Abstract
Rous sarcoma virus (RSV) budding requires an interaction of the L domain within the p2b region of Gag with cellular Nedd4-family E3 ubiquitin protein ligases. Members of our laboratories previously demonstrated that overexpression of a fragment of the chicken Nedd4-like protein (LDI-1 WW) inhibits Gag release in a dominant-negative manner (A. Kikonyogo, F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis, Proc. Natl. Acad. Sci. USA 98:11199-11204, 2001). We have now identified the complete 3′ end of LDI-1 and determined that it has a C-terminal ubiquitin ligase HECT domain, similar to other Nedd4 family members. While overexpression of the full-length LDI-1 clone (LDI-1 FL) had little effect on Gag budding, an LDI-1 FL mutant with a substitution in the HECT domain catalytic site blocked Gag release, similar to LDI-1 WW. The coexpression of Gag and hemagglutinin-tagged ubiquitin (HA-Ub) resulted in the detection of mono- and polyubiquitinated forms of Gag in cells and mostly monoubiquitinated Gag in virus-like particles (VLPs). When the Nedd4-binding site (L domain) was deleted, ubiquitinated Gag was not detected. Interestingly, the release of Gag with ubiquitin covalently linked to the C terminus (Gag-Ub) was still blocked by LDI-1 WW. To understand the mechanism of this inhibition, we examined cells expressing Gag and LDI-1 WW by electron microscopy. In the presence of LDI-1 WW, VLPs were found in electron-dense inclusion bodies in the cytoplasm of transfected cells. In contrast, when cells that coexpressed Gag-Ub and LDI-1 WW were examined, inclusion bodies were detected but did not contain VLPs. These results indicate that the ubiquitination of Gag is dependent upon Nedd4 binding to the L domain and suggest that Nedd4 has additional functions during RSV release besides the ubiquitination of Gag.
The structural proteins of retroviruses are encoded by the gag gene and are translated as a large precursor polyprotein. During or subsequent to the budding process, the Gag polyprotein is cleaved by the viral protease (PR) to release each of the structural proteins. This coincides with maturation of the virus particle and is required for infectivity. The expression of Gag alone is sufficient to lead to the production and release of virus-like particles (VLPs) from transfected cells. Three domains within the Gag polyprotein are required for this process (23). The M domain, found at the N terminus of Gag within the MA protein, is responsible for membrane binding. The late assembly domain (L domain), found in different regions of Gag depending on the retrovirus, is required for particles to bud and separate from the cell. Lastly, the interaction domain (I domain), found within the NC protein coding sequence at the C terminus of Gag, is associated with Gag-Gag interactions during the multimerization process.
The L domain in Rous sarcoma virus (RSV) has been mapped to a proline-rich sequence within the p2b region of Gag and contains the sequence PPPPYV, referred to as the PY motif (22, 24) (see Fig. 1). Deletions or mutations within the PY motif result in a block in particle release and the accumulation of virus particles tethered to the plasma membrane that are unable to completely separate from the cell (14, 22, 24, 27). Members of our laboratories previously reported the identification of late domain interacting protein 1 (LDI-1), which binds to the RSV L domain sequence (9). LDI-1 is a partial clone that contains a C2 domain and four WW domains, and it is a putative member of the Nedd4 family of E3 ubiquitin ligases. The coexpression of RSV Gag and LDI-1 or LDI-1 WW, a truncated version of LDI-1 containing only the four WW domains, inhibits the release of Gag in a dominant-negative manner (9).
FIG. 1.
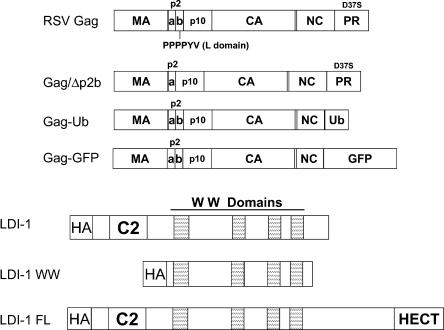
RSV Gag and LDI-1 constructs. RSV Gag is represented by large rectangles. Gag proteins are indicated inside the boxes, with vertical lines representing the boundaries of the viral proteins. Gag/Δp2b lacks the 11-amino-acid p2b region. In the Gag constructs used for this report, the PR subunit contains an inactivating D37S substitution. In the Gag-Ub and Gag-GFP polyproteins, respectively, ubiquitin and GFP were substituted for the last six residues of NC and all of PR. LDI-1 is a partial clone of a putative chicken Nedd4 family member that was described previously (9). LDI-1 WW contains amino acids 132 to 571 of LDI-1. LDI-1 FL is a newly identified clone that contains the complete 3′ end of LDI-1, which includes the catalytic HECT domain of the E3 ubiquitin ligase.
There is accumulating evidence suggesting that the ubiquitination of RSV Gag is an important part of the assembly and budding process. It was shown previously that the treatment of cells with proteasome inhibitors leads to a decrease in RSV Gag release (14). Likewise, the overexpression of ubiquitin or the fusion of a ubiquitin molecule to the C terminus of Gag partially rescues the budding defect caused by proteasome inhibitor treatment (14). It has also been demonstrated that the RSV L domain can mediate the ubiquitination of mini-Gag proteins derived from human immunodeficiency virus type 1 (HIV-1) Gag (17). However, ubiquitinated Gag proteins have not been detected in RSV particles. Interestingly, it has been shown that the concentration of free ubiquitin in RSV particles is higher than that in the cytoplasm of the host cell, suggesting that ubiquitin is somehow recruited into virus particles (16). It is unclear whether free ubiquitin is packaged or if a ubiquitinated viral or cellular protein present in virus particles becomes the substrate for a ubiquitin hydrolase.
Other retroviruses, including human T-cell leukemia virus type 1, Mason-Pfizer monkey virus (MPMV), and murine leukemia virus (MLV), have L domain sequences similar to the PY motif found in RSV Gag and have been shown to bind to Nedd4 family members (1, 4, 25). In addition, enveloped viruses, including the rhabdoviruses vesicular stomatitis virus and rabies virus as well as the filovirus Ebola virus, have amino acid motifs that are similar to the RSV PY motif and bind to Nedd4 family members (5, 6, 18, 26). Furthermore, the overexpression of specific Nedd4 proteins enhances the release of MPMV and Ebola virus (25, 26) particles, and a small amount of Nedd4 has been detected in MPMV particles (4). Multiple groups have demonstrated that the expression of dominant-negative mutants of Nedd4 proteins containing only the WW domain region or an amino acid substitution in the catalytic HECT domain inhibits the release of MPMV, Ebola virus, and human T-cell leukemia virus type 1 (1, 25, 26). Taken together, these results suggest that Nedd4 proteins are involved in the budding process of a variety of viruses and that ubiquitination is a required step in the release process.
