Abstract
Episomal reporter plasmids containing the Epstein-Barr virus (EBV) oriP sequence stably transfected into Akata Burkitt's lymphoma cells were used to analyze EBV lytic cycle gene regulation. First, we found that the Zp promoter of EBV, but not the Rp promoter, can be activated in the absence of protein synthesis in these oriP plasmids, casting doubt on the immediate early status of Rp. An additional level of regulation of Zp was implied by analysis of a mutation of the ZV element. Second, our analysis of late lytic cycle promoters revealed that the correct relative timing, dependence on ori lyt in cis, and sensitivity to inhibitors of DNA replication were reconstituted on the oriP plasmids. Late promoter luciferase activity from oriP plasmids also incorporating replication-competent ori lyt was phosphonoacetic acid sensitive, a hallmark of EBV late genes. A minimal ori lyt, which only replicates weakly, was sufficient to confer late timing of expression specifically on late promoters. Finally, deletion analysis of EBV late promoter sequences upstream of the transcription start site confirmed that sequences between −49 and +30 are sufficient for late gene expression, which is dependent on ori lyt in cis. However, the TATT version of the TATA box found in many late genes was not essential for late expression.
We recently described a system for studying Epstein-Barr virus (EBV) lytic gene regulation with plasmids stably transfected into the Akata Burkitt's lymphoma cell line (3). The plasmids contain the promoter being studied linked to the luciferase reporter gene and also have the EBV oriP plasmid origin of replication and a selectable marker (hygromycin resistance). Cell lines stably maintaining the plasmids, typically at about 10 copies per cell, can readily be isolated. Akata cells containing EBV can be induced into the lytic cycle by treatment with anti-immunoglobulin (anti-Ig), which cross-links the surface B-cell receptor, providing a rapid and physiologically relevant method for studying the EBV lytic cycle in cell culture (28). This system accurately reconstituted the regulation of the Zp immediate early promoter of EBV and was used to study its regulation in detail (3, 5, 14). The stably transfected reporter plasmid has an average copy number similar to that of the endogenous EBV genome present in the same cell, the plasmids are assembled into chromatin like EBV, and the reactivation of endogenous virus provides viral lytic gene products that may act in trans at their normal concentrations. These features make the system a good model for studying viral gene regulation, avoiding artifacts associated with transient transfection and incorrect expression levels of regulatory factors. In this study, we extended the use of the system to investigate the Rp promoter and EBV delayed early and late gene promoters.
Zp and Rp have been considered to be the immediate early promoters of EBV, but controversy has surrounded the status of Rp, partly because of technical difficulties in mapping the 5′ end of the RNA from this promoter. The first exon is very short, and its sequence has a high AT content, hindering RNase protection assay mapping. In earlier work, where immediate early status was deduced for Zp and Rp in Akata cell induction by anti-Ig (21), RNA mapping of Rp was inferred from protection of sequences within BRLF1 rather than direct mapping of the 5′ end of the RNA. Reconstitution of the induction of Zp in a clone of Akata cells lacking EBV (AK31 cells) was a more convincing demonstration (3, 14) that signal transduction from the BCR is able to activate Zp without the involvement of other viral genes (a role for EBNA1 could not be excluded since its presence is required for oriP function). Here we used a similar approach to study Rp regulation directly.
We also extended the system to study delayed early and late genes of EBV. The classification of EBV lytic cycle genes (7) was made on the basis of their sensitivity to inhibitors of DNA replication rather than timing of expression. Herpesvirus true late (γ2) genes depend on viral lytic replication for their expression but are also expressed relatively late in the virus lytic cycle. A core set of early and late genes is conserved in different herpesviruses (reviewed in references 23 and 24). These can be recognized as homologues by sequence or position in the viral genome. The conservation of the mechanism of lytic replication and conservation of many of the late gene coding sequences suggests that the basic mechanism of late gene regulation would also be conserved among herpesviruses. Late gene regulation has been studied in the most detail in herpes simplex virus (HSV) by testing deletion and point mutant forms of late promoters, particularly those for the US11, UL38, UL49.5, and gC genes. These studies (10, 11, 16, 17, 25, 30) showed that the TATA box, sequences around the transcription start site (Inr), and sequences up to about 30 nucleotides downstream of the transcription start site (DAS) are the main determinants of late gene expression in HSV. Some aspects of HSV late gene expression can be reconstituted on small plasmids transiently transfected into cells in which viral replication occurs in trans. Such late promoter activity was dependent on the presence of the viral lytic origin sequence within the plasmid containing the late promoter (15).
Studying EBV late gene expression, Serio et al. (27) showed that lytic-cycle-dependent activation of several EBV late gene promoters could be observed on transiently transfected reporter plasmids. This was dependent on the EBV late lytic replicative cycle occurring in the same cell but, surprisingly, did not require the ori lyt sequence of EBV to be present in cis on the reporter plasmid. These data indicated that a trans-acting factor produced after viral DNA replication is required for EBV late gene expression rather than an ori lyt-dependent function in the plasmid containing the late promoter. Conservation of certain TATA box sequences was noted by these authors in the sequences of EBV late promoters, particularly a T in the fourth position (TATT), and this was shown to be one of the determinants of late promoter activity in EBV (26). Mutation of this T residue in the late promoter for BcLF1 supported its role in late gene regulation (26).
One complication of studying the role of ori lyt in late gene regulation is that it may cause both replication of the template and induction of transcription. It can be difficult to distinguish which of these effects is responsible for activation of late gene expression. In this study, we successfully reconstituted the regulation of delayed early and late genes on oriP plasmids, including the sensitivity of late gene expression to phosphonoacetic acid (PAA). We also tested a smaller version of ori lyt (core ori lyt) that does not cause any measurable increase in plasmid DNA in our assays to study transcription effects on late promoters distinct from a simple increase in template copy number.
In this investigation of EBV delayed early and late promoter transcription on reporter plasmids, we also reconstituted the relative timing of expression of the promoters in the lytic cycle. We observed a specific dependence of late promoters on ori lyt in cis. A previously reported sequence (8) upstream of the TATA box that is conserved in some late promoters was demonstrated to affect the extent of promoter activity but not the temporal lateness or dependence on ori lyt. We also tested the need for the fourth-position T residue in the TATA box sequence for late promoter function in this system.
