Abstract
The enveloped alphavirus Semliki Forest virus (SFV) infects cells via a low pH-triggered membrane fusion reaction mediated by the E1 protein. E1 is a class II fusion protein that contains the hydrophobic fusion peptide loop and converts to a stable homotrimer during the fusion reaction. Intriguingly, the fusion loop is closely associated with a loop connecting the i and j β-strands. This ij loop plays a role in the cholesterol dependence of membrane fusion and is specifically susceptible to proteolysis in the protease-resistant E1 homotrimer. The SFV ij loop contains a histidine residue at position 230. Sequence comparisons revealed that an analogous histidine is completely conserved in all alphavirus and flavivirus fusion proteins. An E1 H230A mutant was constructed using the SFV infectious clone. Although cells infected with H230A RNA produced virus particles, these virions were completely noninfectious and were blocked in both cell-cell fusion and lipid mixing assays. The H230A virions efficiently bound to cell surface receptors and responded to low pH by undergoing acid-dependent conformational changes including dissociation of the E1/E2 dimer, exposure of the fusion loop, association with target liposomes, exposure of acid-conformation-specific epitopes, and formation of the stable E1 homotrimer. Studies with a soluble fragment of E1 showed that the mutant protein was defective in lipid-dependent conformational changes. Our results indicate that the E1 ij loop and the conserved H230 residue play a critical role in alphavirus-membrane fusion and suggest the presence of a previously undescribed late intermediate in the fusion reaction.
A critical step in enveloped virus infection is the fusion of the virus membrane with that of the target cell. Structural and functional studies of virus-membrane fusion have lead to the definition of two classes of fusion proteins (28). Class I fusion proteins include envelope proteins from the Orthomyxovirus, Paramyxovirus, Retrovirus, Filovirus, and Coronavirus genera (reviewed in references 8, 11, and 40). The class II proteins have been defined more recently and to date this class contains the fusion proteins from the Alphavirus and Flavivirus genera (28, 35, 38).
The class I fusion proteins are exemplified by the influenza virus hemagglutinin (HA) (40). HA is composed of a peripheral subunit and a transmembrane subunit containing the viral fusion peptide at its N terminus. Viral HA is organized as a metastable, vertically oriented trimer that refolds to drive the fusion reaction. The final postfusion conformation of HA is a highly stable trimeric hairpin with a central α-helical coiled-coil and the fusion peptide and transmembrane domain at the same end of the molecule. The central coiled-coil appears to be a defining feature of the class I proteins, and indeed computer searches for coiled-coil domains have been used to predict whether a fusion protein falls into class I.
Determination of the neutral pH ectodomain structures of the fusion proteins of the flaviviruses tick-borne encephalitis virus (TBE) (38) and dengue virus (35) and of the alphavirus Semliki Forest virus (SFV) (28) made it clear that there was a striking structural similarity among the class II proteins and a unique structure compared to the class I proteins. The flavivirus E protein and alphavirus E1 are elongated three-domain molecules that lie tangential to the virus membrane and are composed primarily of β-strands. The fusion peptide loop is located in domain II at the membrane distal tip, and the stem and transmembrane regions that connect the ectodomain to the membrane are at the opposite end of the molecule. The class II proteins are synthesized in conjunction with a companion protein, termed E2 for the alphaviruses, which is cleaved by furin during exocytic transport (for reviews, see references 18 and 23). Following processing, alphavirus E1 remains in a heterodimer with E2, while the flavivirus E protein forms a homodimer. A key step in the fusion of the class II viruses is the conversion of the (hetero- or homo-) dimeric fusion protein to a stable target membrane-inserted homotrimer (HT). This is triggered by low pH in the endosome compartment. Recently the three-dimensional structures of the trimeric forms of the fusion protein ectodomains of SFV, dengue virus, and TBE virus were determined (4, 15, 36). Similar to the class I proteins, the class II fusion proteins also convert to a folded-back conformation during fusion, making a trimer of hairpins but without any coiled-coil structure. The class II proteins reorient vertically during target membrane insertion and trimerization, resulting in an orientation similar to that of the class I proteins (13). Thus, although differing structurally, the class I and class II fusion proteins act via a universal overall mechanism of membrane fusion.
In addition to the structural definition of the HT, biochemical and mutational analyses have helped to characterize and order the conformational steps during alphavirus entry and low pH-triggered fusion (for reviews, see references 5 and 23). The E1/E2 heterodimers are organized into 80 trimers on the surface of the virus particle to form a protein shell with T = 4 icosahedral symmetry (34, 50). The E2 protein binds to the virus receptor and mediates endocytic uptake of the virus by the host cell. When the endosome pH reaches the viral fusion threshold of ∼pH 6.2, the E1/E2 dimer dissociates, releasing E1 and allowing subsequent conformational changes (31, 46). The hydrophobic fusion loop of E1 is masked by the E2 protein in the E1/E2 heterodimer at neutral pH, but it becomes exposed when the dimer is dissociated (12, 17). The fusion loop then inserts into membranes in a reaction that, similar to alphavirus fusion, requires both low pH and cholesterol in the target membrane (1, 12). E1 changes conformation as reflected by exposure of specific monoclonal antibody (MAb) epitopes (2), and forms a sodium dodecyl sulfate (SDS)- and trypsin-resistant HT (48) that is critical for SFV fusion (25).
It is clear that the fusion loop interacts with cholesterol-containing target membranes, and that prevention of this interaction by a MAb to the fusion loop prevents E1-membrane association (1, 12). However, data also suggest a possible role for the associated ij loop in the fusion reaction. The crystal structure of the neutral pH E1 protein revealed that domain II is composed of two long insertions between strands of domain I (28). The first insertion encompasses residues 38 to 130. The fusion loop is located at its tip from residues 83 to 100 and connects β-strands c and d. The second insertion includes residues 169 to 273 of domain II, with the ij loop at its tip connecting β-strands i and j. Thus, although the cd and ij loops belong to the two noncontiguous regions that compose domain II, they are closely associated in the three-dimensional structure. This relationship is maintained in the structure of the low-pH-induced E1 HT (15). Proteolytic mapping studies of the E1 HT also indicated a similarity between the protease accessibility of the fusion and ij loops (14). Although the trimer is highly resistant to proteolysis, treatment with β-mercaptoethanol allows cleavage by elastase in the region of the fusion loop and releases the ectodomain HT from the target membrane. The only other region of the HT in which cleavage was detected was within the ij loop, between residues 229 and 230.
An important feature of alphavirus fusion and infection is its promotion by cholesterol and sphingolipid in the target membrane or host cell membrane (23). Cholesterol is also required for insertion of the E1 ectodomain (termed E1*) into the target membrane and for E1* trimerization (1, 12, 27). Three virus mutants with a reduced requirement for cholesterol in fusion have been isolated by their ability to grow in cholesterol-depleted cells (6, 45). These srf mutants (for sterol requirement in function) all have single amino acid changes within domain II of E1. The srf-3 mutant, which was independently isolated more than 10 times, expresses a change of proline 226 to serine within the ij loop. The other two mutants, srf-4 and srf-5, each map to one of the insertions that comprise domain II but are located more distal to the domain II tip. Studies of Sindbis virus have demonstrated that changing the ij loop sequence to that expressed by srf-3 confers cholesterol independence on this alphavirus (32). Mutagenesis studies demonstrated that the overall conformation of the ij loop is involved in the cholesterol requirement, rather than simply the substitution of serine for proline 226 (32). Thus, the structural and functional characteristics of the ij loop together suggest that it may play a part during the alphavirus fusion reaction.
