Abstract
Definition of preleukemia has evolved. It was first used to describe the myelodysplastic syndrome (MDS) with a propensity to progress to acute myeloid leukemia (AML). Individuals with germline mutations of either RUNX1, CEBPA, or GATA2 can also be called as preleukemic because they have a markedly increased incidence of evolution into AML. Also, alkylating chemotherapy or radiation can cause MDS/preleukemia, which nearly always progress to AML. More recently, investigators noted that AML patients who achieved complete morphological remission after chemotherapy often have clonal hematopoiesis predominantly marked by either DNMT3A, TET2 or IDH1/2 mutations, which were also present at diagnosis of AML. This preleukemic clone represents involvement of an early hematopoietic stem cells, which is resistant to standard therapy. The same clonal hematopoietic mutations have been identified in older ‘normal' individuals who have a modest increased risk of developing frank AML. These individuals have occasionally been said, probably inappropriately, to have a preleukemia clone. Our evolving understanding of the term preleukemia has occurred by advancing technology including studies of X chromosome inactivation, cytogenetics and more recently deep nucleotide sequencing.
Preleukemia associated with myelodysplastic syndrome
The term ‘preleukemia' has undergone a major evolution since it was first used in 1953 by Block et al.1 At that time, the term referred to the heterogeneous group of hematopoietic disorders associated with a block in myeloid differentiation and chronic cytopenias. We now call these disorders, myelodysplastic syndromes (MDS).2, 3, 4, 5, 6, 7, 8, 9 Prevalence of these abnormalities is markedly increasing as our population ages; about 75 new patients per 100 000 individuals per year over the age of 65 years develop MDS. About 20–30% of these individuals do indeed progress to acute myeloid leukemia (AML); and thus, these individuals might justifiably be known as preleukemic.
In the late 1960s and 1970s, one of the prominent advances in our understanding of MDS/AML was the concept that the MDS/leukemic event occurred in 1 stem cell, which had a hematopoietic growth advantage causing a clonal population of MDS/leukemic cells. This insight was based on the observation of X-inactivation in females.10, 11 Females have two X chromosomes; between the 4th and 16th cell stage of blastogenesis, each cell stochastically inactivates either its maternal or paternal X chromosome. A number of methods have evolved to determine which X chromosome has been inactivated.12, 13 The initial approach was pioneered by Ernest Beutler who utilized G6PD isoenzymes.14, 15, 16 Over the years, additional methods were developed to identify X-inactivation in females, with one of the most versatile taking advantage of 2 features: cytosine methylation of the inactive X chromosome and different numbers of CAG trinucleotides within the promoter region of the androgen receptor, which resides on the X chromosome.17 This technique allows interrogation of almost all females as opposed to a small population of predominantly African Americans with G6PD isoenzymes. Philip Fialkow's group showed clonal hematopoiesis occurred in a number of hematopoietic diseases including chronic myelogenous leukemia, acute lymphocytic leukemia and AML.18 Also, Fialkow noted that in many cases of AML, the clonality occurred in the myeloid compartment and not the lymphoid compartment. MDS, in contrast, appeared to have an identical clonality of both the myeloid and lymphoid compartments, albeit the myeloid compartment appeared to have a major growth advantage.19 This suggested that the MDS clone initiated in a very early hematopoietic stem cell.
With the era of massively parallel nucleotide sequencing, we have a fairly clear idea of the major mutations that affect patients who have MDS, who evolve from MDS to AML (secondary AML, sAML) and those with de novo AML (Table 1).4, 5, 20, 21, 22, 23, 24, 25, 26, 27, 28 Nine genes occur more frequently in MDS than de novo AML (Table 1) including four spliceosome genes that are mutated in 60–70% of MDS cases, but these latter genes are mutated in fewer than 5–10% of de novo AML cases.29 These mutant genes are involved in ineffective dysplastic hematopoiesis, not frank AML.
Table 1. Exome sequencing: mutational defects in MDS and sAML versus de novo AML.
|
MDS |
sAML |
De novo AML |
|||
|---|---|---|---|---|---|
| Gene | Mutations (%) | Gene | Mutations (%) | Gene | Mutations (%) |
| TET2a | 33 | TET2a | 20 | FLT3b | 28 |
| →SF3B1a | 32 | SF3B1a | 11 | NPM1b | 27 |
| ASXL1a | 23 | ASXL1a | 32 | DNMT3A | 26 |
| →SRSF2a | 17 | SRSF2a | 20 | IDH1/2 | 10 |
| →ZRSR2a | 8 | ZRSR2a | 8 | RUNX1b | 9 |
| EZH2 a | 5 | EZH2a | 9 | TET2 | 9 |
| BCORa | 4 | BCORa | 8 | NRAS/KRASb | 8 |
| →U2AF1a | 8 | U2AF1a | 16 | TP53 | 8 |
| STAG2a | 8 | STAG2a | 14 | CEBPAb | 6 |
| RUNX1 | 10 | RUNX1b | 31 | WT1b | 6 |
| DNMT3A | 13 | NF1b | 6 | PTPN11b | 5 |
| IDH2/IDH1 | 5 | NRAS/KRASb | 31 | KITb | 4 |
| TP53 | 6 | TP53 | 15 | U2AF1 | 4 |
| FLT3 | 5 | IDH1/2 | 20 | KRASb | 4 |
| CBL | 5 | FLT3b | 19 | SMC1A | 4 |
| JAK2 | 5 | DNMT3A | 19 | SMC3 | 4 |
| BCOR | 3 | PTPN11b | 5 | PHF6 | 3 |
| NRAS or KRAS | 7 | CEBPAb | 3 | STAG2 | 2 |
| MPL | 3 | NPM1b | 5 | RAD21 | 2 |
| NF1 | 3 | SMC3 | 2 | FAM5C | 2 |
| ATM | 3 | CBL | 5 | EZH2 | 1 |
Mutations that are often observed in de novo AML are known as Driver or pan-AML mutations including mutations of NPM1, FLT3, IDH1, N/KRAS, RUNX1, CEBPA, WT1, PTPN11, c-KIT. For example, NPM1 is rarely mutated in MDS, but occurs in 27% of de novo AML cases. Likewise, chromosomal translocations (often a fusion of a transcription factor gene with another gene) occur frequently in de novo AML (18%). Examples include fusion of genes such as RUNX1 with ETO, PML with RARA, CBFB with MYHII, as well as fusion of MLL with various partner genes, which only rarely occur in MDS (4%).30 These Driver mutations produce a major block in differentiation providing a proliferative advantage to this clone of cells.30
