Abstract
The intense selection pressure exerted by virus-specific cytotoxic T lymphocytes (CTL) on replicating human immunodeficiency virus and simian immunodeficiency virus results in the accumulation of CTL epitope mutations. It has been assumed that fitness costs can limit the evolution of CTL epitope mutations. However, only a limited number of studies have carefully examined this possibility. To explore the fitness costs associated with viral escape from p11C, C-M-specific CTL, we constructed a panel of viruses encoding point mutations at each position of the entire p11C, C-M epitope. Amino acid substitutions at positions 3, 4, 5, 6, 7, and 9 of the epitope significantly impaired virus replication by altering virus production and Gag protein expression as well as by destabilizing mature cores. Amino acid substitutions at position 2 of the epitope were tolerated but required reversion or additional compensatory mutations to generate replication-competent viruses. Finally, while amino acid substitutions at positions 1 and 8 of the p11C, C-M epitope were functionally tolerated, these substitutions were recognized by p11C, C-M-specific CTL and therefore provided no selection advantage for the virus. Together, these data suggest that limited sequence variation is tolerated by the region of the capsid encoding the p11C, C-M epitope and therefore that only a very limited number of mutations can allow successful viral escape from the p11C, C-M-specific CTL response.
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) escape from dominant epitope-specific cytotoxic T-lymphocyte (CTL) recognition is associated with uncontrolled viral replication and the clinical progression of AIDS. Since virus-specific CTL play a critical role in containing HIV and SIV replication in infected individuals (17, 23, 44), this immune response exerts considerable selection pressure on the replicating virus. The selection pressure leads to an accumulation of CTL epitope mutations (1-4, 6, 8, 10, 14, 19, 23, 29, 33, 34, 53) that shape the evolution of SIV and HIV variation (28). Moreover, an increase in viral replication as a result of these mutational events leads to disease progression in HIV type 1 (HIV-1)-infected humans (4, 13, 15, 21, 39, 40) as well as SIV- and simian-human immunodeficiency virus (SHIV)-infected rhesus monkeys (2, 3). Thus, the mutation of viruses to escape from CTL responses plays a central role in AIDS pathogenesis.
Interestingly, the rate at which particular immunodominant CTL responses select for HIV and SIV mutations can vary dramatically (1, 4, 10, 13, 15, 19, 27, 28, 33, 40). CTL responses to an immunodominant HLA-B44-restricted gp160 epitope or HLA-B8-restricted Nef epitope rapidly select for amino acid substitutions in these epitopes during primary viremia in HIV-1-infected individuals (4, 40). Yet CTL responses to a comparably dominant HLA-B27-restricted HIV-1 Gag epitope select for epitope mutations that can occur after several years of persistent viral replication (15). In addition, the number of residues of a CTL epitope that mutate varies substantially for different epitopes. Viral escape from a dominant Tat epitope-specific CTL response universally occurs within the first 3 weeks of SIVmac239 infection in Mamu-A*01+ rhesus monkeys and occurs by mutation of any one of the nine residues of the epitope. Yet escape from comparably dominant Gag and Env epitope-specific CTL responses occurs more slowly, and mutations are generally restricted to one or two positions of the epitopes (1-3, 7, 33).
While it is assumed that fitness costs for the virus can limit the rate of viral escape from a particular epitope-specific CTL response, this supposition has not been carefully explored (12, 32, 38, 52). The Gag p11C, C-M epitope is extraordinarily dominant in SIV- and SHIV-infected Mamu-A*01+ rhesus monkeys (24). However, viral escape from CTL by mutation of this epitope is slow to evolve and rarely occurs in SIVmac239- and SHIV-89.6P-infected monkeys (2, 3, 33). Moreover, viral escape from p11C, C-M-specific CTL responses occurs at a limited number of residues in the epitope (1-3, 7, 10, 33, 34). For the present study, we sought to determine why viral escape from this immunodominant epitope is infrequent and preferentially occurs at position 2. To this end, we evaluated the functional consequences of individual point mutations in the p11C, C-M epitope and explored the consequences of these mutations on the viability of the virus.
MATERIALS AND METHODS
gag point mutations.
