon FI treatment of FFV-infected cells and that from pFeFV-ARRA particles (lanes 1 and 2) comigrate, supporting the view that both represent the Elp-SU fusion protein.
FI treatment resulted in an about 10-fold decrease of the viral infectivity in the cultures treated with dec-RVKR-cmk. The moderate decrease in titer is probably related to the fact that only a partial inhibition of the furin activity was achieved under the conditions used.
In line with these data and supporting the view that furin itself is responsible for the release of Elp, furin-deficient human LoVo cells (27) exhibit a clearly detectable degree of FFV Elp processing (data not shown) only after overexpression of bovine furin (which is almost identical to human furin) (23, 31).
N118 glycosylation of virion-derived Elp.
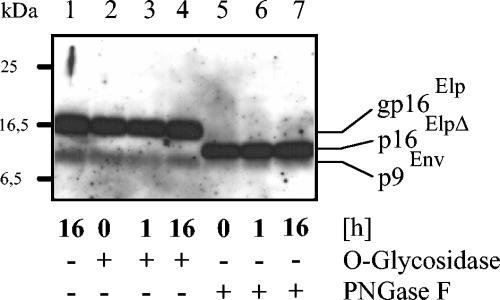
Since we wanted to characterize the C terminus of Elp by MS analysis of proteolytic fragments, it was important to determine whether Elp derived from particles is glycosylated. To this end, enriched FFV particles were lysed in denaturation buffer and then incubated with O-glycosidase (Fig. 6, lanes 2 to 4) and the N-glycosidase PNGaseF (lanes 5 to 7). Only after N-glycosidase treatment did the size of Elp decrease from 16 kDa to about 12 kDa (designated p16ElpΔ), comparable in size to pEnvN* generated by furin digestion of in vitro-synthesized, unglycosylated pEnvN (Fig. 1B and 2B). This result points to a single N-glycosidic modification of the virion-derived mature Elp at residue N118 (Fig. 1A) and to no detectable O-glycosylation. Digestion with N- and O-glycosidases did not affect p9Env; thus, the N-terminal SP of Env is unglycosylated, in line with the lack of glycosylation sites in p9Env. Since the deglycosylated Elp did not comigrate with p9Env, it is unlikely that p9Env represents an unglycosylated Elp, as has been suggested for HFV (15). The MS analysis described below also did not yield further evidence for O-glycosylation or palmitylation at the conserved C60 residue.
FIG. 6.
Deglycosylation of virion-derived Elp. FFV virions were enriched by sedimentation through sucrose, and the viral proteins were denatured for glycosidase digestion as described in Materials and Methods. Aliquots were incubated for 16 h without (−) enzyme (lane 1) and for 0, 1, and 16 h with (+) O-glycosidase (lanes 2 to 4) or PNGaseF (lanes 5 to 7). The reaction products were detected by immunoblotting with the FFV Elp antiserum. The positions of molecular mass markers are marked on the left; the names of the detected FFV proteins are given on the right. Due to the high enzyme concentration used, PNGaseF cleavage had already occurred in the 0-h samples.
MS analysis of virion-derived FFV Elp.
To finally confirm that the FFV Env precursor is proteolytically processed at the furin site between Env residues 127 and 128 in infected cells, FFV particles were purified from about 400 ml of cell culture supernatants of FFV-infected CRFK and deglycosylated with PNGase F. Using preparative SDS-PAGE, the virus-specific band of 16.5 kDa was isolated, subjected to protease digestion, and analyzed by MALDI-TOF (MS). Peptide mass fingerprinting after tryptic and chymotryptic cleavage of the deglycosylated protein band and subsequent analysis by MALDI-MS unequivocally identified the protein as Elp (Fig. 7). None of the identified fragments extended to R127, whereas sequences C-terminal of R127 were not detected, in line with the data that FFV Elp terminates at this residue. Moreover, tryptic cleavage after deglycosylation yielded a peptide at an m/z of 3,138 corresponding to Env 95 to 124. This fragment showed an increase in mass by +1 Da, indicating that one asparagine residue was converted to aspartic acid upon deglycosylation. Sequencing by MALDI-postsource decay analysis mapped the glycosylation to N118, which is in agreement with the predicted position. However, the direct confirmation of the predicted cleavage site after chymotryptic digestion of Elp was not successful, since no chymotryptic fragment with an m/z of 2,319.15 corresponding to the C-terminal Elp peptide 107 to 127 (QVTNSHVTMGGNTSSSSRRRR) could be detected in the mass spectrum.
FIG. 7.