Because cases of sAML evolves from MDS, these cells often contained genes mutated in MDS (Table 1).30 Driver (pan-AML) mutations are gained as MDS evolves to sAML.30 Of interest, individuals with sAML achieve complete remission only about 50% of the time and their event-free survival (EFS) is 4.2 months. In contrast, 92% of de novo AML patients enter a complete remission, and their EFS is ~15.7 months.30
Oligoclonal hematopoiesis: Normal aging versus preleukemic clone
Analysis of X chromosome inactivation discovered skewing of X chromosome inactivation (also known as Lyonization).31, 32, 33, 34 X-inactivation during blastogenesis usually results in equal inactivation of the maternal and paternal X chromosomes in the female. Skewing represents unequal distribution of cells expressing either the paternal and maternal allele of the female X chromosome. It can occur during early embryogenesis, but rarely deviates >10%. However, X chromosomal skewing increases with age approaching 20% of those who are 28–32 years old, and 40% of those⩾60 years.10, 12, 13, 32, 33, 34 A study in normal females ⩾75 of age found that 56% of individuals had skewing of neutrophils and 40% had skewing of T-lymphocytes.34 Thus, older individuals have hematopoietic cells that favor usage of one X allele. This has been explained by unequal exhaustion of hematopoietic stem cells as individuals age. This observation was buttress by genomically marking murine hematopoietic stem cells and using them for complete hematopoietic transplantation. Multiple clones of hematopoiesis initially developed in these transplanted animals; but as the mice aged, oligoclonal hematopoiesis predominated, suggesting that with age, certain hematopoietic clones can predominate.35 Of note, this phenomenon was not noted in humans who underwent hematopoietic transplantation.36
SNP-Chip analysis of peripheral blood of 50 000 normal individuals showed that copy-number changes in the genome increased with increasing age; >0.5% of individuals from birth to 50 years of age had copy number alterations; but after age 50 years, the rate rose to about 2–3%.37 These copy-number changes often occurred in genes that also are abnormal in MDS and AML.37 These individuals with copy-number changes had a 10-fold increased risk of developing MDS or AML.37
Recently, a number of studies using high throughput nucleotide sequencing confirmed what had been suggested by studies of skewing of the inactive X-chromosome in normal elderly individuals. On average, a normal appearing 60-, 70- and 90-year-old individual has a 5%, 10% and 20% incidence, respectively, of clonal hematopoiesis as marked by a mutation.38, 39, 40 A recent study of a normal population of 50–60-year-old individuals found with exceptionally deep sequencing that clonal hematopoiesis occurred in 95% of individuals as marked by a mutation of either DNMT3A or TET2.41 An interesting, unifying study found TET2 mutations in the blood of ten of 182 elderly ‘normal' women who also had chromosomal skewing of the X chromosome. In contrast, 105 normal women of the same age without X-chromosome skewing, had no TET2 mutations.42 Mutation of DNMT3A occurs about fivefold more frequently in the normal elderly individuals than mutations of either TET2, IDH1/2, AXSL1, JAK2, SRSF2 or SF3B1 (to name a few). Many of these mutations of the normal elderly are the same as those in the preleukemic clone identified in complete remission of sAML, or found in the very early hematopoietic stem cells in individuals with either MDS or AML (both to be discussed).38, 39, 40, 43 The base change of these mutations was most commonly cytosine to thymine transition, which is a somatic mutational signature of aging. Usually only one gene and on rare occasions two or three genes are mutated; these additional mutations occur more frequently as the ‘normal' individual becomes older.38, 39, 40 On average, <20% and more often <3% of either the peripheral blood or marrow cells are part of the mutant hematopoietic clone in these elderly individuals (also called variant allele frequency (VAF)).
For lack of a better nomenclature, this hematopoietic clone in the elderly has occasionally been called an age-related preleukemic clone; this is usually a misnomer. About 0.5 to 1% of these individuals per year developed MDS/AML (hazard ratio 11.1–12.9).38, 39 The greater the number of different mutations or the greater the contribution (VAF) of the mutant clone, the greater the chance to evolve into MDS/AML (for example, if the mutant clone constitutes >10% of the hematopoietic population, these individuals have at least a 1% risk per year of developing AML39). Acquisition of a Driver (pan-AML) mutation(s) is required for development of AML in these individuals with morphologically normal marrow cells. These genes behave as the ‘gatekeepers' of leukemogensis providing, when mutated, the needed stimulus for establishment of frank AML.
Thus as humans age, they often develop a hematopoietic clone carrying a mutation of at least either DNMT3A, ASXL1, TET2 or IDH2, but only a small minority of these individuals actually develop myeloid malignancies. These Driver mutations provide a competitive advantage to the hematopoietic stem cells compared with normal hematopoiesis. Whether this mutant stem cell pool is more receptive to secondary mutations requires further study.
Analogous phenomenon occur in the lymphoid system. Monoclonal gammopathy of unknown significance (MGUS) features a small accumulation of plasma cells producing a monoclonal immunoglobulin.44 The aberrant clone of plasma cells infrequently (1% per year) progresses to multiple myeloma. Likewise, a clonal mature B-cell population can develop in older individuals, which rarely progresses to chronic lymphocytic leukemia.45 Incidence of all three of these clonal hematopoietic conditions increase with age. Clonal abnormalities have been noted in other tissue which otherwise appear normal. For example, the normal sun-exposed eyelid frequently has mutations including NOTCH1, NOTCH2, NOTCH3, FAT1, TP53 and RBM10; some of these mutations are associated with clonal expansion of these skin cells.46 Also, we and others noted years ago that the normal appearing colon and lung near the tumor can harbored genomic alterations (for example, aneuploidy, RAS mutations).47, 48 These too represent a precancerous genomic abnormality.