BmgBI and EcoRV restriction sites were introduced flanking the p11C, C-M epitope by PCR mutagenesis with a QuickChange kit (Stratagene, La Jolla, Calif.), with the previously described VRC-WT-gag plasmid as the template (38). The oligonucleotide primers BmgBIF (5′-TCAGGCACGTCCAGAAGGTTGCACC-3′; nucleotides [nt] 1574 to 1598) and BmgBIR (5′-TTCTGGACGTGCCTGAAATCCTGG-3′; nt 1566 to 1589) were used to introduce the BmgBI restriction site. The oligonucleotide primers EcoRVF (5′-TGTGTGGGATATCATCAAGCGGCTATGCAG-3′; nt 1626 to 1655) and EcoRVR (5′-CGCTTGATGATATCCCACACAATTTAACATCTGATTAATGTC-3′; nt 1605 to 1646) were used to introduce the EcoRV restriction site. For the above primers, restriction sites are underlined, mutated nucleotides are shown in bold, and the positions of the oligonucleotide primers are numbered according to the SIV isolate 239 complete proviral genome and flanking sequence (GenBank accession no. M33262) (20). The resulting plasmid, VRC-BEgag, was sequenced to ensure that no additional mutations were introduced. To create the gag point mutations, we digested VRC-BEgag with BmgBI and EcoRV, treated it with calf intestinal alkaline phosphatase, gel purified it, and ligated it to the phosphorylated complementary oligonucleotide pairs shown in Table 1. This process eliminated the newly introduced BmgBI and EcoRV restriction sites, restored these regions of the protein to the wild-type sequence, and introduced the desired p11C, C-M point mutations. In addition, this process changed the aspartic acid coding sequence from GAC to GAT at position 60 of the capsid protein, allowing mutant viruses to be discriminated from wild-type viruses.
TABLE 1.
Phosphorylated primer sequences used to construct SIV capsid mutants
| CA mutant | Forward sequencea (5′-3′) |
|---|---|
| Wild type | TGTCAGAAGGTTGCACCCCCTATGACATTAATCAGATGTTAAATTGTGTGGGAG |
| C46A | TGTCAGAAGGTGCCACCCCCTATGACATTAATCAGATGTTAAATTGTGTGGGAG |
| T47A | TGTCAGAAGGTTGCGCCCCCTATGACATTAATCAGATGTTAAATTGTGTGGGAG |
| P48A | TGTCAGAAGGTTGCACCGCCTATGACATTAATCAGATGTTAAATTGTGTGGGAG |
| Y49A | TGTCAGAAGGTTGCACCCCCGCCGACATTAATCAGATGTTAAATTGTGTGGGAG |
| D50A | TGTCAGAAGGTTGCACCCCCTATGCCATTAATCAGATGTTAAATTGTGTGGGAG |
| I51A | TGTCAGAAGGTTGCACCCCCTATGACGCCAATCAGATGTTAAATTGTGTGGGAG |
| N52A | TGTCAGAAGGTTGCACCCCCTATGACATTGCCCAGATGTTAAATTGTGTGGGAG |
| Q53A | TGTCAGAAGGTTGCACCCCCTATGACATTAATGCCATGTTAAATTGTGTGGGAG |
| M54A | TGTCAGAAGGTTGCACCCCCTATGACATTAATCAGGCCTTAAATTGTGTGGGAG |
| Y49Q | TGTCAGAAGGTTGCACCCCCCAGGACATTAATCAGATGTTAAATTGTGTGGGAG |
| I51L | TGTCAGAAGGTTGCACCCCCTATGACCTTAATCAGATGTTAAATTGTGTGGGAG |
| Q53T | TGTCAGAAGGTTGCACCCCCTATGACATTAATACGATGTTAAATTGTGTGGGAG |
Sequence for nt 1582 to 1635. Bold type indicates the p11C, C-M epitope; underlining indicates mutated codons.
Viruses and cells.
Mutant SIVmac239 proviral plasmids were produced by shuttling gag genes from the VRC-gag plasmids into a full-length wild-type SIVmac239 proviral plasmid. A full-length wild-type SIVmac239 proviral plasmid was obtained as a gift from Heinrich Göttlinger (Dana Farber Cancer Institute, Boston, Mass.). For the production of recombinant viruses, 10-μg samples of wild-type and mutant SIVmac239 proviral plasmid DNAs were transfected into 293T cells by the calcium phosphate method (Invitrogen, Carlsbad, Calif.). Forty-eight hours later, the supernatants were harvested and purified through a 0.45-μm-pore-size filter. Single-round reporter viruses (SIV.GFP) were produced in 293T cells (American Type Culture Collection) by use of the pSIVΔenvGFP plasmid as previously described (37). Viruses were pseudotyped with the vesicular stomatitis virus envelope glycoprotein by the use of previously described plasmids. Forty-eight hours later, the supernatants were harvested and purified through a 0.45-μm-pore-size filter.
Viral replication rates.
Mutant gag genes were shuttled into wild-type p239SpSp5′. p239SpSp5′ was obtained from Ronald Desrosiers through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (20, 41). Recombinant SIVmac239 viruses were produced as previously described (20, 41). Briefly, 5 μg of each proviral half was digested with SphI, extracted with phenol-chloroform, ethanol precipitated, and ligated. The ligation mix was then transfected into CEMx174 cells (American Type Culture Collection) by the DEAE-dextran method. Cultures were monitored for p27 expression and reverse transcriptase (RT) activity by a colorimetric enzyme-linked immunosorbent assay (ELISA) (Roche, Mannheim, Germany).
