Abstract
RNA interference, a natural biological phenomenon mediated by small interfering RNAs (siRNAs), has been demonstrated in recent studies to be an effective strategy against human immunodeficiency virus type 1 (HIV-1). In the present study, we used 21-bp chemically synthesized siRNA duplexes whose sequences were derived from the gp41 gene, nef, tat, and rev regions of viral RNA. These sequences are conserved in select neurotropic strains of HIV-1 (JR-FL, JR-CSF, and YU-2). The designed siRNAs exerted a potent antiviral effect on these HIV-1 strains. The antiviral effect was mediated at the RNA level (as observed by the down-regulation of the HIV-1-specific spliced transcript generating a 1.2-kbp reverse transcription [RT]-PCR product) as well as viral assembly on the cell membrane. Spliced transcripts (apart from the most abundant transcript generating a 1.2-kbp RT-PCR product) arising from an unspliced precursor likely contributed, albeit to a lesser extent, to the antiviral effect. The resultant progeny viruses had infectivities similar to that of input virus. We therefore conclude that these siRNAs interfere with the processing of the unspliced transcripts for the gp41 gene, tat, rev, and nef, eventually affecting viral assembly and leading to the overall inhibition of viral production. Apart from using the gp41 gene as a target, the conservation of each of these targets in the above-mentioned viral strains, as well as several primary isolates, would enable these siRNAs to be used as potent antiviral tools for investigations with cells derived from the central nervous system in order to evaluate their therapeutic potential and assess their utility in inhibiting HIV-1 neuropathogenesis and neuroinvasion.
Vaccines and drugs have thus far been the method of choice for combating viral infections. These agents typically work by inhibiting the function of crucial viral proteins. RNA interference (RNAi) is a newly described natural biological phenomenon mediated by small interfering siRNAs (siRNAs), which target viral mRNAs, rather than the proteins that they encode, for degradation by cellular enzymes. The potential of siRNAs to treat or prevent disease has yet to be proven. However, several proof-of-concept studies have demonstrated the usefulness of siRNAs in controlling a diverse group of human viruses and pathogens (15, 16, 24, 25, 32-35, 38, 40, 50, 51, 53, 55, 70). These data suggest that siRNAs may be widely used as alternate therapeutic agents. RNAi does work effectively as an antiviral agent in cell cultures (25, 33, 50). The emergence of siRNAs as a powerful tool for posttranscriptional gene silencing has given credence to the notion of using siRNAs for certain therapies. In addition, several innovative methods of delivering siRNAs to a wide variety of primary cells are now available (7, 18, 71). These developments are indicative of an increasingly stronger role for siRNAs as a potential therapeutic strategy.
Human immunodeficiency virus (HIV) type 1 (HIV-1) infection is a worldwide disease that requires alternate therapeutic strategies. Despite significant advances, current treatments are limited by toxicity, complexity, cost, and resistance. The last issue is the most critical one, as partial virus suppression allows for the evolution of resistant viruses and the creation of virus reservoirs unaffected by therapeutic agents. The emergence of resistant viruses reduces drug activity and limits future treatment options (48). Of note, HIV-1-specific siRNAs exert potent antiviral effects in a variety of cell culture systems (33, 50). siRNAs containing cognate sequences present within different regions of the HIV-1 genome inhibit infection by specifically degrading genomic HIV-1 RNA, thereby preventing the formation of viral cDNA intermediates (33). HIV-1-specific siRNAs can inhibit infection in permanent cell lines, primary CD4+ T cells, and macrophages (60). The viral life cycle is inhibited after fusion and before reverse transcription (RT) or during the transcription of viral RNA from integrated provirus.
Two main approaches have emerged for siRNA-mediated inhibition of HIV-1 infection. Both viral proteins (15, 16, 33, 38, 50) and the cellular coreceptors (4, 13, 44, 54, 74) used by HIV-1 have been targeted for down-regulation by siRNAs. There are certain perceived shortcomings to both approaches. Targeting viral proteins has been shown to generate escape mutants (10, 17). A combinatorial approach involving targeting several regions of the viral genome may ameliorate the generation of escape mutants, although this theory has not been proven. Chemokines and their receptors play a critical role in host immune surveillance. siRNA-mediated down-regulation of chemokine receptors may compromise important immune functions (23, 75). A series of studies with the mouse nervous system have demonstrated the crucial role played by the chemokine receptor CCR5 in preventing disease progression during the acute and chronic stages of infection of mice with hepatitis virus, which replicates in glial cells and neurons during the acute stage (26-29). Currently, we can only speculate on which approach will become the method of choice (53).
siRNA duplexes targeting the essential Tat and Rev regulatory proteins encoded by HIV-1 can specifically block their expression and function. These same siRNAs can effectively inhibit HIV-1 gene expression and replication in cell cultures, including human T-cell lines and primary CD4+ T lymphocytes (16). siRNAs have also been successfully targeted to block the HIV-1 p24 core antigen (15), gp120 (52), and vif, nef, and the HIV-1 long terminal repeat (LTR) (33). siRNAs against chemokine receptors effectively eliminated their cell surface expression and thereby prevented HIV-1 from entering target cells. siRNAs mediated the silencing of the chemokine receptor genes specifically without influencing CD4 expression. The suppression of HIV-1 coreceptor expression effectively blocked the acute infection of CXCR4+ or CCR5+ U87-CD4+ cells by X4 (NL4-3) or R5 (BaL) HIV-1 strains (44). Of note, in that study (44), viral growth in controls was extremely high, suggesting an unusual level of viral replication. A similar study was conducted with human peripheral blood CD4+ T lymphocytes. A lentivirus-based vector was used to introduce siRNA against CCR5. Substantial protection of lymphocyte populations from CCR5-tropic HIV-1 infection was achieved (54). In another study, Zhou et al. demonstrated selective inhibition of CXCR4-tropic cell-free virus infection of human cells by using siRNA targeting the CXCR4 coreceptor (74).
As noted above, the antiviral effects of HIV-1-specific siRNAs were demonstrated by several research groups (4, 13, 15, 16, 33, 38, 44, 50, 54, 74). However, the targets that were selected in previous studies are not conserved among neurotropic strains of HIV-1. Primary isolates of HIV-1 that utilize the CCR5 coreceptor and that possess a non-syncytium-inducing (NSI) phenotype are widely perceived as neurotropic (59). However, some studies have indicated that microglial tropism is distinct from that for monocyte-derived macrophages (63) and has a decreased dependence on CCR5/CD4 (30).
