Abstract
Lassa virus is endemic to West Africa and causes hemorrhagic fever in humans. To facilitate the functional analysis of this virus, a replicon system was developed based on Lassa virus strain AV. Genomic and antigenomic minigenomes (MG) were constructed consisting of the intergenic region of S RNA and a reporter gene (Renilla luciferase) in antisense orientation, flanked by the 5′ and 3′ untranslated regions of S RNA. MGs were expressed under the control of the T7 promoter. Nucleoprotein (NP), L protein, and Z protein were expressed from plasmids containing the T7 promoter and internal ribosomal entry site. Transfection of cells stably expressing T7 RNA polymerase (BSR T7/5) with MG in the form of DNA or RNA and plasmids for the expression of NP and L protein resulted in high levels of Renilla luciferase expression. The replicon system was optimized with respect to the ratio of the transfected constructs and by modifying the 5′ end of the MG. Maximum activity was observed 24 to 36 h after transfection with a signal-to-noise ratio of 2 to 3 log units. Northern blot analysis provided evidence for replication and transcription of the MG. Z protein downregulated replicon activity close to background levels. Treatment with ribavirin and alpha interferon inhibited replicon activity, suggesting that both act on the level of RNA replication, transcription, or ribonucleoprotein assembly. In conclusion, this study describes the first replicon system for a highly pathogenic arenavirus. It is a tool for investigating the mechanisms of replication and transcription of Lassa virus and may facilitate the testing of antivirals outside a biosafety level 4 laboratory.
Lassa virus is a member of the family of Arenaviridae. This family comprises further human pathogens such as Junin virus, Guanarito virus, Machupo virus, and the prototype arenavirus lymphocytic choriomeningitis virus (LCMV).
The natural hosts of Lassa virus are rodents of the genus Mastomys (43). Transmission of the virus from its reservoir to humans causes Lassa fever, an acute febrile illness associated with bleeding. The disease is endemic to the West African countries of Sierra Leone, Guinea, Liberia, and Nigeria (5, 9, 42, 44), but the virus is probably endemic to larger areas of West Africa (24). Lassa fever is characterized initially by flu-like and gastrointestinal symptoms. Bleeding, organ failure, and shock may occur in the late phase of the disease (36). Death occurs after a mean period of 2 weeks after the onset of illness. The virus can also be transmitted from human to human, giving rise to nosocomial Lassa fever epidemics with fatality rates of up to 65% (17). There is no vaccination available for use in humans. The only drug with a proven therapeutic efficacy in humans with Lassa fever is the broad-spectrum nucleoside analogue ribavirin (35). However, the drug is only effective if given early during the course of disease. Cases of Lassa fever recently imported into Europe show that even state-of-the-art intensive care cannot prevent a fatal outcome (54). Due to the high pathogenicity of Lassa virus and the limitations to the prevention or treatment of infections, the virus is classified as a level 4 pathogen and must be handled under biosafety level 4 (BSL-4) conditions.
Arenaviruses belong to the segmented negative-strand RNA viruses. The genome of Lassa virus, like that of other arenaviruses, consists of two single-stranded RNA segments (33). The small (S) segment is 3.4 kb in length, and the large (L) segment is 7 kb in length. Each segment contains two genes, one in sense orientation and one in antisense orientation, a coding strategy that is called ambisense (4). The S RNA encodes the 75-kDa glycoprotein precursor (GPC) and the 63-kDa nucleoprotein (NP). GPC is posttranslationally cleaved into GP1 and GP2 (8). The L RNA encodes the 11-kDa Z protein, which binds zinc and acts as a matrix protein (47, 55), and the 200-kDa L protein, which is likely to function as the viral polymerase (34, 58). The genes are separated on each RNA segment by an intergenic region (IGR) that predictably folds into a stable secondary structure.
The terminal 19 nucleotides at the 3′ and 5′ ends of the RNA segments are complementary to each other and are highly conserved among all arenaviruses. The termini are essential for replication and transcription (46) and are believed to function as a binding site of the viral polymerase. Replication and transcription of the genome occur in the cytoplasm of an infected cell and both take place within ribonucleoprotein complexes (20). During genome replication, a full-length copy of genomic S and L RNAs is synthesized, yielding the corresponding antigenomic S and L RNAs. Due to the ambisense coding strategy, both genomic and antigenomic RNA serve as templates for the transcription of viral mRNA. The transcripts contain a cap but are not polyadenylated. They terminate within the IGR, suggesting that this element plays a role in transcription termination (37).
Recently, reverse genetics techniques were established for LCMV and Tacaribe virus (31, 32). While these minireplicon systems are still restricted in terms of rescue of genetically modified infectious viruses, they opened the possibility of studying the function of proteins and regulatory elements involved in RNA replication and transcription (30, 46, 48). In particular, the function of Z protein has been studied in detail (12, 13, 27, 47). A sophisticated selection strategy made it possible to generate the first infectious recombinant LCMV with the GPC gene replaced by the glycoprotein gene of vesicular stomatitis virus (49). A reverse genetics system for a highly pathogenic arenavirus like Lassa virus has not yet been published.
In this article, we report the establishment of a minireplicon system for Lassa virus. The system is analogous to the minireplicon systems published for LCMV and Tacaribe virus in gross terms, but it differs in several technical aspects from the other systems. It is also demonstrated that the system is suitable to test antivirals against Lassa virus outside BSL-4 laboratories.
MATERIALS AND METHODS
Virus culture and reverse transcription-PCR.
In the BSL-4 laboratory, Vero cells in several 75-cm2 tissue culture flasks were inoculated with Lassa virus strain AV (24) at a multiplicity of infection of 0.01. After 4 days, the supernatant was cleared by low-speed centrifugation and virus was pelleted from the cleared material by ultracentrifugation with a TST 28.38/17 rotor (Kontron Instruments) at 25,000 rpm at 4°C overnight. The pellets were resuspended in 140 μl of H2O plus 560 μl of buffer AVL (QIAGEN, Hilden, Germany). Virus RNA was purified by using the QIAamp viral RNA kit (QIAGEN) according to the manufacturer's instructions. The purified RNA was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) with virus-specific primers (see below), and the cDNA was amplified with the Expand high-fidelity PCR system (Roche) according to published protocols (24).
Sequencing of Lassa virus genomic ends.
The conserved 5′ and 3′ termini of S RNA of Lassa virus strain AV were sequenced as described previously (37). In brief, purified virus RNA (5 μl) was treated with 5 U of tobacco acid pyrophosphatase (Epicentre) to generate 5′ monophosphorylated termini. These were ligated with the 3′ termini with 10 U of T4 RNA ligase (Biolabs) at 17°C for 16 h. Reverse transcription and PCR were done across the 5′-3′ junction, and the resulting PCR product of about 200 nucleotides was sequenced. The sequence data were sent to GenBank.
Construction of plasmids for protein expression.
Initially, the expression vector pCITE-2/4, containing a T7 RNA polymerase promoter, an internal ribosomal entry site (IRES), a favorable KOZAK sequence with an NcoI restriction site at the translation initiation site, a poly(A) coding sequence, and a T7 RNA polymerase terminator, was constructed by combining the BsaI-SalI fragment of pCITE-2a (Novagen) with the SalI-BsaI fragment of pCITE-4a (Novagen). The control plasmid pCITE-FF-luc was constructed by releasing the firefly luciferase gene from pBI-GL (Clontech) and inserting it into pCITE-2a via the NcoI and XbaI sites.
The Z gene was reverse transcribed and amplified with primers LVLav-Z-N-terminus (GATCTACCCGGGATCCATGGGGAACAGACAAGCCAAAAAT; the AvaI and NcoI sites are underlined, the Z gene start codon is in boldface type) and LVLav-Z-C-terminus (GGAATTCGTCGACTTAGGGTGTGTATGGCGGGGGGTT; the SalI site is underlined) and cloned via the AvaI and SalI sites into pUC18. Several clones were sequenced, and a Z gene exactly matching the Lassa virus AV consensus sequence (GenBank accession no. AY179171) was subcloned via the NcoI and SalI sites into pCITE-2/4, resulting in pCITE-Z.
