Abstract
Although genome-wide association studies (GWASs) have identified thousands of risk loci for many complex traits and diseases, the causal variants and genes at these loci remain largely unknown. Here, we introduce a method for estimating the local genetic correlation between gene expression and a complex trait and utilize it to estimate the genetic correlation due to predicted expression between pairs of traits. We integrated gene expression measurements from 45 expression panels with summary GWAS data to perform 30 multi-tissue transcriptome-wide association studies (TWASs). We identified 1,196 genes whose expression is associated with these traits; of these, 168 reside more than 0.5 Mb away from any previously reported GWAS significant variant. We then used our approach to find 43 pairs of traits with significant genetic correlation at the level of predicted expression; of these, eight were not found through genetic correlation at the SNP level. Finally, we used bi-directional regression to find evidence that BMI causally influences triglyceride levels and that triglyceride levels causally influence low-density lipoprotein. Together, our results provide insight into the role of gene expression in the susceptibility of complex traits and diseases.
Keywords: transcriptome-wide association study (TWAS), genome-wide association study (GWAS), expression quantitative trait loci (eQTLs), susceptibility gene, complex trait, complex disease, genetic covariance, genetic correlation
Introduction
Although genome-wide association studies (GWASs) have identified tens of thousands of common genetic variants associated with many complex traits,1 with some notable exceptions,2, 3 the causal variants and genes at these loci remain unknown. Multiple lines of evidence have shown that GWAS risk variants co-localize with genetic variants that regulate expression—i.e., expression quantitative trait loci (eQTLs).4 This suggests that a substantial proportion of GWAS risk variants influence complex traits by regulating expression levels of their target genes.4, 5, 6, 7 Analyses of genotype, phenotype, and gene expression measurements from multiple tissues in the same set of individuals can directly investigate this plausible chain of causality. However, doing so is challenging because of cost and tissue availability; therefore, GWAS and eQTL datasets remain largely independent (i.e., no overlapping subjects).8, 9 Recent work has shown that one way to integrate GWAS and eQTL data is to predict gene expression levels for GWAS samples and then test for association between the predicted expression and traits.10, 11, 12 This approach, referred to as transcriptome-wide association study (TWAS), can increase power over GWAS when the causal mechanism includes genetic variants that regulate the expression of susceptibility genes. TWAS benefits from a lower multiple-testing burden by probing several thousands of genes, whereas GWAS probes several million SNPs. Although TWAS can also be performed with measured gene expression levels directly, using predicted gene expression has several benefits. First, expression measurements are usually not available in GWAS data. Second, predicted gene expression removes environmental noise by focusing on the genetically regulated component, which can increase statistical power. Third, using the predicted expression to test for association can eliminate potential confounding from reverse causation, where traits affect gene expression levels.10, 11 However, compared with GWAS, TWAS is underpowered when risk is not mediated through expression or when expression data are not available in the right tissue.
In this work, we introduce methods for estimating the genetic correlation between gene expression and a complex trait from summary GWAS and eQTL data. We utilize the local (cis) genetic variation near a gene (i.e., ±0.5 Mb around the transcription start site [TSS]) to estimate the correlation in the genetic effects between gene expression and the trait. We show that under this framework, TWAS can be viewed as a test for non-zero genetic covariance between expression and a trait from summary association data. In addition to identifying susceptibility genes, the predicted expression can also be used for estimating the genome-wide genetic correlation between pairs of complex traits at the level of predicted expression. This is analogous to computing genome-wide genetic correlation between complex traits,13 whereby correlations are determined over predicted gene expression effects rather than SNP effects, and can give insights into the component of genetic correlation mediated through expression. We demonstrate through extensive simulations that our approach is approximately unbiased and well calibrated under the null and slightly conservative when true correlation is near the boundaries. Finally, we utilize estimated effects of predicted expression within a bi-directional regression approach14 to investigate putative causal direction for pairs of complex traits that are genetically correlated.
We analyze summary statistics from 30 GWASs spanning 2.3 million phenotype measurements15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 jointly with 45 expression panels8, 29, 30, 31, 32, 33, 34 sampled from more than 35 tissues to gain insight into the role of expression in the etiology of complex traits. First, we test each gene-tissue pair across 45 panels to perform a multi-tissue TWAS for each of the 30 traits to identify 1,196 gene associations. For example, at four independent loci, we find 11 genes that do not overlap a genome-wide significant SNP for educational years. Notably, all four loci were replicated in a recent, larger GWAS for educational years.35 Second, we identify 43 pairs of traits showing a genome-wide-significant genetic correlation at the level of predicted expression. Overall, the predicted-expression correlation was highly concordant with SNP-level genetic correlation from cross-trait linkage disequilibrium (LD) score regression, which suggests that a large component of genetic correlation between complex traits is driven by local regulation of gene expression. Finally, we use our bi-directional analysis to provide evidence of putative causal effects between pairs of these traits. Overall, our results shed light on shared biological mechanisms responsible for susceptibility to disease and complex traits, as well as potential downstream effects between traits.
Material and Methods
Datasets
We used summary association statistics from 30 large-scale (n = 20,000 subjects) GWASs, including various anthropometric15, 27, 28 (body mass index [BMI], femoral neck bone mineral density [BMD], forearm BMD, lumbar spine BMD, and height), hematopoietic23, 25, 26 (hemoglobin, HbA1c, mean cell hemoglobin [MCH], MCH concentration, mean cell volume, number of platelets, packed cell volume, and red blood cell count), immune-related17, 19 (Crohn disease [OMIM: 266600], inflammatory bowel disease [OMIM: 266600], ulcerative colitis [OMIM: 266600], and rheumatoid arthritis [OMIM: 180300]), metabolic16, 20, 22, 24 (age of menarche, fasting glucose, fasting insulin, high-density lipoprotein [HDL], HOMA-B, HOMA-IR, low-density lipoprotein [LDL], triglycerides [TG], type 2 diabetes [OMIM: 125853], and total cholesterol [TC] levels), neurological18 (schizophrenia [OMIM: 181500]), and social21 (college and educational attainment) phenotypes (see Table S1). We removed SNPs that were strand ambiguous or had a minor allele frequency (MAF) ≤ 1% (see Table S1).
Gene expression data from RNA sequencing data were obtained from the CommonMind Consortium29 (brain, n = 613), the Genotype-Tissue Expression Project8 (GTEx; 41 tissues; see Table S2 for sample size per tissue), and the Metabolic Syndrome in Men study31, 32 (adipose, n = 563). Expression microarray data were obtained from the Netherlands Twins Registry34 (NTR; blood, n = 1,247), and the Young Finns Study30, 33 (YFS; blood, n = 1,264).
Performing TWAS with GWAS Summary Statistics
We estimated SNP heritability for observed expression levels partitioned into cis- (1 Mb region surrounding the TSS) and trans- (rest of genome) components. We used the AI-REML algorithm implemented in Genome-wide Complex Trait Analysis (GCTA),36 which allows estimates to fall outside of the (0, 1) boundaries to maintain unbiasedness. To control for confounding, we included batch variables and the top 20 principal components estimated from genome-wide SNPs. Genes with significant cis-heritability in expression data were used for prediction (cis- p < 0.05 in a likelihood ratio test between the cis-only and joint models). The average number of genes with significant cis- across expression studies was 816 (min = 70 genes from GTEx small intestine samples; max = 3,704 genes from the YFS).
We performed 45 TWASs for each of the 30 GWASs;11 for each trait, we used Bonferroni correction for all gene-tissue pairs tested (see Table S2). In brief, we estimated the strength of association between the predicted expression of a gene and a complex trait (zTWAS) as a function of the vector of GWAS summary Z scores at a given cis-locus, (i.e., vector of SNP association Wald statistics), and the LD-adjusted weight vector learned from the gene expression data, wGE, as
where V is a covariance matrix across SNPs at the locus (i.e., LD). We estimated wGE by using GBLUP37 from eQTL data and computed by using GWAS summary data for all 30 traits and the ∼36,000 gene expression measurements across all studies. We removed all loci in the human leukocyte antigen (HLA) region as a result of complex LD patterns.