Examination of the sequences of the ij loops from alphaviruses and flaviviruses revealed that only one residue, a histidine that lies at position 230 in SFV E1, was completely conserved between both virus families. We have here addressed the role of H230 by replacement of this residue with alanine in the SFV infectious clone. The E1 H230A mutant was completely noninfectious and inactive in both content and lipid-mixing assays of membrane fusion. However, the H230A mutant virus appeared fully active when tested for known responses to acid pH, including membrane insertion and HT formation. Together, our results indicate an important role of H230A and the ij loop in fusion and identify a late stage intermediate in the class II fusion reaction.
(The data in this paper are from a thesis to be submitted by C. Chanel-Vos in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University of Paris VII.)
MATERIALS AND METHODS
Cells.
BHK-21 cells were cultivated in complete BHK medium (Dulbecco's minimal essential medium containing 5% fetal calf serum, 10% tryptose phosphate broth, 100 U of penicillin/ml, and 100 μg of streptomycin/ml) at 37 or 28°C.
Construction of the SFV E1-H230A mutant infectious clone.
Mutagenesis was performed on a pGEM5ZF-based plasmid, DG-1, containing an Nsi1/Spe1 fragment derived from the pSP6-SFV4 wild-type infectious clone (30) and encompassing the C-terminal half of E2 and all of the 6K and E1 proteins. The H230A mutation was introduced into DG-1 by circular mutagenesis, using Pfu Turbo DNA polymerase (Stratagene, Inc., La Jolla, Calif.), methylated DG-1 template, the 5′ primer 5′CTTCACCtGGCATGGTCgcgGTACCGTACACACAGAC3′ and the 3′ primer 5′GTCTGTGTGTACGGTACcgcGACCATGCCaGGTGAAG3′. The mutated Nsi1/Spe1 fragment was then subcloned into pSP6-SFV4 (wt-ic) to generate the H230A infectious clone (H230A-ic). In vitro transcription was used to prepare infectious viral RNA from wt-ic and H230A-ic, and the RNA was electroporated into cells (30) for subsequent analysis. Two independent clones of the H230A mutant construct, obtained from two different starting PCRs, were used to confirm initial results. Both clones were sequenced to confirm the presence of the desired mutation and the absence of other mutations in E1.
Virus assembly assay.
Pulse-chase experiments to evaluate virus assembly were performed essentially as described previously (10). Briefly, BHK cells were electroporated with either wt-ic or H230A-ic RNA, plated in 35-mm diameter dishes for 2 h at 37°C, and further incubated at 37 or 28°C for 4 h or overnight, respectively. The cells were then pulse-labeled with [35S]methionine and cysteine and chased in medium without label, using a 30-min pulse and 2-h chase at 37°C and a 1-h pulse and 5-h chase at 28°C. At the end of the incubation, cell lysates and chase media were immunoprecipitated with a polyclonal antibody against the SFV spike proteins and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Chase medium samples were immunoprecipitated in the absence of detergent to allow capsid recovery.
Electron microscopy of virus-infected cells.
BHK cells were electroporated with wild-type (wt) or mutant RNA, plated on 35-mm diameter dishes, and further incubated at 37 or 28°C for 6 h or overnight, respectively. The cells were then fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 40 min at room temperature, washed in 0.1 M cacodylate buffer, osmicated, dehydrated, and embedded in LX112 (Ladd Research, Burlington, Vt.). Thin sections were stained with uranyl acetate and lead citrate and examined with a 1200EX electron microscope (JEOL, Peabody, Mass.) in the Analytical Imaging Facility of the Albert Einstein College of Medicine.
Preparation of radiolabeled virus and soluble E1 protein.
To prepare 35S-labeled virus, BHK-21 cells were infected by electroporation with RNA, plated at 37°C for 6 h in complete BHK medium, and then radiolabeled at 28°C overnight in methionine- and cysteine-deficient MEM containing 100 μCi of [35S]-methionine and cysteine per ml (PROMIX; Amersham LifeScience) as previously described (10). The virus was purified by banding on a Pfefferkorn gradient (26). The radiolabeled virus was collected relatively early in infection to avoid possible appearance of revertants, and the absence of infectious virus was confirmed for each H230A virus preparation.
The radiolabeled soluble form of the E1 protein, E1s, was prepared essentially as previously described (33). In brief, BHK cells were electroporated with wt-ic or H230A-ic RNA and incubated for 6 h at 37°C or for ∼16 h at 28°C. The cells were then pulse-labeled for 1 h at 37°C or 2 h at 28°C as described above, incubated in chase medium for 30 min at 37°C or 1 h at 28°C, and then incubated in minimal essential medium without bicarbonate plus 10 mM HEPES at pH 6.75 for 90 min at 37°C or 3 h at 28°C. The medium was centrifuged to remove cell debris and virus particles, the E1s released into the medium was concentrated, and the buffer was exchanged to 20 mM morpholine ethanesulfonic acid-130 mM NaCl (pH 8.0), with a microconcentrator with a 30,000-molecular-weight cutoff (Vivascience, Inc.). Similar results were obtained with E1s produced at 28 or 37°C.
Assay of secondary infection.
To assay the ability of wt-ic or H230A-ic mutant viruses to carry out a secondary infection, cells were infected by electroporation with the respective RNA, diluted 1:20 with uninfected cells, and allowed to adhere to 22-mm-square coverslips for 2 h at 37°C. The medium was then changed to complete BHK medium with or without 20 mM NH4Cl, and the cells were incubated overnight at 37 or 28°C. Infected cells were visualized by immunofluorescence with a rabbit antibody to the SFV envelope proteins and photographed with a charge-coupled device camera.
Virus-receptor binding assay.
Virus binding to BHK cells was assayed by incubation of 35S-labeled wt-ic or H230A-ic mutant viruses with BHK cells at the indicated pH for 2 h on ice with shaking. Cells were then scraped and washed twice with ice-cold medium at the indicated pH, followed by quantitation of cell-associated radioactivity, all as previously described (25).
Cell-cell fusion assay.
The cell-cell fusion activity of the wt-ic and H230A-ic E1 proteins was evaluated by infecting cells by RNA electroporation. Electroporated cells were diluted 1:20 with uninfected cells and plated 2 h in complete BHK medium at 37°C, followed by overnight culture at 28°C. The cells were treated with medium at the indicated pH for 3 min at 37°C to trigger cell-cell fusion and incubated for an additional 4 h at 28°C to permit polykaryons to express the viral glycoproteins. The number of nuclei per envelope protein-positive cell was evaluated by fluorescence microscopy, counting at least 200 nuclei per pH point. The fusion index was calculated as [1 − (cells/nuclei)] (29).
Liposome fusion assay.