Preleukemia: germline mutant hematopoietic clone
Twins studies have suggested familial clustering of AML;49, 50 and a large analysis of over 24 000 first-degree relatives of about 7000 patients with AML noted that if the proband develops MDS/AML before the age of 25 years, the first-degree relative has an increased risk of MDS and AML.51 Individuals with inherited germline mutations of one of several transcription factors (RUNX1, GATA2 or CEBPA) have an increased propensity to develop AML over their life.52, 53, 54, 55, 56 Other inherited germline that can also predispose to MDS includes Diamond–Blackfan anemia (20% risk of MDS/AML), dyskeratosis congenita (disorder of telomere maintenance, 30% risk MDS/AML), Fanconi anemia (40% risk), severe congenital neutropenia (20–40% risk), Shwachman-Diamond syndrome (10–35% risk), mutations of the SRP72 gene (increased risk of aplastic anemia or MDS), Bloom syndrome (25% risk for MDS/AML), Li-Fraumeni syndrome (p53 mutation) (5–7% risk) and familial monosomy 7.57, 58, 59 For each of these inherited germline mutation, their progression to frank AML requires the clonal acquisition of addition genetic mutations.60
Familial AML with germline mutant CEBPA (OMIM 11697) is inherited (autosomal dominant) and has nearly complete penetrance for development of AML.54, 56 AML can develop anytime from the age of 1 year to old age. The AML is usually associated with biallelic CEBPA mutations, usually the first mutation is germline and often at the 5′ end of the gene, and the second mutation occurs in the second allele often in the 3′ region of the gene. This is similar to what is observed in those with somatic mutations of CEBPA (de novo AML). CEBPA is critical for myeloid proliferation and granulocytic differentiation. Most of these patients have a normal karyotype when they develop AML; and they have a relatively favorable prognosis with 50–60% that may be cured. The AML is often FAB subtype M1 or M2, with aberrant CD7 expression on the blast cells and often a N-RAS mutation present in the blast cells. As expected, siblings of the proband with the germline CEBPA mutation have a 50% risk of also carrying the mutation.
Familial platelet disorder (FPD), (OMIM 601399) with a propensity to AML is caused by a germline mutation of RUNX1. The RUNX1 gene is important for myeloid proliferation and differentiation; and its alteration in FPD is either a deletion, a loss of function missense mutation or occasionally a dominant-negative missense mutation. These individuals often have a low peripheral blood platelet count, a functional platelet disorder and GCSF hypersensitivity of their myeloid cells.52, 61 The frequency of developing AML with FPD is about 40%, and AML can occur at any age but more often between 40 and 70 years of age.
AML that develops in FPD individuals do not acquire NPM1 or FLT3 mutations, which are frequently found in de novo AML. A recent study found that asymptomatic RUNX1 carriers can harbor additional somatic mutations with a mean VAF varying between 20 and 45%, suggesting that clonal skewed hematopoiesis frequently precedes the development of overt MDS or AML.60 All asymptomatic RUNX1 carriers were less than the age of 50. By the age of 50 years, clonal hematopoiesis was easily found in 80% of RUNX1 carriers.60 One Japanese study reported that these patients had frequent mutations of the CDC25C gene when they progressed to AML.62 However, this mutation could not be detected by other investigators.60, 63 This could represent ethnic differences. The number of mutations in the blast cells found at the time of AML varies between 8 and 18. The VAF of these mutations in both the MDS and AML phases of FPD were comparable (approximately 15–45%).60, 61, 64
Another germline alteration is a heterozygous mutation of the transcription factor GATA2. These individuals often have peripheral blood eosinophilia, deficiency of monocytes, B cells and natural killer cells and an increased incidence of non-tuberculosis mycobacteria and other opportunistic infections including papilloma virus.53, 55, 65, 66, 67 The syndrome is often called (MonoMAC). These patients in addition can develop leg lymphedema and monosomy 7 (Emberger syndrome). The predilection to develop MDS or AML is about 70% with a median age of 29 years, but AML can develop over a wide time span (0.4 to 78 years). Their MDS phase is often characterized as chronic myelomonocytic leukemia. The progression to AML is associated with an initial cytopenias often followed by monosomy 7 (30%) which appears to occur before an ASXL1 mutation (29% of cases). Interestingly, this latter mutation occurs only in females, who are often young; it is associated with a bad prognosis.68, 69 Another frequent secondary mutation is NRAS, but other Driver mutations have been noted including RUNX1, STAG2, IDH2, TP53 and SETDB1. Transformation is often to a hypercellular fibrotic AML.
Clearly, more experience is required to determine at what stage these individual with germline preleukemic mutations need aggressive therapy. Monitoring their marrow for the acquisition of additional Driver mutations might prompt initiation of aggressive therapy such as allogenic hematopoietic stem cell transplant.60 If a sibling allotransplant is contemplated, germline DNA sequencing of the donor for the germline mutation is required because the sibling has a 50% chance of also having the germline mutation. The ability to take fibroblasts from patients with germline mutations, induced dedifferentiation to pluripotent stem cells (iPSC), correct the mutation and then induce these cells to differentiate into functional hematopoietic stem cell, has been done.70, 71 However, to date, full hematopoietic reconstitution in vivo has not been possible using the iPSC technique. Furthermore, gene therapy to repair directly the hematopoietic cells is a distant dream with a multitude of barriers because unlike treating sickle cell anemia or thalassemia, every cell must be corrected.
Therapy-related preleukemia
Several types of therapy-(mutagen)-related MDS/AML have been identified. One that almost always has a preleukemic (MDS) phase is associated with exposure to either alkylating agents (85%), radiation or selected carcinogens, such as benzene.72, 73, 74 Most frequently an individual with Hodgkin disease, non-Hodgkin disease, breast cancer, multiple myeloma and so on receives an alkylating agent and approximately 4 years later develops MDS. The incidence of therapy-related preleukemia varies from <1 to 10% depending on type of agent administered and the intensity of the therapy for the initial cancer.72, 75, 76, 77, 78 Their preleukemic marrow often (>70%) has deletion of either the long arm or the entire chromosome 5 and/or 7; or they develop a complex karyotype (⩾3 abnormalities).75, 79, 80, 81 Some of the frequently mutated genes in this type of preleukemia includes p53, PTPN11 and the ABC family of genes. The p53 mutation often occurs early in the course of MDS. A second group of mutations are commonly associated with secondary AML (Table 1). In contrast to de novo acute leukemia, mutations of NPM1, CEBPA or FLT3 are infrequent.30, 72, 82, 83, 84 The RAEB and RAEB-t subgroups are frequently linked to therapy-related preleukmia (73%).85 These individuals almost always progress to AML with a mean duration of the preleukemic phase of 11.2 months.86, 87, 88 They have a poor prognosis (survival with AML<6–8 months).73, 75, 89 Treatment for these individuals when AML develops has a number of clinical challenges including a greater number of co-morbidities, decreased organ reserve from either previous therapy or primary disease and a higher incidence of unfavorable cytogenetic changes. The key prognostic factors in therapy-related MDS/AML are patient's age, performance status and karyotype. Because of their poor outcome, intensive chemotherapy with allogeneic hematopoietic stem cell transplant should be encouraged.