Estimating the Proportion of Trait Variance Explained by Predicted Expression
We use the LD score regression38, 39 approach described in Guseve et al.11 to quantify the heritability explained by predicted expression for a complex trait (denoted here as ). The expected statistic under a polygenic trait is , where is the number of individuals in the GWAS, M is the number of genes, is the LD score, and is the effect of population structure. We estimate for each gene by predicting expression for 503 European samples in 1000 Genomes40 by using the GBLUP weights (see above) and then computing sample correlation. For each trait, we perform LD score regression by using (which follows a distribution asymptotically) to infer . We estimate heritability for each expression study separately to account for varying sample sizes and repeated gene measurements.
Estimating Genetic Correlation of Expression and Complex Traits from Summary Data
Let expression and traits be modeled as a linear function of the genotypes in a ∼1 Mb locus flanking the gene: and , where is the standardized genotype matrix, and are the standardized effects for expression and traits, respectively, and and are the environmental noise for expression and traits, respectively. The local covariance between expression and complex traits is
where is the LD matrix. If no individuals are shared between studies, then (as in eQTL studies and GWASs). The local genetic correlation between expression and traits can be computed as
where and are the local SNP heritability41 for expression and traits, respectively, estimated at the locus. However, this requires knowledge of the true effect sizes. Given association statistics zT, we estimate an LD-adjusted effect size as . Hence, an estimate of the local genetic covariance42 is given by
where and are the marginal (i.e., LD-unadjusted) standardized effect-size estimates.41, 43 It follows that
We standardize this estimate to obtain our final local genetic correlation estimate as
In practice, we use the variance explained by the local index SNP (i.e., smallest p value) as a proxy for .
Genetic Correlation between Traits at the Level of Predicted Expression
Consider a simple model where the genetic component of a trait can be decomposed into genetic effects that are mediated through cis-gene expressions of k genes plus genetic effects not mediated through expression at other loci in the genome:
where is a vector of genotypes at the cis-locus of gene i, is the casual eQTL effect vector for gene i, is the direct effect of gene expression on a trait, and and refer to the genotype and causal effects, respectively, of variants not mediated through expression. We define the genome-wide genetic correlation at the level of expression between two complex traits as the correlation across the gene effects: . In practice, we do not know , but we can estimate it as
to obtain an estimate of expression correlation by using predicted expression . In practice, we use the standardized estimates of , which are proportional to . Unlike SNP-based genetic correlation , which captures genetic correlation across all common variants in the genome, captures only the component of genetic correlation driven by cis genetic effects on expression (see Figure 1). For instance, a pair of traits with highly correlated effects in cis-regions but weakly correlated effects in trans-regions will result in . In the absence of large trans-eQTL effects, we expect . Furthermore, because accounts for only the shared effect from predicted expression, any genetic effect on a trait not driven through expression in the measured eQTL data will not be represented in . We test for significance by assuming , where is the number of genes and t is the t distribution with M − 2 degrees of freedom. This procedure requires the effects of genes on the trait to be independent, which could be violated in practice; hence, we compute by using one gene per 1 Mb locus.
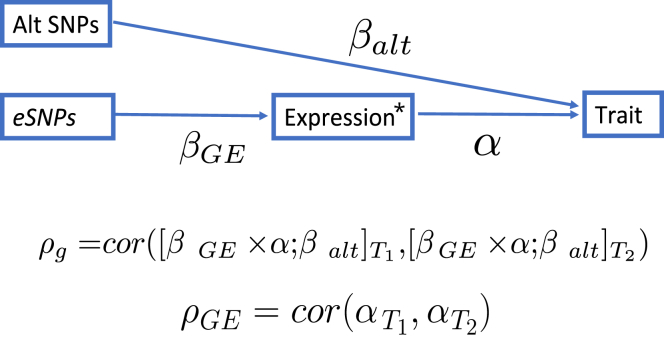
Figure 1.
Causal Diagram Illustrating the Genetic Component of a Trait
The total effect of SNPs on a trait can be partitioned into components that are mediated through cis-regulated (i.e., predicted, indicated by an asterisk) gene expression or through alternative pathways . In contrast to , which quantifies the correlation of the total SNP effects between two traits (; ), focuses exclusively on the effects of cis-regulated gene expression (α).
Estimating Putative Casual Relationships between Pairs of Traits
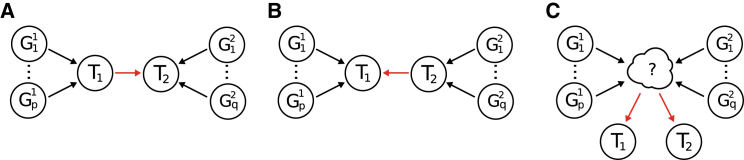
To glean insight into the underlying causal relationship between pairs of traits, we perform a bi-directional regression14 and estimate two different values of by varying gene sets. Before describing the approach, we first review several causal models that explain non-zero between two traits (see Figure 2). Models A and B depict causal relationships in which the effects of a gene set are mediated by one trait on the other. We can formally state model A (without loss of generality for B). Let trait 1 (T1) be defined as , where denotes the matrix of predicted expression at the causal genes, is the effect size, and is environmental noise. We define trait 2 (T2) as
where is the causal effect of on , and are the remaining causal genes and their effects, respectively, for , and is the combined environment component. Under model A, the causal gene set for will have a non-zero effect on (i.e., ); however, if does not cause , this effect will be zero given that unrelated genes have no downstream effect. Bi-directional regression provides a test to distinguish between models A and B by regressing estimated effect sizes for gene sets under model A (i.e., ) and comparing to estimates under model B (i.e., ). Because the causal gene sets for each trait are unknown, we use their identified susceptibility genes as a proxy. We estimate by conditioning on the gene set for trait and denote its value as . We repeat this procedure by ascertaining the gene set for trait to obtain . We perform a Welch’s t test44 to determine whether estimates of and are significantly different, thus providing evidence consistent with a causal direction. To minimize spurious results, we require at least ten genes for estimation in each conditional test. This approach mirrors bi-directional regression analyses of estimated SNP effects on two complex traits.45, 46 We stress that although a bi-directional approach is capable of rejecting model A in favor of model B (or vice versa), it cannot rule out model C, in which a shared pathway (or set of pathways) drives both traits independently (see Figure 2).
Figure 2.
Illustration of Several Causal Models That Explain Expression Correlation for Traits 1 and 2 Given Their Causal Gene Sets
(Model A) Trait 1 directly influences trait 2. In this case, the effect of genes on trait 2 is mediated by trait 1, which implies .
(Model B) Trait 2 directly influences trait 1. Similarly, the effect of genes on trait 1 is mediated by trait 2, which implies .
(Model C) Traits 1 and 2 are influenced independently through an unobserved trait or traits.
Simulation Framework
We simulate gene expression levels by using real genotype data measured in 503 European individuals from the 1000 Genomes Project.40 Given a gene locus, we generate expression levels under the linear model E = Xw + , where E is a gene expression vector of length N, X is the N × 2 mean-centered and variance-standardized genotype matrix over two randomly selected SNPs in the locus, w is the causal effect, and is the environmental noise. We sample effect sizes for i = 1 and 2 and noise from a normal distribution to yield (consistent with what we observe in real gene expression data). We consider only SNPs with a MAF ≥ 0.01 and Hardy-Weinberg equilibrium deviation p ≥ 1 × 10−5. We simulate a complex trait as a linear function of predicted gene expression for k = 100 genes, given by , where Xiwi is the predicted expression of the ith gene with effect sizes . For simulations involving , we simulate the two traits y1 and y2 by using the same process, except effects for the ith gene are drawn from a bivariate normal distribution:
where . Lastly, we perform an association scan on y by using all SNPs at each gene locus to obtain SNP-level Z scores zT.
Results
Accurate Estimation of Expression-Trait Genetic Correlation in Simulations
To validate our statistical framework for estimating , we used real genotype data to perform simulations under various architectures (see Material and Methods). In brief, we simulated gene expression for 100 independent gene loci, which we then used to simulate a complex trait. Using our approach, we performed a GWAS and estimated from TWAS summary statistics (see Material and Methods). We observed unbiased estimates for both when causal variants were typed and when they were masked from the data (see Figure S1). Estimated values of were highly correlated with their true values (r = 0.73; p < 2.2 × 10−16), which indicates that using weights inferred from GBLUP maintains moderate power levels. This slight loss in power extended to estimates, which quantify the total effect of predicted expression on a trait (r = 0.74; p < 6.7 × 10−12; see Table S3). As eQTL datasets increase in sample size, and predictive models become more accurate, we expect this attenuation bias to decrease.