The NP gene was reverse transcribed and amplified with primers LVSav-NP-N-terminus (ATCGTGGATCCAGGTCTCTCATGAGTGCCTCCAAAGAAGTGAAA; the BamHI and BsaI sites are underlined, the NP gene start codon is in boldface type) and LVSav-NP-C-terminus (TCGATCTAGTCGACTTACAGGACGACTCTTGG; the SalI site is underlined) and cloned via the BamHI and SalI sites into pUC18. Several clones were sequenced, and an NP gene exactly matching the Lassa virus AV consensus sequence (GenBank accession no. AF246121) was released from the plasmid by BsaI and SalI digestion and subcloned into the NcoI and SalI sites of pCITE-2/4, resulting in pCITE-NP.
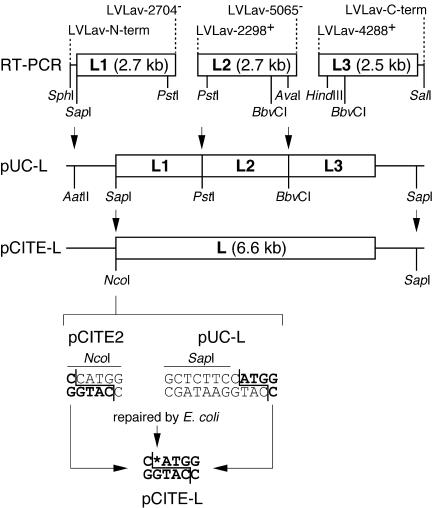
The cloning strategy for the L gene is shown in Fig. 1. The L gene was reverse transcribed and amplified in three overlapping fragments with primers LVLav-N-terminus (GATCTCGCATGCTGGCCAGCTCTTCCATGGAGGAGGACATAGCCTATGT; the SphI and SapI sites are underlined, the L gene start codon is in boldface type) and LVLav2704− (TAATGCTGTGAGTTTGTTGAAGTCATAGTT) for fragment L1, LVLav2298+ (GAGACACCAGATAGACTGACAGATCA) and LVLav5065− (ATTTTCTTTATTTGGCCCCCGGACACATTT) for fragment L2, and LVLav4288+ (TAGATTTGTCGCAGAGTTTAAATCTAGATT) and LVLav-C-terminus (CGGGGTACCGTCGACCTACTCAATGTCCTCAACCTC; the SalI site is underlined) for fragment L3. Fragments L1, L2, and L3 were cloned into pUC18 via the SphI-PstI, PstI-AvaI, and HindIII-SalI sites, respectively. Several clones were sequenced and, if necessary, the sequence was converted to the Lassa virus AV consensus sequence by site-directed mutagenesis with the QuikChange kit (Stratagene). The final clones selected for the assembly of the full-length L gene matched the consensus sequence (GenBank accession no. AY179171) except for one silent G-A exchange at L gene position 1317. Fragments L2 and L3 were assembled by ligation of the AatII-BbvCI fragment of pUC-L2 with the BbvCI-AatII fragment of pUC-L3, resulting in pUC-L2-3. Fragment L1 was assembled with L2-3 by ligation of the AatII-PstI fragment of pUC-L1 with the PstI-AatII fragment of pUC-L2-3, resulting in pUC-L. The complete L gene was subcloned into pCITE-2a by ligation of the SapI-SapI fragment of pUC-L containing the L gene insert with the SapI-NcoI fragment of pCITE-2a. The correct sequence of pCITE-L was ascertained by sequencing.
FIG. 1.
Flowchart of construction of L protein expression plasmid. The L gene was amplified in three fragments by PCR. The primers are indicated above each fragment. Restriction enzyme recognition sites used for cloning are indicated. Sites used for assembly of pUC-L and subcloning of the L gene into pCITE-L are marked by arrows. At the bottom, details of the subcloning of the L gene into the NcoI site of pCITE-L are shown. Note that cloning was possible, although the overhang at the 5′ end of L gene was not fully compatible with an NcoI overhang.
A 3× FLAG sequence was fused with the 3′ ends of the NP and L genes by amplification of the whole pCITE-NP and pCITE-L plasmids, respectively, with the Expand high-fidelity PCR system. The PCR primers bound upstream and downstream of the 3′ end of the gene, were 5′ phosphorylated, and contained heterologous tails coding for a 3× FLAG sequence. Circularization of the PCR fragments by blunt-end ligation of the primer tails resulted in plasmids pCITE-NP_C-flag and pCITE-L_C-flag, expressing NP and L protein, respectively, tagged with three FLAG epitopes at the C terminus. The 5′ ends of the NP and L genes were fused with a 3× FLAG sequence in an analogous way with primers binding upstream and downstream of the 5′ end of the gene. Circularization of the PCR fragments resulted in plasmids pCITE-NP_N-flag and pCITE-L_N-flag, expressing NP and L protein, respectively, tagged with three FLAG epitopes at the N terminus.
Construction of Lassa virus MGs.
Lassa virus minigenomes (MGs) were constructed on the basis of vector pX12ΔT (7) (kindly provided by Ursula Buchholz and Karl-Klaus Conzelmann), which contains a multiple cloning site, the antigenome sequence of the hepatitis delta virus ribozyme (HDR), and a T7 RNA polymerase transcription termination sequence. Based on pX12ΔT, an MG vector with the regulatory elements of antigenomic S RNA of Lassa virus AV, but without the reporter gene, was constructed in several cloning steps. It contained the following elements: T7 RNA polymerase promoter followed by 3 G residues, 3′ untranslated region (UTR) with conserved terminus, cloning site I for fusion of a reporter gene with the start codon of the NP gene (site I, CAACcatgAGGTCTTCCTAAGAAGTGAAGACTGctagCCAG; the NP start codon is in boldface type, BbsI sites are underlined; cleavage with BbsI generates NcoI- and XbaI-compatible overhangs [lowercase letters] for reporter gene insertion), 3′ end of the NP gene (47 nucleotides), complete IGR, 3′ end of the GPC gene (45 nucleotides in antisense orientation), cloning site II for fusion of a reporter gene with the start codon of the GPC gene (site II, AGCCTCTAGAAGAATTCCCATGGTGTG; the GPC start codon [reverse complement] is in boldface type, the XbaI and NcoI sites for reporter gene insertion are underlined), 5′ UTR with conserved terminus, HDR predicted to cleave after the last residue (G) of the conserved 5′ terminus, and T7 terminator. The sequences of the Lassa virus elements in the MG vector exactly corresponded to the consensus sequence (GenBank accession no. AF246121 and sequences of the conserved termini as determined in this study).
The gene of humanized Renilla luciferase (Ren-Luc) was released from plasmid phRL-CMV (Promega) by NcoI and XbaI cleavage and inserted in antisense orientation into the MG vector via cloning site II, resulting in the antigenomic MG plasmid pLAS-agMG. To generate a genomic MG plasmid, the Ren-Luc gene was inserted in sense orientation into cloning site I, and the complete cassette, including the 3′ to 5′ terminal sequences, was amplified by PCR and reinserted in reverse orientation between the T7 promoter and HDR of a pX12ΔT derivative that had been modified to accommodate this insert. This procedure yielded the genomic MG plasmid pLAS-MG. The correct sequence of all functional elements of pLAS-agMG and pLAS-MG was ascertained by sequencing. Plasmids pLAS-agMG and pLAS-MG were further modified by inserting a single C residue between the conserved terminal sequence and the HDR sequence (pLAS-MG-C+ and pLAS-agMG-C+) by site-directed mutagenesis with the QuikChange kit.
For generation of runoff MGs, all functional elements of the MG cassette of pLAS-agMG or pLAS-MG, except for the HDR and T7 terminator sequences, were amplified by PCR with Pwo DNA polymerase (Roche) and a small amount (about 10 pg) of plasmid as a template. The PCR products showed 3′ ends exactly corresponding to the 3′ ends of antigenomic and genomic Lassa virus S RNA, respectively. The unmodified antigenomic runoff MG was amplified from pLAS-agMG with sense primer M13-forward and antisense primer LVS1d+ (CGCACCGGGGATCCTAGGCATTT; the conserved 3′ terminus of antigenomic S RNA is underlined). To generate an antigenomic runoff MG with two G residues following the T7 promoter, the mutagenic primer LVag_T7-GG (TTGTAATACGACTCACTATAGGCGCACAGT; the T7 promoter is underlined with the downstream G residues in boldface type) was used as the sense primer. An antigenomic MG with one G residue was amplified with mutagenic primer LVag_T7-G (same sequence as LVag_T7-GG but with one G residue following the promoter sequence). In an analogous way, the unmodified genomic runoff MG was amplified from pLAS-MG with sense primer M13-forward and antisense primer LVSav3400− (CGCACAGTGGATCCTAGGCTATTGGA; the conserved 3′ terminus of genomic S RNA is underlined). Genomic runoff MGs with two and one G residues following the T7 promoter were generated with sense primers LV_T7-GG (TTGTAATACGACTCACTATAGGCGCACCGG; the T7 promoter is underlined with the downstream G residues in boldface type) and LV_T7-G (same sequence as LV_T7-GG but with one G residue following the promoter sequence), respectively. Amplified DNA fragments were precipitated with ethanol, dissolved in water, and quantified spectrophotometrically.