MATERIALS AND METHODS
Cell culture, transfection of EBV-positive Akata cells, luciferase assays, Southern blot assays, and Western blot assays were done as described previously (3, 5, 14). To maintain a high level of EBV-positive cells, the Akata cells (28) were single cell cloned at various times to give AK6, AK2000, and AK2003 cells, which are all functionally equivalent. AK31 is an EBV-negative clone of Akata cells described previously (3, 5, 14). As in previous luciferase transfection assays (3), for each plasmid, multiple cell lines were assayed in duplicate. The data were corrected for plasmid copy number, and the mean and standard deviations are shown in the graphs.
Plasmids.
All of the EBV genome coordinates in this paper are given as wild-type EBV (EBVwt) genome numbers (6). The EBNA1 expression plasmid puro/oriP-EBNA1 was constructed by cloning the EBNA1 gene under the control of the cytomegalovirus (CMV) immediate early promoter as a SalI/XhoI fragment into the XhoI site of puro/oriP. The puro/oriP plasmid had been assembled by first subcloning the B95-8 EBV oriP with EBV EcoRI and PstI sites into pBluescript KS (Stratagene). The oriP was then excised from this as an EcoRI/BamHI fragment and cloned between the BglII and EcoRI sites of pRG201 (pRG201 contains a puromycin resistance cassette cloned into the polylinker of a pUC vector). The EBNA1 SalI/XhoI fragment was derived by initially cloning the EBNA1 coding sequence as a HindIII fragment from p294 (14) into the HindIII site of pcDNA6/TR (Invitrogen), choosing the orientation that allowed EBNA1 expression from the CMV promoter. The CMV-EBNA1 cassette was then cut out with SalI and XhoI.
The pEH-luc vector was made previously (3) by inserting a luciferase reporter gene into a plasmid similar to pHEBo (32), and the cloning of pEH-luc:ZpXwt has been described previously (14). The Rp promoter was PCR amplified from −1250 to +40 or −446 to +40, tagging the upstream ends with BamHI sites and the +40 end with an XhoI site. These BamHI/XhoI fragments were exchanged with the similar Zp fragment in pEH-luc:ZpXwt to generate pEH-luc:Rp(−1250) and pEH-luc:Rp(−446). The Rp PCR primers were −1250 ATAAGGATCCTGAGGCCGTCAGGGAAA, −446 GATGGATCCTTGTGTGAGGTCTCACCT, and +40 GTACTCGAGTGAGGTGTTGTGTCCTGT.
p294:ZRp-254 (also known as Zp-3241) was constructed by excising the EBV content of pUCIE (14) with XbaI and EcoRI sites in the polylinker of pUC, filling the ends with Klenow DNA polymerase, and then cloning it into the SalI site of p294 (3), which had also been blunted with Klenow. p294:Zp-1276 (also known as Zp-4263) was constructed by first cloning the BamHI R fragment of B95-8 EBV into the BamHI site of p294, choosing a clone with an orientation similar to that of p294:ZRp-254. This plasmid was then cut with SalI, and the larger fragment was ligated to the SalI fragment of pUCIE containing the BZLF1 gene.
p294:ZRp-7101 (also known as RpSal) was made via the intermediate plasmid p294:SalI. p294:SalI was constructed by cloning the XbaI/SalI fragment from pUCIE (EBVwt positions 89825 to 93008) between the NheI and SalI sites of p294. The SalI fragment from M-ABA cosmid CM301-99 (22) corresponding to EBVwt positions 93638 to 100994 was then ligated into the SalI site of p294:SalI to create p294:ZRp-7101.
The core ori lyt fragment (13) was derived from B95-8 EBV BamHI fragment H digested with SacI (EBV position 40098) and NsiI (EBV position 41293) and then cloned into pBK-CMV (Stratagene), which was cut with SacI/PstI. The resulting plasmid was then cut with SacI, treated with T4 DNA polymerase to give blunt ends, and ligated with a BamHI linker (CGCGGATCCGCG). This vector was subsequently cut with BamHI/SalI, and the ori lyt fragment was isolated. pEH-luc was digested with BamHI/SalI and ligated with ori lyt to obtain pEHLyt-luc. Big ori lyt was the BamHI H fragment of B95-8 EBV and was cloned into the BamHI site of pEH-luc. The late promoter regions of BcLF1, BdRF1, and BFRF3, as well as the Cp and BMRF1 promoters (from about −430 to +30 from the transcription start), were amplified by PCR from appropriate cloned fragments of B95-8 EBV. The PCR primers used (and EBVwt sequences cloned) were BcLF1 (cloned positions 125823 to 125362; ACCCAAGCTTGTTTGGCCATGACAGCA and ACCCAAGCTTTCGGCTTCTACTCGGCG), BdRF1 (cloned positions 135931 to 136391; ACCCAAGCTTGTGGGCTCTGAAGTGCC and ACCCAAGCTTCCTCCCTCGAGAACCCA), BFRF3 (cloned positions 48656 to 49116; ACCCAAGCTTGAGCTTGGCGTTGCTGA and ACCCAAGCTTCCCGGATTACCCTCCCT), BMRF1 (cloned positions 67152 to 67612; ACCCAAGCTTATGATCACAAGCAGCAG and ACCCAAGCTTAATAACTACATAAGTAGGG), and Cp (cloned positions 10905 to 11368; ACCCAAGCTTCGGTGTCCTTGTCTCTA and ACCCAAGCTTCCTAGGCCAGCCAGAGA).
The EBVwt coordinates taken to be the transcription starts were as follows: BcLF1, position 125392; BdRF1, position 136361; BFRF3, position 49086; BMRF1, position 67582; Cp, position 11335. Amplified products were either cleaved directly at their terminal HindIII sites or subcloned into TA cloning vector pCR 2.1 (Invitrogen) and then excised with HindIII. Both pEH-luc vectors were cut with HindIII and ligated to the promoter fragments to give the final constructs.
pEHLyt-109(m)Bc/Bd-luc, pEHLyt-49Bc/Bd-luc, and TATA mutants.