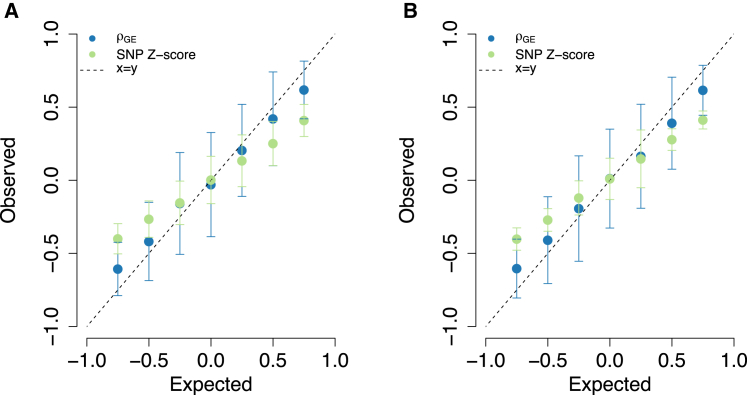
We next performed extensive simulations to validate our procedure for estimating genetic correlation due to predicted expression between pairs of traits. We simulated genetically correlated complex traits from predicted expression by sampling effects from a bivariate normal distribution with correlation (see Material and Methods). We first estimated for each gene-trait pair, which served as input for estimating . Overall, we observed our estimator to be approximately unbiased, with conservative estimates for when its underlying value was near the boundaries (see Figure 3). Importantly, estimates were relatively unbiased when causal variants were untyped in the data. Our method appropriately accounted for LD among variants, resulting in a large improvement over the naive SNP correlation approach (which simply correlates the Z scores by ignoring LD). We also assessed our approach for testing for deviations from = 0 and found estimates consistent with the null distribution with = 0.97 (Jack-knife 95% CI = [0.86, 1.08]; see Figure S2). To measure how sensitive our approach is to estimates of at each gene, we repeated simulations by using variance explained by the top eQTL as a proxy for local heritability. Although estimates were highly similar (r = 0.99; p < 6.6 × 10−7), our approach produced estimates closer to the ground truth (see Figure S3).
Figure 3.
Simulation Results for and Correlation of SNP Z Scores
Each point represents the mean estimate over 100 simulations. Error bars represent the 95% confidence interval estimated by the mean SE across simulations. The dotted line represents the identity line.
(A) Causal SNPs for gene expression are typed in the data.
(B) Causal SNPs are untyped.
TWAS Identifies 1,196 Genes Associated with 30 Complex Traits and Diseases
We integrated GWAS summary data of 30 complex traits with gene expression to identify 1,196 susceptibility genes (i.e., genes with at least one significant trait association), comprising 5,490 total associations (after Bonferroni correction; see Material and Methods). Of these associations, we observed 1,789 distinct gene-trait pairs, of which 783 were found in anthropometric traits, 423 in metabolic traits, 215 in immune-related traits, 213 in hematopoietic traits, 137 in neurological traits (e.g., schizophrenia), and 18 in social traits (see Tables 1, S4, and S5). For example, the 137 susceptibility genes found for schizophrenia included SNX19 (e.g., GTEx cerebellum; p < 2.2 × 10−8) and NMRAL1 (e.g., GTEx skeletal muscle; p < 9.7 × 10−7); this is consistent with a previously reported study12 that used different methods and expression data (see Table S6). We did not find susceptibility genes for forearm BMD, HOMA-B, or MCH concentration, consistent with low GWAS signal for these traits (see Table 1). Indeed, the number of GWAS risk loci strongly correlated with the number of identified susceptibility genes (r = 0.99; p < 2.2 × 10−16). Using the PANTHER database,47 we explored putative molecular function and pathways enriched with identified susceptibility genes but were underpowered to detect molecular function for most individual traits (see Appendix A).
Table 1.
Summary of GWAS and TWAS Results
| Trait | Abbreviation |
Number of GWASs |
Number of Susceptibility Genes |
||||
|---|---|---|---|---|---|---|---|
| Loci | Loci with an eGene | Loci with a Single Susceptibility Gene | Loci with at Least One Susceptibility Gene | Genes Overlapping GWASs | Genes Not Overlapping GWASs | ||
| Age at menarche | AM | 70 | 60 | 14 | 19 | 34 | 9 |
| Body mass index | BMI | 76 | 60 | 10 | 18 | 44 | 11 |
| College | COL | 5 | 5 | 2 | 2 | 1 | 4 |
| Crohn disease | CD | 50 | 48 | 4 | 17 | 65 | 5 |
| Educational years | EY | 7 | 4 | 2 | 2 | 2 | 11 |
| Fasting glucose | FG | 12 | 11 | 2 | 5 | 8 | 1 |
| Fasting insulin | FI | 0 | 0 | 0 | 0 | 0 | 1 |
| Femoral neck bone mineral density | FN | 20 | 20 | 2 | 2 | 2 | 1 |
| Forearm bone mineral density | FA | 3 | 3 | 0 | 0 | 0 | 0 |
| Hemoglobin | HB | 22 | 21 | 2 | 5 | 22 | 3 |
| HbA1c | – | 10 | 10 | 0 | 1 | 4 | 0 |
| Height | – | 482 | 454 | 94 | 225 | 669 | 52 |
| High-density lipoprotein | HDL | 100 | 95 | 11 | 29 | 98 | 4 |
| HOMA-B | – | 4 | 3 | 0 | 0 | 0 | 0 |
| HOMA-IR | – | 0 | 0 | 0 | 0 | 0 | 1 |
| Inflammatory bowel disease | IBD | 63 | 59 | 12 | 23 | 70 | 11 |
| Low-density lipoprotein | LDL | 75 | 72 | 8 | 25 | 84 | 3 |
| Lumbar spine | LS | 24 | 23 | 2 | 3 | 4 | 0 |
| Mean cell hemoglobin concentration | MCHC | 5 | 3 | 0 | 0 | 0 | 0 |
| Mean cell hemoglobin | MCH | 35 | 31 | 5 | 17 | 46 | 7 |
| Mean cell volume | MCV | 43 | 40 | 8 | 20 | 49 | 1 |
| Number of platelets | PLT | 35 | 34 | 6 | 13 | 30 | 8 |
| Packed cell volume | PCV | 14 | 13 | 1 | 3 | 5 | 1 |
| Red blood cell count | RBC | 25 | 21 | 3 | 10 | 35 | 2 |
| Rheumatoid arthritis | RA | 44 | 41 | 7 | 13 | 30 | 5 |
| Schizophrenia | SCZ | 95 | 74 | 15 | 31 | 113 | 24 |
| Total cholesterol | TC | 88 | 85 | 13 | 40 | 117 | 0 |
| Triglycerides | TG | 70 | 67 | 4 | 18 | 59 | 1 |
| Type 2 diabetes | T2D | 12 | 12 | 0 | 1 | 3 | 0 |
| Ulcerative colitis | UC | 37 | 36 | 5 | 9 | 27 | 2 |
| Total | 1,526 | 1,405 | 232 | 551 | 1,621 | 168 | |
The first four numeric columns summarize GWAS risk loci. The last two numeric columns summarize identified TWAS susceptibility genes. The majority (92%) of GWAS risk loci overlap at least one eGene, of which 40% contain at least one susceptibility gene. We report 168 (9%) identified gene-trait pairs that do not overlap a GWAS variant, providing risk loci for follow up.