MG runoff transcripts were synthesized in vitro by using the MEGAscript transcription kit (Ambion) and about 1 μg of runoff MG DNA (i.e., PCR product) as a template. Following DNase I digestion, MG RNA was purified by using the RNeasy kit (QIAGEN) and quantified spectrophotometrically.
Cells and transfections.
BSR T7/5 cells (7) (kindly provided by Ursula Buchholz and Karl-Klaus Conzelmann) stably expressing T7 RNA polymerase and BHK-21 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and antibiotics. Every second passage, 1 mg of G418 per ml of medium was added to the BSR T7/5 cells. One day before transfection, cells were seeded at a density of 5 × 105 cells or 1 × 105 cells per well of a 6- or 24-well plate, respectively. BHK-21 cells were inoculated with modified vaccinia virus Ankara expressing T7 RNA polymerase (MVA-T7) (57) (kindly provided by Gerd Sutter) at a multiplicity of infection of 1 to 5 for 1 h before transfection. DNA and RNA transfections were performed with Lipofectamine 2000 (Invitrogen) (3 μl of DNA or RNA/μg) in DMEM without FCS and antibiotics. The transfection medium was replaced after 4 h with fresh DMEM complemented with FCS and antibiotics. In selected experiments, BSR T7/5 cells were preincubated with different concentrations of natural human alpha interferon (IFN-α) (affinity-purified mixture of different IFN-α species), recombinant human IFN-γ, or recombinant human tumor necrosis factor alpha (Biochrom AG) for 24 h before transfection. Following transfection, the cytokines were added again to the medium. Ribavirin was added to the cells after transfection.
Luciferase assay.
One day after transfection, cells were lysed in 500 or 100 μl of passive lysis buffer (Promega) per well of a 6- or 24-well plate, respectively, and 20 μl of the lysate was assayed for firefly luciferase and Ren-Luc activity by using the Dual-Luciferase Reporter assay system (Promega) as described by the manufacturer.
Antisera.
Lassa virus NP and Z proteins were expressed in bacteria and purified via His tag affinity chromatography as described previously (23). Rabbits were immunized and received boosters three times with the purified proteins with complete and incomplete Freud's adjuvant, respectively.
Immunoblot analysis.
Cells from a well of a six-well plate were harvested and lysed in 100 μl of Triton X-100 lysis buffer 2 days after transfection or 3 days after Lassa virus infection. Nuclei were pelleted by centrifugation, and cytoplasmic lysate was mixed 1:1 with 2× sodium dodecyl sulfate (SDS) lysis buffer (100 mM Tris-HCl [pH 6.8], 20% glycerol, 2% SDS, 0.1% bromophenol blue, and 0.1 M dithiothreitol). Proteins were separated on a 4 to 12% NuPAGE Bis-Tris gradient gel (Invitrogen), transferred to a nitrocellulose membrane (Schleicher & Schuell), and visualized by staining with 0.5% Ponceau red-1% acetic acid for 10 min. Membranes were blocked with 1× Roti-Block (Roth) overnight at room temperature and incubated with rabbit anti-Lassa NP (1:5,000), rabbit anti-Lassa Z (1:1,000), or anti-FLAG M2 monoclonal antibody (1 μg/ml) (Sigma-Aldrich) in Tris-buffered saline-0.2× Roti-Block for 2 h at room temperature. After washing, blots were incubated with horseradish peroxidase-coupled secondary antibodies (1:10,000) (Dianova) for 1 h at room temperature. Protein bands were visualized by chemiluminescence with SuperSignal West Pico substrate (Pierce) and X-ray film (Kodak).
Northern (RNA) blot analysis.
Total RNA of transfected cells was purified by using the RNeasy kit (QIAGEN). RNA (10 μg) was separated on a 1.5% agarose-formaldehyde gel and transferred onto a Hybond N+ membrane (Amersham Pharmacia Biotech). Blots were prehybridized in 50% deionized formamide-0.5% SDS-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5× Denhardt's solution for 1 h at 68°C. Hybridization was done in the same buffer with sense or antisense 32P-labeled riboprobes of the Ren-Luc gene at 68°C for 16 h. Filters were washed several times, and RNA bands were visualized by autoradiography with a PhosphorImager (Typhoon 9210; Amersham Biosciences). Signal intensities were measured with ImageQuant software (Amersham Biosciences).
Nucleotide sequence accession number.
Sequences reported in this article have been sent to GenBank (update to AF24612).
RESULTS
Construction of plasmids for expression of Lassa virus proteins.
Lassa virus strain AV, which had been previously isolated from a patient with fulminant Lassa fever (24), was selected as a basis for the replicon system. Expression plasmids for NP, L protein, and Z protein and genomic and antigenomic MGs containing a reporter gene were constructed in analogy to published replicon systems for arenaviruses and other segmented negative-strand viruses (31, 32, 59). Since Lassa virus replicates in the cytoplasm of a cell, the cytoplasmic T7 RNA polymerase system (16, 19) was chosen for the expression of both virus proteins and the MG. NP and Z genes were cloned as a whole while the large L gene was cloned in three fragments (Fig. 1). The amino acid sequence of the cloned genes exactly corresponded to the previously determined consensus sequence of Lassa virus strain AV. All protein expression plasmids contained an IRES sequence, allowing protein expression in cells that express T7 RNA polymerase from an integrated or transiently transfected gene (16). Thus, protein expression does not depend on recombinant vaccinia virus as a source of T7 RNA polymerase or cytoplasmic capping enzymes (18).
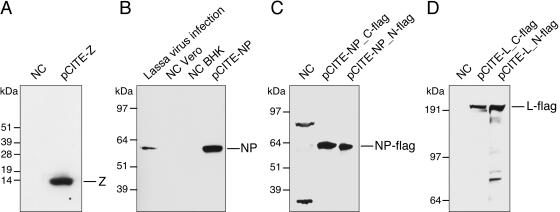
To verify expression of NP, Z protein, and L protein from the plasmids pCITE-NP, pCITE-Z, and pCITE-L (Fig. 2A), respectively, BHK-21 cells were infected with MVA-T7 and transfected with the corresponding plasmid. Z protein and NP were detected by Western blotting with Z protein- and NP-specific antibodies, respectively. Both proteins had the predicted molecular weights, and the electrophoretic mobility of plasmid-expressed NP corresponded to that of NP expressed in Lassa virus-infected cells (Fig. 3A and B). To demonstrate full-length expression of L protein and to substantiate the data obtained for NP, a FLAG sequence was fused to the N and C termini of both proteins. Protein expression from the modified expression plasmids was analyzed by immunoblotting with monoclonal anti-FLAG antibody. N- and C-terminally tagged versions of NP and L protein showed the predicted molecular weights, indicating that both proteins were expressed at their full lengths (Fig. 3C and D).
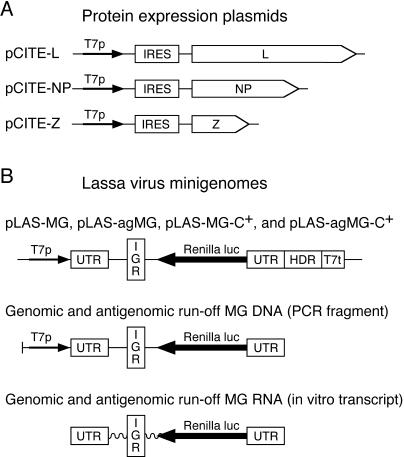
FIG. 2.