The methods to monitor pyrene-labeled SFV fusion with liposomes were essentially as previously described (7). Briefly, pyrene-labeled SFV was prepared by electroporation of infectious RNA into BHK cells that were prelabeled by growth in the presence of 1-pyrenehexadecanoic acid (Molecular Probes, Eugene, Oreg.). Liposomes were prepared with phospholipids from Avanti Polar Lipids (Alabaster, Ala.) and extruded through two 0.2-μm-pore-diameter polycarbonate filters. Complete liposomes contained a 1:1:1:1.5 molar ratio of phosphatidylcholine (PC; from egg yolk), phosphatidylethanolamine (PE, prepared from egg-PC by transphosphatidylation), sphingomyelin (Sph; from bovine brain), and cholesterol. Each fusion assay contained 0.6 μM virus phospholipid (calculated from a virus phospholipid/protein ratio of 0.45 μmol/mg) in a 1.5-ml volume. Fusion was triggered by adjusting the liposome-virus mixture to pH 5.5, and the decrease in pyrene excimer fluorescence was monitored at an excitation wavelength of 343 nm and an emission wavelength of 480 nm with an Aminco-Bowman AB-2 fluorometer (Spectronic Unicam, Rochester, N.Y.) with a thermojacketed cuvette holder and a 470-nm cutoff filter in the emission beam. The 0% fusion level was set to the initial virus excimer fluorescence, and 100% fusion was defined as the background fluorescence of target liposomes.
Precipitation with acid-induced epitope specific antibodies.
To test the accessibility of the fusion peptide in the native or low-pH-triggered virus, 35S-labeled wt-ic or H230A mutant viruses were treated at pH 5.5 or 8.0 for 5 min at 37°C and then adjusted to pH 8.0. Samples were immunoprecipitated in the absence of detergent with mAbE1f, a MAb that maps to residues 85 to 95 of the fusion peptide loop (residues 83 to 100) (12, 17). The bound virus particles were then washed three times in wash buffer (20 mM Tris [pH 8.5], 150 mM NaCl, 1 mM EDTA, 1% aprotinin, 1 mg of bovine serum albumin per ml) to remove unbound antibody, followed by one wash in detergent-containing (radioimmunoprecipitation assay) buffer to disrupt the virus particles and release any E1 that was not specifically antibody bound, followed by a last wash in phosphate-buffered saline. To determine the total amount of E1 protein present, parallel samples were immunoprecipitated in the presence of 1% Triton X-100 with a rabbit polyclonal antibody to the SFV envelope proteins.
Acid-induced conformational changes in E1 were evaluated by treating 35S-labeled wt-ic or H230A-ic mutant viruses at pH 5.5 in the presence of complete liposomes, adjusting to neutral pH, dissolving the samples in lysis buffer, and immunoprecipitating the samples with the acid-conformation specific MAb E1a-1 (2, 24) or with rabbit polyclonal antibody to the SFV envelope proteins to evaluate the total E1. The samples were resuspended in SDS sample buffer and analyzed by electrophoresis on 10% acrylamide gels under nonreducing conditions. Gels were quantitated by phosphorimaging.
Assay of E1 and E1s HT formation.
35S-labeled wt-ic or H230A-ic mutant viruses were incubated with 0.8 mM liposomes (PC/PE/Sph/cholesterol at a ratio of 1:1:1:1.5), treated at the indicated pH for 5 min at 37°C, and adjusted to neutral pH. The samples were solubilized at 30°C for 3 min in SDS sample buffer, and the presence of the HT was analyzed by SDS-PAGE as above. To assess E1s trimerization, 35S-labeled wt or H230A mutant E1s was incubated in the absence or presence of 1 mM liposomes (PC/PE/Sph/cholesterol at a ratio of 1:1:1:3), treated at the indicated pH for 5 min at 37°C, and assayed as above. Alternatively, the E1s HT was assayed by solubilization in 1% Triton X-100 and digestion with 100 μg of trypsin/ml for 10 min at 37°C (14, 33), followed by analysis by SDS-PAGE.
Virus-liposome association.
Virus-liposome binding was measured by cofloatation of radiolabeled virus with liposomes on sucrose step gradients. Virus (30K dpm/sample) was treated for 5 min at 37°C at pH 8.0 or 5.5 in the presence of 0.2 mM complete liposomes or cholesterol-depleted liposomes. The samples were then adjusted to pH 8.0, 40% sucrose, and a volume of 0.45 ml, layered on top of a 60% sucrose cushion; they were then overlaid with 1.1 ml of 25% sucrose and 0.3 ml of 5% sucrose. All sucrose solutions were in 50 mM Tris (pH 8.0)-100 mM NaCl (wt/vol). Gradients were centrifuged for 2 h at 50,000 rpm at 4°C with a TLS-55 rotor. The gradients were then fractionated into seven 0.3-ml fractions, and the proportion of the virus radioactivity in the liposome-containing top three fractions was determined. Recovery of virus radioactivity from the gradients ranged from 80 to 100%.
RESULTS
Sequence comparison of the ij loops of the alphavirus and flavivirus fusion proteins.
The crystal structure of the SFV E1 ectodomain revealed a close interaction between the fusion peptide loop and the ij loop in both the neutral and low pH conformations (15, 28). We compared the sequence of this region from various alphaviruses and flaviviruses, using residues 221 to 233 of SFV E1 and residues 246 to 252 of the TBE E protein as a starting point. Overall, the alphavirus ij loop does not show a high degree of sequence conservation, but a proline at position 224 and a histidine at position 230 are completely conserved among all of the alphavirus sequences from the database, including those of the relatively distant fish alphaviruses salmon pancreas disease virus and sleeping disease virus (Table 1). The analogous ij loop from the flaviviruses is somewhat shorter but also contains a conserved histidine at position 248 followed by a conserved alanine residue. We also analyzed the sequence of Hepatitis C virus (HCV), which is in the genus Hepacivirus of the Flaviviridae family. A model of the HCV E2 protein structure was previously derived based on the TBE E structure, although it has not been experimentally confirmed (49a). Interestingly, the HCV ij loop predicted by this model also contains a histidine. Together, the sequence alignments suggested a potential role for the histidine residue in the ij loop of both alphaviruses and flaviviruses.
TABLE 1.
Sequence alignments of the ij loop of alphavirus and flavivirus fusion proteins
| Virus | ij loopa |
|---|---|
| Alphavirusesb | |
| SFVd | LARPSPGMVHVPY |
| ONNV | -Q--AA-AI---- |
| RRV | -S-----V------ |
| SINV | -LK--AKN----- |
| BFV | -K--AS-N----- |
| EEEV | -Q--QA-I--T-F |
| VEEV | -Q--KA-AI---- |
| WEEV | -LK--VKNI---- |
| NDUV | -D--AS-NI---- |
| MIDV | -A---A-T----- |
| SPDV | VLQ-TNDH---A- |
| SDV | VLQ-TNDH---A- |
| Flavivirusesc | |
| TBE | APHAVKM |
| YFV | P---ATI |
| DE1 | TA--K-Q |
| DE2 | N---K-Q |
| DE3 | NA--K-P |
| DE4 | V---KRQ |
| WN | E---T-Q |
| JE | EA--T-Q |
| Kunjin | E---T-Q |
Completely conserved residues are shown in boldface.