MALDI-TOF (MS) analysis of tryptic and chymotryptic fragments of FFV virion-derived Elp. FFV Elp was purified from FFV particles as described and used for peptide mass fingerprinting to define the C terminus of Elp. (A) The complete sequence of FFV Elp is given with a schematic presentation of the identified tryptic (solid lines above the sequence) and chymotryptic (dashed lines below the sequence) peptide fragments. (B) Presentation of the biophysical features (m/z values) of the Elp-derived tryptic (left column) and chymotryptic (right column) fragments identified by MALDI-TOF (MS) referring to the amino acid positions within Env/Elp. The deglycosylation-related N-to-D modification in the tryptic fragment 95 to 124 is indicated.
DISCUSSION
In the present paper we describe a novel mechanism for the proteolytic release of the N-terminal domain of a surface glycoprotein that is targeted via the ER and Golgi to the cell surface. In many instances, N-terminal signal peptides are required for targeting of secreted or membrane-anchored cellular and viral proteins into the secretory pathway (17). In general, signal peptides are rapidly and efficiently removed from the nascent protein chain by cellular SPases followed by further processing by signal peptide peptidases. In contrast, during synthesis and maturation of FFV Env, this routine pathway of glycoprotein processing is only used to a small extent and yields the minor cleavage product FFV p9Env. This situation appears to be similar to that of human immunodeficiency virus type 1 Env, where SP removal appears to be inefficient and delayed (for a review, see reference 17). In HFV, a smaller Elp form of 14 kDa was proposed to represent the unglycosylated form of the full-length Elp of 18 kDa (15) and not to be an alternative SPase cleavage product, as in FFV. The low-level processing of FFV Env by cellular SPases may be directly related to the fact that algorithms suited for identification of SPase cleavage sites (18) yield only low scores for SPase processing at the p9Env cleavage site of the known FV Env proteins (data not shown).
By cumulative evidence obtained from (i) in vitro cleavages of synthetic peptides and recombinant proteins, (ii) furin inhibitor studies, (iii) transcomplementation studies of furin-deficient LoVo cells, and (iv) functional inactivation of the cleavage site, we identified furin or a related furin-like protease as the major processing enzyme. The direct identification of the Elp C terminus by MS of chymotryptic fragments was not possible, since no fragment with the corresponding mass could be detected in the mass spectra. This may be due to inherent limitations of MALDI-TOF (MS), since this method allows only a partial sequence coverage of the fragments generated (24). Furthermore, tryptic digestion (cleavage C-terminal of R and K) is not suitable to determine whether a furin cleavage does occur within this arginine-rich domain. Determination of the molecular mass of intact Elp by MS without proteolytic digestion could principally be an alternative method for the identification of the C terminus. However, for this approach, substantial amounts of highly purified Elp protein in solution are needed, which is far beyond the amount that could be isolated from FFV particles. Therefore, the predicted C terminus was indirectly confirmed by using a synthetic peptide spanning the predicted cleavage site in Elp and by subsequent analysis of the cleavage products by MALDI-MS.
The furin site separating FFV Elp and SU is conserved in all known FVs. The consensus sequence RXRR of feline, equine, and bovine FVs corresponds to a high-affinity site (28), whereas in the primate FVs, the minimal furin site RXXR is trebled in simian foamy virus type 1, doubled in the prototypic HFV, and present only once in simian foamy virus type 3 (for comparison, see references 29 and 33).
For HFV, processing by an unknown cellular protease at a site about 20 amino acid residues downstream of the furin site identified here had been recently suggested (15). Unpublished data by the same group on HFV now support our finding that furin is responsible for Elp processing (Dirk Lindemann, personal communication). Due to the conservation of the furin sites in all known FVs and since the mature FFV Elp derived from purified particles was not further cleavable by furin under conditions allowing efficient processing of the oversized recombinant pEnvN, we conclude that the FV Elp proteins of 18 kDa (in HFV) (see reference 15) to 16 kDa (in FFV) are exclusively generated by furin or furin-like proteases.