Constructs used for establishment of Lassa virus minireplicon. (A) Plasmids for expression of Lassa virus strain AV proteins. (B) Types of MGs based on Lassa virus S RNA. DNA is indicated by a straight line, and RNA is indicated by a wavy line. Functional elements are abbreviated as follows: T7p, T7 promoter; IRES, internal ribosomal entry site; UTR, untranslated region with conserved termini; IGR, intergenic region; HDR, hepatitis delta ribozyme; T7t, T7 transcriptional terminator; luc, luciferase.
FIG. 3.
Immunoblot analysis of plasmid-expressed Lassa virus proteins. MVA-T7-infected BHK-21 cells were transfected with the constructs as indicated above the blots. Alternatively, Vero cells were infected with Lassa virus (only panel B). Negative control cells (NC) were infected with MVA-T7 and mock transfected. The background bands in the NC lane of panel C result from an overloading of the gel. The blots were incubated with the following antibodies: rabbit anti-Lassa Z protein (A), rabbit anti-Lassa NP (B), and anti-FLAG M2 monoclonal antibody (C and D).
Construction of plasmids for MG expression.
Plasmids pLAS-MG and pLAS-agMG were constructed for expression of genomic and antigenomic Lassa virus S RNA analogues, respectively. Plasmid pLAS-MG contained the following functional elements: T7 promoter followed by three G residues, providing optimal context for transcription initiation (26); 5′ UTR, including conserved terminal sequence; IGR; Renilla luciferase (Ren-Luc) reporter gene in antisense orientation; 3′ UTR, including conserved terminal sequence; HDR; and T7 terminator. Plasmid pLAS-agMG contained the same regulatory Lassa virus elements between the T7 promoter and HDR sequence but in the following order: 3′ UTR, IGR, Ren-Luc gene in antisense orientation, and 5′ UTR. A schematic drawing for the plasmid-integrated MGs is shown in Fig. 2B (top).
The HDR was expected to autolytically cleave the MG RNA after the last G residue of the Lassa virus terminal sequence. To verify HDR activity, both MG plasmids were in vitro transcribed with T7 RNA polymerase and the RNA was analyzed on an agarose gel. However, no RNA processing was observed (data not shown). Therefore, a C residue was inserted at the cleavage site. This modification has been reported to increase HDR activity without affecting LCMV replicon activity (46). Testing of the modified MG plasmids (pLAS-MG-C+ and pLAS-agMG-C+) revealed that 30 to 50% of the in vitro-transcribed RNA was autolytically cleaved (data not shown).
Establishment of Lassa virus replicon system.
Two sets of tests were performed to figure out the optimal experimental setting for the replicon system. Three types of MGs were tested in each set. First, plasmids pLAS-MG-C+ and pLAS-agMG-C+ were directly used for MG expression (Fig. 2B, top). Second, DNA fragments lacking the vector backbone were used for MG expression (Fig. 2B, middle). These runoff MGs were generated by PCR with pLAS-agMG or pLAS-MG as a template. They contained all functional elements of the MG cassette, except for the HDR and T7 terminator sequences, and their 3′ ends exactly corresponded to the 3′ ends of antigenomic and genomic Lassa virus S RNA, respectively. The third type of MG was runoff RNA, synthesized by in vitro transcription of the runoff MG DNA with T7 RNA polymerase (Fig. 2B, bottom).
In the first set of tests, T7 RNA polymerase expressed by MVA-T7 was used to drive the synthesis of virus proteins and MG. BHK-21 cells were infected with MVA-T7 and transfected with pCITE-NP, pCITE-L, and MG either in the form of plasmid, runoff DNA, or runoff RNA. In the second set of tests, the same constructs were transfected in BSR T7/5 cells that stably express T7 RNA polymerase. In control experiments, pCITE-NP or pCITE-L was omitted from the transfection mixture. Ren-Luc activity, indicating transcription of the MG, was measured at 24 h posttransfection.
High levels of Ren-Luc activity and low background in the controls was observed if runoff MGs were transfected in BSR T7/5 cells, independent of whether PCR fragments or in vitro transcripts were used (Fig. 4). Therefore, this experimental setting was chosen for further optimization. In all experiments performed with MVA-T7, a high unspecific Ren-Luc reactivity was observed in the controls. There was no evidence of specific replicon activity with MG plasmid or runoff DNA, whereas some specific activity (eightfold increase compared to the control) was observed after transfection of genomic runoff RNA (data not shown). Similarly, high background and low specific activity, if at all, was seen after transfection of the plasmid-based MGs in BSR T7/5 cells (data not shown).
FIG. 4.
Influence of nontemplated nucleotides at the 5′ end of Lassa virus MGs on replicon activity. Experiments were performed with runoff MGs based on genomic (A) and antigenomic (B) Lassa virus S RNA. The upper panels show schematic drawings of the MG variants tested. The variants were generated by mutagenic PCR and named according to the number of nontemplated G residues at the 5′ end. Functional elements are abbreviated as follows: UTR, untranslated region with conserved termini; IGR, intergenic region; Ren, Renilla luciferase. The lower panels show the replicon activity depending on the presence (+) or absence (−) of NP or L protein and the type of MG. BSR T7/5 cells (5 × 105 cells per well of a six-well plate) were transfected with 2 μg of MG, 1.5 μg of pCITE-NP, and/or 0.5 μg of pCITE-L. Empty pCITE-2 was used to keep the overall DNA amount constant if pCITE-NP or pCITE-L was not included in the transfection mixture. MG variants were transfected as either DNA (PCR fragment) or RNA (in vitro transcript). Ren-Luc activity was measured in RLU 24 h posttransfection. Note the logarithmic scale of the diagrams.
The presence of pCITE-Z was not required for expression of Ren-Luc in the runoff MG/BSR T7/5 replicon system, showing that L protein and NP are the minimal trans-acting factors required for RNA synthesis mediated by Lassa virus polymerase. The minimal cis-acting elements are the 5′ UTR, IGR, and 3′ UTR on both genomic and antigenomic RNA.
Modification of the 5′ end of MG.
The 3′ and 5′ terminal sequences are highly conserved among all arenaviruses, and minor modification of these sequences can dramatically influence the transcription and replication activity of the LCMV replicon system (46). While the runoff MGs have authentic 3′ ends, their 5′ ends do not correspond to the termini of Lassa virus RNA because of the 3 G residues that were attached to the 5′ end to enhance T7 promoter activity. In fact, one nontemplated G residue is often present at the 5′ end of genomic or antigenomic RNA of different arenaviruses, including Lassa virus (3, 21, 38, 39, 50, 51).
To convert the 5′ end stepwise into the authentic sequence, one or two of the G residues were removed from the runoff MGs by mutagenesis. According to T7 promoter mapping studies (26), the resulting sequences at the initiation site of the T7 RNA polymerase (G+1GC and G+1CG after removal of one and two G residues, respectively) are still compatible with high promoter activity. Complete removal of all nontemplated G residues would generate an initiation sequence (C+1GG) that is no longer compatible with an active T7 promoter (26).
Genomic and antigenomic runoff MGs with one to three nontemplated G residues at the 5′ end were generated by PCR (Fig. 4, upper panels) and transfected directly as DNA fragments or after in vitro transcription into BSR T7/5 cells, together with the expression plasmids for NP and L protein. Removal of G residues from the 5′ end of the genomic MG enhanced specific Ren-Luc expression by 1 to 2 log units, irrespective of whether MG DNA or RNA was transfected (Fig. 4A). The effect was also observed if the antigenomic MG was transfected as RNA (Fig. 4B), indicating a positive effect of the mutations on the Lassa virus promoter. However, the effect was not observed if antigenomic DNA fragments were used (Fig. 4B). The removal of the G residues probably downregulates the T7 promoter on the antigenomic DNA construct, and this adverse effect obscures the positive effect of the mutations on the Lassa virus promoter. In summary, these results indicate that nontemplated residues at the 5′ ends of genomic and antigenomic RNA decrease the activity of the Lassa virus promoter. The highest Ren-Luc activity was observed with the genomic runoff MG with one nontemplated 5′ G residue (Fig. 4A, construct 1G). This MG was used in all further experiments (called genomic MG).
Optimal ratios of MG, NP, and L protein constructs.