Residue numbers in SFV sequence are 221 to 233.
Residue numbers in TBE sequence are 246 to 252.
Virus abbreviations: ONNV, O'nyong nyong virus; RRV, Ross River virus; SINV, Sindbis virus; BFV, Barmah Forest virus; EEEV, Eastern equine encephalitis virus; VEEV, Venezuelan EEV; WEEV, Western EEV; NDUV, Ndumu virus; MIDV, Middelburg virus; SPDV, salmon pancreas disease virus; SDV, sleeping disease virus; YFV, yellow fever virus; DE, Dengue virus; WN, West Nile virus; JE, Japanese encephalitis virus.
Generation and initial characterization of an SFV E1 H230A mutant.
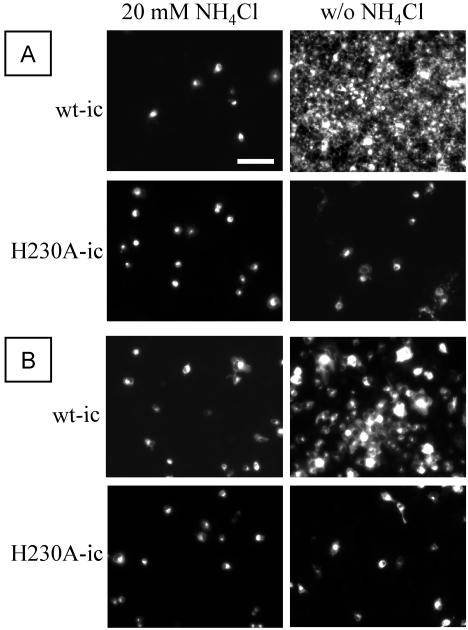
To better understand the role played by the ij loop histidine during the SFV life cycle, we constructed a mutant of the SFV infectious clone containing a substitution of alanine for E1 histidine 230 (H230A-ic). The mutant infectious clone was used as template for in vitro transcription of viral RNA, and the phenotype of the H230A mutant was studied by electroporation of the RNA into BHK cells. To test the ability of the mutant RNA to support primary and secondary viral infection, electroporated cells were mixed in a 1:20 ratio with nonelectroporated cells and incubated overnight at either 37 or 28°C in the presence or absence of 20 mM NH4Cl. Infected cells were identified by immunostaining. NH4Cl blocks SFV infection by raising the endosomal pH above the virus fusion threshold (19). Thus, cultures incubated in the presence of NH4Cl reflect primary virus infection due to RNA electroporation, while in the absence of NH4Cl virus can spread by secondary infection to nonelectroporated cells (25). At 37°C, the initial wt virus infection spread to almost every cell in the culture (Fig. 1A). In contrast, although cells became infected by electroporation of H230A RNA, there was no increase in the number of infected cells in cultures incubated without NH4Cl (Fig. 1A). The infectivity assay was also performed at 28°C, since several mutants with an assembly defect at 37°C have been shown to assemble more efficiently at 28°C (6, 10). Although the spread of infection was slower than at 37°C, the wt virus showed secondary infection at 28°C, while the H230A mutant again showed no secondary infection (Fig. 1B).
FIG. 1.
Infectivity of wild-type and H230A viruses on BHK cells. BHK cells were electroporated with H230A-ic or wt-ic RNA, diluted 1:20 with nonelectroporated cells, and plated on duplicate coverslips. Cells were allowed to adhere for 2 h at 37°C, at which point half of the coverslips were switched to medium containing 20 mM NH4Cl to prevent secondary infection. After overnight incubation at either 37°C (A) or 28°C (B), cells were fixed with methanol and labeled with a rabbit polyclonal antibody against the SFV envelope glycoproteins, followed by fluorescein-conjugated goat anti-rabbit antibody. Samples were photographed by fluorescence microscopy, and data shown are a representative example of three experiments. Bar, 100 μm.
H230A mutant protein expression and assembly.
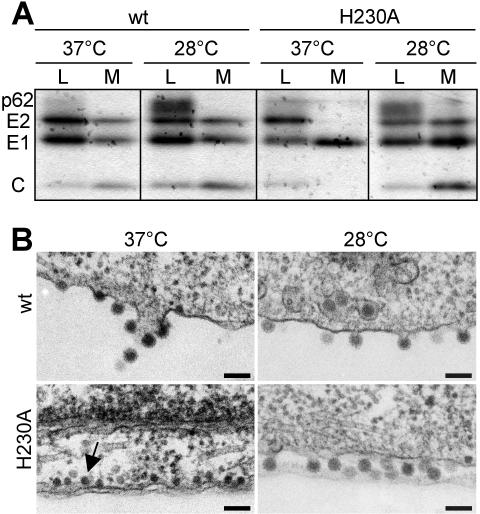
To determine if the lack of secondary infection by H230A was due to misfolding of the mutant E1 protein, cell surface expression was tested by immunostaining of BHK cells electroporated with wt or H230A RNA. The wt and mutant E1 and E2 proteins were efficiently expressed at the cell surface after culture of the cells at either 37 or 28°C (data not shown). Virus assembly was then evaluated by pulse-chase analysis of wt and mutant-infected BHK cells (Fig. 2A). H230A-infected cells released only a soluble truncated form of E1 when incubated at 37°C, similar to several other E1 mutants that are temperature sensitive for virus assembly (6, 10, 33). In contrast, radiolabeled H230A virus particles were efficiently released when cells were incubated at 28°C (Fig. 2A). In keeping with the pulse-chase results, electron microscopy of H230A-infected cells incubated at 37°C revealed an accumulation of viral nucleocapsids underneath the plasma membrane, but no budding virus particles (Fig. 2B). H230A-infected cells incubated at 28°C showed budding of morphologically normal virus particles (Fig. 2B).
FIG. 2.
Assembly of wt SFV and H230A mutant in BHK cells. BHK cells were electroporated with wt-ic or H230A-ic RNA and incubated at 37°C for 2 h. Cells were further incubated for 4 h at 37°C or overnight at 28°C before analysis as described. (A) Protein expression and virus assembly. Cells were pulse-labeled with [35S]methionine and cysteine and chased at 37 or 28°C. SFV proteins in the cell lysates (L) and medium (M) were immunoprecipitated with a polyclonal antibody to E1 and E2, and samples were analyzed by SDS-PAGE. Chase medium samples were immunoprecipitated in the absence of detergent to allow recovery of intact virus particles containing the viral nucleocapsid. The position of the different viral proteins is indicated (C, capsid). Data shown are representative of three experiments. (B) Electron microscopy of wt or H230A virus-infected-cells. The arrow indicates the presence of viral nucleocapsids underneath the plasma membrane. Bar, 0.1 μm.
Together, these H230A results are in contrast to prior results with the G91A fusion loop mutant and the double srf-4-srf-5 mutant (6, 25). Both of these mutants are temperature sensitive for assembly but are infectious and fusion active at either temperature once assembled at 28°C. The complete absence of secondary infection of the H230A mutant at 28°C thus suggests that the mutant virus particles have a defect in entry into host cells. We therefore analyzed the steps in virus entry and fusion for the H230A mutant.