Next, we quantified the overlap of susceptibility genes and GWAS signals. Of the 1,789 identified gene-trait pairs, 168 (9%) were not proximal (more than 0.5 Mb from the TSS) to any genome-wide-significant SNP for that respective trait (see Table 2). This measure was robust to increases in window size, such that 140 (8%) gene-trait pairs did not overlap a genome-wide-significant SNP within 1 Mb of the TSS. We observed increased SNP association statistics at these genes (mean χ2 = 6.5; see Figure S4), which suggests that GWASs with an increased sample size will discover genome-wide-significant SNPs nearby. We tested this hypothesis by assessing the new TWAS loci for educational years21 (n = 126,599) in a recent, much larger GWAS for educational years35 (n = 293,723). All four independent loci contained a genome-wide-significant SNP in the larger GWAS (see Table S7). Of the 1,526 GWAS risk loci, 1,405 (92%) overlapped at least one eGene (i.e., a gene with heritable expression levels in at least one of the considered expression panels), and 551 (36%) overlapped at least one susceptibility gene (see Table 1). Focusing on the 1,621 TWAS associations that overlapped a genome-wide-significant SNP, we observed 1,350 (83%) genes that were not the closest, suggesting that the traditional heuristic of prioritizing genes closest to GWAS SNPs is typically not supported by evidence from eQTL data48 (see Figure S5). This is also supported by the mean χ2 association statistics for genes closest to index SNPs (χ2 = 43.9) and the top association (χ2 = 72.9; see Figure S6). In addition, lead GWAS SNPs typically have a weaker eQTL effect for the proximal gene than for the TWAS-implicated gene in 1,088 of 1,350 TWAS associations. This result, consistent with earlier reports,11, 12 highlights the importance of utilizing the entire locus and estimates of LD to prioritize genes.
Table 2.
Susceptibility Genes That Do Not Overlap a Genome-wide Significant SNP within 0.5 Mb of the Transcription Start and End Sites for Each Trait
| Trait | Genes |
|---|---|
| AM | CCDC65, COG6, INO80E, NUCKS1, PMS2P5, RAB7L1, SLC26A9, STAG3L2, and TMEM180 |
| BMI | CDK5RAP3, CERCAM, DHRS11, GGNBP2, INO80E, RP11-6N17.10, RP11-6N17.9, SLC27A4, STAG3L1, TUBA1C, and URM1 |
| CD | CCDC88B, CISD1, PPP1R14B, RIT1, and SMIM19 |
| COL | ABCB9, AC091729.9, AFF3, and RNF123 |
| EY | ABCB9, EIF3CL, MIR4721, MPHOSPH9, NFATC2IP, RP11-1348G14.4, SDCCAG8, SH2B1, STK24, SULT1A1, and TUFM |
| FG | MAPRE3 |
| FI | KNOP1 |
| FN | FGFRL1 |
| HB | CCDC117, UBE2Q2, and WNT3 |
| HDL | HRAS, KNOP1, RETSAT, and TYRO3 |
| HEIGHT | ARL17A, ATF1, ATP5J2, C20orf194, C9orf156, CCDC116, CNIH4, COX6B1, CRELD1, CRHR1, DAB2IP, DESI1, DLG5, DUS3L, ECHDC2, FAM35A, FUCA2, H2AFJ, HIBADH, INO80E, IQGAP1, KANSL1, LBX2-AS1, LRRC37A2, MAPT, MAT2A, MED4, MEGF9, MGMT, MORC2-AS1, MSRB2, P4HTM, PHF19, PLEKHA1, PSMD5, PSMD5-AS1, RP11-173M1.8, RP11-455F5.3, RP11-4O1.2, RP11-67A1.2, RP13-39P12.3, RP4-612B15.3, RRN3, SFTPD, SH3YL1, SUSD1, TMEM128, UBE2L3, UTP18, WDR60, YPEL3, and YWHAB |
| HOMA-IR | KNOP1 |
| IBD | ADCY3, CCDC88B, FAM189B, GBA, GBAP1, HCN3, PPP1R14B, RMI2, SATB2, TMEM180, ZFP90 |
| LDL | DHRS13, ERAL1, and WDR25 |
| MCH | AP003419.16, GSTP1, PABPC4, PTPRCAP, RP11-69E11.4, RP1-18D14.7, and RPS6KB2 |
| MCV | COX4I2 |
| PCV | PLEKHH2 |
| PLT | ACTR1A, BAZ2A, CCDC17, IPP, MUTYH, PRIM1, TESK2, and TMEM180 |
| RA | METTL21B, RNF40, RPS26, SLC26A10, and SUOX |
| RBC | COX4I2 and FBXL20 |
| SCZ | ALMS1P, ARL14EP, CAD, CBR3, CEBPZ, CORO7, CPNE7, DND1, EMB, ENDOG, EPN2, GRAP, IK, NMRAL1, NRBP1, PCNX, PFDN1, PRR12, PRRG2, RNF112, RP11-135L13.4, SEPT10, SRA1, and TMCO6 |
| TG | L3MBTL3 |
| UC | SATB2 and TNPO3 |
For details on individual genes, expression studies, and association statistics, see Table S4. Genome-wide significance: p < 5 × 10−8.
Although GWAS SNPs provide the majority of the power in this approach, the flexibility of TWASs to leverage allelic heterogeneity provides a significant gain.11 We found 219 instances across 19 traits where association signal was stronger (20% higher χ2 statistics on average) in TWASs than in GWASs. For example, predicted expression in CCDC88B (OMIM: 611205; a gene involved in T cell maturation and inflammation49) exhibited strong association with Crohn disease (pTWAS = 6.32 × 10−8), whereas the index SNP (i.e., top overlapping GWAS SNP) at site rs11231774 was only suggestive (pGWAS = 2.47 × 10−6). This effect was most dramatic for height, such that 108 susceptibility genes had a stronger signal than GWAS index SNPs. We observed that the χ2 statistics for predicted expression in CRELD1 (OMIM: 607170; pTWAS = 1.55 × 10−10) were 2.6× higher than those for the index SNP rs1473183 (pGWAS = 6.33 × 10−5).
Recent work50 applied a similar approach12 that used summary eQTLs from blood and GWAS data to identify 71 genes for 28 complex traits.50 Of the investigated traits, 12 overlapped those in our study. Overall, whereas that study reported 63 genes for these traits, we identified 564 genes. Surprisingly, despite using independent methods and expression data, we replicated 40 out of 51 associations for genes assayed in both studies (see Table S8). This increase in power can be attributed to two reasons. First, we integrated many more expression panels sampled from many tissues, leading to many more genes for the assay. Second, we used a method that jointly tests the entire locus rather than the index SNPs. We have shown that many identified susceptibility genes contain signals of allelic heterogeneity; therefore, using individual SNPs will decrease power.
Genes Associated with Multiple Traits
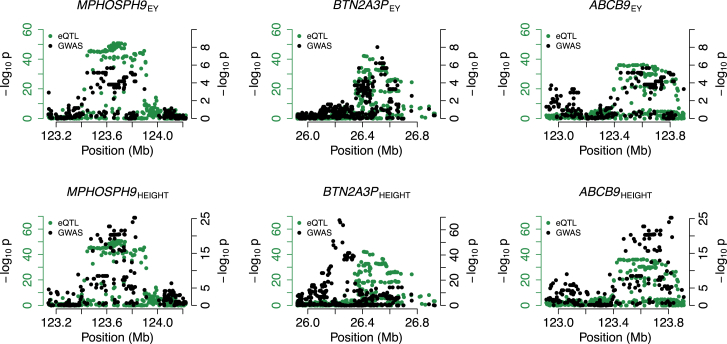
We investigated the degree of pleiotropic susceptibility genes (i.e., genes associated with more than one trait) in our data and found 380 (32%) genes associated with multiple traits (see Figure S7). For example, IKZF3 (OMIM: 606221) displayed strong associations with Crohn disease (NTR; p = 1.6 × 10−9), HDL levels (NTR; p = 6.6 × 10−15), inflammatory bowel disease (NTR; p = 7.9 × 10−16), rheumatoid arthritis (NTR; p = 6.0 × 10−8), and ulcerative colitis (NTR; p = 9.2 × 10−10). Indeed, IKZF3 has been shown to influence lymphocyte development and differentiation.51, 52 These traits are known to have a strong autoimmune component;53 hence, association with predicted IKZF3 expression levels is consistent with a model where cis-regulated variation in IKZF3 product levels contributes to risk. Similarly, we observed three susceptibility genes shared between educational years (EY) and height (see Figure 4): ABCB9 (OMIM: 605453; GTEx heart left ventricle; pheight = 1.38 × 10−15; pEY = 1.28 × 10−6), BTN2A3P (OMIM: 613592; GTEx subcutaneous adipose; pheight = 3.82 × 10−12; pEY = 1.90 × 10−7), and MPHOSPH9 (OMIM: 605501; GTEx thyroid; pheight = 5.84 × 10−18; pEY = 1.30 × 10−6). Although not direct evidence of co-localization of educational years and height at these loci, this result is consistent with a recent study13 that reported a non-zero genetic correlation between height and educational years ( = 0.13; p = 3.82 × 10−6).