In the present study, we provide new insights into how LDI-1 WW inhibits RSV Gag budding, which involves the formation of VLPs in cytoplasmic inclusion bodies. In addition, we have determined that ubiquitination is dependent upon a functional L domain. Moreover, the presence of ubiquitin at the C terminus of Gag alters the assembly and budding process but does not replace the requirement for an interaction between Gag and Nedd4 family members. Finally, we have identified the complete 3′ end of LDI-1 and determined that the overexpression of an LDI-1 mutant with an amino acid substitution in the catalytic HECT domain (LDI-1 FL C/A) has a severe inhibitory effect on Gag release. These results indicate that Nedd4-dependent ubiquitination of Gag is required for the budding of RSV and that Nedd4-like proteins may have multiple functions during the release process.
MATERIALS AND METHODS
Cell culture.
293/E cells are derived from human embryonic kidney cells and stably express the EBNA-1 protein from Epstein-Barr virus (9). 293/E and COS-1 cells were maintained in Dulbecco's modified Eagle medium (Gibco, Carlsbad, Calif.) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, Calif.), penicillin (1,000 U/ml), and streptomycin (1,000 μg/ml; Gibco). In all experiments, 293/E and COS-1 cells were transfected with the Fugene 6 transfection reagent according to the manufacturer's instructions (Roche, Indianapolis, Ind.).
Plasmids.
p2036-Gag, p2036-LDI-1, and p2036-LDI-1 WW have been described previously (9). p2036-Gag/Δp2b is identical to p2036-Gag except that it lacks the nucleotide sequence coding for the PPPPYV amino acid sequence within the p2b region of Gag. pGag-Ub and pGag-GFP were obtained from John Wills (Pennsylvania State University). For the generation of p2036-Gag-Ub, the Gag-ubiquitin (Gag-Ub) coding region was subcloned from pGag-Ub (14) and inserted into p2036. The Gag-Ub coding sequence was amplified by PCR to introduce a KpnI site at the 5′ end and a BsrGI site at the 3′ end, allowing for insertion into p2036. pGag-GFP encodes the RSV Gag protein minus the last six residues of NC and all of PR fused to enhanced green fluorescent protein (EGFP) in the pEGFP-N2 backbone (14). pGag/Δp2b-GFP lacks the nucleotide sequence coding for the PPPPYV sequence within p2b as well as the last six residues of NC and all of PR. The Gag/Δp2b sequence was amplified from p2036-Gag/Δp2b by PCR to produce a HindIII site at the 5′ end and a KpnI site at the 3′ end to allow for insertion into pEGFP-N2, with Gag/Δp2b placed in frame with EGFP. pMT123 (19) encodes a hemagglutinin (HA)-tagged ubiquitin molecule and was obtained from Dirk Bohmann (University of Rochester Medical Center). The 3′ end of LDI-1 was determined by use of a Smart RACE cDNA amplification kit (BD Biosciences, Palo Alto, Calif.) and was amplified from chicken liver cDNA (BD Biosciences) by the use of primers complementary to the previously identified LDI-1 sequence (9). After identification of the 3′ end, p2036-LDI-1 FL was constructed by amplification of the LDI-1 coding sequence from chicken liver cDNA by PCR to introduce a KpnI site at the 5′ end and an MfeI site at the 3′ end to enable insertion into p2036. The LDI-1 coding sequence within p2036-LDI-1 FL was amplified beginning at nucleotide position 1 of the previously identified partial LDI-1 clone (9) and ending at the newly identified stop codon at the 3′ end. p2036-LDI-1 FL(C/A) was constructed by PCR-based mutagenesis and contains the LDI-1 full-length coding sequence with a cysteine-to-alanine amino acid substitution in the catalytic site of the HECT domain (Cys932). Similar to the previously described LDI-1 constructs, LDI-1 FL and LDI-1 FL C/A were expressed as fusion proteins with HA tags (YPYDVPDYA) at their N termini. AGP5 is a simian virus 40 large T antigen expression vector that has been described previously (9). pcDNA3.1 (Invitrogen, Carlsbad, Calif.) and p2036 were used as filler DNAs to normalize the amount of DNA transfected in each experiment.
Antibodies.
Rabbit anti-Nedd4 was raised against WW domain 2 of rat Nedd4 and was purchased from Upstate Biotechnology (Lake Placid, N.Y.). An anti-avian myeloblastosis virus (AMV) p19 (MA) monoclonal antibody developed by David Boettiger was obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa (Iowa City). A rabbit anti-RSV polyclonal antiserum was produced after the immunization of rabbits with detergent- and heat-denatured RSV. Rabbit and mouse anti-influenza virus HA antibodies were purchased from Covance (Berkeley, Calif.). Goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (HRP) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Fluorescently tagged mouse anti-rabbit (tetramethylrhodamine isocyanate [TRITC]) and rabbit anti-mouse (fluorescein isothiocyanate [FITC]) secondary antibodies were obtained from Zymed Laboratories Inc. (San Francisco, Calif.).
Budding assay.
Six-well plates of 293/E cells were transfected with 0.5 μg of p2036-Gag or p2036-Gag-Ub, 0.5 to 1.5 μg of p2036-LDI-1, p2036-LDI-1WW, p2036-LDI-1 FL, or p2036-LDI-1 FL(C/A), and 0.01 μg of AGP5. The total DNA transfected per well was normalized to 2 μg with pcDNA3.1. At 48 h posttransfection, the cells were labeled with [35S]Met and [35S]Cys, and Gag proteins were immunoprecipitated from the medium and lysate fractions by use of a rabbit anti-RSV polyclonal antiserum as described previously (9). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and dried. Gels were analyzed with a Storm imaging system or a PhosphorImager (Amersham Biosciences, Piscataway, N.J.). The budding level was determined by dividing the signal of the Gag band in the medium fraction by the sum of the signals of the Gag bands in the medium and lysate fractions. The level of Gag budding was set to 100%.
Coimmunoprecipitation assay.