Virus-receptor binding.
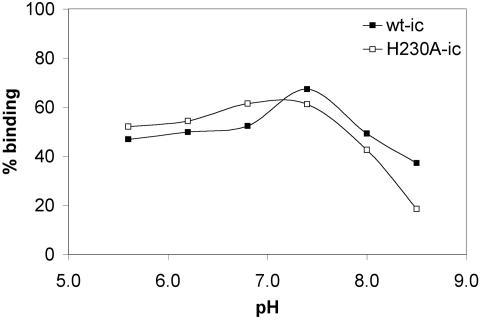
The first step in SFV entry is the binding of virus to receptors on the surface of the target cell, mediated by the E2 protein (43). It was possible that a change in the conformation of the E1 protein could affect the conformation of its E2 dimeric partner and thus interfere with virus-receptor binding. 35S-labeled wt and H230A mutant viruses were produced from RNA-transfected cells incubated at 28°C. The radiolabeled H230A virus preparations were noninfectious, as confirmed by plaque assays. Radiolabeled viruses were incubated on ice with BHK cells at the indicated pH, and the bound virus was quantitated (Fig. 3). Both wt and H230A mutant viruses showed efficient binding, with maximal levels of binding at pH 7.4 and somewhat reduced binding at more basic pH values. Thus, the lack of H230A infectivity was not due to a defect in virus-receptor interaction. We also confirmed that the receptor-bound mutant was efficiently endocytosed by BHK cells (data not shown).
FIG. 3.
pH dependence of binding of wt SFV and H230A mutant to BHK cells. 35S-labeled wt-ic or H230A-ic viruses were bound to duplicate 35-mm plates containing BHK cells at the indicated pH for 2 h on ice with shaking. Cells were scraped and washed, and the bound radioactivity was quantitated by scintillation liquid counting. Data shown are a representative example of three experiments.
Virus fusion activity.
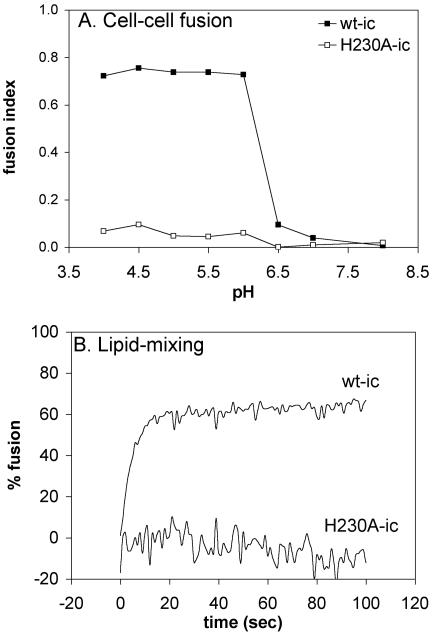
We then tested the capacity of the H230A mutant to fuse with a target lipid bilayer. Cells were electroporated with wt or mutant RNA, and cell-cell fusion was triggered by a 3-min low-pH treatment of the virus-infected cells at either 37 or 28°C (25, 49). Polykaryon formation was quantitated (Fig. 4A). The wt-infected cells fused efficiently with a pH threshold of ∼pH 6.2. In contrast, the mutant-infected cells showed no fusion above background at either temperature, even after treatment as low as pH 4.0.
FIG. 4.
Membrane fusion activity of wt SFV and H230A mutant. (A) Low-pH-dependent polykaryon formation in wt and H230A-infected cells. BHK cells were electroporated with wt-ic or H230A-ic mutant RNA, diluted 1:20 with nonelectroporated BHK cells, plated on duplicate coverslips for 2 h at 37°C, and then cultured overnight at 28°C. The cells were treated at the indicated pH for 3 min at 37°C to induce cell-cell fusion, recultured at 28°C for 3 to 4 h, and fixed. Cells were stained with an antibody to the SFV spike proteins, and nuclei were stained with propidium iodide. The number of nuclei per expressing cell was evaluated by fluorescence microscopy, and the fusion index was determined. The data shown are the average of two experiments. (B) Low-pH-dependent liposome fusion activity. Fusion of pyrene-labeled wt-ic and H230A-ic viruses with unlabeled liposomes was analyzed. Pyrene-labeled wt-ic or H230A-ic viruses (0.6 μM) were mixed with unlabeled complete liposomes (200 μM) and preequilibrated for 5 min at 28°C. The pH was adjusted to 5.5 at time zero, and the fluorescence signal was recorded in real time. Data shown are a representative example of three experiments.
Both virus infection and E1-induced cell-cell fusion are content-mixing assays that detect complete fusion between lipid bilayers. It was possible that the H230A mutant was blocked in full fusion but could mediate the initial mixing of the outer leaflets of the virus and target membranes, a process known as hemifusion (22). We therefore performed a real-time lipid-mixing assay using pyrene-labeled wt and H230A mutant viruses produced at 28°C, following the decrease of the pyrene excimer peak upon fusion with unlabeled liposomes (Fig. 4B). The wt virus showed rapid and efficient fusion when treated at pH 5.5 at 28°C in the presence of cholesterol-containing liposomes, with maximal fusion in less than 20 s. No dilution of the pyrene probe was observed for the H230A mutant even after treatment as low as pH 5.0. Addition of detergent to the H230A virus preparation confirmed that dilution of the pyrene probe would be easily detected for the mutant (data not shown). Thus, the H230A mutant virus was completely impaired for membrane fusion, even in the initial step of lipid mixing.
Dimer dissociation and fusion peptide exposure.
Since the mutant is blocked in fusion, we set out to determine the step in the fusion pathway that is inhibited by the H230A substitution. The first conformational change that is biochemically detectable following treatment of SFV at acid pH is the dissociation of the E1-E2 heterodimer (21, 46). Dimer dissociation exposes the previously masked fusion loop on the E1 protein (12), and mutants that are blocked in dimer dissociation are blocked in the subsequent conformational changes in E1 and in membrane fusion (39, 51). We used MAb E1f, which maps to the E1 fusion loop, to assess the masking of the fusion loop by dimer association and its exposure by dimer dissociation (12, 17). The wt and mutant virus particles were not efficiently recognized by MAb E1f when tested at pH 8.0 (Fig. 5). Following treatment at pH 5.5, the fusion loop epitope became exposed in both wt and H230A viruses. The proportion of the total E1 recognized by MAb E1f after low pH treatment was about 40% for both viruses. The wt and H230A mutant thus show similar dimer association in the virus particle, similar dimer dissociation at acid pH, and comparable properties of fusion loop exposure.
FIG. 5.
Low-pH-triggered exposure of the fusion peptide in wt SFV and H230A mutant. Purified 35S-labeled wt-ic or H230A-ic viruses were treated at the indicated pH for 5 min at 37°C, the pH was adjusted to pH 8.0, and incubation continued for 1 h on ice with a polyclonal antibody against the SFV envelope proteins (Rab) or with the fusion peptide-specific MAb E1f. The amount of E1 specifically precipitated by the antibody was determined by SDS-PAGE and quantitation. The graph represents the average of three experiments.