Furin and furin-like proteases are ubiquitously expressed, and often, more than a single furin family member is expressed in a given cell (28). For instance, furin-deficient LoVo cells (27) release small amounts of infectious FFV upon transfection with wt proviral DNA clones, indicating that the essential SU-TM cleavage had been executed by another furin-like protease. The involvement of alternative proteases (e.g., SPases) or the utilization of other furin-like proteases with reduced susceptibility toward the inhibitor used and/or relaxed cleavage site specificity may be the reasons for the remaining infectivity detected upon mutagenesis of the cleavage site in FFV and HFV (Dirk Lindemann, personal communication). Alternatively, release of Elp/p9Env may not be required for FV infectivity, and the Elp-SU fusion protein may support viral infectivity. The assumption that proteolytic processing of the N terminus of FFV Env by furin (and to a much lower degree by SPases) is not necessary for virus replication appears unlikely, since Env of released FFV particles is fully processed at both the Elp-SU and SU-TM sites (35). However, the residual, poorly efficient processing of FFV Env by cellular SPases (see also reference 11) may be sufficient to maintain a reduced level of infectivity seen when the major cleavage site used by furin had been inactivated by mutagenesis. In line with the latter argument, our analyses show that complete proteolytic processing of Env is not a prerequisite for particle release (Fig. 4 and 5), and it is thus possible that particles containing only some properly processed Env proteins display even almost wt infectivity.
As reported previously (11) and in agreement with the data presented here, FV particles contain high concentrations of the 16-kDa Elp but low levels of the 9-kDa FFV Env SP. The N-terminal sequences extending into the cytosol before budding and thereafter into the virion are identical for both proteins. Thus, the N-terminal Env sequences interacting with Gag during budding are present in both proteins (35). However, the ectodomain of Elp is significantly larger than that of the SP, and its absence may contribute to the reduced infectivity. The membrane-proximal Elp ectodomains of the known FVs show some sequence homology, in particular, two conserved Trp residues may be of functional importance. The external part of Elp could, for instance, be important for the proper assembly and arrangement of the Env trimers on virus particles (34).
From the data presented in this study, it is not clear why, in contrast to the Orthoretrovirinae, the primary N-terminal processing of FV Env is executed by furin or a furin-like protease and not by a canonical SPase. However, according to published data, the N terminus of FV Env does not only function as a signal peptide but also contributes to morphogenesis and particle release (15, 35). This gain of function may have led to an increased size of Elp far exceeding that of canonical SPs. In consequence, as Env processing by cellular SPases became inefficient, alternative processing by furin proteases allowed release of Elp and circumvented signal peptide peptidase-mediated destruction of this essential protein. The utilization of furin for the processing of Elp may have been favored by the fact that furin or related proteases were already engaged in Env processing, obviating the need to establish new targeting and trafficking mechanisms.
Acknowledgments
We thank Helmut Bannert for excellent technical assistance; Wolfgang Garten for providing the furin expression plasmid, the furin antiserum, and valuable suggestions; Dirk Lindemann for sharing results prior to publication; Hans-Richard Rackwitz (Peptide Specialty Laboratories GmbH) for providing peptides; Helmut Fickenscher for LoVo cells; Yong-Boum Kim for support in immunoblotting; Valerie Bosch for critically reading the manuscript; and Lutz Gissmann for support.
REFERENCES
- 1.Alke, A., A. Schwantes, K. Kido, M. Flötenmeyer, R. M. Flügel, and M. Löchelt. 2001. The bet gene of feline foamy virus is required for virus replication. Virology 287:310-320. [DOI] [PubMed] [Google Scholar]
- 2.Alke, A., A. Schwantes, M. Zemba, R. M. Flügel, and M. Löchelt. 2000. Characterization of the humoral immune response and virus replication in cats experimentally infected with feline foamy virus. Virology 275:170-176. [DOI] [PubMed] [Google Scholar]
- 3.Bansal, A., K. L. Shaw, B. H. Edwards, P. A. Goepfert, and M. J. Mulligan. 2000. Characterization of the R572T point mutant of a putative cleavage site in human foamy virus Env. J. Virol. 74:2949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak, A., M. Zhong, J. Munzer, M. Chretien, and N. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornsen, K. 2000. Influence of salts, buffers, detergents, solvents, and matrices on MALDI-MS protein analysis in complex mixtures. Methods Mol. Biol. 146:387-404. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1847. In B. N. Fields, D. M. Knipe, and P. Howley (ed.), Virology. Raven Press, New York, N.Y.