Culture dishes (100-mm diameter) containing 293/E cells were transfected with 10 μg of p2036-Gag, p2036-Gag/Δp2b, or pcDNA3.1. Forty-eight hours after transfection, the cells were washed two times with phosphate-buffered saline (PBS) and lysed with RIPA buffer (1% Igepal CA-630, 0.5% sodium deoxycholate, and 0.1% SDS in 1× PBS) containing protease inhibitor cocktail tablets (Roche). After passage through a 21-gauge needle, the lysate was incubated on ice for 60 min and the cellular debris was pelleted at 10,000 × g for 10 min at 4°C. The supernatant was precleared with 0.5 μg of normal mouse IgG (Santa Cruz Biotechnology) and 20 μl of protein A agarose (Invitrogen) for 30 to 60 min at 4°C with rocking. The precleared lysate was incubated on ice for 1 h with an anti-AMV MA monoclonal antibody. Twenty microliters of protein A agarose was then added, and the complexes were incubated overnight at 4°C with rocking. Immunoprecipitated complexes were washed three or four times with RIPA buffer and suspended in 2× loading buffer (225 mM Tris [pH 6.8], 5% SDS, 50% glycerol, 5% β-mercaptoethanol, and 0.05% bromophenol blue). Immunoprecipitated proteins were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.). After blocking of the membrane with wash buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.1% Tween 20) containing 5% nonfat dry milk, coimmunoprecipitated Nedd4 proteins were detected with a rabbit anti-Nedd4 antibody and an anti-rabbit IgG-HRP secondary antibody by enhanced chemiluminescence (ECL; Amersham Biosciences). To verify that similar amounts of Gag and Gag/Δp2b were immunoprecipitated, we reprobed the membrane with an anti-AMV MA monoclonal antibody and an anti-mouse IgG-HRP secondary antibody. Gag proteins were detected by ECL.
Ubiquitination assay.
Dishes (100-mm diameter) of 293/E cells were transfected with 5 μg of p2036-Gag or p2036-Gag/Δp2b and 2.5 μg of pMT123. The amount of DNA transfected per plate was normalized to 7.5 μg with pcDNA3.1. Forty-eight hours after transfection, VLPs were harvested from the medium. Cellular debris was pelleted by centrifugation at 3,000 × g for 20 min at 4°C. The supernatant was layered onto a 20% sucrose cushion in 5TE (50 mM Tris-HCl [pH 8.0], 1 mM EDTA), and VLPs were pelleted by centrifugation at 76,000 × g for 45 min at 4°C. The pellet was suspended in RIPA buffer containing protease inhibitor cocktail tablets. For harvest of the cell lysate, the cells were washed two times with PBS and lysed with RIPA buffer containing protease inhibitor cocktail tablets. After passage through a 21-gauge needle, the lysate was incubated on ice for 60 min and the cellular debris was pelleted at 10,000 × g for 10 min at 4°C. Gag proteins were immunoprecipitated with an anti-AMV MA monoclonal antibody on ice for 1 h. Twenty microliters of protein A agarose was then added, and the complexes were incubated overnight at 4°C with rocking. The immunoprecipitated proteins were washed three times with RIPA buffer and then suspended in 2× loading buffer. The immunoprecipitated proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. After blocking of the membrane with wash buffer containing 5% nonfat dry milk, ubiquitinated Gag proteins were detected with a mouse or rabbit anti-HA antibody and an anti-mouse or anti-rabbit IgG-HRP secondary antibody by ECL.
Confocal microscopy.
COS-1 cells in six-well plates (seeded at 0.5 × 105 cells/ml and grown to 60% confluence) were transfected with 3 μg of pGag-GFP, pGag/Δp2b-GFP, or p2036-Gag-Ub and 7 μg of p2036LDI-1 WW. At 48 h posttransfection, the cells were washed once with PBS and fixed in 4% formaldehyde (Fisher, Pittsburgh, Pa.) in Ca2+-free, Mg2+-free PBS for 20 min. The samples were then washed three times for a total of 5 min with PBS, permeabilized with 0.1% Triton X-100 for 5 min, and washed three times again with PBS. After being blocked for 10 min in PBS containing 1% bovine serum albumin, the cells were incubated with a primary antibody for 1 h at 37°C, rinsed with PBS, and then incubated with a fluorescently (TRITC or FITC) tagged secondary antibody for 30 min at 37°C. When indicated, a nuclear Hoechst stain (Molecular Probes, Eugene Oreg.) was added for the last 10 min. After being rinsed, the cells were mounted with p-phenylenediamine and Immunomount. Confocal images were captured with an inverted fluorescence-differential interference contrast Zeiss Axiovert 200 M microscope equipped with an AxioCam HRm camera (Zeiss, Thornwood, N.Y.) and a mercury arc lamp light source using a 63× Plan-Apochromat (NA 1.40) oil objective and operating with AxioVision, version 3.1 (Zeiss), software. More than 30 cell images were examined for each experiment, and experiments were repeated three times. Twenty to thirty optimal sections along the z axis were acquired in increments of 0.4 μm. Figures show the central image or, when indicated, z sections from the adherent surface through the nucleus. The fluorescent data sets were deconvolved by the constrained iterative method (AxioVision, version 3.1). The following excitation and emission wavelengths were used for imaging: for Hoechst, λex was 360 ± 20 nm and λem was 460 ± 25 nm; for FITC (to see GFP), λex was 480 ± 20 nm and λem was 535 ± 25 nm; and for TRITC (to see Texas Red), λex was 560 ± 25 nm and λem was 645 ± 35 nm.
Electron microscopy.
Sixty-millimeter-diameter dishes of 293/E cells were transfected with 0.5 μg of p2036-Gag or p2036-Gag-Ub, 1.5 μg of p2036-LDI-1, p2036-LDI-1 WW, p2036-LDI-1 FL, or p2036-LDI-1 FL(C/A), and 0.01 μg of AGP5. The total DNA transfected per dish was normalized to 2 μg with p2036. At 48 h posttransfection, the cells were washed with PBS at room temperature and fixed in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4, at 4°C for 30 min. The cells were scraped from the tissue culture dish and pelleted at 1,000 × g for 10 min at 4°C. The cell pellets were fixed an additional 2 h in 2.5% glutaraldehyde and then postfixed for 1 h with osmium tetroxide. The cell pellets were dehydrated in a series of alcohol washes and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and examined by use of a Zeiss 900 electron microscope.
Nucleotide sequence accession number.
The GenBank sequence for LDI-1 has been updated with the LDI-1 FL sequence (accession number AF412121).