Acid-induced conformational changes in the E1 protein.
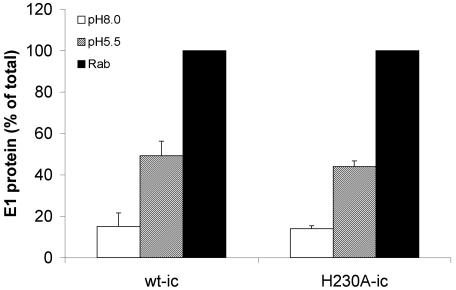
We then analyzed the specific response of the wt and mutant E1 proteins to low pH. Following dimer dissociation, the E1 protein undergoes conformational changes that expose acid-conformation-specific epitopes. One such epitope is recognized by MAb E1a-1, which we have mapped to the region of residue G157 in domain I of the E1 protein (2). The neutral pH forms of the wt and H230A E1 proteins were not recognized by MAb E1a-1, but following treatment at acid pH both the mutant and wt E1 proteins were immunoprecipitated by MAb E1a-1 (Fig. 6A). A similar efficiency of the E1 conformational change was observed for the wt and mutant, with about 40% of the total E1 recognized by MAb E1a-1 for each virus.
FIG. 6.
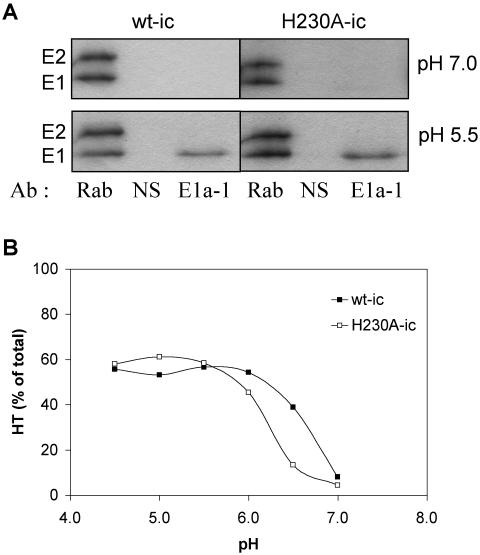
Analysis of low-pH-dependent E1 conformational changes in wt SFV and H230A mutant. Purified 35S-labeled wt-ic or H230A-ic virus was preincubated with complete liposomes for 3 min at 37°C, acid treated at the indicated pH for 5 min at 37°C, returned to pH 8.0 on ice, and tested for acid-specific epitope exposure and E1 HT formation. (A) Samples were immunoprecipitated with either a polyclonal antibody against the SFV envelope proteins (Rab), a nonspecific antibody (NS), or an E1 acid-specific MAb (E1a-1) and analyzed by SDS-PAGE. Shown is a representative example of three experiments. (B) Formation of the E1 HT was determined by solubilizing samples in SDS sample buffer at 30°C and carrying out analysis by SDS-PAGE. The data shown are the average of two experiments.
A key conformational change in E1 during fusion is the formation of a very stable E1 HT that is resistant to trypsin digestion and to dissociation by SDS at 30°C (14, 47, 48). The HT is critical for virus fusion, as shown by the phenotype of the E1 G91D mutant (25). This lethal mutation within the fusion loop completely inhibits membrane fusion and blocks E1 HT formation. We tested the ability of wt and H230A virus to form the E1 HT by treating virus at a pH range from pH 4.5 to 7.0 in the presence of complete liposomes. HT formation was assessed by quantitation of the SDS-resistant trimer band in SDS-PAGE (Fig. 6B). Both wt and mutant formed the E1 HT at comparable efficiency, indicating that the H230A E1 HT, similar to the wt, was stable to SDS treatment. The H230A HT also showed resistance to trypsin digestion similar to that of the wt (data not shown), suggesting that its overall structure resembled that of the wt HT. Interestingly, although the H230A mutation replaces a residue that is presumably titratable at acid pH, the pH dependence of E1 trimerization was comparable between the two viruses.
Virus-lipid bilayer interaction.
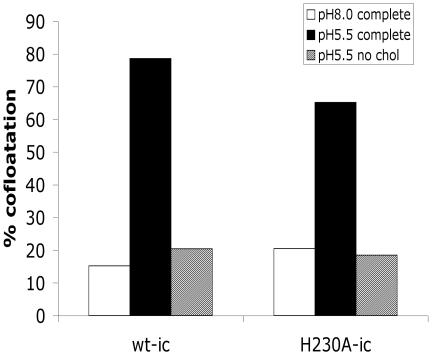
The dual interaction of E1 with both the virus and target membranes and the refolding of E1 to the HT conformation are believed to drive the alphavirus fusion reaction. However, both wt and H230A viruses have the capacity to form a stable E1 HT even in absence of a target lipid bilayer (14, and data not shown). Thus, it was critical to determine the ability of the H230A E1 protein to interact with a target membrane. Radiolabeled wt or mutant virus was mixed with cholesterol-containing liposomes and treated at pH 8.0 or 5.5. The association of the virus with the target membrane was then tested by sucrose gradient floatation analysis (Fig. 7). Low pH triggered the association of >65% of the wt and H230A virus with target liposomes. This interaction was dependent on low-pH treatment, since much less cofloatation was observed when the virus-liposome mixture was maintained at pH 8.0.
FIG. 7.
Low-pH-dependent liposome association of wt SFV and H230A mutant. Purified 35S-labeled wt-ic or H230A-ic viruses were mixed with 0.2 mM complete or cholesterol-depleted (no chol) liposomes. The samples were treated at the indicated pH for 5 min at 37°C and adjusted to neutral pH. Virus-liposome association was then determined by cofloatation analysis on discontinuous sucrose gradients as described in Materials and Methods. The pH 8.0 and 5.5 samples were treated in presence of complete liposomes, and the cholesterol-depleted sample was treated at pH 5.5. The data shown are a representative example of three experiments.
We also analyzed the cholesterol dependence of the virus-membrane interaction since we have previously shown that the ij loop, particularly residue 226, is important in the cholesterol dependence of the alphavirus fusion (7, 32, 45). Little binding was observed when either wt or mutant virus was treated at pH 5.5 in the presence of cholesterol-depleted liposomes (Fig. 7). Thus, the H230A mutant was able to carry out the initial interaction of E1 with the target membrane. Similar to the wt virus, this membrane interaction was dependent on both low pH and cholesterol.
Soluble E1 HT formation.