Figure 4.
Susceptibility Genes Shared for Educational Years and Height
We indicate –log10 p values for eQTLs in green and trait-specific GWASs in black on separate axes to simplify illustration.
The Effect of cis Expression on Traits Is Consistent across Tissues
Having established the importance of individual predicted gene expression levels for these traits, we next estimated the amount of trait variance explained by predicted expression by using all examined genes, including those not significantly associated, and an LD score regression approach (see Material and Methods). We found 108 tissue-trait pairs across 17 traits and 33 tissues where the cumulative effect of all measured genes on the trait was significantly greater (p < 0.05/45) than for the significant-only set (see Table S9). For example, in height we estimated = 0.07 (Jack-knife SE = 0.02; p = 5.6 × 10−4) by using all 3,733 measured genes in YFS and = 0.015 (Jack-knife SE = 6.9; p = 0.03) by using only the 169 YFS susceptibility genes (pall>sig = 5.6 × 10−3). This suggests that height has additional susceptibility genes, which we are underpowered to detect. Strikingly, the predicted expression from all YFS genes accounts for 12% of SNP heritability measured in height.54 However, for most trait-tissue pairs, we did not observe a significant difference at our given sample sizes. Indeed, we measured a significant association between expression-study sample size and number of eGenes (r = 0.73; SE = 0.10; p = 1.3 × 10−8), which indicates that smaller studies lack power to find eGenes and thus underestimate the total .
We next asked whether any tissues are burdened with increased levels of risk for a given trait. To test this hypothesis, we examined the difference between estimated trait variance explained per gene and the average. Our results did not suggest tissue-specific enrichment at the current sample sizes (see Table S10). We observed a significant correlation between gene expression sample size and tissue enrichment estimates (p = 62.4 × 10−6). One explanation for this relationship is that the number of eGenes identified per study increases with sample size, which increases estimates. Given no observable difference in tissue-specific risk, we expect local estimates of genetic correlation to be highly similar across tissues. When estimating , we observed consistent effect-size estimates in both sign and magnitude estimates across tissues (mean tissue-tissue r = 0.82; see Figure 5). These results are compatible with earlier work that found that cis effects on expression are largely consistent across tissues.55 To obtain a meta-estimate of local genetic correlation for gene-trait pairs with measurements in multiple tissues, we used the mean genetic correlation across all expression panels in all of the following analyses.
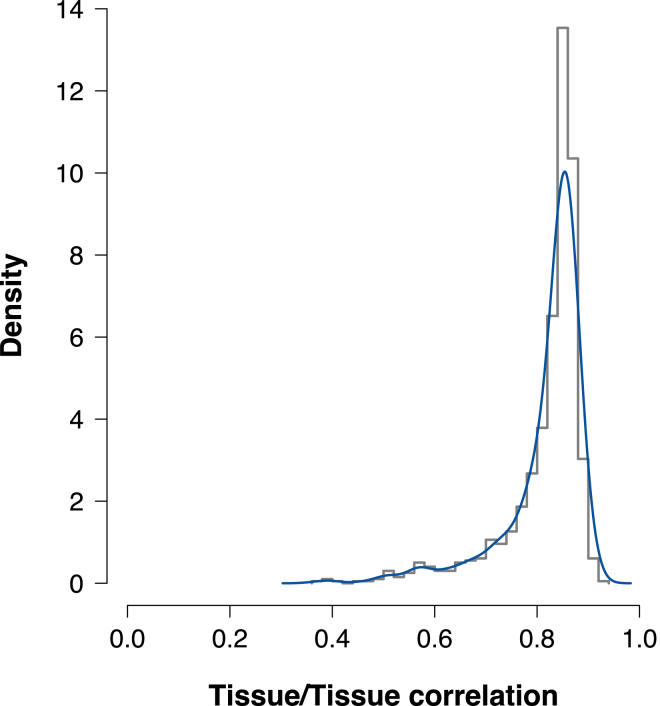
Figure 5.
Histogram and Density Estimate for Correlation of across Tissues
We computed the correlation across pairs of different tissues by using local estimates of genetic correlation between expression and traits. Most tissues exhibited a high correlation over the underlying gene effects on traits with an estimated mean of r = 0.82.
Genetic Correlation between Traits at the Level of Predicted Expression
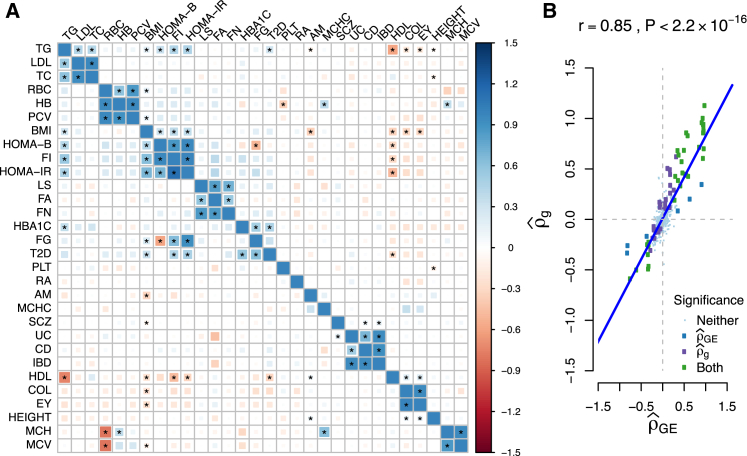
To evaluate the shared contribution of predicted expression on pairs of traits, we used nominally significant (p < 0.05) genes to compute the genome-wide genetic correlation at levels of predicted expression (see Material and Methods). For 435 distinct pairs, we discovered 43 significant expression correlations, 22 of which had previously reported non-zero genetic correlations13 (see Figure 6 and Table 3). For example, age of menarche and BMI had = −0.32 (95% CI = [−0.32, −0.21]; p = 7.97 × 10−8). This negative correlation is consistent with estimates published in epidemiological studies,56 in addition to studies probing genetic correlation across complex traits.13 To determine whether estimates were sensitive to changes in scale, we recomputed by using the top eQTL as a proxy for local heritability of gene expression and observed similar results (r = 0.99; p = 2.2 × 10−16; see Figure S8). Results were also robust to increasing window size for gene pruning, such that there was no significant difference in estimates between 2 and 4 Mb windows (r2Mb = 0.99; r4Mb = 0.98). Using estimates of , we clustered traits and observed groups forming naturally in the trait-trait matrix (see Figure 6). Interestingly, BMI clustered with insulin-related traits (HOMA-B, HOMA-IR, and fasting insulin). Our estimates were highly consistent with the results of LD score regression (see Figure 6 and Table S11). Out of 435 pairs of traits, 35 demonstrated significance for and , whereas 8 and 27 were exclusive to and , respectively. Given the high degree of concordance between estimates, we tested for significant differences and found four insulin-related pairs of traits and three blood-related pairs with more extreme values for (see Table S11). Differences for these pairs of traits can be partially explained by overconfident standard errors for (see Table S12). Overall, we found to explain most of the variation in (r2 = 0.72). We compared this to the naive approach of computing the correlation of SNP Z scores across susceptibility gene loci and observed a much smaller proportion of variance explained in (r2 = 0.46). This reinforces that, compared to the naive approach, our method incorporates LD to aggregate signal.
Figure 6.
Estimates of Genetic Correlation Obtained from LD Scores versus Estimates of Expression Correlation from Nominally Significant TWAS Results
(A) Correlation matrix for 30 traits. The lower triangle contains , and the upper triangle contains estimates. Correlation estimates that are significantly non-zero (p < 0.05/435) are marked with an asterisk (∗). The strength and direction of correlation are indicated by size and color. We found 43 significantly correlated traits by using predicted expression and 62 by using genome-wide SNPs.
(B) Linear relationship between estimates of and . We indicate whether individual estimates were significant in either approach by color. Non-significant trait pairs are reduced in size for visibility.
Table 3.