In summary, the development of unexplained pancytopenia and the finding of specific karyotypic abnormalities (loss of chromosome 5 and/or 7 or their long arms in the marrow) in patients who received chemotherapy and/or radiation therapy for another disease about 4 years earlier is pathognomonic of preleukemia. Evolution to overt leukemia is almost universal if the preleukemic individual survives the complications of hemorrhage and infection. Therapy preleukemia can be viewed as an early phase of therapy AML in which the malignant hematopoietic clone is established and becomes predominant.
The second type of therapy-related AML develops in individuals whose initial malignancy was treated with a topoisomerase II inhibitor (for example, etoposide, doxorubicin, mitoxantrone).72, 90, 91 Typical latency period from therapy to AML is a median of 33 months but can be as short as 12 months. The AML often has a chromosomal translocation especially at 11q23 (MLL gene) or chromosome 21q22 (RUNX1). The morphologic phenotype often is FAB M4 or M5. These individuals usually do not have a MDS (preleukemic) phase. Overall survival of these individuals after development of AML is equal to that of a patient with de novo leukemia having the same cytogenetic abnormalities in their AML cells.
Preleukemia in morphologic complete remission after AML chemotherapy
During the 1980s and the early 1990s, studies by Phil Fialkow and Claus Bartram noted that about ¼ of AML patients who achieved complete morphologic remission by chemotherapy, continued to have an abnormal clone as measured by X-inactivation.31 This has been buttress by studies showing strong association between persistent karyotypic markers of AML in first complete morphologic remission. Hence, the term ‘preleukemic clone' has been used to describe the loss of the major phenotype of AML (Driver mutations associated with AML blast cells), but where the clonal hematopoiesis persisted, and could evolve into relapse AML. Thus, the term preleukemia is used in a very different context than utilized in the 1950s and 1960s.
High throughput sequencing of AML samples reinforced the concept that a preleukemic clone can exist at the time of complete morphologic remission after standard aggressive chemotherapy for AML. Individuals whose AML cells at diagnosis had mutations of either DNMT3A, IDH2, TET2, UAF1 or SRSF2 often had the same alteration at complete morphologic remission.92, 93, 94, 95, 96, 97, 98 In contrast, more classical Driver (pan-AML) mutations such as FLT3, RUNX1, RAS, NPM1, CEBPA and WT1 genes, although present at diagnosis of AML, were no longer found in complete remission. Dr. Ley's group reported that 21/37 individuals whose AML cells carried DNMT3A, TET2 or IDH1/2 mutations at diagnosis, continued to have the same mutation in >5% of the hematopoietic cells at complete morphologic remission; but the classical Driver mutations (for example, NPM1, FLT3 and RAS) found at diagnosis, were not detectable at remission.94 Of note, if the individual had a persistence of a mutation at complete remission (day 30 after induction therapy), their event-free survival was 6 months and their overall survival was 10 months as compared with an event-free survival of 18 months and an overall survival of 42 months in individuals with no mutation in the 30-day complete remission marrow.
Another study focused on DNMT3A mutations. The investigators noted that the DNMT3A mutation was often present in the marrow cells in complete morphologic remission at an allele frequency of 1-50% and could be detected even 8 years after initial complete remission of AML with no other molecular markers of AML. Likewise, the mutations could be found at a lower allele frequency in T and B lymphocytes in the same patients.97 The authors concluded that DNMT3A mutations precede NPM1 and FLT3-ITD mutations in AML pathogenesis; and the fact that this mutation can be found in B and T cells suggest that it involves a very early hematopoietic stem cell that can differentiate both into lymphoid and myeloid cells.93, 97
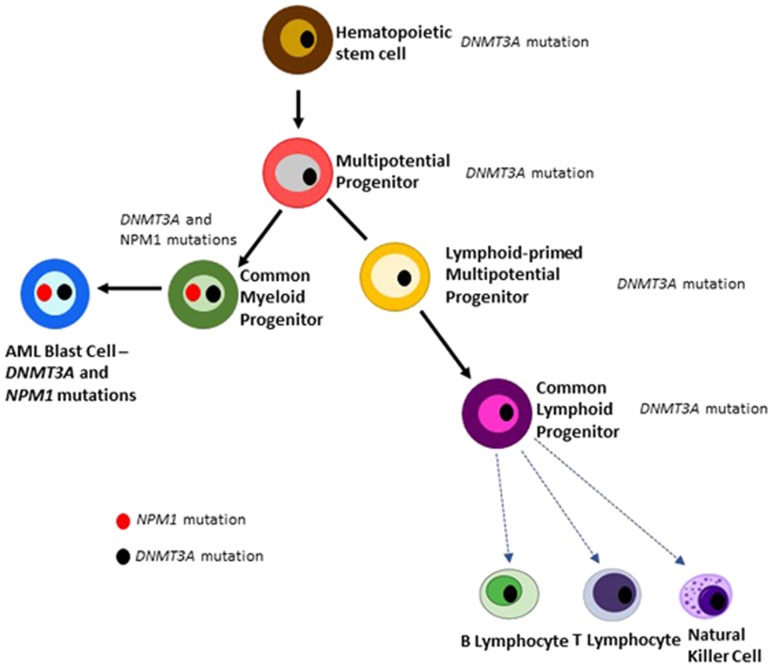
Dr. Dick's group studied a cohort of individuals whose AML blast cells had mutations of DNMT3A and NPM1 at both diagnosis and relapse.93 In morpholoigc complete remission, only the DNMT3A mutation could be found. In a second part of the study, human bone marrow cells at diagnosis of AML (264 samples) were injected into immunodeficient mice (NSG mice). A total of 37% of samples did not engraft, 40% of samples recapitulated the AML blasts in the mice; but most interestingly, 23% of the samples produced multi-lineage engraftment that appeared normal, but continued to have mutations of either DNMT3A or IDH1/2.93 This suggests that even at diagnosis of AML, a preleukemic clone exists. Interestingly, the same group also purified different stages of hematopoietic cells at diagnosis of AML whose blasts cells were marked with both DNMT3A and NPM1 mutations.93 The very early common hematopoietic progenitors carried the mutation of DNMT3A, but not NPM1. In contrast, the more mature myeloid progenitor cells (granulocyte-monocyte progenitors), as well as the AML blast cells had both the DNMT3A and NPM1 mutations (Figure 1 summarizes these studies).