In the current study, nine siRNAs were designed to contain cognate sequences present within the coding regions for the gp41 gene, tat, rev, and nef. Of these nine siRNAs, three siRNAs, two of which are derived from the gp41 gene and one of which is derived from nef, have been studied in detail. We designed our HIV-1-specific siRNAs by ensuring that the sequences are conserved among viral isolates that meet the criteria which define a neurotropic strain (R5 tropic and NSI [R5/NSI]). The strains of HIV-1 selected for this investigation, apart from possessing an R5/NSI phenotype, were isolated from brain tissues or cerebrospinal fluid. In addition to being conserved among select neurotropic strains, the cognate sequences of these siRNAs are also relatively well conserved among several primary isolates. A novel aspect of this study was the demonstration of an antiviral effect of the siRNA derived from the HIV-1 gp41 gene. These HIV-1-specific siRNAs targeting transcripts for the gp41 gene, tat, rev, and nef will degrade both unspliced and spliced transcripts in these regions of the HIV-1 genome (21, 67). The present study enabled us to determine whether these siRNAs can be used as antiviral tools for investigations with cells derived from the central nervous system (CNS) in order to evaluate their therapeutic potential and assess their utility in inhibiting HIV-1 neuropathogenesis and neuroinvasion.
MATERIALS AND METHODS
Cell lines.
The HeLaCD4-LTR-β-Gal cell line was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (catalog number 1470). HeLaCD4-LTR-β-Gal, HeLaCD4, and 293T cell lines were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 2 mM l-glutamine, 50 U of penicillin G/ml, and 50 μg of streptomycin/ml. Additionally, the culture medium for the HeLaCD4-LTR-β-Gal cell line was supplemented with 0.2 mg of G418/ml and 0.1 mg of hygromycin B/ml. For HeLaCD4 cells, the culture medium was supplemented with 0.5 mg of G418/ml. Human monocytes were isolated by positive selection of CD11b+ cells from peripheral blood mononuclear cells obtained from healthy, HIV-1-seronegative donors. CD11b magnetic beads (Miltenyi Biotec) were used to identify the positive population, which was subsequently isolated by Automacs (Miltenyi Biotec). Monocytes (5 × 106 cells/well in six-well plates) were allowed to adhere and differentiate into monocyte-derived macrophages (MDMs) for 7 to 10 days prior to infection with cell-free HIV-1. Monocytes were differentiated in DMEM containing 10% heat-inactivated FBS, 10% heat-inactivated horse serum, 2 mM l-glutamine, 50 U of penicillin G/ml, 50 μg of streptomycin/ml, 0.5 ng of granulocyte-macrophage colony-stimulating factor/ml, and 0.5 ng of macrophage colony-stimulating factor/ml.
Plasmid constructs.
Plasmids pNL4-3, pYU-2, and pYK-JR-CSF, encoding HIV-1 clones NL4-3, YU-2, and JR-CSF, respectively, were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (catalog numbers 114, 1350, and 2708, respectively) and have been described in detail elsewhere (2, 14, 42).
Viral stocks.
HIV-1 NL4-3 stocks were prepared by transfecting 293T cells with pNL4-3. Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer's instructions. Essentially, lipid complexes were generated by mixing pNL4-3 and Lipofectamine 2000 in Optimem-I reduced serum medium (Invitrogen). 293T cells in Optimem-I (70 to 90% confluent) on poly-d-lysine-coated plates (Becton Dickinson) were incubated with lipid complexes for 5 h. The medium was changed to Optimem-I containing 10% heat-inactivated FBS. At 48 h posttransfection, the supernatant containing viral particles was harvested, clarified of cellular debris by centrifugation at 10,000 × g for 10 min, and filtered through 0.22-μm-pore-size polyvinylidene difluoride membranes. Viral titers were determined by the p24 antigen enzyme-linked immunosorbent assay (ELISA) with an Alliance HIV-1 p24 ELISA kit (Perkin-Elmer).
siRNA selection and preparation.
Web-based tools at the Ambion, Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology, and National Center for Biotechnology Information websites were used for the selection of siRNA sequences and for BLAST searches. Apart from the Web-based tools, Jellyfish 3.0 (LabVelocity) was used to establish conservation among select primary viral isolates obtained from CNS-based cells or cerebrospinal fluid.
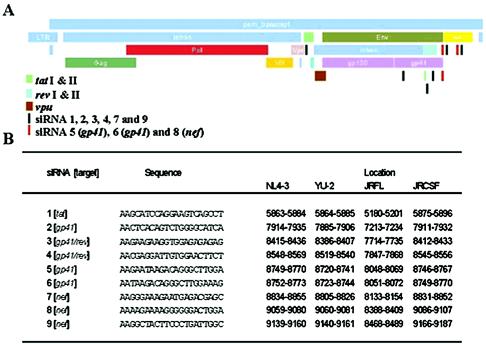
siRNA targets were selected from within the region of the HIV-1 NL4-3 genome containing the gp41 gene, tat, rev, and nef. Twenty-one-base-pair stretches downstream of AA repeats and with a GC content of 40 to 50% were initially selected (20, 31). siRNA sequences were BLAST searched either in the expressed sequence tag (EST)-human database or databases containing viral nucleic acid sequences. In either database, BLAST searches were for “short nearly exact matches. ” Sequences that were conserved among select neurotropic strains, YU-2, JR-CSF, and JR-FL, were BLAST searched against the EST-human database to ensure the absence of homology to human transcripts. Sequences thus selected were further BLAST searched against primary isolates of HIV-1. Nine sequences showing maximal conservation among a wide variety of primary isolates were tested for their antiviral efficacy (Fig. 1 and 2).
FIG. 1.
HIV-1-specific siRNA sequences and their relative positions in select neurotropic strains. Twenty-one-base-pair siRNA duplexes were selected from within a region of the HIV-1 NL4-3 transcripts for the gp41 gene, tat, rev, and nef. (A) The positions of the siRNA cognate sequences in HIV-1 NL4-3 are shown by narrow rectangular bars (red bars indicate the positions of the three most potent siRNAs). These siRNA cognate sequences are conserved in NL4-3 and select neurotropic strains of HIV-1. (B) Relative positions of these sequences in NL4-3, YU-2, JR-FL, and JR-CSF. These sequences are also well conserved in a wide variety of primary isolates but lack significant homologies in the EST-human database.