RESULTS
Gag interacts with endogenous Nedd4 proteins in an L domain-dependent manner.
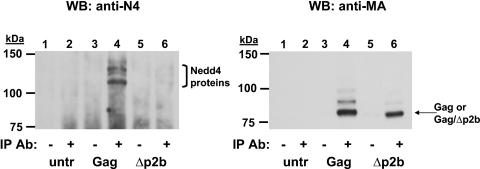
Peptides containing the PY motif (L domain) of RSV Gag (Fig. 1) bind to Nedd4-like proteins containing WW motifs. In addition, the overexpression of fragments of the chicken Nedd4-like protein (LDI-1) inhibits RSV Gag release in a dominant-negative manner (9). To determine if endogenous Nedd4 proteins bind to RSV Gag in an L domain-dependent fashion, we performed a series of coimmunoprecipitation experiments. As shown in Fig. 2, endogenous Nedd4 proteins were coimmunoprecipitated from the lysate of cells transfected with p2036-Gag but not from that of cells transfected with p2036-Gag/Δp2b. The interaction between Gag and Nedd4 was specific because Nedd4 was not immunoprecipitated in the absence of Gag expression or without the anti-AMV MA antibody (Fig. 2, left panel, lanes 1 to 3). As a control, the blot was reprobed with the same anti-AMV MA antibody that was used for immunoprecipitation. Equal amounts of Gag and Gag/Δp2b were detected in the immunoprecipitates (Fig. 2, right panel, lanes 4 and 6). The slower migrating Gag bands seen in lane 4 (right panel) may represent ubiquitinated forms of Gag. The Gag constructs used in all of the experiments described here contained a D37S mutation in the PR coding sequence that inactivates this enzyme (Fig. 1). As a consequence, only Pr76Gag is detected in the gels.
FIG. 2.
Coimmunoprecipitation of endogenous Nedd4 proteins with RSV Gag. RSV Gag or Gag/Δp2b was expressed in 293/E cells and immunoprecipitated with an anti-AMV MA monoclonal antibody as indicated. After separation by SDS-PAGE, immunoprecipitated proteins were detected by use of an anti-Nedd4 antibody (left). The membrane was then reprobed with an anti-AMV MA antibody (right) as described in Materials and Methods. Untransfected cells, lanes 1 and 2; cells transfected with p2036-Gag, lanes 3 and 4; cells transfected with p2036-Gag/Δp2b, lanes 5 and 6. The plus and minus symbols denote which samples were incubated with the anti-AMV MA antibody used for immunoprecipitation. IP, immunoprecipitation; WB, Western blot.
Overexpression of full-length LDI-1 does not enhance RSV Gag budding.
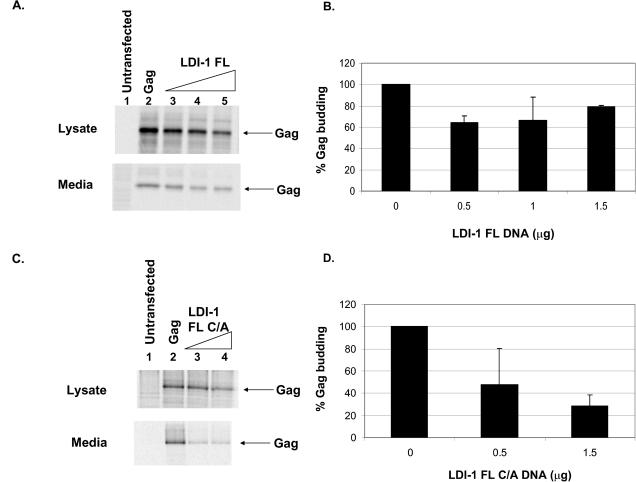
We identified the complete 3′ end of LDI-1 by random amplification of cDNA ends and designated the full-length clone LDI-1 FL. In addition to containing a C2 domain and four WW domains, LDI-1 FL contains a ubiquitin ligase HECT domain (Fig. 1). The domain structure of LDI-1 FL is similar to that of other Nedd4 family members, providing further evidence that LDI-1 is a chicken Nedd4 protein. We tested the effect of overexpression of LDI-1 FL on the budding of Gag VLPs. We found that the overexpression of LDI-1 FL had a small effect on Gag release (Fig. 3). The signals of Pr76Gag in the medium and lysate fractions in the presence of increasing amounts of LDI-1 FL are shown in Fig. 3A. Note that the level of Gag in the lysate decreased in the presence of increasing amounts of LDI-1 FL. However, quantitation of the signals of Gag in the medium and lysate fractions indicated that the level of Gag release in the presence of LDI-1 FL was 70 to 80% the level of Gag budding in the absence of LDI-1 FL (Fig. 3B). Although there was little effect on Gag release, the expression of LDI-1 FL was verified by Western blot analysis and by the appearance of inclusion bodies in thin-section electron micrographs (data not shown).
FIG. 3.
Effect of overexpression of full-length LDI-1 (LDI-1 FL) or a HECT domain mutant of LDI-1 (LDI-1 FL C/A) on release of RSV Gag from 293/E cells. (A) RSV Gag was expressed in 293/E cells in the absence or presence of increasing amounts of p2036-LDI-1 FL. The amounts of Gag in the lysate and medium fractions were determined as described in Materials and Methods. Untransfected cells, lane 1; cells transfected with 0.5 μg of p2036-Gag, lane 2; cells cotransfected with 0.5 μg of p2036-Gag and 0.5 μg of p2036-LDI-1 FL (lane 3); cells cotransfected with 0.5 μg of p2036-Gag and 1 μg of p2036-LDI-1 FL (lane 4); or cells cotransfected with 0.5 μg of p2036-Gag and 1.5 μg of p2036-LDI-1 FL (lane 5). (B) Graphical representation of relative amounts of Gag released into the medium in the presence of LDI-1 FL (derived from two independent experiments). (C) RSV Gag was expressed in 293/E cells in the absence or presence of increasing amounts of p2036-LDI-1 FL C/A. Untransfected cells, lane 1; cells transfected with 0.5 μg of p2036-Gag, lane 2; cells cotransfected with 0.5 μg of p2036-Gag and 0.5 μg of p2036-LDI-1 FL C/A, lane 3; cells cotransfected with 0.5 μg of p2036-Gag and 1.5 μg of p2036-LDI-1 FL C/A, lane 4. (D) Graphical representation of relative amounts of Gag released into the medium in the presence of LDI-1 FL C/A (derived from two independent experiments).