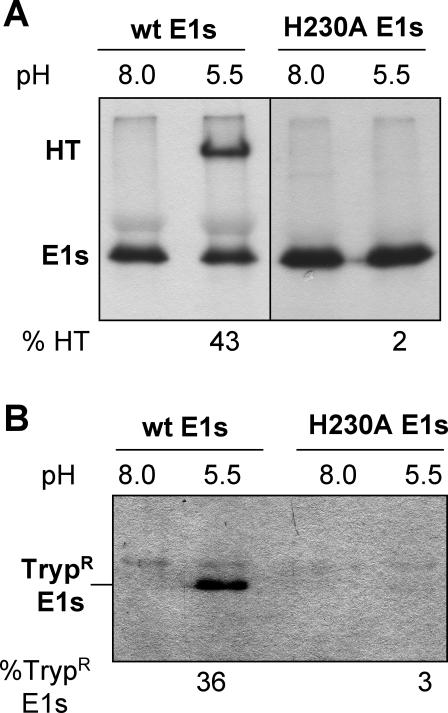
While full-length virus E1 can form the HT in the absence of a target membrane, a truncated monomeric form of E1 missing the transmembrane domain does not trimerize unless treated at low pH in the presence of a cholesterol-containing target membrane. This lipid dependence is observed both for soluble E1 generated by in vitro protease cleavage (E1*) (12, 27) and for the soluble E1s fragment produced during virus assembly and budding (33). We therefore took advantage of the E1s system to assay the productive interaction of H230A E1 with target membranes at low pH. Radiolabeled E1s was prepared from wt or H230A-infected BHK cells at 28°C. The proteins were mixed with cholesterol-containing liposomes or with buffer alone, and treated at pH 8.0 or 5.5. In keeping with previous results, wt E1s did not form a HT at neutral pH or at low pH in the absence of liposomes (Fig. 8A and data not shown). However, when incubated with complete liposomes and treated at pH 5.5, over 40% of the wt E1s formed the SDS-resistant HT, confirming the lipid dependence of the E1s conformational change. In contrast, H230A E1s did not form significant HT when treated at acid pH in the presence of cholesterol-containing liposomes, showing on average only ∼2% trimerization (Fig. 8A). Detection of the HT by the trypsin-resistance assay confirmed these results, showing efficient trimerization of the wt E1s protein at low pH in the presence of complete liposomes and inefficient H230A trimer formation under the same conditions (Fig. 8B). Thus, the lack of detection of the H230A HT in SDS-PAGE was not due to a decrease in the SDS stability of the trimer, as was previously observed for the srf-4 and srf-5 mutants (6), but reflects a reduction in HT formation. The inhibition of H230A E1s trimerization did not appear to be due to a lower E1s concentration, since H230A and wt E1s were prepared in parallel from cells electroporated at high efficiency and yielded equivalent amounts of radiolabeled E1s. Mutant trimerization efficiency was not increased when tested at a 5- to 20-fold-higher protein concentration (data not shown). Thus, the fusion defect of the H230A virus correlates with impaired activity of the soluble form of the H230A E1 protein.
FIG. 8.
HT formation by the soluble E1 fragment from wt SFV and H230A mutant. 35S-labeled wt or H230A E1s was preincubated with complete liposomes for 3 min at 37°C, treated at the indicated pH for 5 min at 37°C, and adjusted to neutral pH. Quantitation of the low-pH-induced HT was performed by two methods, and the average percentage of HT formation (% HT) from three experiments is shown below each gel. (A) HT formation was evaluated by solubilization in SDS sample buffer at 30°C, followed by SDS-PAGE. (B) HT formation was evaluated by trypsin resistance. Parallel aliquots from the samples shown in panel A were subjected to trypsin digestion, followed by trichloroacetic acid precipitation. Trypsin-resistant E1s (TrypR E1s) was detected by SDS-PAGE.
DISCUSSION
The E1 ij loop region was previously implicated in membrane fusion because of its observed role in alphavirus cholesterol dependence (7, 32, 45) and specific susceptibility to proteolysis in the E1 HT (15). We showed here that mutation of the highly conserved ij loop residue histidine 230 to alanine blocked SFV fusion and infection and that the initial stage of lipid mixing was also blocked. Intriguingly, however, although virus fusion activity was completely inhibited, tests of the known steps in the alphavirus fusion pathway revealed that H230A behaved in a manner similar to that of the wt virus. The mutant virus bound efficiently to the cellular receptor and showed a similar pH dependence of receptor binding. The mutant E1 interacted with E2 in a heterodimer, which masked the fusion loop at neutral pH and dissociated to expose the fusion loop after acid treatment. Following dimer dissociation, H230A E1 responded to low pH as detected by liposome association, acid-specific antibody binding, and formation of a stable, SDS- and trypsin-resistant HT. Thus, the phenotype of the H230A mutant suggests the presence of a previously undescribed late intermediate in the alphavirus membrane fusion reaction.
In contrast, all other reported alphavirus membrane fusion mutants and inhibitors have identified steps that appear to lie upstream of the H230A defect, such as dimer dissociation, membrane interaction, or E1 HT formation. For example, a number of studies have shown that blocking furin processing of p62 inhibits fusion because the low pH-triggered dissociation of the E1/p62 dimer is resistant to the normal physiological pH range of the endosome (39, 41, 51). Treatment of SFV with Zn2+ inhibits SFV fusion (9). In the presence of Zn2+, dimer dissociation and virus-target membrane interaction occur, but HT formation is strongly reduced. A G91D mutation in the SFV fusion loop inhibits fusion and infection by completely blocking formation of the E1 HT (25). In the flavivirus system, a similar mutation in the fusion loop, L107D, inhibits fusion by blocking virus-membrane interaction, although trimerization is unaltered (3). The novel phenotype of the H230A mutant, considered in the context of our knowledge of E1 structure and function, suggests several possible explanations for the mechanism of the fusion block.
The only biochemical assay of the H230A mutant that showed a significant difference from that of the wt was the activity of the truncated E1s form of the fusion protein. Mutant E1s had greatly reduced HT formation when acid treated in the presence of cholesterol-containing liposomes. These E1s results suggest that some property of E1-membrane interaction or its consequences is aberrant in the mutant. Unlike the full-length proteins, the truncated forms of both the alphavirus and flavivirus fusion proteins require target membranes for trimerization (27, 42). While the mechanism of this membrane requirement is not clear, it may reflect a specific role of cholesterol/target membrane in the E1 conformational change, a role of the membrane in increasing the local concentration of E1 and correctly orienting each monomer with its partners during trimerization, or some combination of these factors (for discussion, see references 12 and 42). Thus, an alteration in the membrane interaction of the mutant protein could be responsible for the observed defect in E1s trimerization and might also be the cause of the virus fusion defect.
What is known about the interaction of E1 with the target membrane? An antibody to the fusion loop blocks E1*-membrane insertion, suggesting that this part of the protein is the first or dominant portion that interacts with the membrane (12). The structure of the fusion loop in the low-pH-induced HT suggests that it does not undergo a major conformational change upon membrane insertion and that the loop does not insert deeply into the hydrocarbon portion of the lipid bilayer (15). The E1 fusion loop is known to preferentially interact with detergent-resistant, cholesterol-enriched membrane domains, perhaps reflecting an interaction of the protein with cholesterol (1). Alteration of this interaction could decrease the local concentration of the protein and inhibit E1s trimerization, and an altered cholesterol interaction might also inhibit fusion. In contrast with H230A E1s, the H230A virus efficiently bound cholesterol liposomes at low pH, demonstrating that even in the absence of fusion the initial association of mutant E1 with the target membrane was stable. However, there are many unanswered questions about E1-membrane insertion, since the structure of the E1* HT was determined after membrane solubilization and lipid removal. It is possible that there are several steps in membrane insertion, with the ij loop playing a role. For example, multiple steps in membrane insertion have been observed for the bacterial pore-forming toxin perfringolysin O (44). This cholesterol-dependent cytolysin contains four protein domains. Initial membrane interaction is via a superficial insertion of domain 4, which interestingly contains a conserved histidine suggested to be important in pore formation (44). Subsequently the toxin oligomerizes, and portions of domain 3 insert into the membrane via a cholesterol-dependent step to form the lytic pore (16, 20). While the structures of the membrane-inserted regions of perfringolysin O and E1 are clearly different (15), the toxin does suggest a precedent for the interaction of multiple protein regions with the membrane during membrane insertion.