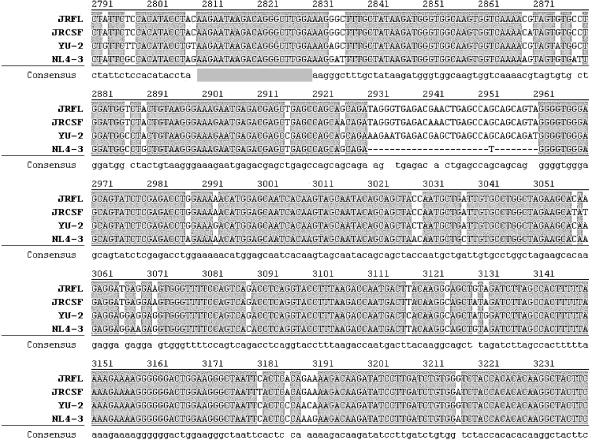
FIG. 2.
Conservation of target sequences for the most potent siRNAs (5, 6, and 8) in select neurotropic strains of HIV-1 and NL4-3. Shown is an alignment of the select neurotropic strains in the region where the cognate sequences for siRNAs 5, 6, and 8 are present. Viral sequences are numbered with reference to the sense primer (HIV-5964-5983-F) for the NL4-3 genome and are from within a region of the transcripts for the gp41 gene, tat, rev, and nef. The primer nomenclature indicates the location of the sequence in the NL4-3 genome.
Chemically synthesized siRNAs were used (Dharmacon). siRNA duplexes with 3′ AA overhangs were prepared, desalted, purified, and 2′ deprotected. siRNA duplexes (20 μM) were stored in 6 mM HEPES-KOH buffer (pH 7.5) containing 20 mM KCl and 0.2 mM MgCl2 at −20°C. Scrambled (negative control) siRNA (Ambion) was used to ensure the specificity of the observed effects. This negative control siRNA lacks any sequence homology to transcripts in the human genome.
Antiviral assays.
HeLaCD4 cells were transfected with chemically synthesized siRNA duplexes and subsequently infected with HIV-1 NL4-3. To each well of a six-well plate were added 0.6 × 106 cells in DMEM containing 10% heat-inactivated FBS and 2 mM l-glutamine. Approximately 18 h after plating, when the cells had reached 30% confluence, siRNA (10 pmol/well)-siPort lipid (Ambion) complexes in Optimem-I were added for transfection for 5 h. Lipid complexes were generated according to the manufacturer's instructions. Subsequently, HeLaCD4 cells were infected with HIV-1 NL4-3 (multiplicity of infection [MOI], 0.1 pg of p24/cell) for 2 h. At the end of the 2-h period, cells were washed with phosphate-buffered saline to remove input virus. Cells were cultured in Optimem-I without any supplements. HIV-1 p24 antigen levels in cell extracts and culture supernatants were measured by an ELISA. The infectivity of progeny virions in culture supernatants was determined by multinuclear activation in a galactosidase indicator assay (MAGI assay) with a HeLaCD4-LTR-β-Gal cell line as described previously (36).
293T cells and HeLaCD4 cells were used in cotransfection experiments, wherein the antiviral efficacy of the siRNA was tested against a range of plasmid DNAs for NL4-3, YU-2, and JR-CSF. To each well of a six-well plate were added 106 cells in DMEM containing 10% heat-inactivated FBS and 2 mM l-glutamine. Approximately 18 h after plating, when the cells had reached at least 50% confluence, cells were transfected with siRNA and plasmid DNA was complexed with Lipofectamine 2000 (for 293T cells) or siPort lipid (for HeLaCD4 cells). In either setting, lipid complexes in Optimem-I were generated according to the manufacturer's instructions. Cells were incubated in the same medium during transfection. For each of the transfections, 10 pmol of each of three different siRNA duplexes (5, 6, and 8) was used along with plasmid DNAs (62.5 to 1,000 ng) for the above-mentioned HIV-1 strains. At the end of a 5-h transfection period, the culture medium was replaced with Optimem-I containing 10% heat-inactivated FBS. HIV-1 p24 antigen levels in culture supernatants were measured by an ELISA.
MDMs were transfected with chemically synthesized siRNA duplexes and subsequently infected with HIV-1 NL4-3. At the time of transfection, macrophages were 50% confluent. For each of the transfections, 10 pmol of each of three different siRNA duplexes (5, 6, and 8) was used. siRNA-siPort lipid complexes in Optimem-I were generated according to the manufacturer's instructions. MDMs were incubated in Optimem-I for the duration of transfection and infection. MDMs were transfected with siRNA-lipid complexes for 5 h. Subsequently, they were infected with HIV-1 YU-2 (MOI, 0.1 pg of p24/cell) for 2 h. At the end of the 2-h period, cells were washed with phosphate-buffered saline to remove input virus. Cells were cultured in DMEM containing 10% heat-inactivated FBS. HIV-1 p24 antigen levels in culture supernatants were measured by an ELISA at 2 and 9 days postinfection (dpi).
RT-PCR and Southern blotting.
Total RNA was extracted from HeLaCD4 cells (transfected with siRNA and infected with HIV-1) at 2 dpi with an RNeasy minikit (Qiagen). RNA was incubated with DNase I (amplification grade; Invitrogen) to remove residual genomic DNA. RT was performed by SuperScript one-step RT-PCR with Platinum Taq (Invitrogen) and intron-spanning primers for HIV-1 NL4-3 to detect a spliced product(s) resulting from processing of the transcripts for the gp41 gene, tat, rev, and nef. The sense-antisense primer pair was HIV-5964-5983-F (5′-CTCCTATGGCAGGAAGAAGC-3′) and HIV-9486-9505-R (5′-TATATGCAGCATCTGAGGGC-3′); the numbers indicate the location of the sequence (or its reverse complement) in the NL4-3 genome. For RT of β-globin transcripts, previously described primer pair sets were used (56). Reaction products were analyzed on 1% agarose gels. RT-PCR products were blotted onto Hybond N (Amersham) nylon membranes according to the manufacturer's instructions. Three internal probes corresponding to target sequences for siRNAs 5, 6, and 8 were used (Fig. 1). Gene Images AlkPhos direct labeling and a CDP-Star detection system (Amersham) were used to label probes (directly with alkaline phosphatase) and for chemiluminescence detection of hybridized probes.