Figure 1.
Schematic diagram of the hematopoietic tree with stem cells having a preleukemic mutation (DNMT3A) and blast cells acquiring a Driver (pan-AML) mutation (NPM1 mutation), and retaining the DNMT3A mutations. Black dot, DNMT3A mutation; Red dot, NPM1 mutation. Figure summarizes data by John Dick's lab.93
A very detailed study focused on IDH2 found that about 22% of individuals with an IDH2 mutant allele at diagnosis of sAML continued to have the mutational clone in complete morphologic remission after chemotherapy; but with marrow transplant, these aberrant alleles were destroyed.98 In addition, the VAF of the mutant IDH2 clone exceeded the VAF of the NPM1 or FLT3 clones in frank AML, and the NPM1 and FLT3 clones disappeared with complete remission. Colony assays of CD34+ hematopoietic stem cells and progenitor cells were done in complete remission and 96% and 50% of the erythroid and granulocyte macrophage colonies, respectively, had only the IDH2 mutation. Further, colony and single-cell-nucleotide sequencing also confirmed that DNMT3A, TET2 and ASXL1 were early clonal mutations during evolution to AML.99, 100
Interestingly, recent studies found a rapid expansion of a pre-existing non-leukemic/preleukemic mutant hematopoietic clone occurred following induction therapy for de novo AML.101, 102 Investigators discovered that at diagnosis of AML, a small, barely detectable, non-leukemic clone was overshadowed by the leukemic clone. After remission inducing chemotherapy, the non-leukemic/preleukemic clone markedly expanded (VAF of 20–40%) compared with the normal hematopoietic population. Thus, the non-leukemic/preleukemic mutant clones had a competitive advantage over the normal hematopoietic cells. These studies highlight the importance to distinguish Driver-related leukemia versus non-leukemic/preleukemic mutant populations when the physician assesses by DNA sequencing the response to induction chemotherapy.
Preleukemic stem cells and therapeutic implications
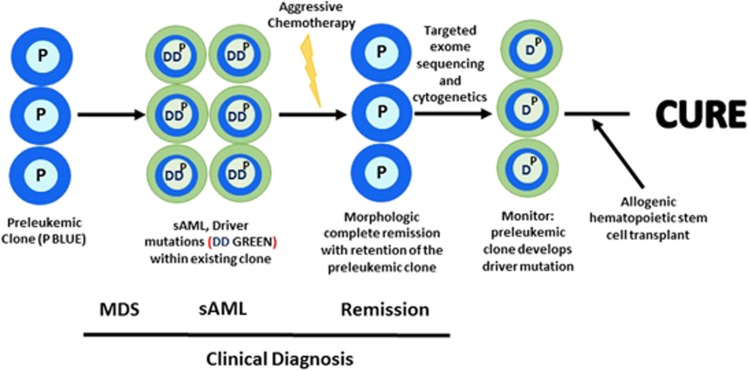
A long-lasting preleukemic (MDS) clone often marked by mutations of either DNMT3A, TET2 IDH1/2 or a spliceosome gene, can have a competitive growth advantage over the normal hematopoietic cells. These mutations can cause ineffective hematopoiesis. This can be followed by a subclone that develops a major Driver mutation (for example, NPM1) or fusion gene, resulting in frank AML with blast cells. These Driver (pan-AML) mutations block differentiation providing these cells with a proliferative advantage and causing life-threatening peripheral blood cytopenias. After ablative chemotherapy accompanied by a complete morphologic remission, the preleukemic clone may persist suggesting that these cells are resistant to standard chemotherapy, but are still able to differentiate to normal-appearing mature blood cells. If frank AML reoccurs, the original or new blastic AML subclone emerges from the preleukemic clone. Event-free survival of individuals with a preleukemic clone at morphologic complete remission, is inferior to those with no preleukemic clone during complete remission.30, 94 Perhaps, therapeutic intervention might be considered while these patients are in complete morphologic remission if nucleotide monitoring shows the acquisition of a Driver mutation.103 This might prompt hematopoietic transplantation before frank AML develops (Figure 2). Timing will be critical because reports have found that the preleukemic clone may be present for 8–10 years without emergence of relapsed blastic AML. Repeat targeted nucleotide sequencing of the marrow may be no more expensive than cytogenetic examination; and in the future sequencing may supplant cytogenetics in define circumstances.
Figure 2.
Evolution to MDS→secondary (s) AML (sAML)→remission and potential therapeutic intervention.
IDH2 mutant AML cells can generate D-2-hydroglycolate (D-2-HG) as a marker of the mutation.98 The plasma level of this compound is high in AML patients; however, those in complete morphologic remission with a mutant IDH2 clone may also have elevated serum D-2-HG. Therefore, measuring D-2-HG is not a good surrogate for making clinical decisions.98 Specific pharmacologic inhibitors of IDH2 have been developed. IDH2 mutation is often in the founder mutant clone; this clone acquires a Driver mutation to develop frank AML. The ability of the IDH2 inhibitor to eliminate the AML or founder clone requires further study.
Of interest, mathematic modeling noted a good correlation between the lifetime risk of developing cancer and the number of stem cell division that specific tissue undergoes during an individual's lifespan.104, 105 Mutations are passed to daughter cells only at cell division. Normal cells undergo about 1 mutation per 108 cell divisions. An estimated 11 000 to 22 000 early hematopoietic stem cells are present in an individual. This number decreases with age. Indeed, a likely hypothesis is the fewer hematopoietic stem cells that are present as an individual ages, the greater their proliferative pool to maintain normal peripheral blood counts; hence, the greater chance for a mutational event.104, 105 Many of the random mutations that occur in these cells affect non-critical genes (passenger genes) providing no growth advantage to these stem cells. Thus, ‘bad luck' happens when a mutation occurs in a gene that provides a proliferative advantage to the hematopoietic cell. The resulting clone of cells has a greater chance to acquire additional mutations that might evolve into frank AML.