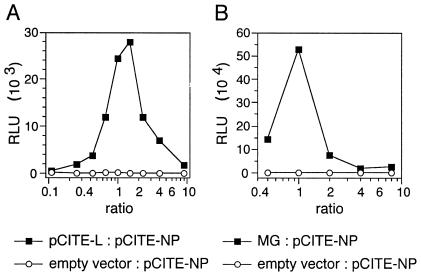
To further maximize replicon activity, the optimal ratios of the components, i.e., genomic MG, pCITE-NP, and pCITE-L, were determined. First, variable ratios of pCITE-NP to pCITE-L were cotransfected with a fixed amount of genomic MG DNA. The maximum activity was observed if both expression plasmids were present in equal amounts, reflecting a molar ratio of pCITE-NP to pCITE-L of about 2:1 (Fig. 5A). Second, changing the ratio of MG DNA to protein expression plasmids revealed maximum Ren-Luc activity at a ratio of MG to each expression plasmid of 1:1 (Fig. 5B). Taking both experiments together, replicon activity is maximal at a ratio of MG DNA to pCITE-NP to pCITE-L of 1:1:1. The same optimal ratios were obtained when MG RNA was used (data not shown).
FIG. 5.
Determination of the optimal ratios between MG, pCITE-NP, and pCITE-L. (A) Titration of ratio between pCITE-NP and pCITE-L. BSR T7/5 cells (5 × 105 cells per well of a six-well plate) were transfected with 2 μg of genomic MG DNA (PCR fragment of construct 1G in Fig. 4A) and a total amount of 2 μg of pCITE-L and pCITE-NP in different ratios (filled squares). In negative-control experiments, pCITE-L was replaced by empty pCITE-2 (open circles). Ren-Luc activity was measured in RLU 24 h posttransfection. (B) Titration of ratio between MG and protein expression plasmids. BSR T7/5 cells (105 cells per well of a 24-well plate) were transfected with MG and a 1:1 mixture of pCITE-NP and pCITE-L (filled squares). The ratio of MG to pCITE-NP (identical to the ratio of MG to pCITE-L) differed in each experiment, but the total amount of transfected DNA was kept constant at 800 ng. In negative-control experiments, MG DNA was replaced by empty pCITE-2 (open circles).
Time kinetics of replicon activity.
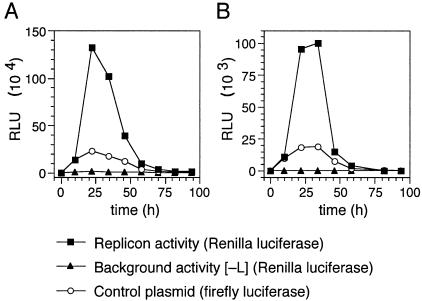
To determine the time kinetics of replicon activity, pCITE-NP, pCITE-L, and genomic MG DNA were transfected in several wells and one well was harvested every 12 h over a time period of 96 h. A sharp maximum of Ren-Luc activity was reached 24 to 36 h after transfection; thereafter, the activity returned to background values (Fig. 6A). Essentially, the same kinetics were observed after transfection of MG RNA (Fig. 6B). The expression of firefly luciferase from the T7 RNA polymerase-driven control plasmid pCITE-FF-luc also followed these kinetics. This suggests that a decrease in T7 RNA polymerase-mediated expression of the replicon components accounts for the rapid decrease of replicon activity after 36 h.
FIG. 6.
Time kinetics of replicon activity. (A) Time kinetics with MG DNA. BSR T7/5 cells (105 cells per well of a 24-well plate) were transfected with 270 ng of genomic MG DNA (PCR fragment of construct 1G in Fig. 4A), 270 ng of pCITE-NP, 270 ng of pCITE-L, and 10 ng of pCITE-FF-luc. Cells were lysed at different times posttransfection, and Renilla (filled squares) and firefly luciferase activities (open circles) were measured in RLU. In negative-control experiments, pCITE-L was replaced by empty pCITE-2 (filled triangles). (B) Time kinetics with MG RNA. Experiments were done as described above except that 270 ng of genomic MG RNA (in vitro transcript of construct 1G in Fig. 4A) was used.
Northern blot analysis of MG transcription and replication.
To provide more evidence for replicon functionality, the synthesis of genomic RNA, Ren-Luc mRNA, and antigenomic RNA was measured by RNA blot analysis during the maximum of Ren-Luc activity. BSR T7/5 cells were transfected with genomic MG DNA, pCITE-NP, and pCITE-L. In control experiments, empty pCITE vector was transfected instead of pCITE-L. Cells were harvested for isolation of total cellular RNA at 0, 6, 24, and 36 h posttransfection. Genomic RNA was detected with a Ren-Luc sense probe, and Ren-Luc mRNA and antigenomic RNA were detected with a Ren-Luc antisense probe.
Figure 7 shows the blots and the quantitative evaluation of the experiments. Synthesis of Ren-Luc mRNA and antigenomic RNA was observed if pCITE-L was present in the transfection mixture but not in the control experiments (Fig. 7A), demonstrating that the Lassa virus polymerase has synthesized this RNA. Ren-Luc mRNA and antigenomic RNA could not be distinguished on the blot, as both RNA species hardly differ in length. The first RNA signal was already seen at 6 h after transfection (Fig. 7A, insert). The time kinetics of Ren-Luc mRNA and antigenomic RNA steady-state levels with a maximum at 24 h faithfully reflected the expression levels of Ren-Luc as measured in the luciferase assay (compare Ren-Luc mRNA and antigenomic RNA levels in Fig. 7C with Ren-Luc levels in Fig. 6A). This is consistent with the assumption that a substantial fraction of the RNA corresponds to Ren-Luc mRNA.
FIG. 7.
Northern blot analysis of Lassa virus replicon. (A) Detection of antigenomic RNA/Ren-Luc mRNA. BSR T7/5 cells (1.2 × 106 cells per 50-mm-diameter dish) were transfected with 2.7 μg of genomic MG DNA (PCR fragment of construct 1G in Fig. 4A), 2.7 μg of pCITE-NP, 2.7 μg of pCITE-L, and 100 ng of pCITE-FF-luc (lanes +). In control experiments, pCITE-L was replaced by empty pCITE-2 (lanes −). Total RNA was isolated at different time posttransfection, and Northern blot hybridization was performed with a 32P-labeled antisense probe of the Ren-Luc gene. The insert in the lower left corner shows the signal area of lanes 0 h and 6 h with enhanced contrast. The methylene blue-stained 28S rRNA is shown below the blot as a semiquantitative marker for gel loading and RNA transfer. (B) Detection of genomic RNA. Experiments were done as described above except that a 32P-labeled sense probe of the Ren-Luc gene was used for Northern blot hybridization. (C) Quantification of the RNA signals in panels A (lanes +, filled squares) and B (lanes +, open squares; lanes −, open circles). Arrows indicate the fraction of genomic RNA resulting from MG replication by L protein.
The synthesis of genomic RNA by Lassa virus polymerase could be measured only indirectly. The reason for this is that the genomic RNA in cells with an active replicon is a mixture of molecules. One fraction is synthesized from the runoff MG by T7 RNA polymerase (input RNA), and the other fraction results from the replication of the antigenomic RNA by Lassa virus polymerase. Control cells lacking L protein solely contain input RNA (Fig. 7B, lanes −). A quantitative evaluation of the blot shows that the levels of genomic RNA in cells with L protein were higher than in control cells at all time points (Fig. 7C). Apparently, the Lassa virus polymerase has synthesized a large fraction of the RNA in cells with L protein. Genomic RNA produced by L protein already appeared 6 h after transfection, which is consistent with the detection of the corresponding template RNA, i.e., antigenomic RNA, at this time point (Fig. 7A, insert). The difference from the control cells (Fig. 7C) was most obvious 36 h after transfection. At this time, a high level of genomic RNA was detected in cells with L protein while input RNA in the control cells was close to the background (Fig. 7B, compare lanes + and − at 36 h). This suggests that the genomic RNA detected at this time point almost exclusively originated from the replication of antigenomic RNA. The disappearance of the input RNA at 36 h after transfection is consistent with the disappearance of firefly luciferase expressed from control plasmid pCITE-FF-luc at 60 h after transfection (Fig. 6). Considering the delayed synthesis of protein relative to RNA, the observations suggest that T7 RNA polymerase-mediated transcription ceases 24 to 36 h after transfection of BSR T7/5 cells while L protein-mediated transcription and replication continue until the concentrations of L protein and NP drop below the critical level. In conclusion, RNA blot analysis and the Ren-Luc assay provide evidence that the Lassa virus replicon system is capable of performing both transcription and replication of the MG.