Statistical analysis.
Statistically significant differences between control and test groups were determined by Student's t tests. P values of ≤0.05 were considered significant.
RESULTS
HIV-1-specific siRNAs inhibit HIV-1 NL4-3 infection.
To test whether the designed HIV-1-specific siRNAs (Fig. 1) exerted an antiviral effect, we transfected HeLaCD4 cells with these moieties. The transfected cells then were infected with HIV-1 NL4-3 (MOI, 0.1 pg of p24/cell), and the HIV-1 p24 antigen levels in culture supernatants were measured. On day 1, siRNAs 1, 3, and 6 exerted a statistically significant antiviral effect (P ≤ 0.003). In HeLaCD4 cells transfected with siRNA 6, HIV-1 p24 antigen levels were 49.5% those in the control (HeLaCD4 cells infected with HIV-1 NL4-3) (Fig. 3). On day 2, all nine siRNAs exerted an antiviral effect ranging from 66.18 to 24.16% that seen in the control. The most potent effect was exerted by siRNA 5 (31.55% that seen in the control), siRNA 6 (24.17% that seen in the control), and siRNA 8 (37.04% that seen in the control) (P ≤ 0.001). There was a significant decrease in the antiviral effect from day 1 to day 2 for siRNAs 1, 3, and 4 (P ≤ 0.001). siRNAs whose cognate sequence was exclusively from within the gp41 coding region did not show this decrease in the antiviral effect. This result is in contrast to the results obtained with siRNAs 3 and 4, the cognate sequences of which overlap in the gp41 and rev coding regions. As noted above, both siRNAs 3 and 4 exhibited a gradual decrease in the antiviral effect. Hence, from our initial screening experiments, four of the nine siRNAs (2, 5, 6 and 8) seemed to be candidates for further analysis. siRNAs 2 and 8 did not exert a statistically significant antiviral effect at 2 dpi (P = 0.462). Among siRNAs 2, 5, and 6, the cognate sequences of which are within the gp41 coding region, siRNAs 5 and 6 had a stronger effect than siRNA 2. In addition, siRNAs 5, 6, and 8 continued to exert superior antiviral effects (20.46, 20.87, and 33.19% that seen in the control, respectively) compared with siRNA 2 (data not shown) at 3 dpi (P ≤ 0.001). Thus, we chose siRNAs 5 (gp41 gene), 6 (gp41 gene), and 8 (nef) for further investigations.
FIG. 3.
Antiviral efficacy of HIV-1-specific siRNAs for HIV-1 NL4-3 infection in HeLaCD4 cells. HeLaCD4 cells were transfected with siRNAs and then infected with HIV-1 NL4-3 (MOI, 0.1 pg of p24/cell). HIV-1 p24 antigen levels in culture supernatants were measured at 1 and 2 dpi. The means of three experiments and the standard deviations are indicated. siRNAs 5, 6, and 8 exerted the most potent inhibition of NL4-3 infection.
HIV-1-specific siRNAs induce inefficient viral assembly.
The HIV-1-specific siRNAs appeared to inhibit viral replication by affecting viral assembly (and/or release) at the cellular membrane. An experiment similar to that described above (Fig. 3) was performed, except that HIV-1 p24 antigen levels in both cell extracts and culture supernatants were measured from 1 to 3 dpi (Fig. 4). When cell-free (virion-associated) p24 antigen is expressed as a percentage of intracellular p24 antigen, the result essentially is a reflection of the efficiency of viral production. For the control infection, with an ongoing duration of infection, viral production became increasingly inefficient. However, with each of the HIV-1-specific siRNAs, viral production was found to be most inefficient at 2 dpi. Significantly, viral production became relatively more efficient (P ≤ 0.008) at 3 dpi. This type of increase was not observed either in the control infection (P = 0.078) or when cells were transfected with nonspecific, scrambled siRNA (P = 0.305). The inefficiency of viral production seemed to be correlated with the time at which most siRNAs would exert maximal interference in such transient assays. The inefficient viral production at 2 dpi likely was the result of altered stoichiometries of viral components (at least some of which are encoded by the transcripts for the gp41 gene, tat, rev, and nef) at the cell membrane.
FIG. 4.
HIV-1-specific siRNAs affect viral assembly. Intracellular levels of HIV-1 p24 antigen are not down-regulated in a significant manner. When extracellular p24 antigen levels (progeny virion-associated p24) are analyzed as a percentage of intracellular p24 antigen levels, the efficiency of viral production can be assessed. Infection in the absence of HIV-1-specific siRNAs becomes increasingly inefficient. However, infection in cells transfected with HIV-1-specific siRNAs exhibits maximal inefficiency at 2 dpi and then increases in a statistically significant manner. The inefficiency of viral production seems to be correlated temporally with maximal interference by siRNAs. Bars: NL4-3, HeLaCD4 cells not transfected with siRNA but infected with HIV-1 NL4-3; Scrambled, HeLaCD4 cells transfected with a nonspecific siRNA and infected with HIV-1 NL4-3; 5, 6, and 8, HeLaCD4 cells transfected with HIV-1-specific siRNAs 5, 6, and 8, respectively, and infected with HIV-1 NL4-3; Multiplexed, HeLaCD4 cells transfected simultaneously with HIV-1-specific siRNAs 5, 6, and 8 and infected with HIV-1 NL4-3. The means of three experiments and the standard deviations are indicated.
HIV-1-specific siRNAs do not alter the infectivity of progeny virions.
As noted above, the cognate sequences for at least siRNAs 5 and 6 are within the gp41 coding region. Hence, there was a possibility that progeny virions could have reduced infectivity. Supernatants (at 2 dpi) from the experiment described in Fig. 4 were tested in a MAGI assay (Fig. 5). In all of the samples, the infectivity of the progeny virions varied from 0.7 to 3.14 infectious units per pg of HIV-1 p24 antigen. These values were statistically similar to the infectivity of the input virus (0.93 infectious unit per pg of HIV-1 p24 antigen) (P = 0.433). It is likely that viral particles with aberrant stoichiometries of viral components failed to assemble and bud from the cell; this scenario would be one of the mechanisms for reduced viral production from cells possessing the above-described siRNAs.
FIG. 5.