Alternatively, the critical effect of the H230A mutation on fusion could be due to alterations in the interaction of the stem with the core HT. It has been shown for the class I influenza fusion protein HA that fusion can be blocked by mutations that affect the interaction of the C-terminal leash region with the N terminus of the central coiled-coil (37). These leash mutations appear to act via a late stage in fusion and in formation of the final hairpin structure. From the structure of the SFV E1 low-pH-induced trimer, the C-terminal stem region could act analogously to the HA C-terminal leash. The E1 stem follows a groove situated between two adjacent E1 molecules, with the ij loop oriented towards this groove (4, 15). It is possible that H230A changes the ij loop, impairs the interaction of the membrane-proximal stem with domain II of the same E1 subunit, and thus inhibits lipid mixing. In contrast to the HA leash mutants, the mutation in H230A is located on the core portion of the trimer instead of the leash. To address this model more fully, we need to know more about E1 stem-trimer interaction, particularly for the stem region not represented in the trimer structure, and to understand potential alterations in the H230A trimer that are not detectable by analysis of overall trimer stability to SDS and protease.
A third explanation for the effect of H230A on fusion could lie in the observed cooperative interactions between HTs. Electron microscopy revealed that membrane insertion was highly cooperative and that the membrane-inserted proteins were associated in rings of five or six trimers (13). The crystal structure of the low-pH-induced trimer showed interactions between the fusion loops of trimers, which could be propagated to give rings of five or six trimers (15). Due to the proximity of the fusion peptide loop and ij loop within each E1 protein, it is possible that the ij loops play a role in regulating the formation of such oligomeric complexes. If such cooperation is critical for fusion, its inhibition could be the mechanism of the H230A mutant block.
It will be important to carefully evaluate the exact portions of E1 that insert into the target membrane, the mechanism of membrane insertion, the effect of stem-trimer interaction in mediating the final steps of membrane merger, and the importance of intertrimer cooperativity during fusion. The involvement of the ij loop in these processes has not been explored. Further analysis of the sequence requirements at position 230 and of mutations that can compensate for the lethal H230A substitution may begin to bring these many unknowns closer to understanding.
Acknowledgments
We thank Brigid Reilly and Zoya Niazova for expert technical assistance, Alyson Urian for her significant contributions to the virus-liposome binding studies, Don Gibbons for providing the DG-1 construct, and the members of the Kielian lab for helpful discussions and critical reading of the manuscript. We also thank Félix Rey for his insightful comments on the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM57454) and by Cancer Center Core Support grant NIH/NCI P30-CA13330.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, A., M. R. Klimjack, P. K. Chatterjee, and M. Kielian. 1999. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J. Virol. 73:10029-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, R., J. M. Wahlberg, H. Garoff, and J. Wilschut. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, P. K., C. H. Eng, and M. Kielian. 2002. Novel mutations that control the sphingolipid and cholesterol dependence of the Semliki Forest virus fusion protein. J. Virol. 76:12712-12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 9.Corver, J., R. Bron, H. Snippe, C. Kraaijeveld, and J. Wilschut. 1997. Membrane fusion activity of Semliki forest virus in a liposomal model system: Specific inhibition by Zn2+ ions. Virology 238:14-21. [DOI] [PubMed] [Google Scholar]
- 10.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 69:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, M. Liao, L. Hammar, R. H. Cheng, and M. Kielian. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 78:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., I. Erk, B. Reilly, J. Navaza, M. Kielian, F. A. Rey, and J. Lepault. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114:573-583. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons, D. L., M.-C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 16.Giddings, K. S., A. E. Johnson, and R. K. Tweten. 2003. Redefining cholesterol's role in the mechanism of the cholesterol-dependent cytolysins. Proc. Natl. Acad. Sci. USA 100:11315-11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammar, L., S. Markarian, L. Haag, H. Lankinen, A. Salmi, and H. R. Cheng. 2002. Prefusion rearrangements resulting in fusion peptide exposure in Semliki forest virus. J. Biol. Chem. 17:17. [DOI] [PubMed] [Google Scholar]
- 18.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius, A., J. Kartenbeck, K. Simons, and E. Fries. 1980. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 84:404-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuck, A. P., R. K. Tweten, and A. E. Johnson. 2003. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J. Biol. Chem. 278:31218-31225. [DOI] [PubMed] [Google Scholar]
- 21.Justman, J., M. R. Klimjack, and M. Kielian. 1993. Role of spike protein conformational changes in fusion of Semliki Forest virus. J. Virol. 67:7597-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 23.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit: examples from the alphaviruses, p. 409-455. In H. Hilderson and S. Fuller (ed.), Subcellular biochemistry, vol. 34. Fusion of biological membranes and related problems. Plenum Publishers, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 24.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielian, M. C., S. Keränen, L. Kääriäinen, and A. Helenius. 1984. Membrane fusion mutants of Semliki Forest virus. J. Cell Biol. 98:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 29.Levy-Mintz, P., and M. Kielian. 1991. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 65:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobigs, M., and H. Garoff. 1990. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J. Virol. 64:1233-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, Y. E., T. Cassese, and M. Kielian. 1999. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 73:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, Y. E., C. H. Eng, S. G. Shome, and M. Kielian. 2001. In vivo generation and characterization of a soluble form of the Semliki forest virus fusion protein. J. Virol. 75:8329-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 35.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 37.Park, H. E., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048-1053. [DOI] [PubMed] [Google Scholar]
- 38.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 39.Salminen, A., J. M. Wahlberg, M. Lobigs, P. Liljeström, and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 41.Smit, J. M., W. B. Klimstra, K. D. Ryman, R. Bittman, R. E. Johnston, and J. Wilschut. 2001. PE2 cleavage mutants of Sindbis virus: correlation between viral infectivity and pH-dependent membrane fusion activation of the spike heterodimer. J. Virol. 75:11196-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tweten, R. K., M. W. Parker, and A. E. Johnson. 2001. The cholesterol-dependent cytolysins. Curr. Top. Microbiol. Immunol. 257:15-33. [DOI] [PubMed] [Google Scholar]
- 45.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlberg, J. M., W. A. M. Boere, and H. Garoff. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlberg, J. M., R. Bron, J. Wilschut, and H. Garoff. 1992. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 66:7309-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahlberg, J. M., and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza and vesicular stomatitis viruses. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, X., M. Fugere, R. Day, and M. Kielian. 2003. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 77:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]