5′ primers specific for −109 and −49 and 3′ primers as used for construction of pEHLyt-BcLF1-luc and pEHLyt-BdRF1-luc (see above), both incorporating flanking HindIII sites, were used for PCR from the respective pEHLyt-luc vectors. The amplified regions and pEHLyt-BdRF1-luc were cut with HindIII and ligated. The correct orientation in pEHLyt-109Bc/Bd-luc and pEHLyt-49Bc/Bd-luc was identified by PCR. The −109 promoter fragments were then subcloned into pBluescript as HindIII fragments. −109mut, which contains a change of the conserved region in BcLF1 and BdRF1 −81 to −73 (AGACTCTGA to AGAATTCGA) and an introduced EcoRI site, was generated by site-directed mutagenesis in pBS-109BcLF1 and pBS-109BdRF1. The −109mut fragments were then cloned back into pEHLyt-BcLF1-luc and pEHLyt-BdRF1-luc, respectively, by HindIII digestion to generate pEHLyt-109mBcLF1-luc and pEHLyt-109mBdRF1-luc.
The TATA box mutants pEHLyt-BcLF1-luc and pEHLyt-BdRF1-luc were made by site-directed mutagenesis with the QuikChange kit (Stratagene). In each case, a single nucleotide was altered to convert TATT to TATA. The promoter regions of all plasmids were sequenced to ensure that no unwanted mutations had been introduced in the PCR and cloning procedures.
5′ RACE analysis.
The 5′ RACE (rapid amplification of cDNA ends) kit (Life Technologies) was used to synthesize cDNA with the primer Z5234. After removal of the RNA with RNase and purification of the cDNA, a polymeric dC tail was added with terminal deoxynucleotidyl transferase. Amplification of the RACE products was done in two rounds with nested PCR primers. The first-round PCR was done with Z7675 and the 5′ RACE abridged-anchor primer (Life Technologies), followed by a second amplification with the nested Z5235 primer and the abridged universal amplification primer (Life Technologies). The primers used were Z5234 (TAACACGTAGCGCATCACTA), Z7675 (CGGAATTCGGCATTCTCAGCCCGTCTT), and Z5235 (CGGAATTCAGACCAGAGAGCCCAGCTGC). Amplified products were gel purified, cloned into pCR2.1 with the TA cloning kit (Invitrogen), and sequenced.
RESULTS
Rp functions as a delayed early promoter but does not respond directly to BCR signaling in an episomal luciferase plasmid.
The Zp immediate early promoter was previously mapped with RNA from Akata cells treated with anti-Ig and protein synthesis inhibitors for 4 h (21). However, for Rp, the short, AT-rich first exon makes it difficult to do RNase protection assay analysis of the 5′ end, so this has not been mapped directly (Fig. 1A and B). RNAs containing BRLF1 were first characterized (20) by sequencing cDNA clones of RNA from Raji cells that had been treated with tetradecanoyl phorbol acetate for 48 h, long after addition of the inducer. In herpesvirus saimiri orf50 (the equivalent of the gene for BRLF1), there is a change in promoter usage during infection (12, 31) so it was not certain that the previously established Rp would necessarily be the immediate early promoter.
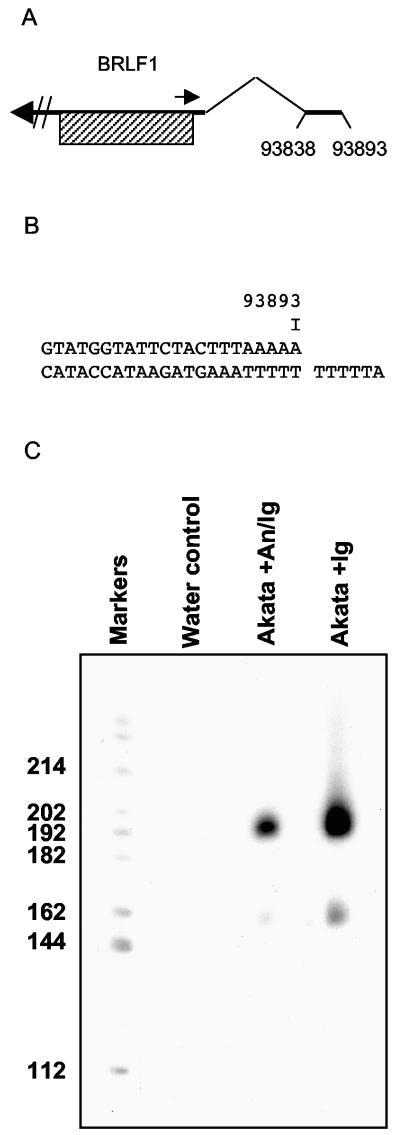
FIG. 1.
(A) Structure of the 2.8-kb RNA encoding BRLF1; coordinates of the first exon are numbered. The small rightward arrow shows the position of the 5′ RACE primers. The BRLF1 open reading frame is shaded. (B) EBV genome sequence up to position 93893, the start of the 2.8-kb mRNA. The additional 5′ sequence (TTTTTA in two clones and TTTTTTA in one clone sequenced) present in the 5′ RACE cDNAs is shown. (C) Polyacrylamide gel electrophoresis of the 5′ RACE products, showing the 192- and 162-nucleotide products. Akata cells were treated with anti-Ig with (An/Ig) or without (Ig) anisomycin to inhibit protein synthesis. The values on the left are nucleotides.
To check that Rp is the promoter for BRLF1 mRNA detected in Akata cells under immediate early conditions, a 5′ RACE analysis was performed on RNA from Akata cells treated with anti-Ig and anisomycin (to inhibit protein synthesis). Two species of RNA were amplified (Fig. 1C). These were sequenced and found to correspond to the structures described previously (20). The major structure corresponding to the 192-nucleotide RACE product starts at position 93893, and the minor product starts at about position 93860 (Fig. 1B and C). The minor product corresponds to the Z8 cDNA (20), subsequently renamed RAZ (9), and its expression was almost completely prevented by inhibition of protein synthesis. The results thus support the established Rp as the promoter for BRLF1 RNA present under immediate early conditions.
The 5′ RACE products from Rp starting at position 93893 all had an additional ATTTTT sequence at the 5 ′ end (Fig. 1B) not encoded in the EBV genome (this part of the Akata EBV DNA was sequenced to confirm that it does not contain the additional ATTTTT sequence). This ATTTTT sequence is most likely an artifact of the RACE PCR procedure in the AT-rich sequence, but the possibility of a novel 5′-end structure on the mRNA should be noted.