Infectivity of progeny virions generated from HeLaCD4 cells treated with HIV-1-specific siRNAs. Supernatants containing progeny virions generated from HeLaCD4 cells transfected with HIV-1-specific siRNAs at 2 dpi were tested to determine the infectivity of the viral particles in a MAGI assay. HeLaCD4-LTR-β-Gal indicator cells were stained for β-galactosidase enzyme activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The data represent the infectivity of progeny virions generated from HeLaCD4 cells transfected with no siRNA (N/2dpi), scrambled (negative control) siRNA (S/2dpi), siRNA 5 (5/2dpi), siRNA 6 (6/2dpi), siRNA 8 (8/2dpi), and siRNAs 5, 6, and 8 (multiplexed) (M/2dpi). I.U., infectious units. The means of three experiments and the standard deviations are indicated.
HIV-1-specific siRNAs down-regulate the major spliced transcript.
Alterations in the stoichiometries of viral components likely are the result of the siRNAs tested here clearing the spliced transcripts arising from the unspliced transcripts for the gp41 gene, nef, tat, and rev. To investigate this possibility, we performed RT-PCR of total RNA obtained from HeLaCD4 cells transfected with siRNAs 5, 6, and 8 and infected with HIV-1 NL4-3 (Fig. 6). The major spliced transcript generated a 1.2-kbp RT-PCR product. The identity of the RT-PCR product was verified by Southern blotting. Oligonucleotides possessing cognate sequences for the three siRNAs were used as probes. Both the control infection and infection in the presence of the scrambled siRNA had similar levels of the major spliced transcript generating the 1.2-kbp RT-PCR product. However, with each of the siRNAs (individually or multiplexed), this spliced transcript was down-regulated. For each of the RNA samples tested above, similar quantities of β-globin transcripts were detected. RT-PCR without reverse transcriptase did not generate any products, indicating the absence of genomic DNA contamination. Thus, the down-regulation of the major spliced transcript likely is a factor contributing to the antiviral effect described above.
FIG. 6.
HIV-1-specific siRNAs down-regulate the major spliced transcript. (A) Southern blot hybridization of RT-PCR products with three internal probes (used simultaneously) corresponding to the siRNA 5, 6, and 8 target sequences. (B) RT-PCR products generated by intron-spanning primers within the unspliced transcripts for the gp41 gene, tat, rev, and nef in the HIV-1 NL4-3 genome. The major spliced transcript generated a 1.2-kbp RT-PCR product observed on agarose gels or by hybridization. (C) RT-PCR products generated by β-globin primers. Lanes for panels A to C: lane 1, uninfected; lane 2, HeLaCD4 cells infected with NL4-3; lane 3, HeLaCD4 cells transfected with scrambled (negative control) siRNA and infected with NL4-3; lanes 4, 5, and 6, HeLaCD4 cells transfected with siRNAs 5, 6, and 8, respectively, and infected with NL4-3; lane 7, HeLaCD4 cells transfected with siRNAs 5, 6, and 8 simultaneously (multiplexed) and infected with NL4-3. (D) Schematic representation of the HIV-1 NL4-3 genome. Red bars indicate the positions of the oligonucleotide probes used for Southern blotting in panel A. Light-blue bars indicate the positions of intron-spanning primers. The unspliced transcript would generate a 4.5-kbp RT-PCR product. The major spliced transcript would generate a 1.2-kbp RT-PCR product.
HIV-1-specific siRNAs exert a potent antiviral effect on HIV-1 strains YU-2, JR-CSF, and NL4-3.
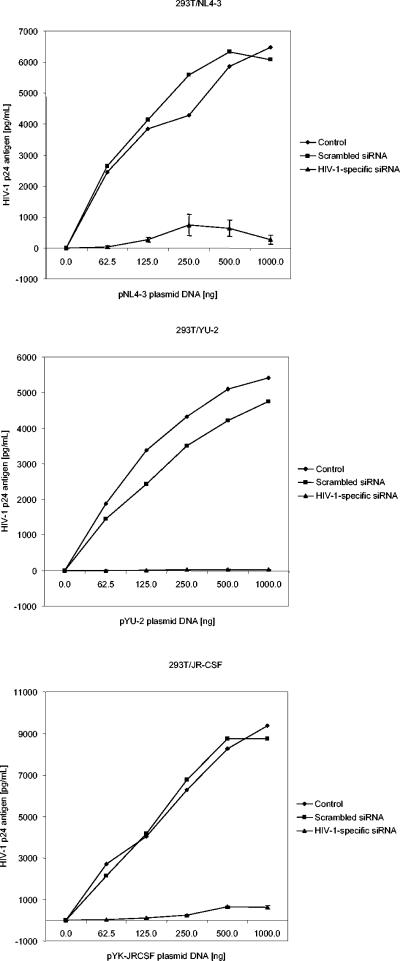
Having established the antiviral effect of the siRNAs tested here, we next wished to test their efficacy in inhibiting the replication of YU-2, JR-CSF, and NL4-3. We performed cotransfection experiments with both 293T cells (Fig. 7) and HeLaCD4 cells (data not shown). In either setting, various amounts of plasmid DNAs (for these viral strains) were cotransfected with siRNAs 5, 6, and 8 (multiplexed). Scrambled (negative control) siRNA was also used to verify the specificity of inhibition. In both 293T and HeLaCD4 cells, the most potent antiviral effect was observed with HIV-1 YU-2. The decrease in the production of HIV-1 YU-2 in 293T cells varied from 1,040- to 242-fold that seen in the control. The decrease in HeLaCD4 cells varied from 331- to 6-fold that seen in the control. The decrease in the production of HIV-1 JR-CSF was between those for YU-2 and NL4-3. In 293T cells, the decrease was 121- to 18-fold that seen in the control, while in HeLaCD4 cells, there was a slightly more potent decrease, varying from 225- to 15-fold that seen in the control. In 293T cells, the decrease in HIV-1 NL4-3 infection varied from 100- to 7-fold that seen in the control. The decrease in HeLaCD4 cells varied from 29- to 6-fold that seen in the control. In each of these settings, the strongest inhibition was observed with the smallest amount of plasmid DNA (62.5 ng), and there was an almost proportional decrease in the levels of inhibition with increasing concentrations of plasmid DNAs. Cells cotransfected with scrambled (negative control) siRNA had levels of viral production similar to those of cells transfected with plasmid DNA alone (P = 0.77). The results demonstrate that the HIV-1-specific siRNAs used in these experiments do indeed exert strong antiviral effects on these select neurotropic strains of HIV-1 and on strain NL4-3.
FIG. 7.