Influence of Z protein expression on replicon activity.
An inhibitory effect of Z protein was previously demonstrated with the LCMV and Tacaribe virus replicon systems (12, 32). We therefore wondered whether a similar effect is also observed in the Lassa virus replicon system. Various amounts of pCITE-Z were cotransfected with pCITE-NP, pCITE-L, and genomic MG DNA or RNA. As little as 16 ng of pCITE-Z caused a significant reduction of Ren-Luc activity, and higher amounts of pCITE-Z inhibited the replicon activity by up to 3 orders of magnitude. The effect was independent of whether MG DNA or RNA was transfected (Fig. 8 and data not shown). Thus, Lassa virus Z protein has an inhibitory effect on transcription and/or replication of Lassa virus within the replicon context.
FIG. 8.
Inhibitory effect of Lassa virus Z protein on minireplicon activity. BSR T7/5 cells (105 cells per well of a 24-well plate) were transfected with 200 ng of genomic MG DNA (PCR fragment of construct 1G in Fig. 4A), 200 ng of pCITE-NP, 200 ng of pCITE-L, and 10 ng of pCITE-FF-luc. Various quantities of pCITE-Z were cotransfected. The total amount of transfected DNA was kept constant by the addition of pCITE-2. Cells were lysed 24 h posttransfection, and Renilla (filled squares) and firefly luciferase activities (open circles) were measured in RLU. Note the logarithmic scale of the diagram.
Inhibition of the replicon system by ribavirin.
Ribavirin is currently the only available drug with proven therapeutic efficiency in patients with Lassa fever (35). However, the mode of action of the drug against Lassa virus is still not known. In a first attempt to narrow down the steps in the virus life cycle targeted by the drug, we tested whether ribavirin inhibits the Lassa virus replicon system. BSR T7/5 cells were transfected with genomic MG DNA, pCITE-NP, and pCITE-L, and ribavirin was added to the cells 4 h after transfection. Plasmid pCITE-FF-luc expressing firefly luciferase under the control of the T7 promoter was cotransfected as an internal reference. The Ren-Luc values of the replicon were standardized with the firefly luciferase values of the internal reference to correct for variations in transfection efficacy and unspecific effects of the drug. Ribavirin specifically inhibited the activity of the replicon by 2 log units at concentrations of 50 μg/ml (Fig. 9A). Therefore, it is likely that the drug acts at the level of replication or transcription of Lassa virus.
FIG. 9.
Inhibition of replicon activity by ribavirin and IFN-α. (A) Effect of ribavirin. BSR T7/5 cells (105 cells per well of a 24-well plate) were transfected with 270 ng of genomic MG DNA (PCR fragment of construct 1G in Fig. 4A), 270 ng of pCITE-NP, 270 ng of pCITE-L, and 10 ng of pCITE-FF-luc. Different concentrations of ribavirin were added 4 h after transfection. Cells were lysed 24 h posttransfection, and Renilla and firefly luciferase activities were measured. Ren-Luc levels were corrected with the firefly luciferase levels (standardized RLU). The number of RLU in untreated cells was defined as 1. (B) Effect of IFN-α. Experiments were done as described above except that cells were treated with human IFN-α for 24 h before transfection. The same concentrations were added again 4 h after transfection. Note the logarithmic scale of the diagrams.
Inhibition of the replicon system by IFN-α.
Recently, it has been demonstrated that Lassa virus replication can be inhibited by IFN-α in vitro (2). To test whether the Lassa virus replicon system is also susceptible to this cytokine, BSR T7/5 cells were preincubated with human IFN-α for 24 h and then transfected with the replicon components as well as with the internal reference plasmid. The cytokine was again added after transfection. IFN-α specifically inhibited replicon activity by up to 2 log units at concentrations of 1,000 U/ml (Fig. 9B), suggesting that it interferes with Lassa virus replication or transcription. The data obtained with ribavirin and IFN-α also demonstrate that the Lassa virus replicon system may be used for testing of antivirals outside BSL-4 laboratories.
DISCUSSION
This study describes the establishment of a replicon system for Lassa virus. It was based on T7 RNA polymerase-driven expression of MG and virus proteins. The MG contained Ren-Luc as a reporter gene, providing a broad dynamic range for measurement of replicon activity. The use of IRES sequences for protein expression rendered the system independent of vaccinia virus. The replicon was functional with genomic and antigenomic runoff MGs. It was optimized with respect to the ratios of the main components and by modifying the 5′ end of the MG. The signal-to-background ratio was 2 to 3 log units. Maximum activity was observed during a period 24 to 36 h after transfection. Northern blot analysis provided evidence that the system is capable of replicating and transcribing the MG. Z protein strongly inhibited replicon activity. Both ribavirin and IFN-α interfered with the reporter gene expression of the replicon.
Replicon systems for the arenaviruses LCMV and Tacaribe have been previously established (31, 32). The system for Lassa virus as described here has many aspects in common with the other systems, but there are also differences. For example, the Lassa virus replicon was functional without vaccinia virus infection, which was required for the first LCMV system and the Tacaribe virus system. More recently, LCMV RNA analogs were rescued by using an RNA polymerase I-based system, which also circumvents the use of vaccinia virus (48). Vaccinia virus expresses an array of enzymes in the cytoplasm. While some of these enzymes, for example those involved in mRNA capping and polyadenylation (18), are important for protein expression in some replicon systems, others may adversely affect replicon functions or obscure effects in mutational studies (53). This is excluded in the Lassa virus system as well as in the RNA polymerase I-based LCMV system.
Another difference from existing systems concerns the expression of the MG. The Lassa virus replicon was only functional with runoff MG transfected as DNA or RNA. This strategy of MG expression was rather less successful in reverse genetics projects in the past (11). In this study, the runoff MGs were generated and transfected as PCR fragments, a strategy that obviates cloning and can facilitate MG modification by PCR in future mutagenesis studies. Although the plasmid-expressed MG RNA was processed by HDR, at least after insertion of a C residue as recommended by a previous study (46), no specific replicon activity was observed with these constructs. This contrasts with the LCMV replicon, which was more active with the plasmid-expressed MG than the runoff MG (31). Technical variations such as the use of an LCMV-specific ribozyme rather than HDR for cleavage may account for the differences. A possible disadvantage of the runoff method compared to ribozyme cleavage is microheterogeneity at the 3′ end of the MG input RNA. T7 RNA polymerase is known to add a nontemplated nucleotide, preferably A or C, to the 3′ end of about half of the RNA transcripts (28, 41). This effect has been observed with cell-free reactions and is likely to occur also during RNA polymerization within cells. In model assays, this effect can be efficiently suppressed by using chemically modified nucleotides at the 5′ end of the DNA template strand (28). Experiments are under way to elucidate whether this approach can further enhance the activity of the Lassa virus replicon.
A major problem was the high unspecific luciferase activity observed after infection with MVA-T7. Our data do not exclude that MVA-T7-expressed T7 RNA polymerase supports transcription and replication of the MG. Indeed, we found some activity with the genomic runoff RNA. However, with the plasmid- and PCR fragment-based MGs, the Ren-Luc levels in the absence of L protein were so high that specific signals could not be distinguished from unspecific activity. Similar problems with this recombinant virus were reported previously in the establishment of reverse genetic systems for filoviruses (45). The simplest explanation for these observations would be that MVA expresses enzymes that are capable of transcribing RNA from hidden promoters on the transfected MG constructs. Furthermore, besides its DNA-dependent activity, T7 RNA polymerase is also able to transcribe or even replicate RNA molecules (6, 10). In conjunction with MVA-specific enzymes for capping and polyadenylation, this RNA-dependent RNA polymerase activity may have contributed to the high unspecific expression level of Ren-Luc mRNA. Noteworthy, the LCMV and Tacaribe virus replicon systems (31, 32) were established using a different vaccinia virus as a source for T7 RNA polymerase expression, namely vTF7-3, which is based on the Western Reserve strain (19). In contrast to the Western Reserve strain, MVA is highly attenuated and contains several deletions that make up 15% the genome size (40). It is likely that both virus strains differ with respect to many biological parameters that are also relevant to reverse genetics systems. For example, MVA-T7, but not vTF7-3, interferes with Marburg virus glycoprotein processing (53).