Having established that Rp is the start detected for BRLF1 mRNA under immediate early conditions, we tested whether activation of Rp in response to BCR cross-linking by anti-Ig could be reconstituted in EBV-negative Akata cells, as we have reported previously for Zp (3). An oriP plasmid containing Rp linked to the luciferase reporter in pEH-luc was transfected into EBV-positive Akata cells or EBV-negative Akata cells that already contained an EBNA1 expression plasmid, the EBNA1 allowing episomal maintenance of the Rp reporter plasmid (Fig. 2A). Treatment with anti-Ig resulted in a strong activation of the Rp reporter in the EBV-positive cells but no activation in the EBV-negative Akata cells (Fig. 2B). In the same cell background, the Zp reporter was activated by anti-Ig in both EBV-positive and EBV-negative cells (Fig. 2B). It was also noticeable that the initial induction of Rp-luciferase was delayed relative to that of Zp luciferase (Fig. 2B). It therefore seems that the Rp promoter plasmids tested, which are highly active under delayed early conditions, are not functional in the absence of EBV; i.e., Rp does not behave as an immediate early promoter in this assay.
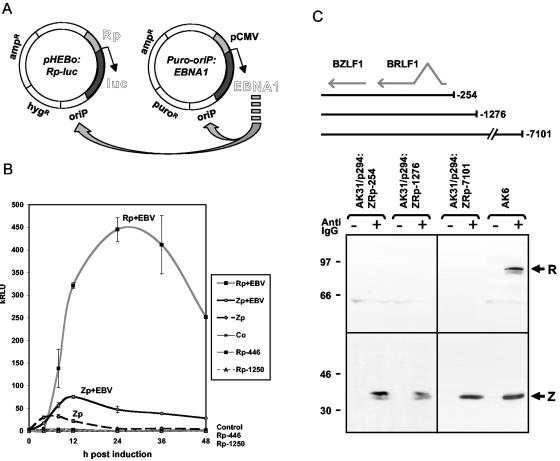
FIG. 2.
(A) Schematic diagram of the pHEBo:Rp-Luc reporter plasmid and the EBNA1 expression plasmid puro/oriP-EBNA1. (B) Luciferase reporter assays following induction with anti-Ig. Rp+EBV and Zp+EBV were in AK2003 cells, and Rp and Zp were in EBV-negative AK31 cells. Co is the pHEBo-luc vector control in AK2003 cells. (C) Western blotting assay to detect induction of BZLF1 (BZ-1 antibody) or BRLF1 (8C12 antibody, 1:100) in EBV-negative AK31 cell lines containing p294:ZRp-254, p294:Zp-1276, or p294:ZRp-7101 compared to the positive control AK6 cells.
Although it is a less sensitive method than the luciferase assay, we also tested larger fragments of the EBV genome in oriP plasmids in EBV-negative AK31 cells for anti-Ig induction of BRLF1 by Western blotting. These EBV genomic fragments included the BZLF1 gene, BRLF1, and various amounts of EBV upstream of Rp, as far as 7.1 kb upstream. Although BZLF1 was induced by anti-Ig from all of these plasmids, no BRLF1 could be detected (Fig. 2C). To ensure that this was not due to a trivial defect in the BRLF1 open reading frame in the plasmids, the BRLF1 open reading frame was PCR amplified from p294:ZRp-254 and p294:Zp-1276, cloned, and translated in vitro. BRLF1 was efficiently translated in vitro from these clones and the in vitro translated protein was readily detected by Western blotting (data not shown). It was surprising that the BZLF1 protein induced by anti-Ig did not activate Rp to express BRLF1 in these assays, raising the possibility that an additional level of control on Rp is present in the whole EBV genome but lacking in these plasmids.
Mutation of the ZV element in Zp has little effect on production of BZLF1 protein.
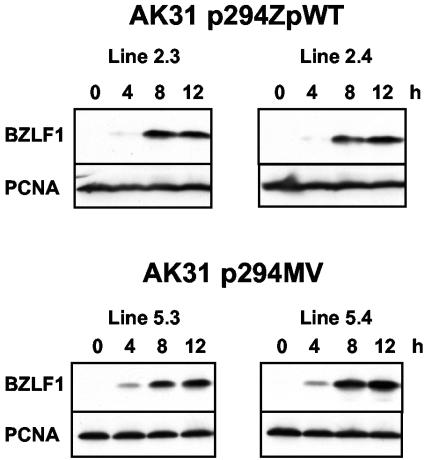
Previous site-directed mutagenesis of the Zp promoter has identified several sequences that bind transcription factors that are important regulators of Zp and viral reactivation. These include the ZI, ZII, and ZIII elements, which have been tested in both luciferase and chloramphenicol acetyltransferase reporter systems and with the complete BZLF1 gene (3, 14). Mutation of an additional element known as ZV was found to cause a large increase in the response of the Zp promoter to either tetradecanoyl phorbol acetate or BCR signal transduction (3, 18, 19). This ZV mutation had the largest effect of any Zp mutation tested in luciferase reporter assays, giving an about 8- to 10-fold increase in Zp activity after anti-Ig induction but not making the promoter constitutively active. We therefore tested whether the same mutation of ZV would increase the production of BZLF1 protein when the mutation was introduced into the complete BZLF1 gene, expecting much higher expression of BZLF1 protein than from the wild-type gene. In fact there was little difference in BZLF1 expression between the wild-type and ZV mutant promoters in response to anti-Ig induction of BCR signaling (Fig. 3). The examples shown in Fig. 3 have the largest MV increase in BZLF1 expression observed; other, similar experiments showed no difference between the wild type and MV (data not shown). There thus appears to be an additional level of control of BZLF1 expression that limits the production of BZLF1 protein even when ZV is mutated. This indicates that the signal transduction already described through the ZI, ZII, and ZIII elements is the more quantitatively important pathway by which reactivation occurs in Akata cells.
FIG. 3.
Western blot assays of BZLF1 and PCNA at the indicated times after induction with anti-Ig in AK31 cells containing Zp-82LF1 plasmids. The upper panels are wild-type Zp, and the lower panels are the MV mutant form of Zp. Protein samples were electrophoresed on 12.5% SDS gels, blotted to nitrocellulose, and probed with either BZ-1 (1:500 dilution) or PCNA antibody PC10 (1:1,000), followed by enhanced-chemiluminescence detection (Amersham).
Regulation of delayed early and late genes can also be reconstituted on oriP plasmids in the presence of the whole EBV genome.