Pairs of Traits with Significant Estimates of
| Trait 1 | Trait 2 |
All Nominally Significant Genes |
|||
|---|---|---|---|---|---|
| 95% CI | M | ||||
| AM | BMI | −0.33 | −0.43 | −0.21 | 257 |
| BMI | COL | −0.31 | −0.44 | −0.18 | 190 |
| BMI | EY | −0.31 | −0.43 | −0.18 | 210 |
| BMI | FI | 0.39 | 0.25 | 0.51 | 164 |
| BMI | HDL | −0.34 | −0.45 | −0.23 | 256 |
| BMI | HOMA-B | 0.31 | 0.17 | 0.44 | 168 |
| BMI | HOMA-IR | 0.36 | 0.22 | 0.49 | 162 |
| BMI | TG | 0.29 | 0.17 | 0.41 | 233 |
| CD | IBD | 0.93 | 0.91 | 0.94 | 366 |
| CD | UC | 0.51 | 0.41 | 0.60 | 218 |
| COL | EY | 0.95 | 0.94 | 0.96 | 363 |
| FA | FN | 0.57 | 0.44 | 0.67 | 149 |
| FA | LS | 0.60 | 0.49 | 0.69 | 170 |
| FG | FI | 0.65 | 0.53 | 0.74 | 133 |
| FG | HOMA-B | −0.60 | −0.70 | −0.47 | 125 |
| FG | HOMA-IR | 0.92 | 0.89 | 0.94 | 136 |
| FI | HDL | −0.31 | −0.44 | −0.17 | 168 |
| FI | HOMA-B | 0.97 | 0.96 | 0.98 | 243 |
| FI | HOMA-IR | 0.99 | 0.99 | 0.99 | 383 |
| FI | TG | 0.57 | 0.45 | 0.66 | 152 |
| FN | LS | 0.86 | 0.83 | 0.89 | 264 |
| HB | MCH | 0.37 | 0.23 | 0.50 | 156 |
| HB | MCHC | 0.40 | 0.23 | 0.55 | 105 |
| HB | PCV | 0.97 | 0.96 | 0.97 | 338 |
| HB | PLT | −0.36 | −0.49 | −0.20 | 141 |
| HB | RBC | 0.95 | 0.94 | 0.96 | 260 |
| HbA1c | T2D | 0.46 | 0.30 | 0.59 | 110 |
| HbA1c | TG | 0.37 | 0.21 | 0.50 | 137 |
| HDL | HOMA-IR | −0.32 | −0.46 | −0.18 | 159 |
| HDL | T2D | −0.32 | −0.45 | −0.19 | 186 |
| HDL | TG | −0.74 | −0.79 | −0.69 | 274 |
| HOMA-B | HOMA-IR | 0.97 | 0.96 | 0.98 | 227 |
| HOMA-B | TG | 0.43 | 0.27 | 0.56 | 127 |
| HOMA-IR | TG | 0.48 | 0.34 | 0.60 | 138 |
| IBD | UC | 0.96 | 0.95 | 0.96 | 415 |
| LDL | TC | 0.97 | 0.96 | 0.97 | 452 |
| LDL | TG | 0.54 | 0.44 | 0.63 | 231 |
| MCH | MCHC | 0.63 | 0.51 | 0.72 | 127 |
| MCH | MCV | 0.96 | 0.95 | 0.97 | 320 |
| MCH | RBC | −0.81 | −0.85 | −0.76 | 207 |
| MCV | RBC | −0.80 | −0.85 | −0.75 | 208 |
| PCV | RBC | 0.96 | 0.95 | 0.97 | 278 |
| TC | TG | 0.61 | 0.53 | 0.68 | 248 |
Estimates were computed with pruned genes that were nominally significant (p < 0.05) in both traits.
Bi-directional Regression Suggests Putative Causal Relationships
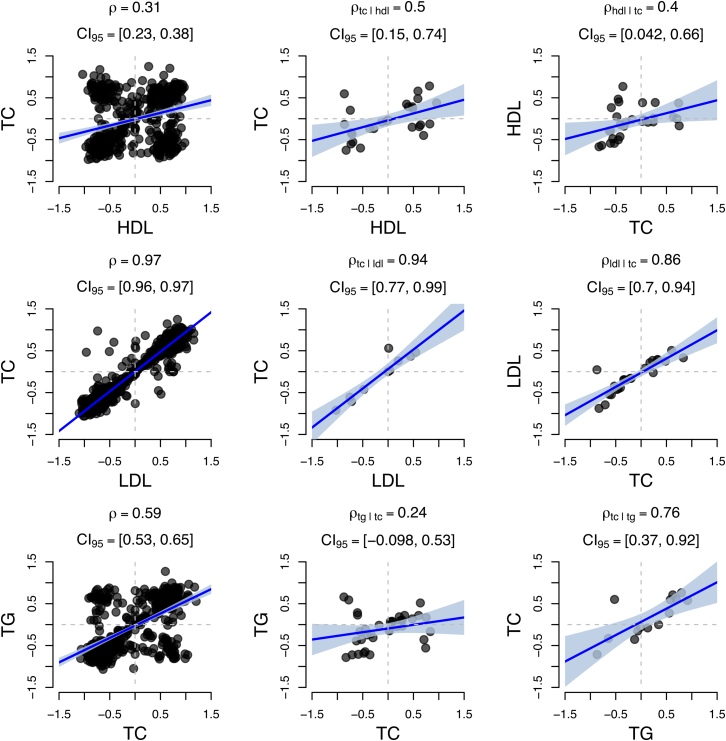
Given pairs of traits with significant estimates of , we aimed to distinguish among possible causal explanations by performing bi-directional regression analyses (see Material and Methods). To empirically validate our approach, we regressed HDL, LDL, and TG with TC. TC is the direct consequence of summing over TG, HDL, and LDL levels, so we expected to observe higher signal for than for . Of these three, we found evidence that TG influences TC (p = 2.34 × 10−3). We observed consistent, but not significant, evidence for the effects of LDL on TC (p = 0.07) and HDL on TC (p = 0.55; see Figure 7). These results suggest that point estimates from the bi-directional approach favor the correct model but might not have adequate power required for significance.
Figure 7.
Estimates of Expression Correlation between TC and HDL, LDL, and TG
(Left column) Estimates of with the use of nominally significant genes (p < 0.05).
(Middle column) We repeated the analysis by using only susceptibility genes found in the x axis trait but not found in the y axis trait.
(Right right) Same analysis as in the middle column but with the other trait’s susceptibility genes.
All three analyses resulted in stronger point estimates for when conditioning on HDL, LDL, and TG genes than for ; however, significance was observed only for (p = 2.34 × 10−3). Shaded regions indicate the estimated 95% confidence interval for the regression line.
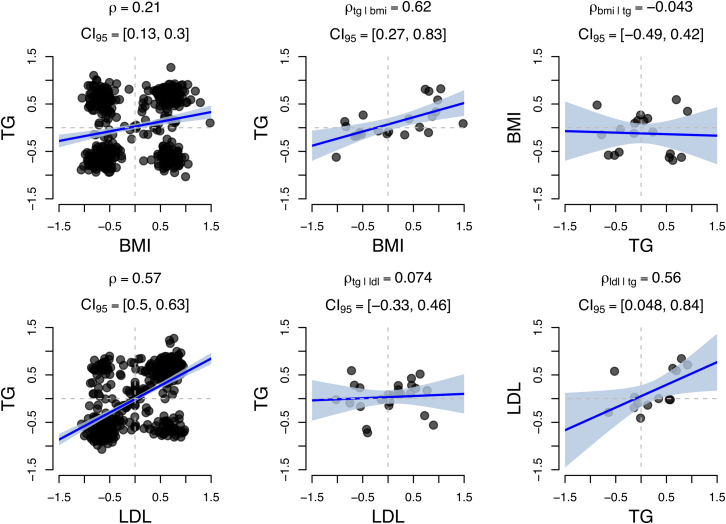
We tested the 43 pairs of traits identified above (see Table 3) while ascertaining susceptibility genes and observed asymmetric effects at p < 0.05 for BMI-TG and LDL-TG (see Figure 8 and Table 4). For example, in the bi-directional analysis on BMI and TG, we observed a significant effect for = 0.62 (95% CI = [0.27, 0.83]; p = 2.06 × 10−3). By contrast, the reverse analysis estimate overlapped 0 at (95% CI = [−0.49, 0.42]; p = 0.86). Individual estimates for and were significantly different (p = 0.01, Welch’s t test), which is consistent with a model where BMI directly influences TG levels. In practice, we used susceptibility genes found through a TWAS (p ∼ 1 × 10−6), but this could be too strict an inclusion threshold for genes for which we lack power to detect. We conducted analyses with weaker thresholds and observed similar results (see Tables S13 and S14). Our results reinforce previous estimates of putative causal effects where BMI influences TG levels.45, 57
Figure 8.