We also noticed Ren-Luc activity of 103 to 104 relative light units (RLU) in BSR T7/5 cells containing runoff MG but lacking L protein and NP. This background is still 10- to 100-fold above the cutoff of the luciferase assay as determined with nontransfected cells. Cellular enzymes as well as the above mentioned RNA-dependent activity of T7 RNA polymerase (6, 10) are presumably involved in this unspecific Ren-Luc expression. Importantly, in contrast to the much higher background produced by MVA-T7, this unspecific activity did not obscure the specific activity of the replicon which showed a sharp peak at 24 to 36 h after transfection with 106 to 107 RLU. Thereafter, the activity decreased. Since the expression of the T7 RNA polymerase-driven control plasmid exactly followed these kinetics, it is likely that the decrease in activity is not the result of a regulated process but reflects the degradation of input DNA and RNA that served as the templates for expression of Lassa virus proteins and MG.
The data obtained with the replicon system indicate that the minimal cis-acting elements required for Lassa virus replication and transcription are the 5′ and 3′ UTRs and the IGR. The minimal trans-acting factors are L protein and NP, whereas Z protein has a strong inhibitory effect. This is in agreement with the studies of LCMV and Tacaribe virus (31, 32), supporting the notion that arenaviruses share a common replication and transcription mechanism, at least in gross terms. The number of nontemplated nucleotides at the 5′ end of the Lassa virus MG was inversely correlated with replicon activity. This is consistent with the high degree of conservation of the genomic ends and their importance in replication and transcription of the LCMV replicon (46). Since arenavirus RNA species with nontemplated 5′ nucleotides are generated during infection (3, 21, 38, 39, 50, 51), it may be that the addition of nontemplated nucleotides represents a means of regulation of promoter strength.
The Northern blot data provide evidence for replication and transcription being performed by the replicon. A quantitative analysis of the genomic RNA level 36 h after transfection indicates a 10- to 20-fold excess of genomic RNA in cells with an active replicon compared to control cells. This suggests a single input RNA molecule made by T7 RNA polymerase had been replicated 10 to 20 times by the Lassa virus polymerase. This estimate is larger than that for MG replication by the initial vaccinia virus-based LCMV replicon (less than 2-fold) and in the same order of magnitude as the estimation made for the LCMV replicon based on cellular RNA polymerase I expression (70-fold) (48). Due to its characteristics, the Lassa virus replicon system may facilitate studies with the aim of dissecting the molecular mechanisms of replication and transcription.
In the past, the testing of antiviral compounds against Lassa virus was greatly hampered by its classification as a level 4 pathogen. Most antiviral tests were performed with related, less-pathogenic arenaviruses (1). The replicon system offers an alternative for testing antiviral compounds against Lassa virus outside the BSL-4 laboratory. However, the system is only suitable for the testing of compounds that interact at the level of ribonucleoprotein assembly, RNA replication, or transcription. Since it is independent of vaccinia virus infection, possible confounding drug effects on vaccinia virus replication or gene expression need not be taken into account. Inclusion of a T7 promoter-driven control plasmid expressing firefly luciferase served as an internal reference for potential effects of the drug on T7 RNA polymerase or cell viability. Replicon activity (Ren-Luc) and internal reference (firefly luciferase) display a dynamic range over several orders of magnitude, and both can be measured simultaneously with commercially available reagents.
The suitability of the system for testing antivirals has been exemplified with ribavirin and IFN-α. Inhibition of replicon activity by ribavirin was also observed with the LCMV replicon using the vaccinia virus expression system (52). Our data extend these findings to Lassa virus and indicate that the drug directly acts on the replicon because inhibition of vaccinia virus by ribavirin (29) cannot play a role. In an in vitro infection system, ribavirin inhibited replication of Lassa virus strain AV with a 50% inhibitory concentration of 9 μg/ml and a 90% inhibitory concentration of 14 μg/ml (25). Both values are well in agreement with the inhibition kinetics that were observed with the strain AV-based replicon system, suggesting that the replicon system indeed mirrors the effect of the drug on infectious virus.
Ribavirin is a nucleoside analogue, and it was recently proposed to act by inducing an error catastrophe in some virus infections. When incorporated into the progeny RNA, as shown with poliovirus, it induces mutations that eventually render the virus population inviable (called lethal mutagenesis) (14, 15). Although it cannot be excluded that short cis-acting viral elements are affected, it is more likely that mutations interfere with protein functions. However, in the replicon system, the proteins are synthesized from the plasmid and they cannot undergo mutation. Therefore, the data shown here do not support the hypothesis that ribavirin inhibits Lassa virus via lethal mutagenesis, which is also in agreement with the observation that ribavirin does not increase the mutation frequency during LCMV multiplication in cell culture (52). The mode of action remains speculative. According to the replicon data, the drug probably interferes with steps during replication and transcription, either directly (22) or by inhibition of cellular enzymes important in nucleotide metabolism (56).
The second substance that was tested in the replicon system was IFN-α. This cytokine suppresses multiplication of Lassa virus by about 1 log unit in vitro (2). A similar degree of inhibition was observed in the replicon system, substantiating the previous observations and the rationale for designing antiviral strategies against Lassa fever with the aim of enhancing the IFN response. The fact that natural human IFN-α (a mixture of different IFN-α species) was obviously active on a rodent (hamster) cell line (BSR T7/5 is derived from a BHK-21 subclone) (7) was somewhat surprising and indicates extensive cross-reactivity of human IFN-α. IFN-γ and tumor necrosis factor alpha were also tested, but no effect was observed. It is unclear whether the hamster cell line is refractory to these cytokines or whether the replicon is resistant to the antiviral state induced by these cytokines in these cells.
In conclusion, this study describes the first replicon system for a highly pathogenic arenavirus, namely Lassa virus. It will serve as a tool to gain insights into the mechanisms of replication and transcription of this pathogen and facilitate the testing of antivirals outside a BSL-4 laboratory.
Acknowledgments
We thank Ursula Buchholz and Karl-Klaus Conzelmann for providing vector pX12ΔT and BSR T7/5 cells, Gerd Sutter for providing MVA-T7, Simon Vieth for help with sequencing the S RNA termini of Lassa virus AV, Stephan Becker and Friedemann Weber for many helpful discussions, and Herbert Schmitz for continuous support of the work.