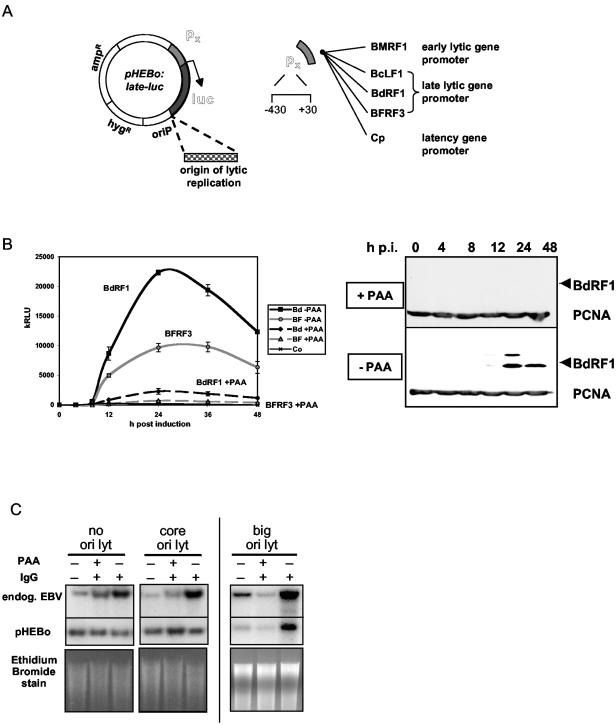
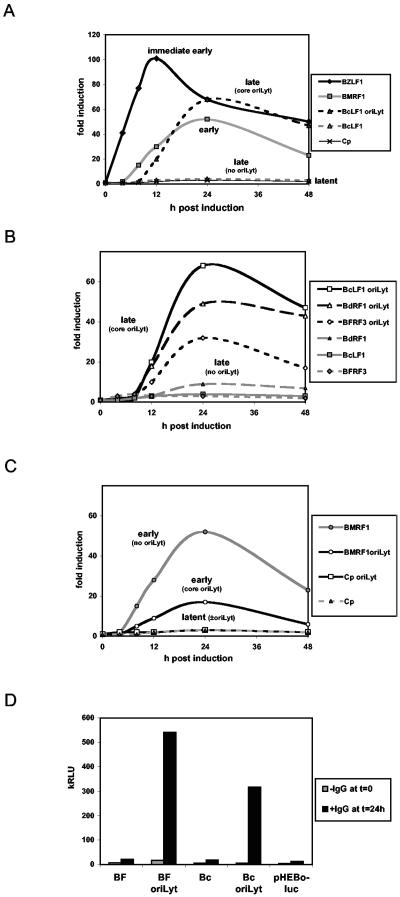
Luciferase reporter plasmids were created in the pEH-luc vector for a delayed early promoter, a latent cycle promoter (Cp), and several late lytic cycle EBV gene promoters (Fig. 4). The late promoters were cloned with or without ori lyt (13) in the plasmid (in this case, the EBV sequence encompassing positions 36560 to 42565, known as big ori lyt). The transcription start sites for all of the EBV promoters tested had previously been mapped accurately (4, 7, 8) so that a TATA box at about −30 relative to the transcription start could be identified. Sequences from about −430 to +30 relative to the transcription starts were cloned for each promoter (Fig. 4A) in front of the luciferase reporter gene. These plasmids were then transfected into EBV-positive Akata cells and stably transfected cell lines were isolated by selection with hygromycin. As in our previous studies (3), several lines were isolated for each promoter, the transfected cell lines were treated with anti-Ig to induce the EBV lytic cycle, and extracts were assayed for luciferase activity. The late promoters for BdRF1 and BFRF3 were only slightly affected by anti-Ig treatment of the cells unless ori lyt was also present in the plasmid. With big ori lyt, there was a very large induction of the late promoter and that induction was mostly prevented by treatment of the cells with PAA (Fig. 4B), a characteristic of true late promoters of herpesviruses. Either orientation of ori lyt in the plasmids functioned in this way (data not shown). The timing of expression of the late promoters (e.g., that for BdRF1) in the reporter plasmids matched the timing of expression of the BdRF1 late lytic cycle protein from the EBV genome in Akata 6 cells (Fig. 4B).
FIG. 4.
(A) Schematic diagram of EBV promoter in pHEBo-luc, with or without ori lyt. Late promoters were those for BcLF1, BdRF1, and BFRFC) Similar plasmids were made with the promoter for BMRF1 (delayed early) and the Cp latent promoter. (B) Luciferase reporter assays following anti-Ig induction of late promoter luciferase plasmids in AK2003 cells, with or without PAA treatment (0.85 mM). The Western blot assay of the BdRF1 protein demonstrates inhibition of the late lytic cycle by PAA and timing of BdRF1 expression. PCNA was also blotted as a loading control. kRLU, 103 relative light units; p.i., postinfection. (C) Southern blotting of 2 μg of EcoRI-digested total DNA from AK2003 cells containing the promoter for BdRF1 with no ori lyt or with the core ori lyt or the big ori lyt. The probe was an equal mixture of the B95-8 EcoRI I fragment (for EBV) and the pGL2 vector (Promega) for the luciferase plasmid, labeled by random priming. Restriction fragments corresponding to the endogenous (endog.) EBV or the pHEBo plasmid are shown above an ethidium bromide stain of the DNA on the gel as a loading control.
In our previous analyses of Zp and Rp, template copy number could be controlled very accurately because the plasmids did not support lytic replication. The replication of plasmids containing big ori lyt made it difficult to accurately quantitate how much of the induction of the late promoter was due to an increase in template copy number and how much was due to gene activation. To estimate the effect of replication, we measured the plasmid copy number in the presence and absence of PAA for pEH-luc plasmids containing no ori lyt, a replication-incompetent ori lyt (see below), and the big ori lyt with Southern blot assays (Fig. 4C). These data and the magnitude of the inhibition by PAA suggested that, of the 100-fold increase in luciferase caused by big ori lyt on the late promoter in response to anti-Ig, about 10-fold was due to replication causing an increase in the template copy number. So the remaining 10-fold is presumably due to transcription effects specific for the late promoters.
Core ori lyt confers late timing of induction on late promoters in the absence of detectable DNA replication.