Estimates of for TG with BMI and for TG with LDL
We present results for pairs of traits that displayed a significant difference (p < 0.05, Welch’s t test) in their conditional estimates. These results are consistent with a causal model where BMI influences TG and TG influences LDL. Shaded regions indicate the estimated 95% confidence interval for the regression line.
Table 4.
Bi-directional Estimates of Genome-wide Genetic Correlation at the Level of Predicted Expression
| Trait 1 | Trait 2 |
Results when Ascertaining for Trait 1 |
Results when Ascertaining for Trait 2 |
Test for Difference |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SE | p | M | SE | p | M | t | p | ∼M | ||||
| BMI | TG | 0.62 | 0.10 | 2.06 × 10−3 | 22 | −0.04 | 0.22 | 8.62 × 10−1 | 19 | 2.74 | 1.12 × 10−2 | 25 |
| LDL | TG | 0.07 | 0.19 | 7.25 × 10−1 | 25 | 0.56 | 0.13 | 3.55 × 10−2 | 14 | −2.17 | 3.69 × 10−2 | 36 |
| TC | TG | 0.24 | 0.14 | 1.63 × 10−1 | 36 | 0.76 | 0.08 | 1.79 × 10−3 | 14 | −3.22 | 2.34 × 10−3 | 47 |
We denote the number of ascertained genes used in the test as M. We tested for a difference as a t statistic, where and df is the approximate degrees of freedom determine by the Welch-Satterthwaite equation.
Discussion
In this work, we described an approach to estimate the local genetic covariance and correlation between gene expression and complex traits by using GWAS summary data. We also introduced a method of estimating genome-wide genetic correlation between complex traits at the level of predicted expression. Using simulations, we demonstrated that both approaches are relatively unbiased under realistic scenarios. We used GWAS summary statistics from 30 complex traits and diseases jointly with expression data collected across 45 expression panels to identify 1,196 susceptibility genes for complex traits. Interestingly, susceptibility genes that were identified for educational years and not proximal to a genome-wide significant SNP were validated in a much larger GWAS.35 We leveraged estimates of local genetic correlation between gene expression and traits to compute for 435 trait pairs. This quantified the shared effect of predicted expression levels between two complex traits. To provide evidence of possible causal direction, we adapted a recently proposed causality test45 to operate at the level of predicted gene expression. Our results suggest that TG influences LDL and that BMI influences TG. As more GWAS and eQTL summary results become publicly available, we expect additional studies to integrate cross-trait information to make inferences about mechanistic bases for complex traits. Indeed, recent work has combined chromatin phenotypes with alternatively spliced introns and total gene expression (the latter of which overlaps expression used in this study) to identify regulatory mechanisms for schizophrenia.58
Under the assumption that gene expression mediates the effect of genetics on complex traits, testing for association between predicted gene expression and traits is equivalent to a two-sample Mendelian randomization test for a causal effect of expression on a trait.59, 60 This test for causality is valid if SNPs do not exhibit pleiotropic effects, which is difficult to prove; therefore, TWAS associations do not provide direct evidence of causal relationships between gene expression and complex traits but rather reflect associations between expression levels and traits. This set of assumptions extends to our bi-directional approach to inferring causal direction. A bi-directional regression is capable of distinguishing between directions of effect but cannot rule out pleiotropy. Therefore, our results show consistency with a putative causal mechanism and should not be interpreted as direct proof of causality.
We conclude with several caveats. First, we note that using estimates of genetic correlation to find susceptibility genes could still be biased as a result of confounding. The expression weights used for TWASs could tag variants that are causal through other genes or non-genic mechanisms. In principle, this can be partially remedied by joint testing of multiple genes and a trait. In this work, we combined estimates across tissues by taking the mean effect to compute the genetic correlation between traits and expression. This approach is unbiased but could be inefficient. Recent work61 has described a random-effect model that combines estimates across tissues to increase power. Finally, our method of estimating correlation between traits by using the genetically predicted component of gene expression makes several simplifying assumptions. First, we remedied the non-independence of genes by sampling single genes within a 1 Mb region, an approach that has been used previously.46 However, a more powerful approach could take correlations across genes into account. Second, we limited predictive models to the local (or cis) effects on gene expression, which ignores distal (or trans) effects that regulate gene expression. Although the predictive accuracy of models for gene expression used in this study can account for most of the variation due to genetics,11 we believe that incorporating additional sources of genomic information (e.g., functional priors on SNP effects39, 62, 63) could make additional refinement possible.
Acknowledgments
We would like to thank Valerie Arboleda, Robert Brown, Kathy Burch, and Malika Kumar for helpful discussions and feedback. We also thank Dr. Nicole Soranzo for sharing summary data for the platelet traits. This research was funded in part by NIH awards GM105857, GM053275, and HG009120. G.K. is supported by the Biomedical Big Data Training Program (NIH-NCI T32CA201160). CMC data were generated as part of the CommonMind Consortium, supported by funding from Takeda Pharmaceuticals, F. Hoffman-La Roche, and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881, R37MH057881S1, HHSN271201300031C, AG02219, AG05138, and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the National Institute of Mental Health (NIMH) Human Brain Collection Core. CommonMind Consortium leadership includes Pamela Sklar, Joseph Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Keisuke Hirai, Hiroyoshi Toyoshiba (Takeda Pharmaceuticals), Enrico Domenici, Laurent Essioux (F. Hoffman-La Roche), Lara Mangravite, Mette Peters (Sage Bionetworks), Thomas Lehner, Barbara Lipska (NIMH).
Published: February 23, 2017
Footnotes
Supplemental Data include 12 figures and 14 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.01.031.
Contributor Information
Nicholas Mancuso, Email: nmancuso@mednet.ucla.edu.
Bogdan Pasaniuc, Email: bpasaniuc@mednet.ucla.edu.
Appendix A: Pathway Analysis
We used the PANTHER database47 to explore putative molecular function and pathways enriched with identified susceptibility genes. Using all susceptibility genes across all traits, we found 13 significantly enriched categories, of which seven were related to binding functions. Catalytic activity exhibited the strongest enrichment at 1.3× (GO: 0003824; p = 5.17 × 10−9; see Figure S9). We next focused on individual traits (see Figure S10); however, most individually tested gene sets did not indicate significant enrichment, except for height, LDL, and TC. For example, height had a significant enrichment of genes with catalytic activity (1.31×; p = 4.77 × 10−4). We next looked at biological processes and found TWAS genes enriched at 1.2× for metabolic processes (GO: 0008152; p = 7.29 × 10−11) and 1.57× cellular catabolic processes (GO: 0044248; p = 2.51 × 10−2; see Figures S11 and S12). Enrichment was most pronounced in susceptibility genes specific to height (1.3×; p = 1.03 × 10−6).
Web Resources
CommonMind Consortium, https://www.synapse.org
FUSION software package, http://gusevlab.org/projects/fusion/
Gene Ontology, http://www.geneontology.org/
GTEx Portal, http://www.gtexportal.org/home/
OMIM, http://www.omim.org
RhoGE software, https://github.com/bogdanlab/RHOGE
Supplemental Data
Sample size and the number of genes tested for association with trait by training set. The per-trait Bonferroni correction factor used to determine significance with traits is listed in the “Total Tests” column. Some GWAS did not have overlapping SNPs to perform expression imputation and thus individual expression/trait association was absent.
The “Expression Study” column denotes the original study used to fit weights. The p values are based on a two-tailed Z score test determined from the TWAS test.
Comparison of association strength for genes shared with Zhu et al.12 The “TWAS Z” and “TWAS P” columns indicate the association strength determined using the TWAS expression study weights. PSMR and PHEIDI correspond to the Zhu et al. findings.
Comparison of association strength for genes and traits shared with Pavlides et al.50
Heritability explained by gene expression computed using all genes measured versus reported susceptibility genes only for each tissue.
Mean variance explained in trait per gene. Each estimate was tested for against the cross-tissue mean using a one-sided Z-test.