Ubiquitin ligase activity is required for RSV Gag budding.
After the identification of the 3′ end of LDI-1 and the determination that LDI-1 has a HECT domain, we tested whether the ubiquitin ligase activity of LDI-1 FL is required for Gag budding. To do this, we introduced a cysteine-to-alanine amino acid substitution into the conserved active site of the HECT domain of LDI-1 FL (Cys 932). The active-site cysteine of LDI-1 FL was identified by an alignment of the LDI-1 FL HECT domain sequence with the sequences of the HECT domains of other Nedd4 family members. When 293/E cells were cotransfected with this construct (LDI-1 FL C/A) and p2036-Gag, we found that Gag budding was significantly inhibited in an LDI-1 FL C/A dose-dependent manner (Fig. 3C). In the presence of 1.5 μg of p2036-LDI-1 FL C/A, the level of Gag release was reduced to 25% the level of Gag release in the absence of LDI-1 FL C/A (Fig. 3D). This result was in contrast to the results shown in Fig. 3A and B for wild-type LDI-1 FL. It should be noted that the expression levels of LDI-1 FL and LDI-1 FL C/A in lysates of 293/E cells were similar, as determined by Western blot analysis (data not shown). These data clearly demonstrate that the ubiquitin ligase activity of Nedd4 proteins is required for the RSV budding process.
Gag VLPs are found in inclusion bodies in cells expressing LDI-1 WW, but not in cells expressing LDI-1 FL C/A.
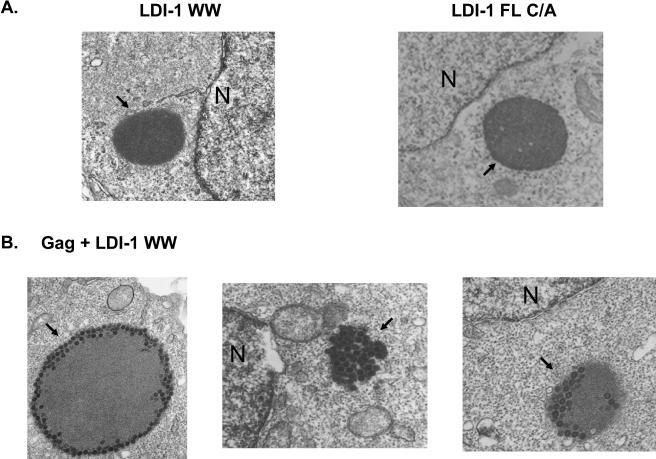
In order to better understand the mechanism by which LDI-1 WW and LDI-1 FL C/A inhibit Gag budding, we examined cells that were transfected with p2036-Gag and p2036-LDI-1 WW or p2036-LDI-1 FL C/A by thin-section electron microscopy. Electron-dense granules lacking membranes were detected in the cytoplasm of cells expressing LDI-1 WW and LDI-1 FL C/A (Fig. 4A). These granules were indicative of inclusion body-like structures and were not detected in untransfected cells or cells expressing Gag alone. When Gag was coexpressed with LDI-1 WW, VLPs accumulated in the cytoplasm in these electron-dense inclusion body structures (Fig. 4B; n = 20 cells). The inclusion bodies varied in size and in the numbers of particles that they contained. Some contained particles on their outer edges, while others appeared to be completely filled. VLPs were not regularly detected anywhere else in the cell. In contrast, VLPs were not detected in the inclusion bodies in cells that were cotransfected with p2036-Gag and p2036-LDI-1 FL C/A (data not shown), nor were they detected elsewhere in these cells. Taken together, these results suggest that LDI-1 WW and LDI-1 FL C/A block Gag assembly and release at different stages. These experiments were repeated four times, and approximately 200 cells were examined in each experiment.
FIG. 4.
Effect of LDI-1 WW expression on RSV Gag release from 293/E cells. (A) Cells transfected with p2036-LDI-1 WW (left) or p2036-LDI-1 FL C/A (right). Magnification, ×12,000. (B) Cells cotransfected with p2036-LDI-1 WW and p2036-Gag. Magnification, ×20,000. The arrows denote the inclusion bodies found in the cytoplasm. N, nucleus.
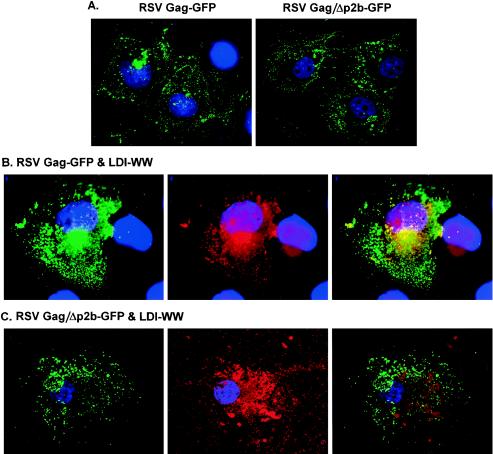
We next examined the localization of Gag and LDI-1 WW by performing confocal microscopy (Fig. 5). To do this, we transfected COS-1 cells with pGag-GFP or pGag/Δp2b-GFP alone (A, left and right panels, respectively), with p2036-LDI-1 WW and pGag-GFP (B), or with pGag/Δp2b-GFP (C). In both the absence and presence of LDI-1 WW, a subpopulation of Gag protein was found in a perinuclear region of the cell, while the rest of the Gag protein appeared to be distributed throughout the cytoplasm. Examinations of sections taken throughout the z plane of the cell indicated that the latter Gag population was actually localized at the plasma membrane (data not shown). In cells that coexpressed Gag and LDI-1 WW, the majority of LDI-1 WW was found in the perinuclear region (B, middle panel) and was colocalized with Gag (B, right panel). The colocalized perinuclear staining of Gag and LDI-1 WW was suggestive of the presence of inclusion bodies. In contrast, in cells that coexpressed LDI-1 WW and Gag/Δp2b, LDI-1 WW was distributed throughout the cytoplasm, and colocalization with Gag/Δp2b was not detected (C, right panel). This observation indicates that the association of Gag and LDI-1 WW in the perinuclear region of the cell was a direct result of L domain recruitment of LDI-1 WW to the perinuclear site and suggests that LDI-1 WW was also present in the inclusion bodies that contained Gag VLPs (Fig. 4B). It should be noted that LDI-1 WW also inhibits Gag budding in COS-1 cells (data not shown).
FIG. 5.