Summary
In summary, the term ‘preleukemia' has evolved over the years. Initially in the 1950s and 1960s, preleukemia was viewed as MDS. We now know that the clinical course as well as chromosomal and mutational changes of MDS are often markedly different from those present in de novo AML. Individuals with an inherited mutation of RUNX1, GATA2 or C/EBP in their germline have a very high chance of developing AML; and thus have a preleukemic mutation. Also, previous treatment of a primary malignancy with either an alkylating drug or radiation therapy can result in a preleukemia that almost always progresses to AML. As outlined in Figure 3, very early hematopoietic stem cells can develop mutations that results in clonal hematopoiesis especially in those >60 years of age. In hematopoietically normal older individuals, this clone probably should not be called preleukemic. The incidence of evolution to AML in these individuals requires further study, but may be 0.5 to 1% per year. Quizzically, the mutations associated with MDS are often the same as those observed both in the normal elderly individual as well as the preleukemic clone in AML patients who are in morphologic complete remission. Development of overt AML requires Driver mutations. X-inactivation and DNA-sequencing studies showed that AML patients that obtained a complete morphologic remission can retain a preleukemic clone suggesting that these cells are resistant to normal ablative chemotherapy perhaps because they involve quiescent hematopoietic stem cells not easily killed by our standard chemotherapy. Patients whose leukemia evolves from an ancestral preleukemic clone, often will require therapy that exceeds our standard chemotherapy and perhaps monitoring of the preleukmic clone for acquisition of Driver mutations during remission may help to determine when aggressive therapy should be initiated. In conclusion, preleukemia might be defined as a condition that has modifying mutations in the bone marrow that either cause MDS or cause clonal hematopoietic expansion initially without disease but associated with progression to AML.
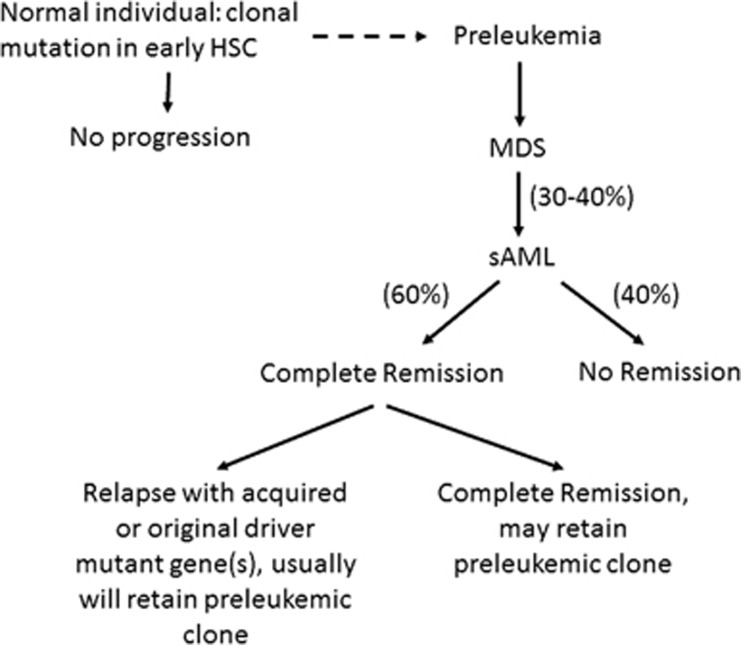
Figure 3.
Hematopoiesis: Normal elderly individual±evolution to sAML. Normal elderly individuals can develop clonal hemtopoiesis marked by a mutation in their early hematopoietic stem cells (HSC). Some of these individuals will develop myelodysplastic syndrome (MDS); and with the acquisition of a Driver mutation, they can develop secondary AML (sAML). These patients may go into complete morphologic remission but often retain their preleukemic clone. With relapse, they can either retain the original Driver mutation or acquire a new Driver mutation. Dotted line suggests that some normal elderly individuals with clonal hematopoiesis may develop MDS and sAML.
Acknowledgments
We thank the Melamed Family and Reuben Yeroushalmi for their generous support and Tim Babich for motivation to write this review. This work was also funded by the Leukemia Lymphoma Society of America, the Singapore Ministry of Health's National Medical Research Council (NMRC) under its Singapore Translational Research (STaR) Investigator Award to H. Phillip Koeffler (NMRC/STaR/0021/2014), Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2013-T2-2-150), the NMRC Centre Grant awarded to National University Cancer Institute of Singapore (NMRC/CG/012/2013) and the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiatives.
Footnotes
The authors declare no conflict of interest.
References
- Block M, Jacobson LO, Bethard WF. Preleukemic acute human leukemia. J Am Med Assoc 1953; 152: 1018–1028. [DOI] [PubMed] [Google Scholar]
- Koeffler HP. Myelodysplastic syndromes (preleukemia). Semin Hematol 1986; 23: 284–299. [PubMed] [Google Scholar]
- Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997; 89: 2079–2088. [PubMed] [Google Scholar]
- Brunning ROA, Germing U, Beau MM, Porwit A, Baumann I et al Myelodysplastic syndromes. In: Swerdlow SH International Agency for Research on Cancer, World Health Organization (eds). WHO classification of Tumours of Haematopoietic and Lymphoid Tissues; World Health Organization Classification of Tumours, 4th edn. International Agency for Research on Cancer: Lyon, France, 2008; p 439. [Google Scholar]
- Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med 2009; 361: 1872–1885. [DOI] [PubMed] [Google Scholar]
- Cazzola M, Della Porta MG, Travaglino E, Malcovati L. Classification and prognostic evaluation of myelodysplastic syndromes. Semin Oncol 2011; 38: 627–634. [DOI] [PubMed] [Google Scholar]
- Malcovati L, Hellstrom-Lindberg E, Bowen D, Ades L, Cermak J, Del Canizo C et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European Leukemia Net. Blood 2013; 122: 2943–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122: 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014; 28: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, West C, Johnson C. Involvement of the erythroid series in acute myeloid leukemia. Blood 1979; 53: 1203–1205. [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet 1962; 14: 135–148. [PMC free article] [PubMed] [Google Scholar]
- Busque L, Gilliland DG. X-inactivation analysis in the 1990 s: promise and potential problems. Leukemia 1998; 12: 128–135. [DOI] [PubMed] [Google Scholar]
- Gale RE, Linch DC. Clonality studies in acute myeloid leukemia. Leukemia 1998; 12: 117–120. [DOI] [PubMed] [Google Scholar]
- Beutler E, Yeh M, Fairbanks VF. The normal human female as a mosaic of X-chromosome activity: studies using the gene for C-6-PD-deficiency as a marker. Proc Natl Acad Sci USA 1962; 48: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Collins Z, Irwin LE. Value of genetic variants of glucose-6-phosphate dehydrogenase in tracing the origin of malignant tumors. N Engl J Med 1967; 276: 389–391. [DOI] [PubMed] [Google Scholar]
- Linder D, Gartler SM. Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas. Science 1965; 150: 67–69. [DOI] [PubMed] [Google Scholar]
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Fialkow PJ. Use of genetic markers to study cellular origin and development of tumors in human females. Adv Cancer Res 1972; 15: 191–226. [DOI] [PubMed] [Google Scholar]