Comparisons between LD score estimates of genetic correlation (ρg) versus genetic correlation using predicted expression (ρGE). LD-Score was run genome-wide using the same summary data. The threshold for significance was p < 0.05 / 435, where 435 is the total number of pairwise combinations. Significant estimates are denoted with an asterisk (∗).
Comparisons between LD score estimates of genetic correlation (ρg) versus genetic correlation using predicted expression (ρGE). Standard error estimates for ρGE were obtained by jack-knifing over genes.
Bi-directional estimates for all pairs of traits. The number of genes used in the calculation is indicated in the M column. Gene sets included genes that attained p < 1 × 10−6 exclusively for that trait. We pruned genes by selecting a single gene per 1 Mb locus.
The analyses in Table S13 were repeated with relaxed gene sets that attained p < 1 × 10−3.
References
- 1.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claussnitzer M., Dankel S.N., Kim K.-H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 6.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 8.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. advance online publication. [DOI] [PubMed] [Google Scholar]
- 13.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.-R., Duncan L., Perry J.R., Patterson N., Robinson E.B., ReproGen Consortium. Psychiatric Genomics Consortium. Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H.F., Forgetta V., Hsu Y.H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., AOGC Consortium. UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry J.R.B., Day F., Elks C.E., Sulem P., Thompson D.J., Ferreira T., He C., Chasman D.I., Esko T., Thorleifsson G., Australian Ovarian Cancer Study. GENICA Network. kConFab. LifeLines Cohort Study. InterAct Consortium. Early Growth Genetics (EGG) Consortium Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rietveld C.A., Medland S.E., Derringer J., Yang J., Esko T., Martin N.W., Westra H.-J., Shakhbazov K., Abdellaoui A., Agrawal A., LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., WTCCC Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Harst P., Zhang W., Mateo Leach I., Rendon A., Verweij N., Sehmi J., Paul D.S., Elling U., Allayee H., Li X. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raitakari O.T., Juonala M., Rönnemaa T., Keltikangas-Järvinen L., Räsänen L., Pietikäinen M., Hutri-Kähönen N., Taittonen L., Jokinen E., Marniemi J. Cohort profile: the cardiovascular risk in Young Finns Study. Int. J. Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 31.Stancáková A., Civelek M., Saleem N.K., Soininen P., Kangas A.J., Cederberg H., Paananen J., Pihlajamäki J., Bonnycastle L.L., Morken M.A. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 2012;61:1895–1902. doi: 10.2337/db11-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stancáková A., Javorský M., Kuulasmaa T., Haffner S.M., Kuusisto J., Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuotio J., Oikonen M., Magnussen C.G., Jokinen E., Laitinen T., Hutri-Kähönen N., Kähönen M., Lehtimäki T., Taittonen L., Tossavainen P. Cardiovascular risk factors in 2011 and secular trends since 2007: the Cardiovascular Risk in Young Finns Study. Scand. J. Public Health. 2014;42:563–571. doi: 10.1177/1403494814541597. [DOI] [PubMed] [Google Scholar]
- 34.Wright F.A., Sullivan P.F., Brooks A.I., Zou F., Sun W., Xia K., Madar V., Jansen R., Chung W., Zhou Y.-H. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.-B., Emilsson V., Meddens S.F.W., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Los Campos G., Vazquez A.I., Fernando R., Klimentidis Y.C., Sorensen D. Prediction of complex human traits using the genomic best linear unbiased predictor. PLoS Genet. 2013;9:e1003608. doi: 10.1371/journal.pgen.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi H., Kichaev G., Pasaniuc B. Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet. 2016;99:139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi H., Mancuso N., Spendlove S., Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. bioRxiv. 2016 doi: 10.1016/j.ajhg.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., Genetic Investigation of ANthropometric Traits (GIANT) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch B.L. The generalisation of student’s problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 45.Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Do R., Willer C.J., Schmidt E.M., Sengupta S., Gao C., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy J.M., Fodil N., Torre S., Bongfen S.E., Olivier J.-F., Leung V., Langlais D., Meunier C., Berghout J., Langat P. CCDC88B is a novel regulator of maturation and effector functions of T cells during pathological inflammation. J. Exp. Med. 2014;211:2519–2535. doi: 10.1084/jem.20140455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pavlides J.M.W., Zhu Z., Gratten J., McRae A.F., Wray N.R., Yang J. Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med. 2016;8:84. doi: 10.1186/s13073-016-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosokawa Y., Maeda Y., Takahashi E.-i., Suzuki M., Seto M. Human aiolos, an ikaros-related zinc finger DNA binding protein: cDNA cloning, tissue expression pattern, and chromosomal mapping. Genomics. 1999;61:326–329. doi: 10.1006/geno.1999.5949. [DOI] [PubMed] [Google Scholar]
- 52.Quintana F.J., Jin H., Burns E.J., Nadeau M., Yeste A., Kumar D., Rangachari M., Zhu C., Xiao S., Seavitt J. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farh K.K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J.H., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Bakshi A., Zhu Z., Hemani G., Vinkhuyzen A.A.E., Lee S.H., Robinson M.R., Perry J.R.B., Nolte I.M., van Vliet-Ostaptchouk J.V., LifeLines Cohort Study Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez-Arcelus M., Ongen H., Lappalainen T., Montgomery S.B., Buil A., Yurovsky A., Bryois J., Padioleau I., Romano L., Planchon A. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 2015;11:e1004958. doi: 10.1371/journal.pgen.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons T.J., Power C., Logan S., Summerbell C.D. Childhood predictors of adult obesity: a systematic review. Int. J. Obes. Relat. Metab. Disord. 1999;23(Suppl 8):S1–S107. [PubMed] [Google Scholar]
- 57.Fall T., Hägg S., Mägi R., Ploner A., Fischer K., Horikoshi M., Sarin A.-P., Thorleifsson G., Ladenvall C., Kals M., European Network for Genetic and Genomic Epidemiology (ENGAGE) consortium The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gusev A., Mancuso N., Finucane H.K., Reshef Y., Song L., Safi A., Oh E., McCaroll S., Neale B., Ophoff R. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. bioRxiv. 2016 doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pickrell J. Fulfilling the promise of Mendelian randomization. bioRxiv. 2015 [Google Scholar]
- 60.Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Gamazon E.R., Pierce B.L., Stranger B.E., Im H.K., Gibbons R.D., Cox N.J., Nicolae D.L., Chen L.S. Imputing Gene Expression in Uncollected Tissues Within and Beyond GTEx. Am. J. Hum. Genet. 2016;98:697–708. doi: 10.1016/j.ajhg.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kichaev G., Yang W.-Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample size and the number of genes tested for association with trait by training set. The per-trait Bonferroni correction factor used to determine significance with traits is listed in the “Total Tests” column. Some GWAS did not have overlapping SNPs to perform expression imputation and thus individual expression/trait association was absent.
The “Expression Study” column denotes the original study used to fit weights. The p values are based on a two-tailed Z score test determined from the TWAS test.
Comparison of association strength for genes shared with Zhu et al.12 The “TWAS Z” and “TWAS P” columns indicate the association strength determined using the TWAS expression study weights. PSMR and PHEIDI correspond to the Zhu et al. findings.
Comparison of association strength for genes and traits shared with Pavlides et al.50
Heritability explained by gene expression computed using all genes measured versus reported susceptibility genes only for each tissue.
Mean variance explained in trait per gene. Each estimate was tested for against the cross-tissue mean using a one-sided Z-test.
Comparisons between LD score estimates of genetic correlation (ρg) versus genetic correlation using predicted expression (ρGE). LD-Score was run genome-wide using the same summary data. The threshold for significance was p < 0.05 / 435, where 435 is the total number of pairwise combinations. Significant estimates are denoted with an asterisk (∗).
Comparisons between LD score estimates of genetic correlation (ρg) versus genetic correlation using predicted expression (ρGE). Standard error estimates for ρGE were obtained by jack-knifing over genes.
Bi-directional estimates for all pairs of traits. The number of genes used in the calculation is indicated in the M column. Gene sets included genes that attained p < 1 × 10−6 exclusively for that trait. We pruned genes by selecting a single gene per 1 Mb locus.
The analyses in Table S13 were repeated with relaxed gene sets that attained p < 1 × 10−3.