HIV-1-specific siRNAs inhibit the replication of HIV-1 NL4-3, YU-2, and JR-CSF. To assess the potency of HIV-1-specific siRNAs in inhibiting viral replication, siRNAs (multiplexed) were cotransfected into 293T cells along with various quantities (62.5 to 1,000 ng) of plasmid DNAs for NL4-3, YU-2, and JR-CSF. HIV-1 p24 antigen levels in culture supernatants were measured at 48 h posttransfection. Parallel experiments were also performed with HeLaCD4 cells. Control data are for 293T cells transfected with plasmid DNA forHIV-1 alone (one set). Scrambled siRNA data are for 293T cells cotransfected with scrambled (negative control) siRNA and plasmid DNA for HIV-1 (one set). HIV-1-specific siRNA data are for 293T cells cotransfected with HIV-1-specific siRNAs (multiplexed) and plasmid DNA for HIV-1 (in triplicate; the means and standard deviations are indicated).
HIV-1-specific siRNAs inhibit HIV-1 strain YU-2 infection in macrophages.
Having established that the HIV-1-specific siRNAs tested here possess potent antiviral activity against the neurotropic isolates YU-2 and JR-CSF, we next proceeded to verify whether these siRNAs have the ability to similarly inhibit viral infection in primary human macrophages. Since maximal inhibition was observed against HIV-1 YU-2, we tested the antiviral efficacy of these siRNAs in macrophages (Fig. 8). HIV-1 YU-2 infection was inhibited and the antiviral effect was sustained over a prolonged duration, as previously observed in a similar study (60). At 2 dpi, no inhibitory effect was obvious; however, by 9 dpi, the siRNAs inhibited HIV-1 YU-2 infection significantly, to 46.12% that seen in the control (P = 0.037).
FIG. 8.
Antiviral efficacy of HIV-1-specific siRNAs for HIV-1 YU-2 infection in human MDMs. Primary human macrophages were transfected with siRNAs and then infected with HIV-1 YU-2 (MOI, 0.1 pg of p24/cell). HIV-1 p24 antigen levels in culture supernatants were measured at 2 and 9 dpi. The data show the levels of viral production in macrophages infected with HIV-1 YU-2 and not transfected with siRNA (YU-2) and in macrophages infected with HIV-1 YU-2 and transfected with HIV-1-specific siRNAs 5, 6, and 8 (YU-2 + HIV-1-specific siRNA). The means of three experiments and the standard deviations are indicated.
DISCUSSION
These data demonstrate that siRNAs that target the region of the HIV-1 genome which contain regulatory as well as structural genes and in which transcripts undergo complex splicing events leading to the generation of multiple spliced transcripts are effective antiviral tools (21, 62, 67). Our results demonstrate that disruption of these splicing events leads to the degradation of at least some of the spliced transcripts. These HIV-1-specific siRNAs significantly inhibited viral assembly without altering the infectivity of the progeny virions. A connection between these two observations may be related to stoichiometric alterations of viral components.
We did not observe any aberrant transcriptional products in our analyses by both RT-PCR and Southern blotting. The fate of a transcript degraded by an siRNA is not clearly understood. Is there a simple down-regulation leading to reduced amounts of full-length transcripts, or do aberrant partially degraded transcripts exist? More importantly, are partially degraded transcripts translatable? It can only be speculated whether reduced quantities of viral proteins or aberrant viral proteins contribute to the observed antiviral effects. Although the former situation is clear, several studies have suggested how aberrant viral proteins affect the assembly process. For example, mutations in the HIV-1 gp41 cytoplasmic tail are known to drastically reduce the incorporation of gp120 in some cell types (3). Such mutations can also affect oligomerization in a manner distinct from the incorporation of gp120 (8). It is also likely that aberrant progeny virions (if present) did not form a significant majority of the viral population and hence had no influence on their overall infectivity. Nef expression is correlated with an increase in viral infectivity, at least in part by enhancing the quantity of Env products incorporated into viral particles (57). There are no studies supporting the reverse, i.e., the consequence of Nef down-regulation on viral assembly and infectivity. Again, although we found that siRNA with a cognate sequence from the Nef coding region led to the inhibition of viral production, a concomitant decrease in viral infectivity was not observed.
Our ability to inhibit infection by neurotropic strains of HIV-1 (YU-2 and JR-CSF) along with X4-tropic strain NL4-3 is a key feature of this investigation. These strains appear to be potent antiviral tools for dissecting HIV-1 neuropathogenesis and neuroinvasion. Viral reservoirs, especially those associated with the CNS, are difficult to eliminate. The unique dynamics of HIV-1 replication in macrophages and the relative lack of cytopathic effects ensure sustained survival after infection, eventually contributing to making macrophages a formidable viral reservoir (5). Macrophages represent a key target of HIV-1 in vivo. They are also the primary targets of HIV-1 in the nervous system. Infection of neurons and astrocytes was demonstrated recently by laser capture microdissection (68), although this aspect of HIV-1 pathogenesis is still highly controversial. One of the prevalent hypotheses suggests that productive infection of macrophages or microglia leads to altered neuronal function and damage, which ultimately lead to HIV-associated dementia. Neurologic dysfunction in HIV-associated dementia appears to be a consequence of microglial infection and activation. Several neurotoxic immunomodulatory factors are released from infected and activated microglia, leading to altered neuronal function, synaptic and dendritic degeneration, and eventual neuronal apoptosis (22, 64, 72). Hence, the ability to control infection by neurotropic strains of HIV-1 (R5/NSI) in macrophages or microglia is likely to enable interdiction of this crucial reservoir and possibly ameliorate the associated dysfunction.
MDMs have been shown to be able to be transfected with chemically synthesized siRNA duplexes to achieve sustained effects up to 7 days after transfection (60). Additionally, several developments to efficiently deliver siRNAs make the prospect of sustained siRNA-mediated antiviral effects in these cells feasible. Several studies have attempted the in vivo delivery of siRNAs (9, 39, 46, 60, 61). More importantly, a variety of vector-based delivery systems have been tested. The earliest developments involved plasmid vectors (12), and now there are lentiviral vectors (1, 6, 37, 41, 45, 54, 71), retroviral vectors (7, 19, 43), and adenoviral vectors (11, 58, 66, 73) for delivering siRNAs into a wide variety of cells. Most of these vector systems use the U6 promoter (47) or the HI promoter (49). Inducible plasmid vectors (69) and inducible lentiviral vectors (65, 71) are among the more complex recent additions. The availability of a wide variety of methods for delivering siRNAs and our HIV-1-specific siRNAs should enable investigators to obtain insights into developing RNAi-based therapeutic agents for altering certain viral reservoir sites.