- Janssen JW, Buschle M, Layton M, Drexler HG, Lyons J, van den Berghe H et al. Clonal analysis of myelodysplastic syndromes: evidence of multipotent stem cell origin. Blood 1989; 73: 248–254. [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011; 365: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 2013; 27: 1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012; 366: 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 2011; 364: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 2012; 44: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih LY, Huang CF, Wang PN, Wu JH, Lin TL, Dunn P et al. Acquisition of FLT3 or N-ras mutations is frequently associated with progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia 2004; 18: 466–475. [DOI] [PubMed] [Google Scholar]
- Dicker F, Haferlach C, Sundermann J, Wendland N, Weiss T, Kern W et al. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia 2010; 24: 1528–1532. [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016; 374: 2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015; 125: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialkow PJ, Janssen JW, Bartram CR. Clonal remissions in acute nonlymphocytic leukemia: evidence for a multistep pathogenesis of the malignancy. Blood 1991; 77: 1415–1417. [PubMed] [Google Scholar]
- Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996; 88: 59–65. [PubMed] [Google Scholar]
- Gale RE, Wheadon H, Goldstone AH, Burnett AK, Linch DC. Frequency of clonal remission in acute myeloid leukaemia. Lancet 1993; 341: 138–142. [DOI] [PubMed] [Google Scholar]
- Gale RE, Fielding AK, Harrison CN, Linch DC. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol 1997; 98: 512–519. [DOI] [PubMed] [Google Scholar]
- Williams DA, Lemischka IR, Nathan DG, Mulligan RC. Introduction of new genetic material into pluripotent haematopoietic stem cells of the mouse. Nature 1984; 310: 476–480. [DOI] [PubMed] [Google Scholar]
- Nash R, Storb R, Neiman P. Polyclonal reconstitution of human marrow after allogeneic bone marrow transplantation. Blood 1988; 72: 2031–2037. [PubMed] [Google Scholar]
- Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012; 44: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 2015; 10: 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016; 7: 12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012; 44: 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N Engl J Med 2002; 346: 564–569. [DOI] [PubMed] [Google Scholar]
- Strati P, Shanafelt TD. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood 2015; 126: 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015; 348: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker M, Lee S, Spira S, Takeuchi S, Koeffler HP. DNA aneuploidy in morphologically normal colons from patients with colon cancer. Mod Pathol 1995; 8: 360–365. [PubMed] [Google Scholar]
- Yakubovskaya MS, Spiegelman V, Luo FC, Malaev S, Salnev A, Zborovskaya I et al. High frequency of K-ras mutations in normal appearing lung tissues and sputum of patients with lung cancer. Int J Cancer 1995; 63: 810–814. [DOI] [PubMed] [Google Scholar]
- Cannon-Albright LA, Thomas A, Goldgar DE, Gholami K, Rowe K, Jacobsen M et al. Familiality of cancer in Utah. Cancer Res 1994; 54: 2378–2385. [PubMed] [Google Scholar]
- Kerber RA, O'Brien E. A cohort study of cancer risk in relation to family histories of cancer in the Utah population database. Cancer 2005; 103: 1906–1915. [DOI] [PubMed] [Google Scholar]
- Goldin LR, Kristinsson SY, Liang XS, Derolf AR, Landgren O, Bjorkholm M. Familial aggregation of acute myeloid leukemia and myelodysplastic syndromes. J Clin Oncol 2012; 30: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 1999; 23: 166–175. [DOI] [PubMed] [Google Scholar]
- Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, Li XC et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 2011; 43: 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Cavenagh JD, Lister TA, Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med 2004; 351: 2403–2407. [DOI] [PubMed] [Google Scholar]
- Wlodarski MW, Hirabayashi S, Pastor V, Stary J, Hasle H, Masetti R et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 2016; 127: 1387–1397. [DOI] [PubMed] [Google Scholar]
- Sellick GS, Spendlove HE, Catovsky D, Pritchard-Jones K, Houlston RS. Further evidence that germline CEBPA mutations cause dominant inheritance of acute myeloid leukaemia. Leukemia 2005; 19: 1276–1278. [DOI] [PubMed] [Google Scholar]
- Liew E, Owen C. Familial myelodysplastic syndromes: a review of the literature. Haematologica 2011; 96: 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels EM, Soodalter J, Churpek JE, Godley LA. Recognizing familial myeloid leukemia in adults. Ther Adv Hematol 2013; 4: 254–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Bessler M. Genetic predisposition syndromes: when should they be considered in the work-up of MDS? Best Pract Res Clin Haematol 2015; 28: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood 2015; 126: 2484–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin DW, Sakurai M, Nah GS, Du L, Jacob B, Yokomizo T et al. RUNX1 haploinsufficiency results in granulocyte colony-stimulating factor hypersensitivity. Blood Cancer J 2016; 6: e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi A, Toya T, Kawazu M, Ueno T, Tsukamoto A, Iizuka H et al. Recurrent CDC25C mutations drive malignant transformation in FPD/AML. Nat Commun 2014; 5: 4770. [DOI] [PubMed] [Google Scholar]
- Shiba N, Hasegawa D, Park MJ, Murata C, Sato-Otsubo A, Ogawa C et al. CBL mutation in chronic myelomonocytic leukemia secondary to familial platelet disorder with propensity to develop acute myeloid leukemia (FPD/AML). Blood 2012; 119: 2612–2614. [DOI] [PubMed] [Google Scholar]
- Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 2004; 104: 3565–3572. [DOI] [PubMed] [Google Scholar]
- Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 2011; 118: 2653–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 2011; 118: 2656–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 2011; 43: 929–931. [DOI] [PubMed] [Google Scholar]
- Wang X, Muramatsu H, Okuno Y, Sakaguchi H, Yoshida K, Kawashima N et al. GATA2 and secondary mutations in familial myelodysplastic syndromes and pediatric myeloid malignancies. Haematologica 2015; 100: e398–e401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Hsu AP, Holland SM, Cuellar-Rodriguez J, Hickstein DD. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica 2014; 99: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JP, Kwon EM, Gao Y, Trivedi NS, Elkahloun AG, Horwitz MS et al. Targeted correction of RUNX1 mutation in FPD patient-specific induced pluripotent stem cells rescues megakaryopoietic defects. Blood 2014; 124: 1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka H, Kagoya Y, Kataoka K, Yoshimi A, Miyauchi M, Taoka K et al. Targeted gene correction of RUNX1 in induced pluripotent stem cells derived from familial platelet disorder with propensity to myeloid malignancy restores normal megakaryopoiesis. Exp Hematol 2015; 43: 849–857. [DOI] [PubMed] [Google Scholar]