Confocal images of COS-1 cells expressing LDI-1 WW and RSV Gag or Gag/Δp2b. (A) Cells expressing Gag-GFP (left) or Gag/Δp2b-GFP (right) alone. (B) Cells coexpressing LDI-1 WW and Gag-GFP. (C) Cells coexpressing LDI-1 WW and Gag/Δp2b-GFP. Gag-GFP and Gag/Δp2b-GFP were visualized by green fluorescence (left panels). LDI-1 WW (red) was visualized by indirect immunofluorescence using a rabbit anti-HA primary antibody and a TRITC-tagged mouse anti-rabbit secondary antibody (middle panels). Nuclei (blue) were stained with Hoechst dye. The colocalization of Gag and LDI-1 WW was detected in the merged images by the yellow areas (right panels).
Gag is ubiquitinated in an L domain-dependent manner.
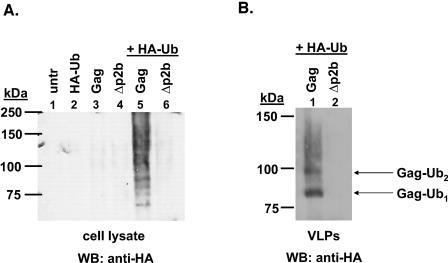
Since RSV Gag interacts with endogenous Nedd4 proteins, we wanted to determine whether Gag is ubiquitinated in 293/E cells and whether the Gag protein incorporated into VLPs is ubiquitinated. Although it was shown previously that the L domain of RSV Gag mediates the ubiquitination of mini-Gag molecules derived from HIV-1 Gag (17), this has not been determined in the context of full-length RSV Gag. To do this, we transfected 293/E cells with p2036-Gag or p2036-Gag/Δp2b and a vector expressing an HA-tagged ubiquitin molecule. HA-tagged ubiquitin was used to enhance the detection of ubiquitinated Gag with an anti-HA antibody. There was a ladder (or smear) of bands detected in the cell lysate, indicating that Gag was mono- and polyubiquitinated (Fig. 6A, lane 5). In contrast, when a Gag molecule containing an L domain deletion was used, ubiquitinated forms of Gag were not detected (lane 6). The binding of the anti-HA antibody to the bands in lane 5 represented specific ubiquitination of Gag and not cross-reactive binding of the anti-HA antibody to Gag because no bands were detected in the absence of expression of HA-tagged ubiquitin (lane 3). Additionally, the sizes of the anti-HA-reactive bands detected in lane 5 corresponded approximately to the expected masses of ubiquitinated Gag proteins.
FIG. 6.
Ubiquitination of RSV Gag in 293/E cells and released VLPs. RSV Gag or Gag/Δp2b was expressed in 293/E cells in the presence or absence of HA-Ub. (A) Gag proteins were immunoprecipitated from cell lysates with an anti-AMV antibody and were separated by SDS-PAGE. Ubiquitinated Gag proteins were detected by Western blotting with an anti-HA antibody as described in Materials and Methods. Untransfected cells, lane 1; cells transfected with pMT123, lane 2; cells transfected with p2036-Gag, lane 3; cells transfected with p2036-Gag/Δp2b, lane 4; cells transfected with p2036-Gag and pMT123, lane 5; and cells transfected with p2036-Gag/Δp2b and pMT123, lane 6. (B) VLPs were harvested from the medium fraction for the cells in lanes 5 and 6 (panel A) and were separated by SDS-PAGE. Ubiquitinated Gag proteins in the VLPs were detected by Western blotting with an anti-HA antibody (lanes 1 and 2). WB, Western blot.
When we examined VLPs released from cells (Fig. 6B), we detected two prominent bands representing ubiquitinated Gag (lane 1). In contrast to the Gag proteins in the cell lysate, the ubiquitinated Gag proteins present in VLPs (lane 1) were mainly monoubiquitinated. This was indicated by the dark band migrating at approximately 84 kDa, which is the expected molecular mass for a monoubiquitinated Gag protein. There was also another slower migrating band that may represent a Gag protein that was di-ubiquitinated or monoubiquitinated at two different residues. Together, these results indicate that Gag is ubiquitinated in the cell lysate and that ubiquitinated Gag proteins incorporated into VLPs are mainly monoubiquitinated. This suggests that the ubiquitination of Gag serves a trafficking or signaling function and is not a mechanism for the degradation of Gag. After the membrane shown in Fig. 6B was reprobed with an anti-AMV MA antibody, we estimated that <10% of the Gag protein incorporated into VLPs was ubiquitinated (data not shown), similar to what has been described for HIV-1 and MLV (12, 13).
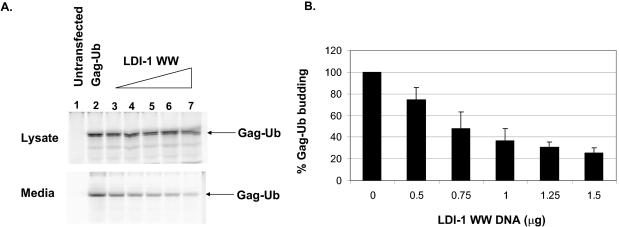
Gag-Ub budding is inhibited by overexpression of LDI-1 WW.
Since RSV Gag interacts with Nedd4 ubiquitin ligases and is ubiquitinated, we wanted to determine whether the sole function of the interaction of Gag and Nedd4 proteins during budding is to ubiquitinate Gag. Therefore, we tested whether the dominant-negative mutant LDI-1 WW would still inhibit Gag release if a ubiquitin molecule were fused to the C terminus of Gag (Gag-Ub). We found that, similar to the case for Gag, the budding of Gag-Ub was inhibited by the overexpression of LDI-1 WW (Fig. 7). The expression level of Gag-Ub was relatively constant in the lysate, while the level of Gag-Ub detected in the medium decreased in a dose-dependent fashion as the amount of p2036-LDI-1 WW increased (Fig. 7A). Quantitation of the intensities of the Gag-Ub bands in the medium and lysate fractions indicated that the budding of Gag-Ub decreased at least 75% in the presence of LDI-1 WW (Fig. 7B). This is similar to the level of inhibition of Gag budding in the presence of LDI-1 WW (9).
FIG. 7.