Acknowledgments
We thank Jianhua Fang for critical suggestions, Michelle Vanella-Kudenko for laboratory support, and Rita M. Victor and Brenda O. Gordon for excellent secretarial assistance.
This work was supported in part by USPHS grants NS27405, MH58526, NS41864, and NS44513 to R.J.P.
REFERENCES
- 1.Abbas-Terki, T., W. Blanco-Bose, N. Deglon, W. Pralong, and P. Aebischer. 2002. Lentiviral-mediated RNA interference. Hum. Gene Ther. 13:2197-2201. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akari, H., T. Fukumori, and A. Adachi. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74:4891-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J., A. Banerjea, V. Planelles, and R. Akkina. 2003. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res. Hum. Retrovir. 19:699-706. [DOI] [PubMed] [Google Scholar]
- 5.Aquaro, S., R. Calio, J. Balzarini, M. C. Bellocchi, E. Garaci, and C. F. Perno. 2002. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 55:209-225. [DOI] [PubMed] [Google Scholar]
- 6.Banerjea, A., M. J. Li, G. Bauer, L. Remling, N. S. Lee, J. Rossi, and R. Akkina. 2003. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 8:62-71. [DOI] [PubMed] [Google Scholar]
- 7.Barton, G. M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 99:14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein, H. B., S. P. Tucker, S. R. Kar, S. A. McPherson, D. T. McPherson, J. W. Dubay, J. Lebowitz, R. W. Compans, and E. Hunter. 1995. Oligomerization of the hydrophobic heptad repeat of gp41. J. Virol. 69:2745-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrand, J. R., M. Pottier, A. Vekris, P. Opolon, A. Maksimenko, and C. Malvy. 2002. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 296:1000-1004. [DOI] [PubMed] [Google Scholar]
- 10.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boden, D., O. Pusch, F. Lee, L. Tucker, and B. Ramratnam. 2004. Efficient gene transfer of HIV-1-specific short hairpin RNA into human lymphocytic cells using recombinant adeno-associated virus vectors. Mol. Ther. 9:396-402. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 13.Butticaz, C., A. Ciuffi, M. Munoz, J. Thomas, A. Bridge, S. Pebernard, R. Iggo, P. Meylan, and A. Telenti. 2003. Protection from HIV-1 infection of primary CD4 T cells by CCR5 silencing is effective for the full spectrum of CCR5 expression. Antiviral Ther. 8:373-377. [PubMed] [Google Scholar]
- 14.Cann, A. J., J. A. Zack, A. S. Go, S. J. Arrigo, Y. Koyanagi, P. L. Green, S. Pang, and I. S. Chen. 1990. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J. Virol. 64:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 16.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, A. T., T. R. Brummelkamp, E. M. Westerhout, M. Vink, M. Madiredjo, R. Bernards, and B. Berkhout. 2004. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 78:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dave, R. S., and R. J. Pomerantz. 2003. RNA interference: on the road to an alternate therapeutic strategy. Rev. Med. Virol. 13:373-385. [DOI] [PubMed] [Google Scholar]
- 19.Devroe, E., and P. A. Silver. 2004. Therapeutic potential of retroviral RNAi vectors. Expert Opin. Biol. Ther. 4:319-327. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 21.Furtado, M. R., R. Balachandran, P. Gupta, and S. M. Wolinsky. 1991. Analysis of alternatively spliced human immunodeficiency virus type-1 mRNA species, one of which encodes a novel tat-env fusion protein. Virology 185:258-270. [DOI] [PubMed] [Google Scholar]
- 22.Garden, G. A. 2002. Microglia in human immunodeficiency virus-associated neurodegeneration. Glia 40:240-251. [DOI] [PubMed] [Google Scholar]
- 23.Garred, P., J. Eugen-Olsen, A. K. Iversen, T. L. Benfield, A. Svejgaard, B. Hofmann, et al. 1997. Dual effect of CCR5 delta 32 gene deletion in HIV-1-infected patients. Lancet 349:1884. [DOI] [PubMed] [Google Scholar]
- 24.Ge, Q., M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 26.Glass, W. G., B. P. Chen, M. T. Liu, and T. E. Lane. 2002. Mouse hepatitis virus infection of the central nervous system: chemokine-mediated regulation of host defense and disease. Viral Immunol. 15:261-272. [DOI] [PubMed] [Google Scholar]
- 27.Glass, W. G., and T. E. Lane. 2003. Functional analysis of the CC chemokine receptor 5 (CCR5) on virus-specific CD8+ T cells following coronavirus infection of the central nervous system. Virology 312:407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass, W. G., and T. E. Lane. 2003. Functional expression of chemokine receptor CCR5 on CD4+ T cells during virus-induced central nervous system disease. J. Virol. 77:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glass, W. G., M. T. Liu, W. A. Kuziel, and T. E. Lane. 2001. Reduced macrophage infiltration and demyelination in mice lacking the chemokine receptor CCR5 following infection with a neurotropic coronavirus. Virology 288:8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harborth, J., S. M. Elbashir, K. Vandenburgh, H. Manninga, S. A. Scaringe, K. Weber, and T. Tuschl. 2003. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 13:83-105. [DOI] [PubMed] [Google Scholar]
- 32.Hu, W. Y., C. P. Myers, J. M. Kilzer, S. L. Pfaff, and F. D. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301-1311. [DOI] [PubMed] [Google Scholar]
- 33.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia, Q., and R. Sun. 2003. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 77:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, M., and J. Milner. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041-6048. [DOI] [PubMed] [Google Scholar]
- 36.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, M. T., G. A. Coburn, M. O. McClure, and B. R. Cullen. 2003. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 77:11964-11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, D. L., J. E. Hagstrom, A. G. Loomis, J. A. Wolff, and H. Herweijer. 2002. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 32:107-108. [DOI] [PubMed] [Google Scholar]