- Iwanski GBTN, Park DJ, Koeffler HP Therapy-related acute myelogenous leukemia. In: Wiernik PH GJ, Dutcher JP, Kyle R ed. Neoplastic Diseases of the Blood, 5th edition. Springer: New York, NY, USA, Science+Business Media 2013; pp 455–486. [Google Scholar]
- Pedersen-Bjergaard J, Philip P, Mortensen BT, Ersboll J, Jensen G, Panduro J et al. Acute nonlymphocytic leukemia, preleukemia, and acute myeloproliferative syndrome secondary to treatment of other malignant diseases. Clinical and cytogenetic characteristics and results of in vitro culture of bone marrow and HLA typing. Blood 1981; 57: 712–723. [PubMed] [Google Scholar]
- Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A et al The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes Blood 2009; 114: 937–951. [DOI] [PubMed]
- Rowley JD, Golomb HM, Vardiman JW. Nonrandom chromosome abnormalities in acute leukemia and dysmyelopoietic syndromes in patients with previously treated malignant disease. Blood 1981; 58: 759–767. [PubMed] [Google Scholar]
- Kantarjian HM, Keating MJ, Walters RS, Smith TL, Cork A, McCredie KB et al. Therapy-related leukemia and myelodysplastic syndrome: clinical, cytogenetic, and prognostic features. J Clin Oncol 1986; 4: 1748–1757. [DOI] [PubMed] [Google Scholar]
- Gundestrup M, Klarskov Andersen M, Sveinbjornsdottir E, Rafnsson V, Storm HH, Pedersen-Bjergaard J. Cytogenetics of myelodysplasia and acute myeloid leukaemia in aircrew and people treated with radiotherapy. Lancet 2000; 356: 2158. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J. Insights into leukemogenesis from therapy-related leukemia. N Engl J Med 2005; 352: 1591–1594. [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Albain KS, Larson RA, Vardiman JW, Davis EM, Blough RR et al. Clinical and cytogenetic correlations in 63 patients with therapy-related myelodysplastic syndromes and acute nonlymphocytic leukemia: further evidence for characteristic abnormalities of chromosomes no. 5 and 7. J Clin Oncol 1986; 4: 325–345. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Rowley JD. The balanced and the unbalanced chromosome aberrations of acute myeloid leukemia may develop in different ways and may contribute differently to malignant transformation. Blood 1994; 83: 2780–2786. [PubMed] [Google Scholar]
- Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: overview report. Genes Chromosomes Cancer 2002; 33: 331–345. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J, Andersen MT, Andersen MK. Genetic pathways in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Hematol Am Soc Hematol Educ Program 2007, 392–397. [DOI] [PubMed]
- Ok CY, Patel KP, Garcia-Manero G, Routbort MJ, Fu B, Tang G et al. Mutational profiling of therapy-related myelodysplastic syndromes and acute myeloid leukemia by next generation sequencing, a comparison with de novo diseases. Leuk Res 2015; 39: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015; 518: 552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian HM, Estey EH, Keating MJ. Treatment of therapy-related leukemia and myelodysplastic syndrome. Hematol Oncol Clin North Am 1993; 7: 81–107. [PubMed] [Google Scholar]
- Dreyfus B. Preleukemic states. I. Definition and classification. II. Refractory anemia with an excess of myeloblasts in the bone marrow (smoldering acute leukemia). Nouv Rev Fr Hematol Blood Cells 1976; 17: 33–55. [PubMed] [Google Scholar]
- Koeffler HP, Golde DW. Human myeloid leukemia cell lines: a review. Blood 1980; 56: 344–350. [PubMed] [Google Scholar]
- Saarni MI, Linman JW. Preleukemia. The hematologic syndrome preceding acute leukemia. Am J Med 1973; 55: 38–48. [DOI] [PubMed] [Google Scholar]
- Churpek JE, Larson RA. The evolving challenge of therapy-related myeloid neoplasms. Best Pract Res Clin Haematol 2013; 26: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Behm FG, Raimondi SC, Dodge RK, George SL, Rivera GK et al. Secondary acute myeloid leukemia in children treated for acute lymphoid leukemia. N Engl J Med 1989; 321: 136–142. [DOI] [PubMed] [Google Scholar]
- Ratain MJ, Kaminer LS, Bitran JD, Larson RA, Le Beau MM, Skosey C et al. Acute nonlymphocytic leukemia following etoposide and cisplatin combination chemotherapy for advanced non-small-cell carcinoma of the lung. Blood 1987; 70: 1412–1417. [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA 2014; 111: 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014; 506: 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 2015; 314: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 2015; 126: 2491–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QY, Ding LW, Tan KT, Chien W, Mayakonda A, Lin DC et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia 2016; e-pub ahead of print 8 July 2016 doi:10.1038/leu.2016.160. [DOI] [PMC free article] [PubMed]
- Ploen GG, Nederby L, Guldberg P, Hansen M, Ebbesen LH, Jensen UB et al. Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol 2014; 167: 478–486. [DOI] [PubMed] [Google Scholar]
- Wiseman DH, Williams EL, Wilks DP, Sun Leong H, Somerville TD, Dennis MW et al. Frequent reconstitution of IDH2R140Q mutant clonal multilineage hematopoiesis following chemotherapy for acute myeloid leukemia. Leukemia 2016; 30: 1946–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun 2016; 7: 12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Magrini V, Demeter R, Miller CA, Fulton R, Fulton LL et al. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet 2014; 10: e1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Miller CA, Klco JM, Petti A, Demeter R, Helton NM et al. Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood 2016; 127: 893–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Miller CA, Griffith OL, Krysiak K, Skidmore ZL, Ramu A et al. Optimizing cancer genome sequencing and analysis. Cell Syst 2015; 1: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey A, Hills RK, Simpson MA, Jovanovic JV, Gilkes A, Grech A et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med 2016; 374: 422–433. [DOI] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015; 347: 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]