Effect of overexpression of LDI-1 WW on release of RSV Gag-Ub from 293/E cells. (A) RSV Gag-Ub was expressed in 293/E cells in the absence or presence of increasing amounts of p2036-LDI-1 WW. The amounts of Gag in the lysate and medium fractions were determined as described in Materials and Methods. Lane 1, untransfected cells; lane 2, cells transfected with 0.5 μg of p2036-Gag-Ub; lanes 3 to 7, cells transfected with 0.5 μg of p2036-Gag-Ub and with p2036-LDI-1 WW (0.5 μg, lane 3; 0.75 μg, lane 4; 1.0 μg, lane 5; 1.25 μg, lane 6; 1.5 μg, lane 7). (B) Graphical representation of the relative amounts of Gag-Ub released into the medium in the presence of LDI-1 WW (derived from two independent experiments).
Gag-Ub VLPs are not detected in inclusion bodies in cells expressing LDI-1 WW.
To attempt to better understand the Gag-Ub budding defect caused by LDI-1 WW, we examined cells that were transfected with p2036-Gag-Ub and p2036-LDI-1 WW by thin-section electron microscopy. Similar to the case for cells transfected with p2036-Gag and p2036-LDI-1 WW, electron-dense inclusion bodies were detected in the cytoplasm of cells expressing Gag-Ub and LDI-1 WW (data not shown). However, in contrast to the results shown in Fig. 4B, VLPs were not detected in any of the observed inclusion bodies. Furthermore, VLPs could not be detected anywhere else in the cells, suggesting that the ubiquitin molecule fused to the C terminus of Gag altered the assembly process. Although Gag-Ub VLPs were not detected in these cells, we believe that this was not due to an expression defect or stability problem of Gag-Ub. The Gag-Ub construct used in this study was as stable as wild-type Gag and exhibited a similar budding efficiency (compare the results in Fig. 3A and C, lane 2, to that in Fig. 7A, lane 2). The microscopy experiment was repeated three times, and approximately 200 cells were examined in each experiment.
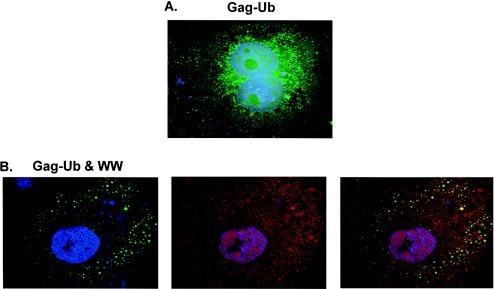
It was unclear whether Gag-Ub was present in the inclusion bodies and unable to form VLPs or was absent from these structures. To try to clarify this problem, we examined cells that were transfected with p2036-Gag-Ub and p2036-LDI-1 WW by using confocal microscopy (Fig. 8). Similar to Gag, Gag-Ub appeared to be distributed throughout the cytoplasm when it was expressed alone (Fig. 8A). In addition, examinations of sections taken throughout the z plane of the cells indicated that most of the Gag-Ub protein was located at the cell periphery on the plasma membrane (data not shown). However, in contrast to Gag, whose overall cytoplasmic distribution was not extensively perturbed by coexpression with LDI-1 WW, the distribution of peripherally located Gag-Ub was significantly altered (B, left panel). Instead of the fine punctate distribution observed when Gag-Ub was expressed alone, the peripheral population of Gag-Ub was dispersed in many larger vesicle-like structures in the presence of LDI-1 WW (B, left panel). The fluorescence signal of LDI-1 WW was also broadly dispersed, and although some of it was located in the vesicles containing Gag-Ub, colocalization of the two proteins was not apparent (B, right panel). Moreover, 90% of the cells lacked the perinuclear subpopulation of Gag-Ub or LDI-1 WW (Fig. 8B; n = 15 cells) that was seen with Gag and LDI-1 WW (Fig. 5). Interestingly, in the few cases in which Gag-Ub was detected in the perinuclear region of the cell (data not shown) (n = 1 to 2 cells), LDI-1 WW was also observed near the nucleus and the two proteins were colocalized. These results suggest that Gag-Ub does not have the same perinuclear localization as Gag and that the mechanism by which LDI-1 WW blocks Gag-Ub assembly and release is different from the way it inhibits Gag assembly and release.
FIG. 8.
Confocal images of COS-1 cells expressing RSV Gag-Ub and LDI-1 WW. (A) Cells expressing Gag-Ub alone. (B) Cells coexpressing Gag-Ub and LDI-1 WW. Gag-Ub (green) was visualized by indirect immunofluorescence using a mouse anti-AMV MA primary antibody and a FITC-tagged rabbit anti-mouse secondary antibody. HA-tagged LDI-1 WW (red) was also detected by indirect immunofluorescence using a rabbit anti-HA primary antibody and a TRITC-tagged mouse anti-rabbit secondary antibody. Nuclei (blue) were stained with Hoechst dye. A merged image of Gag-Ub and LDI-1 WW is shown in the right panel (B).
DISCUSSION
The results of this study demonstrate that the interaction between Nedd4 proteins and RSV Gag is dependent upon the L domain found within the p2b region of Gag. Previous work from our laboratory (9) has shown that RSV Gag interacts with endogenous Nedd4 proteins, and we have now determined that this interaction is dependent solely on the L domain sequence within full-length Gag. Furthermore, we have now shown that the ubiquitination of Gag is dependent upon the presence of the L domain. Together, these findings suggest that the ubiquitination of Gag is mediated by the Nedd4 family of ubiquitin ligases. With the isolation of a full-length clone of LDI-1 which contains a HECT domain at its C terminus, we have confirmed the identity of LDI-1 as a chicken Nedd4 protein and have shown that Nedd4 proteins have a role in RSV budding.
The overexpression of certain Nedd4 family members has previously been shown to lead to an enhancement of MPMV and Ebola VLP release (25, 26). However, we did not detect an increase in the level of RSV Gag release from 293/E cells after the overexpression of LDI-1 FL. It is possible that Nedd4 proteins are not limiting in the budding process in these cells, thereby explaining why the overexpression of LDI-1 FL did not enhance the level of Gag release. It is also possible that although Nedd4 proteins are required during RSV budding, we have not identified the specific Nedd4 protein that is involved in Gag release. However, a mutation of the conserved cysteine residue in the catalytic HECT domain of LDI-1 FL led to a severe decrease in Gag budding. The release defect caused by overexpression of the HECT domain mutant (LDI-1 FL C/A) was similar to the effect caused by the overexpression of LDI-1 or LDI-1 WW (9). This result clearly demonstrates the requirement of ubiquitin ligase activity for RSV release. We have also determined that LDI-1 FL and LDI-1 FL C/A were expressed to similar levels in 293/E cells, as estimated by the appearance of comparable numbers of inclusion bodies by electron microscopy and by Western blot analyses of 293/E cell lysates (data not shown).