In an attempt to separate the DNA replication effects of ori lyt from potential late gene regulation effects, a smaller version of ori lyt, known here as core ori lyt, was substituted for the big ori lyt. Core ori lyt has only about 1% of the replication activity of big ori lyt (13). The latent cycle, delayed early, and late promoter pEH-luc plasmids were all cloned with or without core ori lyt. As before, these plasmids were transfected into EBV-positive Akata cells and stably transfected cell lines were isolated by selection with hygromycin. Several lines were isolated for each promoter, and the time course of the response to anti-Ig induction was tested (Fig. 5A to C).
FIG. 5.
(A) Luciferase reporter assays for different classes of EBV promoter in AK2000 cells to show the relative timing after anti-Ig treatment and dependence of the late promoter on ori lyt. BZLF1, immediate early; BMRF1, delayed early; BcLF1, late; Cp, latent. Data are expressed as fold induction above the level of activity without anti-Ig treatment because of the different inherent strengths of the promoters. (B) All three of the late promoters tested in AK2000 cells require ori lyt for induction in response to anti-Ig and show late timing. (C) Core ori lyt reduces the lytic cycle activity of the delayed early promoter (BMRF1) in AK2000 cell lines and has no effect on the latent cycle promoter (Cp) in luciferase reporter assays after anti-Ig treatment. (D) Luciferase reporter assays with AK2000 cell lines for the BFRF3 and BcLF1 promoters with or without core ori lyt, compared to empty vector pHEBo-luc without ori lyt. Relative light unit (kRLU, 103 relative light units) data are shown as averages of duplicate determinations.
Because the promoters have different inherent strengths, the data in Fig. 5 are expressed as fold induction above the promoter activity in uninduced cells. Inducibility of the late promoters was dependent on the presence of ori lyt in the plasmid (Fig. 5A). The late promoters were also activated later than the immediate early Zp and the delayed early promoter for BMRF1. BcLF1 promoter activity was only measurable later than 8 h after anti-Ig induction, while immediate early BZLF1 promoter (Zp) activity increased almost instantly after anti-Ig induction and early BMRF1 activity preceded late BcLF1 activity by about 4 h, as measured by luciferase. This temporal spacing was seen with all three of the late promoters tested (Fig. 5B). Consistent with the previously reported very low replication activity of core ori lyt (13), Southern blotting analysis of the plasmid DNA did not detect any increase in template copy number (Fig. 4C, core ori lyt) and PAA had no effect on the induction of the late promoters caused by core ori lyt (data not shown). The core ori lyt did not increase the activity of the latent cycle or delayed early promoters—there was a decrease in delayed early promoter activity (Fig. 5C), and the latent cycle promoter Cp was little affected by any of the treatments (Fig. 5C). It thus appears that in this assay a contribution that ori lyt can make to lytic cycle expression of these EBV late gene promoters on the small plasmids is distinguished from the component of replication that can be inhibited by PAA. The core ori lyt conferred late timing of expression specifically on the late promoters without detectable DNA replication and only activated the late promoters.
The much greater anti-Ig-dependent activity of late promoters with core ori-lyt in cis is shown in the primary luciferase values given in Fig. 5D for the BFRF3 and BcLF1 promoters. Late regulation of the promoters acting in trans would induce in the absence of ori-lyt (27), but the small induction observed (Fig. 5D) did not differ significantly from that of the empty reporter vector lacking the late promoter and was insignificant relative to the values obtained with core ori-lyt (or big ori lyt [Fig. 4B]) in this system.
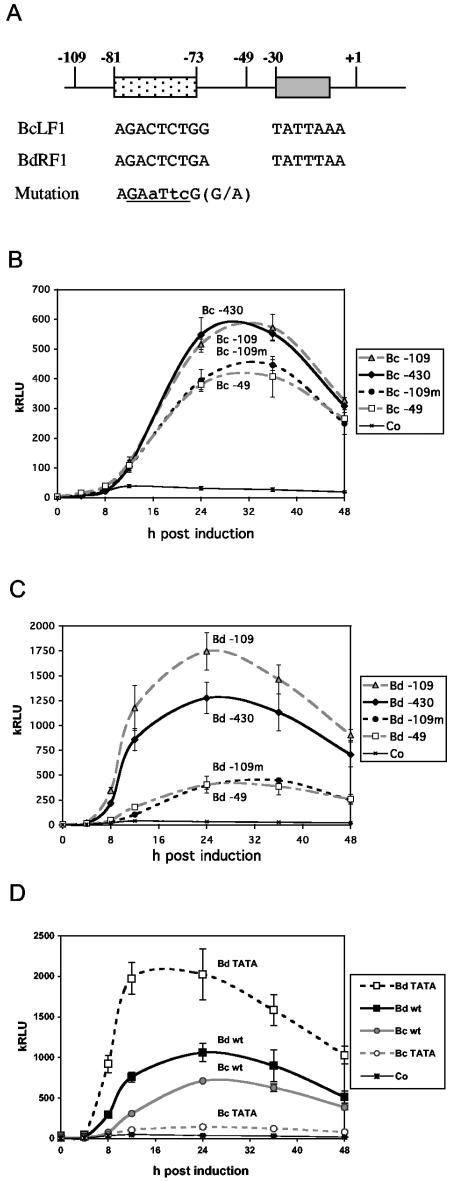
Conserved sequences upstream of the TATA box affect promoter strength but not lateness.
Previous studies of EBV promoter sequences noted a GACTCTG motif that was precisely conserved upstream of the TATA box at positions −73 to −81 in promoters of several EBV late genes (8). Deleting upstream sequences that contained the GACTCTG motif caused a loss of constitutive promoter activity in a Xenopus oocyte microinjection assay (8). To test the role of sequences upstream of the TATA box in the Akata cell assay described here, various 5′ deletions of the promoters for the BcLF1 and BdRF1 late genes were constructed in the pEHLyt-luc vector together with site-directed mutations of the GACTCTG motif (summarized in Fig. 6A). Figure 6B shows that 5′ deletion of the promoter for BcLF1 to position −109 made no difference from the −430 construct and deletion to position −49 caused only a slight decrease in inducibility. Mutation of the GACTCTG motif in the −109 background gave activity similar to that of the −49 plasmid. It therefore appears that sequences between positions −49 and +30 are sufficient for the core ori lyt-dependent promoter activity in the EBV late promoters tested and that the GACTCTG motif contributes to the increase in activity of the promoter but does not determine its late timing. Similar results were found when the corresponding mutations were analyzed in the BdRF1 promoter (Fig. 6C), where the magnitude of the contribution of the GACTCTG motif was greater and there was a small effect of the sequences between positions −109 and −430.