- 40.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 41.Li, M. J., G. Bauer, A. Michienzi, J. K. Yee, N. S. Lee, J. Kim, S. Li, D. Castanotto, J. Zaia, and J. J. Rossi. 2003. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol. Ther. 8:196-206. [DOI] [PubMed] [Google Scholar]
- 42.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, C. M., D. P. Liu, W. J. Dong, and C. C. Liang. 2004. Retrovirus vector-mediated stable gene silencing in human cell. Biochem. Biophys. Res. Commun. 313:716-720. [DOI] [PubMed] [Google Scholar]
- 44.Martinez, M. A., A. Gutierrez, M. Armand-Ugon, J. Blanco, M. Parera, J. Gomez, B. Clotet, and J. A. Este. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS 16:2385-2390. [DOI] [PubMed] [Google Scholar]
- 45.Matta, H., B. Hozayev, R. Tomar, P. Chugh, and P. M. Chaudhary. 2003. Use of lentiviral vectors for delivery of small interfering RNA. Cancer Biol. Ther. 2:206-210. [DOI] [PubMed] [Google Scholar]
- 46.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 47.Miyagishi, M., and K. Taira. 2002. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20:497-500. [DOI] [PubMed] [Google Scholar]
- 48.Montaner, J. S., and J. W. Mellors. 1999. Effective salvage therapy for HIV-1 infection—an unmet challenge. Antiviral Ther. 4:59-60. [PubMed] [Google Scholar]
- 49.Myslinski, E., J. C. Ame, A. Krol, and P. Carbon. 2001. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 29:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 51.Olson, K. E., Z. N. Adelman, E. A. Travanty, I. Sanchez-Vargas, B. J. Beaty, and C. D. Blair. 2002. Developing arbovirus resistance in mosquitoes. Insect Biochem. Mol. Biol. 32:1333-1343. [DOI] [PubMed] [Google Scholar]
- 52.Park, W. S., M. Hayafune, N. Miyano-Kurosaki, and H. Takaku. 2003. Specific HIV-1 env gene silencing by small interfering RNAs in human peripheral blood mononuclear cells. Gene Ther. 10:2046-2050. [DOI] [PubMed] [Google Scholar]
- 53.Pomerantz, R. J. 2002. RNA interference meets HIV-1: will silence be golden? Nat. Med. 8:659-660. [DOI] [PubMed] [Google Scholar]
- 54.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saiki, R. K., S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich, and N. Arnheim. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350-1354. [DOI] [PubMed] [Google Scholar]
- 57.Schiavoni, I., S. Trapp, A. C. Santarcangelo, V. Piacentini, K. Pugliese, A. Baur, and M. Federico. 2004. HIV-1 Nef enhances both membrane expression and virion incorporation of Env products. A model for the Nef-dependent increase of HIV-1 infectivity. J. Biol. Chem. 279:22996-23006. [DOI] [PubMed] [Google Scholar]
- 58.Shen, C., A. K. Buck, X. Liu, M. Winkler, and S. N. Reske. 2003. Gene silencing by adenovirus-delivered siRNA. FEBS Lett. 539:111-114. [DOI] [PubMed] [Google Scholar]
- 59.Smit, T. K., B. Wang, T. Ng, R. Osborne, B. Brew, and N. K. Saksena. 2001. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology 279:509-526. [DOI] [PubMed] [Google Scholar]
- 60.Song, E., S. K. Lee, D. M. Dykxhoorn, C. Novina, D. Zhang, K. Crawford, J. Cerny, P. A. Sharp, J. Lieberman, N. Manjunath, and P. Shankar. 2003. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77:7174-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song, E., S. K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Lieberman. 2003. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9:347-351. [DOI] [PubMed] [Google Scholar]
- 62.Sonza, S., H. P. Mutimer, K. O'Brien, P. Ellery, J. L. Howard, J. H. Axelrod, N. J. Deacon, S. M. Crowe, and D. F. Purcell. 2002. Selectively reduced tat mRNA heralds the decline in productive human immunodeficiency virus type 1 infection in monocyte-derived macrophages. J. Virol. 76:12611-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strizki, J. M., A. V. Albright, H. Sheng, M. O'Connor, L. Perrin, and F. Gonzalez-Scarano. 1996. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J. Virol. 70:7654-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swindells, S., J. Zheng, and H. E. Gendelman. 1999. HIV-associated dementia: new insights into disease pathogenesis and therapeutic interventions. AIDS Patient Care STDS 13:153-163. [DOI] [PubMed] [Google Scholar]
- 65.Tiscornia, G., V. Tergaonkar, F. Galimi, and I. M. Verma. 2004. CRE recombinase-inducible RNA interference mediated by lentiviral vectors. Proc. Natl. Acad. Sci. USA 101:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomar, R. S., H. Matta, and P. M. Chaudhary. 2003. Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene 22:5712-5715. [DOI] [PubMed] [Google Scholar]
- 67.Tornatore, C., K. Meyers, W. Atwood, K. Conant, and E. Major. 1994. Temporal patterns of human immunodeficiency virus type 1 transcripts in human fetal astrocytes. J. Virol. 68:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trillo-Pazos, G., A. Diamanturos, L. Rislove, T. Menza, W. Chao, P. Belem, S. Sadiq, S. Morgello, L. Sharer, and D. J. Volsky. 2003. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 13:144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu, Y., J. Kulkosky, E. Acheampong, G. Nunnari, J. Sullivan, and R. J. Pomerantz. 2004. HIV-1-mediated apoptosis of neuronal cells: proximal molecular mechanisms of HIV-1-induced encephalopathy. Proc. Natl. Acad. Sci. USA 101:7070-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, J., O. Yamada, T. Sakamoto, H. Yoshida, T. Iwai, Y. Matsushita, H. Shimamura, H. Araki, and K. Shimotohno. 2004. Down-regulation of viral replication by adenoviral-mediated expression of siRNA against cellular cofactors for hepatitis C virus. Virology 320:135-143. [DOI] [PubMed] [Google Scholar]
- 74.Zhou, N., J. Fang, M. Mukhtar, E. Acheampong, and R. J. Pomerantz. Inhibition of HIV-1 fusion with small interfering RNAs targeting the chemokine coreceptor CXCR4. Gene Ther., in press. [DOI] [PubMed]
- 75.Zhou, Y., T. Kurihara, R. P. Ryseck, Y. Yang, C. Ryan, J. Loy, G. Warr, and R. Bravo. 1998. Impaired macrophage function and enhanced T cell-dependent immune response in mice lacking CCR5, the mouse homologue of the major HIV-1 coreceptor. J. Immunol. 160:4018-4025. [PubMed] [Google Scholar]