- 7.Cullen, B. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46:973-982. [DOI] [PubMed] [Google Scholar]
- 8.Einfeld, D. 1996. Maturation and assembly of retroviral glycoproteins. Curr. Top. Microbiol. Immunol. 214:133-176. [DOI] [PubMed] [Google Scholar]
- 9.Garten, W., S. Hallenberger, D. Ortmann, W. Schäfer, M. Vey, H. Angliker, E. Shaw, and H. D. Klenk. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76:217-225. [DOI] [PubMed] [Google Scholar]
- 10.Garten, W., A. Stieneke, E. Shaw, P. Wikstrom, and H. Klenk. 1989. Inhibition of proteolytic activation of influenza virus hemagglutinin by specific peptidyl chloroalkyl ketones. Virology 172:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiselhart, V., A. Schwantes, P. Bastone, M. Frech, and M. Löchelt. 2003. Features of the Env leader protein and the N-terminal Gag domain of feline foamy virus important for virus morphogenesis. Virology 310:235-244. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, I., M. Schnölzer, I. Kaufmann, and W. Franke. 2002. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13:1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 14.Lindemann, D., and P. A. Goepfert. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111-129. [DOI] [PubMed] [Google Scholar]
- 15.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maley, F., R. Trimble, A. Tarentino, and T. Plummer. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195-204. [DOI] [PubMed] [Google Scholar]
- 17.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Pancino, G., H. Ellerbrok, M. Sitbon, and P. Sonigo. 1994. Conserved framework of envelope glycoproteins among lentiviruses. Curr. Top. Microbiol. Immunol. 188:77-105. [DOI] [PubMed] [Google Scholar]
- 20.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poland, J., A. Urbani, H. Lage, M. Schnölzer, and P. Sinha. 2004. Study of the development of thermoresistance in human pancreatic carcinoma cell lines using proteome analysis. Electrophoresis 25:173-183. [DOI] [PubMed] [Google Scholar]
- 22.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer, W., A. Stroh, S. Berghofer, J. Seiler, M. Vey, M. L. Kruse, H. F. Kern, H. D. Klenk, and W. Garten. 1995. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 14:2424-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheler, C., S. Lamer, Z. Pan, X. Li, J. Salnikow, and P. Jungblut. 1998. Peptide mass fingerprint sequence coverage from differently stained proteins on two-dimensional electrophoresis patterns by matrix assisted laser desorption/ionization-mass spectrometry (MALDI-MS). Electrophoresis 19:918-927. [DOI] [PubMed] [Google Scholar]
- 25.Shaw, K. L., D. Lindemann, M. J. Mulligan, and P. A. Goepfert. 2003. Foamy virus envelope glycoprotein is sufficient for particle budding and release. J. Virol. 77:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha, P., J. Poland, M. Schnölzer, and T. Rabilloud. 2001. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics 1:835-840. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi, S., K. Kasai, K. Hatsuzawa, N. Kitamura, Y. Misumi, Y. Ikehara, K. Murakami, and K. Nakayama. 1993. A mutation of furin causes the lack of precursor-processing activity in human colon carcinoma LoVo cells. Biochem. Biophys. Res. Commun. 195:1019-1026. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tobaly-Tapiero, J., P. Bittoun, M. Neves, M. C. Guillemin, C. H. Lecellier, F. Puvion-Dutilleul, B. Gicquel, S. Zientara, M. L. Giron, H. de The, and A. Saib. 2000. Isolation and characterization of an equine foamy virus. J. Virol. 74:4064-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umemoto, J., V. Bhavanandan, and E. Davidson. 1977. Purification and properties of an endo-alpha-N-acetyl-D-galactosaminidase from Diplococcus pneumoniae. J. Biol. Chem. 252:8609-8614. [PubMed] [Google Scholar]
- 31.Vey, M., W. Schafer, S. Berghofer, H. D. Klenk, and W. Garten. 1994. Maturation of the trans-Golgi network protease furin: compartmentalization of propeptide removal, substrate cleavage, and COOH-terminal truncation. J. Cell Biol. 127:1829-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, A., A. Doerks, M. Aboud, A. Alonso, T. Tokino, R. M. Flügel, and M. Löchelt. 2000. Induction of cellular genes is mediated by the Bel1 transactivator in foamy virus-infected human cells. J. Virol. 74:4441-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, G., and M. J. Mulligan. 1999. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J. Gen. Virol. 80:245-254. [DOI] [PubMed] [Google Scholar]
- 34.Wilk, T., F. de Haas, A. Wagner, T. Rutten, S. Fuller, R. M. Flügel, and M. Löchelt. 2000. The intact retroviral Env glycoprotein of human foamy virus is a trimer. J. Virol. 74:2885-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler, I., J. Bodem, L. Haas, M. Zemba, H. Delius, R. Flower, R. M. Flügel, and M. Löchelt. 1997. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J. Virol. 71:6727-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zemba, M., A. Alke, J. Bodem, I. G. Winkler, R. L. Flower, K. Pfrepper, H. Delius, R. M. Flügel, and M. Löchelt. 2000. Construction of infectious feline foamy virus genomes: cat antisera do not cross-neutralize feline foamy virus chimera with serotype-specific Env sequences. Virology 266:150-156. [DOI] [PubMed] [Google Scholar]