FIG. 6.
(A) Schematic diagram of the conserved −73 to −81 sequence present upstream of some late promoters and the mutation introduced. For the TATA box mutation, TATT was changed to TATA. (B) Luciferase reporter assays of the late promoter for BcLF1 in response to anti-Ig treatment in AK2000 cells. 5′ deletions were to position −430, −109, or −49, and −109m has the −81 to −73 sequence mutated in the −109 background. (C) Similar to panel B but with the late promoter for BdRF1. (D) TATT-to-TATA mutations of the TATA box in promoters for BdRF1 and BcLF1 in the −109 plasmids. kRLU, 103 relative light units; wt, wild type.
Role of the fourth-position T in the TATA box of late promoters.
Several lines of evidence point to a role for the TATA box in determining the activity of herpesvirus late promoters, and a T in the fourth position of the TATA box was found to be important for the activity of an EBV late promoter in a previous report (26). To test this in our system, we mutated the TATT sequence of the BcLF1 and BdRF1 late promoters to TATA, making them identical to the TATA box in several early EBV promoters. EBV-positive Akata cell lines containing these plasmids were prepared, and they were assayed as before. Similar to the results of Serio et al. (26), mutation of the TATT sequence in the promoter for BcLF1 to TATA caused a substantial reduction in promoter activity, about fourfold in our system (Fig. 6D). In contrast, the TATT mutation in the late promoter for BdRF1 caused a twofold increase in anti-Ig-inducible promoter activity (Fig. 6D).
DISCUSSION
In this study, we have expanded the use of our episomal oriP-based vector system (3, 5, 14) to investigate the regulation of the lytic cycle gene BRLF1 and late genes, exemplified by BcRF1, BdLF1, and BFRF3. With this system, the relative timing of promoter activity can be studied in the lytic cycle genes of this virus. By using stably transfected plasmids and the endogenous signaling of the Akata cell response to anti-Ig stimulation of the BCR, the results should accurately reflect the physiological regulation of EBV gene expression.
Although Zp clearly responds directly to signaling from the BCR in Akata cells, we have not been able to find any evidence of a direct response of Rp. For these studies, we tested sequences from positions +40 to −1250 in a luciferase reporter system (Fig. 2) and also tested larger fragments of the EBV genome with BRLF1 protein expression as a readout. It remains possible that we have not included the correct sequences in the promoter constructs or that there is a level of control in the whole EBV genome that is not reconstituted in the oriP plasmids we have used, but these new data argue against Rp being an immediate early promoter of EBV. Because of the feedback activation of BZLF1 and BRLF1 on Zp and Rp, there is much less of these RNAs expressed when this activation is prevented by protein synthesis inhibitors, so distinguishing whether the residual level of RNA (as detected in the 5′ RACE experiment) represents immediate early activity or escape from the inhibitor of protein synthesis is difficult. The ability of BRLF1 to activate the EBV lytic cycle in epithelial cells (33), the constitutive expression of BRLF1 by rearranged defective EBV present in some P3HR1 cell cultures (2), and the importance of BRLF1 homologues in reactivation of other gamma herpesviruses all indicate a key role for BRLF1 in EBV reactivation, but in the Akata cell response to anti-Ig cross-linking of the BCR, activation of Rp does not appear to work without another EBV function. In this respect, the failure of the BZLF1 induced from the pZRp-7101 plasmid to induce BRLF1 (Fig. 2C) and the additional ATTTTT sequence on the 5′ end of the BRLF1 mRNA remain to be explained fully. The possibility that there is a specific mutation or defect in the AK31 cell line that prevents activation of Rp in response to BCR signal transduction cannot be excluded but seems unlikely since all three arms of the signal transduction were shown to function normally at the biochemical level in an earlier analysis (5). The kinetics of induction of the Rp luciferase reporter in a different clone of EBV-positive Akata cells also showed a clear delay relative to the induction of Zp (Fig. 2B), supporting a mainly indirect activation of Rp.
The published data on the ZV element of Zp (3, 18, 19) indicate a large (8- to 10-fold) increase in Zp promoter induction in response to anti-Ig when ZV is mutated to MV. These data from luciferase reporter constructs in both transient and stable transfection systems were not born out when the altered promoter was expressing BZLF1 (Fig. 3). It is not clear whether this is a consequence of a further promoter element within the BZLF1 coding sequence that prevents the effects of MV or whether the local increase in BZLF1 that might result from the MV plasmid inhibits Zp activity. The low-affinity binding site for BZLF1 in Zp (ZIIIA) might be a candidate for mediating such feedback, although this would be difficult to test experimentally because of the role of ZIIIA in the response of Zp to BCR signaling (3).
Some recent publications on late gene regulation of EBV have concluded that late promoters are induced in trans by the late lytic cycle without the need for ori lyt in cis in the plasmids containing the late promoter being tested (26, 27). In contrast, with the Akata oriP plasmid system described here, we found a requirement for ori lyt in cis, specifically for late promoter induction. Some of the increase in luciferase activity could be accounted for by an increase in template copy number as the big ori lyt plasmids replicated, but there was also some late regulation of the late promoter; this was dependent on ori lyt in cis and occurred late in the lytic cycle. There might be subpopulations of EBV genomes during the replicative phase of the lytic cycle with different activity, but that would make it even more difficult to account for late gene induction by an increase in genome copy number and reinforce the case for transcriptional regulation. It is likely that at least three effects contribute to late gene regulation in EBV. The replication of the plasmid increases the copy number and may expose the late promoters that were previously masked in the unreplicated viral chromatin. Second, replication occurs at nuclear sites localized near PML domains (1), so plasmids for replication may have to migrate to these sites and we speculate that this location may favor transcription from late promoters in the late lytic cycle. The obvious potential mechanism for this would be mediated by ori lyt in the plasmid, and we suggest that this might be the effect we observed with the core ori lyt. Nonspecific enhancer effects of BZLF1 and BRLF1 via ori lyt have been reported in various transient assay systems, but it is important to note the specificity of the core ori lyt induction for late promoters in our system. Finally, there may also be a transacting factor that helps to activate late genes, comparable to the IVa2 protein in late gene regulation of adenovirus (29), as indicated for EBV by the results of Serio et al. (26, 27).