This work was supported by grant E/B41G/1G309/1A403 from the Bundesamt für Wehrtechnik und Beschaffung. The Bernhard-Nocht-Institut is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
REFERENCES
- 1.Andrei, G., and E. De Clercq. 1990. Inhibitory effect of selected antiviral compounds on arenavirus replication in vitro. Antivir. Res. 14:287-299. [DOI] [PubMed] [Google Scholar]
- 2.Asper, M., T. Sternsdorf, M. Hass, C. Drosten, A. Rhode, H. Schmitz, and S. Günther. 2004. Inhibition of different Lassa virus strains by alpha and gamma interferons and comparison with a less pathogenic arenavirus. J. Virol. 78:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auperin, D. D., D. R. Sasso, and J. B. McCormick. 1986. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology 154:155-167. [DOI] [PubMed] [Google Scholar]
- 4.Auperin, D. D., V. Romanowski, M. Galinski, and D. H. Bishop. 1984. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J. Virol. 52:897-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bausch, D. G., A. H. Demby, M. Coulibaly, J. Kanu, A. Goba, A. Bah, N. Conde, H. L. Wurtzel, K. F. Cavallaro, E. Lloyd, F. B. Baldet, S. D. Cisse, D. Fofona, I. K. Savane, R. T. Tolno, B. Mahy, K. D. Wagoner, T. G. Ksiazek, C. J. Peters, and P. E. Rollin. 2001. Lassa fever in Guinea. I. Epidemiology of human disease and clinical observations. Vector Borne Zoonotic Dis. 1:269-281. [DOI] [PubMed] [Google Scholar]
- 6.Biebricher, C. K., and R. Luce. 1996. Template-free generation of RNA species that replicate with bacteriophage T7 RNA polymerase. EMBO J. 15:3458-3465. [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchmeier, M. J., P. J. Southern, B. S. Parekh, M. K. Wooddell, and M. B. Oldstone. 1987. Site-specific antibodies define a cleavage site conserved among arenavirus GP-C glycoproteins. J. Virol. 61:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey, D. E., G. E. Kemp, H. A. White, L. Pinneo, R. F. Addy, A. L. Fom, G. Stroh, J. Casals, and B. E. Henderson. 1972. Lassa fever. Epidemiological aspects of the 1970 epidemic, Jos, Nigeria. Trans. R. Soc. Trop. Med. Hyg. 66:402-408. [DOI] [PubMed] [Google Scholar]
- 10.Cazenave, C., and O. C. Uhlenbeck. 1994. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 91:6972-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conzelmann, K. K. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu. Rev. Genet. 32:123-162. [DOI] [PubMed] [Google Scholar]
- 12.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornu, T. I., and J. C. de la Torre. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J. Virol. 76:6678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 16.Elroy-Stein, O., and B. Moss. 1990. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA 87:6743-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher-Hoch, S. P., O. Tomori, A. Nasidi, G. I. Perez-Oronoz, Y. Fakile, L. Hutwagner, and J. B. McCormick. 1995. Review of cases of nosocomial Lassa fever in Nigeria: the high price of poor medical practice. BMJ 311:857-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst, T. R., and B. Moss. 1989. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells. Importance of the 5′ untranslated leader. J. Mol. Biol. 206:333-348. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller-Pace, F. V., and P. J. Southern. 1989. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J. Virol. 63:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcin, D., and D. Kolakofsky. 1990. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J. Virol. 64:6196-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goswami, B. B., E. Borek, O. K. Sharma, J. Fujitaki, and R. A. Smith. 1979. The broad spectrum antiviral agent ribavirin inhibits capping of mRNA. Biochem. Biophys. Res. Commun. 89:830-836. [DOI] [PubMed] [Google Scholar]
- 23.Günther, S., O. Kühle, D. Rehder, G. N. Odaibo, D. O. Olaleye, P. Emmerich, J. ter Meulen, and H. Schmitz. 2001. Antibodies to Lassa virus Z protein and nucleoprotein co-occur in human sera from Lassa fever endemic regions. Med. Microbiol. Immunol. (Berlin) 189:225-229. [DOI] [PubMed] [Google Scholar]
- 24.Günther, S., P. Emmerich, T. Laue, O. Kühle, M. Asper, A. Jung, T. Grewing, J. ter Meulen, and H. Schmitz. 2000. Imported Lassa fever in Germany: molecular characterization of a new Lassa virus strain. Emerg. Infect. Dis. 6:466-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Günther, S., M. Asper, C. Röser, L. K. S. Luna, C. Drosten, B. Becker-Ziaja, P. Borowski, H.-M. Chen, and R. S. Hosmane. 2004. Application of real-time PCR for testing antiviral compounds against Lassa virus, SARS coronavirus, and Ebola virus in vitro. Antivir. Res. 63:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imburgio, D., M. Rong, K. Ma, and W. T. McAllister. 2000. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry 39:10419-10430. [DOI] [PubMed] [Google Scholar]
- 27.Jacamo, R., N. Lopez, M. Wilda, and M. T. Franze-Fernandez. 2003. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J. Virol. 77:10383-10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, C., M. Zheng, and S. Rudisser. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5:1268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz, E., E. Margalith, and B. Winer. 1976. Inhibition of vaccinia virus growth by the nucleoside analogue 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole, ribavirin). J. Gen. Virol. 32:327-330. [DOI] [PubMed] [Google Scholar]
- 30.Lee, K. J., M. Perez, D. D. Pinschewer, and J. C. de la Torre. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J. Virol. 76:6393-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, K. J., I. S. Novella, M. N. Teng, M. B. Oldstone, and J. C. de La Torre. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J. Virol. 74:3470-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez, N., R. Jacamo, and M. T. Franze-Fernandez. 2001. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J. Virol. 75:12241-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukashevich, I. S., T. A. Stelmakh, V. P. Golubev, E. P. Stchesljenok, and N. N. Lemeshko. 1984. Ribonucleic acids of Machupo and Lassa viruses. Arch. Virol. 79:189-203. [DOI] [PubMed] [Google Scholar]
- 34.Lukashevich, I. S., M. Djavani, K. Shapiro, A. Sanchez, E. Ravkov, S. T. Nichol, and M. S. Salvato. 1997. The Lassa fever virus L gene: nucleotide sequence, comparison, and precipitation of a predicted 250 kDa protein with monospecific antiserum. J. Gen. Virol. 78:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCormick, J. B., I. J. King, P. A. Webb, C. L. Scribner, R. B. Craven, K. M. Johnson, L. H. Elliott, and R. Belmont-Williams. 1986. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 314:20-26. [DOI] [PubMed] [Google Scholar]
- 36.McCormick, J. B., I. J. King, P. A. Webb, K. M. Johnson, R. O'Sullivan, E. S. Smith, S. Trippel, and T. C. Tong. 1987. A case-control study of the clinical diagnosis and course of Lassa fever. J. Infect. Dis. 155:445-455. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, B. J., and P. J. Southern. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J. Virol. 67:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer, B. J., and P. J. Southern. 1994. Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J. Virol. 68:7659-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer, B. J., and P. J. Southern. 1997. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol. 71:6757-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72:1031-1038. [DOI] [PubMed] [Google Scholar]
- 41.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monath, T. P., M. Maher, J. Casals, R. E. Kissling, and A. Cacciapuoti. 1974. Lassa fever in the Eastern Province of Sierra Leone, 1970-1972. II. Clinical observations and virological studies on selected hospital cases. Am. J. Trop. Med. Hyg. 23:1140-1149. [DOI] [PubMed] [Google Scholar]
- 43.Monath, T. P., V. F. Newhouse, G. E. Kemp, H. W. Setzer, and A. Cacciapuoti. 1974. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science 185:263-265. [DOI] [PubMed] [Google Scholar]
- 44.Monath, T. P., P. E. Mertens, R. Patton, C. R. Moser, J. J. Baum, L. Pinneo, G. W. Gary, and R. E. Kissling. 1973. A hospital epidemic of Lassa fever in Zorzor, Liberia, March-April 1972. Am. J. Trop. Med. Hyg. 22:773-779. [DOI] [PubMed] [Google Scholar]
- 45.Mühlberger, E., B. Lotfering, H. D. Klenk, and S. Becker. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72:8756-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez, M., and J. C. de la Torre. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 77:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinschewer, D. D., M. Perez, and J. C. de la Torre. 2003. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 77:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinschewer, D. D., M. Perez, A. B. Sanchez, and J. C. de la Torre. 2003. Recombinant lymphocytic choriomeningitis virus expressing vesicular stomatitis virus glycoprotein. Proc. Natl. Acad. Sci. USA 100:7895-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polyak, S. J., S. Zheng, and D. G. Harnish. 1995. 5′ termini of Pichinde arenavirus S RNAs and mRNAs contain nontemplated nucleotides. J. Virol. 69:3211-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raju, R., L. Raju, D. Hacker, D. Garcin, R. Compans, and D. Kolakofsky. 1990. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology 174:53-59. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz-Jarabo, C. M., C. Ly, E. Domingo, and J. C. de la Torre. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37-47. [DOI] [PubMed] [Google Scholar]
- 53.Sänger, C., E. Mühlberger, H. D. Klenk, and S. Becker. 2001. Adverse effects of MVA-T7 on the transport of Marburg virus glycoprotein. J. Virol. Methods 91:29-35. [DOI] [PubMed] [Google Scholar]
- 54.Schmitz, H., B. Köhler, T. Laue, C. Drosten, P. J. Veldkamp, S. Günther, P. Emmerich, H. P. Geisen, K. Fleischer, M. F. Beersma, and A. Hoerauf. 2002. Monitoring of clinical and laboratory data in two cases of imported Lassa fever. Microbes Infect. 4:43-50. [DOI] [PubMed] [Google Scholar]
- 55.Strecker, T., R. Eichler, J. ter Meulen, W. Weissenhorn, H. D. Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Streeter, D. G., J. T. Witkowski, G. P. Khare, R. W. Sidwell, R. J. Bauer, R. K. Robins, and L. N. Simon. 1973. Mechanism of action of 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. USA 70:1174-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 58.Vieth, S., A. E. Torda, M. Asper, H. Schmitz, and S. Günther. 2004. Sequence analysis of L RNA of Lassa virus. Virology 318:153-168. [DOI] [PubMed] [Google Scholar]
- 59.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]