We believe that our results, in combination with the findings of others, suggest that Nedd4 proteins have multiple roles in RSV budding. At least one of the roles of Nedd4 family members in budding is to mediate the ubiquitination of Gag. We estimate that <10% of the Gag molecules found in VLPs are ubiquitinated. This suggests that not every Gag molecule needs to be ubiquitinated for budding to occur, with the one caveat that ubiquitin may have been removed from some Gag proteins by a ubiquitin hydrolase. Patnaik et al. demonstrated that RSV Gag release is sensitive to proteasome inhibitor treatment. The block in budding due to proteasome inhibitor treatment was partially rescued by the fusion of ubiquitin to the C terminus of Gag. More specifically, it was demonstrated previously that the fusion of ubiquitin to the C terminus of Gag rescued budding only when the L domain sequence was also present within Gag (14). It should be noted that the Gag-Ub construct used for this study was the same one used by Patnaik and coworkers and was shown to be as stable as wild-type Gag and to have a similar budding efficiency (14). These results suggest that in addition to ubiquitination, the Nedd4 binding site (L domain) within Gag needs to be accessible to mediate VLP release. Similarly, we found that the budding of Gag-Ub was inhibited by LDI-1 WW, further suggesting that Nedd4 has functions during Gag release in addition to the ubiquitination of Gag.
The observations that VLPs were present in inclusion bodies in cells expressing Gag and LDI-1 WW, but not in cells expressing Gag and LDI-1 FL C/A or Gag-Ub and LDI-1 WW, are interesting. These findings suggest that when Gag interacts with LDI-1 WW and becomes trapped in inclusion bodies, it is still able to form recognizable VLPs. The colocalization of LDI-1 WW and Gag, but not Gag/Δp2b, further supports the conclusion that both LDI-1 WW and Gag are present in these inclusion bodies. The fluorescence microscopy results also indicate that Gag modulates the organization or distribution of LDI-1 WW. The structures detected by confocal microscopy that contained Gag-Ub and LDI-1 WW were not detected when LDI-1 WW was coexpressed with Gag. It is therefore possible that the ubiquitin moiety on the C terminus of Gag interfered with the transport of Gag-Ub and LDI-1 WW to specific sites on the plasma membrane or to the perinuclear region, where some event necessary for VLP assembly may occur. Although we have not shown that the effect of LDI-1 WW on Gag-Ub localization is mediated through an interaction with the L domain, we speculate that this is the case because of the results presented in Fig. 2.
Further experiments will be needed to determine whether Gag is present in the inclusion bodies found in cells expressing Gag and LDI-1 FL C/A. It is likely that LDI-1 FL C/A is present in the inclusion bodies because these structures formed in cells that were transfected with LDI-1 FL C/A alone and because immunofluorescence studies indicated that LDI-1 WW was recruited to the perinuclear location by the L domain within Gag. In addition, confocal microscopy experiments are under way to determine the localization of Gag in the presence of LDI-1 FL C/A. If Gag is present in these granules, it will suggest that the full-length Nedd4 protein inhibits Gag from forming VLPs, while the truncated version (LDI-1 WW) does not. It is possible that LDI-1 FL C/A is able to interact with other proteins that might be involved in the budding process and thereby lead to the formation of a larger protein complex in the inclusion bodies. An aggregated protein complex may be more likely to inhibit Gag VLP formation than LDI-1 WW. Alternatively, proteins recruited by the full-length Nedd4 protein might impose temporal or spatial controls on the site where the budding complex is formed, preventing assembly within the inclusion bodies. Cells expressing LDI-1 FL also contained granular inclusion body-like structures (data not shown) similar to those in cells expressing LDI-1 WW or LDI-1 FL C/A. It is possible that the small decrease in Gag budding caused by the overexpression of LDI-1 FL was indirectly related to the formation of these inclusion bodies.
Accumulating evidence suggests that retroviruses make use of the cellular protein-sorting pathway leading to the multivesicular body (MVB) to bud particles from infected cells. The L domain of HIV-1 Gag binds to Tsg101, which is a component of the ESCRT-1 endosomal protein sorting complex and is involved in targeting monoubiquitinated proteins to the MVB (2, 3, 7, 10, 20). Furthermore, mutants of the Vps4 ATPase inhibit the release of HIV-1, MLV, and RSV, suggesting that these viruses all take advantage of proteins associated with the MVB pathway to exit the cell (3, 21, 27). We speculate that RSV Gag is ubiquitinated by Nedd4 proteins in order to target an association with proteins of the MVB sorting pathway. This hypothesis is supported by the fact that the Saccharomyces cerevisiae Nedd4 homolog, Rsp5, is required for the ubiquitination and sorting of proteins into the MVB pathway in yeast (8, 11). We believe that the data presented in this paper indicate that during the budding process, Nedd4 proteins are needed for functions in addition to mediating Gag ubiquitination. For example, the Nedd4 proteins may act as ligands for members of the ESCRTI-III complex to promote assembly and trafficking of the budding complex. It is also possible that Nedd4 proteins are needed to ubiquitinate other proteins in the MVB pathway (15). It will be interesting to elucidate the steps in the RSV budding pathway between the required Nedd4 and Vps4 functions.
Acknowledgments
This work was supported by U.S. Public Health Service grants CA52047 (to J.L.) and GM 48294 (to C.C.) and by the Cancer Biology Fellowship Program, Chicago Baseball Cancer Charities, from the Robert H. Lurie Comprehensive Cancer Center (to M.L.V.).
We thank John Wills for the Gag-Ub and Gag-GFP vectors. We thank Stephen T. Oh for critically reading the manuscript.
REFERENCES
- 1.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 4.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 8.Katzmann, D. J., S. Sarkar, T. Chu, A. Audhya, and S. D. Emr. 2004. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell 15:468-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 11.Morvan, J., M. Froissard, R. Haguenauer-Tsapis, and D. Urban-Grimal. 2004. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic 5:383-392. [DOI] [PubMed] [Google Scholar]
- 12.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 13.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polo, S., S. Sigismund, M. Faretta, M. Guidi, M. R. Capua, G. Bossi, H. Chen, P. De Camilli, and P. P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451-455. [DOI] [PubMed] [Google Scholar]
- 16.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 17.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 19.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 20.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 24.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y., and J. Leis. Unpublished data.