By using the core ori lyt to separate late promoter induction from the increase in template copy number due to replication, we analyzed the roles of upstream sequences and the TATA box in late promoters. Although there is conservation of certain sequences upstream of some late promoters, these contributed to promoter activity but not lateness. The fourth-position T is conserved in the TATA box of many late promoters. In one late promoter, we confirmed the previously published importance of this T residue, but in another, late promoter mutation of this T residue did not reduce the late promoter activity, perhaps consistent with the fact that not all late promoters have the fourth-position T in the TATA box. Our findings on the requirement for ori lyt in cis and the relatively small region of DNA that constitutes a late promoter help to bring EBV late gene regulation more in line with the published data on late promoters in HSV, as might have been predicted from the homology of many late genes in herpesviruses.
Finally, the comparison of immediate early, delayed early, and late promoters in the oriP plasmid luciferase reporter assay showed that the correct sequence of expression of these classes of promoter was reconstituted. Previously, EBV genes have been classified on the basis of their response to inhibitors of DNA replication, but now we can include the relative timing of expression in the lytic cycle as a property of their promoters.
With EBV, it is easy to obtain a biologically relevant latent infection that can be studied in cell culture. Investigation of the Akata system provided the first evidence that the transition between latency and reactivation in this herpesvirus involves acetylation of histones in the nucleosomes close to the Zp immediate early promoter (14). The further clarification provided in this paper of the role of Rp and the regulation of delayed early and late genes will help to provide a coherent understanding of the mechanism of reactivation and lytic cycle gene expression of this herpesvirus.
Acknowledgments
We thank Evelyne Manet for the 8C12 antibody to BRLF1, Kenzo Takada for the Akata cell line, Jaap Middeldorp for the BdRF1 antibody, and Richard Greaves for the pRG201 plasmid.
REFERENCES
- 1.Bell, P., P. M. Lieberman, and G. G. Maul. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800-11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggin, M., M. Bodescot, M. Perricaudet, and P. Farrell. 1987. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J. Virol. 61:3120-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binné, U., W. Amon, and P. Farrell. 2002. Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76:10282-10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodescot, M., M. Perricaudet, and P. J. Farrell. 1987. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J. Virol. 61:3424-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, H., and P. Farrell. 2002. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 76:10290-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jesus, O., P. R. Smith, L. Spender, C. Elgueta Karstegl, H. Niller, D. Huang, and P. J. Farrell. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 84:1443-1450. [DOI] [PubMed] [Google Scholar]
- 7.Farrell, P. J. 1989. The Epstein-Barr virus genome, p. 103-132. In I. G. Klein (ed.), Advances in viral oncology. Raven Press, New York, N.Y.
- 8.Farrell, P. J., A. Bankier, C. Seguin, P. Deininger, and B. G. Barrell. 1983. Latent and lytic cycle promoters of Epstein-Barr virus. EMBO J. 2:1331-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furnari, F. B., V. Zacny, E. B. Quinlivan, S. Kenney, and J. S. Pagano. 1994. RAZ, an Epstein-Barr virus transdominant repressor that modulates the viral reactivation mechanism. J. Virol. 68:1827-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzowski, J. F., J. Singh, and E. K. Wagner. 1994. Transcriptional activation of the herpes simplex virus type 1 UL38 promoter conferred by the cis-acting downstream activation sequence is mediated by a cellular transcription factor. J. Virol. 68:7774-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 67:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, K. T., A. J. Stevenson, D. J. Goodwin, P. C. Gibson, A. F. Markham, and A. Whitehouse. 1999. The activation domain of herpesvirus saimiri R protein interacts with the TATA-binding protein. J. Virol. 73:9756-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins, P., U. Binné, and P. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, P. A., and R. D. Everett. 1986. The control of herpes simplex virus type-1 late gene transcription: a ‘TATA-box’/cap site region is sufficient for fully efficient regulated activity. Nucleic Acids Res. 14:8247-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kibler, P. K., J. Duncan, B. D. Keith, T. Hupel, and J. R. Smiley. 1991. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J. Virol. 65:6749-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, D. B., S. Zabierowski, and N. A. DeLuca. 2002. The initiator element in a herpes simplex virus type 1 late-gene promoter enhances activation by ICP4, resulting in abundant late-gene expression. J. Virol. 76:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus, R., J. G. Perrigoue, and J. Mertz. 2003. ZEB negatively regulates the lytic-switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 77:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, R. J., S. J. Mirocha, H. M. Stephany, J. R. Puchalski, and J. E. Mertz. 2001. Identification of a novel element involved in regulation of the lytic switch BZLF1 gene promoter of Epstein-Barr virus. J. Virol. 75:867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manet, E., H. Gruffat, M. C. Trescol-Biemont, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Packham, G., M. Brimmell, D. Cook, A. Sinclair, and P. Farrell. 1993. Strain variation in Epstein-Barr virus immediate early genes. Virology 192:541-550. [DOI] [PubMed] [Google Scholar]
- 22.Polack, A., G. Hartl, U. Zimber, U. K. Freese, G. Laux, K. Takaki, B. Hohn, L. Gissmann, and G. W. Bornkamm. 1984. A complete set of overlapping cosmid clones of M-ABA virus derived from nasopharyngeal carcinoma and its similarity to other Epstein-Barr virus isolates. Gene 27:279-288. [DOI] [PubMed] [Google Scholar]
- 23.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2425-2426. In D. Knipe and P. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 24.Roizman, B., and P. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2388-2394. In D. Knipe and P. Howley (ed.), Fields virology, fourth ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 25.Romanelli, M. G., P. Mavromara-Nazos, D. Spector, and B. Roizman. 1992. Mutational analysis of the ICP4 binding sites in the 5′ transcribed noncoding domains of the herpes simplex virus 1 UL49.5 γ2 gene. J. Virol. 66:4855-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serio, T. R., N. Cahill, M. E. Prout, and G. Miller. 1998. A functionally distinct TATA box required for late progression through the Epstein-Barr virus life cycle. J. Virol. 72:8338-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serio, T. R., J. L. Kolman, and G. Miller. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71:8726-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weir, J. P., and P. R. Narayanan. 1990. Expression of the herpes simplex virus type 1 glycoprotein C gene requires sequences in the 5′ noncoding region of the gene. J. Virol. 64:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 33.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]