Core stability assay.
The stabilities of wild-type and mutant cores were determined as previously described (11). Briefly, cell-free virus was concentrated by ultracentrifugation (100,000 × g for 4 h at 4°C) through a cushion of 20% (wt/vol) sucrose in STE buffer (10 mM Tris-HCl [pH 7.3], 100 mM NaCl, 1 mM EDTA). Virions were resuspended in 200 μl of STE buffer, loaded onto a linear sucrose gradient (10 ml of 30 to 70% sucrose in STE buffer) with or without a layer of detergent (1 ml of 15% sucrose-0.5% NP-40 in STE buffer), and then ultracentrifuged (100,000 × g for 20 h at 4°C). Fractions (1 ml) were collected from the top and assayed for SIV p27 by a colorimetric ELISA (Coulter, Miami, Fla.). To control for variations in the amount of virus loaded onto the linear sucrose gradient, we calculated the percent yield of cores for each mutant virus as follows: SIV p27 in core fractions/total SIV p27 in all fractions.
Protein quantification and Western blots.
293T cells were transfected with 10 μg of full-length proviral plasmid DNA by the calcium phosphate method (Invitrogen). Forty-eight hours later, the cell supernatants were collected and the cells were lysed for 20 min with a solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and protease inhibitors (Roche). The cell lysates were centrifuged for 20 min at 16,000 × g, and the soluble fraction was separated from the insoluble pellet. A SIV core antigen assay (Coulter) was used to quantify the concentration of SIV p27 in the supernatants and soluble cell lysates. Supernatants and soluble cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes for Western blotting. An anti-SIVmac p27 monoclonal antibody (55-2F12) was obtained from Niels Pederson through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH (16). The primary detection antibody 55-2F12 (anti-SIVmac p27) and anti-HSP-90 (BD Biosciences, San Diego, Calif.) were used at a 1:2,000 dilution and were detected with a SuperSignal West Pico mouse IgG detection kit (Pierce, Rockford, Ill.).
Virus sequencing.
Virus sequence analyses of 500-bp regions of gag were performed as described below. Viruses were isolated from cell-free supernatants by use of a viral RNA isolation kit (Qiagen, Valencia, Calif.). The positions of the oligonucleotide primers are numbered according to the SIV isolate 239 complete proviral genome and flanking sequence (accession no. M33262). Viral sequences were amplified with a OneStep RT-PCR kit (Qiagen) and the primers gag-fwd (5′-CTTTCGGTCTTAGCTCCATTAGTGCC-3′; nt 1233 to 1258) and gag-rev (5′-TGTCTGTTCTGCTCTTAAGCTTTTGTAG-3′; nt 1955 to 1982). RT-PCR products were purified in agarose gels, the 749-bp fragment was TA cloned into the pCR4-TOPO sequencing vector (Invitrogen), and individual transformed colonies were subjected to T7/SP6 dideoxy sequencing with the oligonucleotide gag-seq (5′-CTTTCGGTCTTAGCTCCATTAGTG-3′; nt 1233 to 1256).
CTL assays.
Functional CTL assays were performed as previously described (42). Lymphocytes were isolated from monkey peripheral blood by Ficoll-diatrizoate gradient centrifugation. Cultures were started with 4 × 106 peripheral blood lymphocytes (PBL) per ml in RPMI 1640 containing 12% fetal calf serum, antibiotics, and 1 μM wild-type p11C, C-M peptide. On day 3 of culture, 20 U of recombinant interleukin-2 per ml was added. On days 12 to 17 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr-release CTL assay. Target cells were major histocompatibility complex (MHC) class I-deficient Mamu-A*01 transfectants of either the B-lymphoblastoid cell line C1R (42) or the cell line 721.221, kindly provided by D. I. Watkins (Wisconsin National Primate Research Center, Madison, Wis.), that had been incubated for 1.5 h or overnight with the peptide at various concentrations as well as with 75 to 100 μCi of sodium [51Cr]chromate. After being washed, 1,000 to 2,000 target cells per well were added to 96-well U-bottomed plates. Effectors were added at various effector-to-target cell ratios in a final volume of 200 μl of RPMI 1640 with 12% fetal calf serum and were incubated for 4 h at 37°C. Next, 50 μl of the supernatant was added to 200 μl of scintillation fluid, and the mixture was analyzed in a 1450 Microbeta liquid scintillation counter. All experimental wells were set up in duplicate, and maximal release and spontaneous release wells were set up in quadruplicate. Maximal release was calculated by incubating labeled targets with 1% Triton X-100, and spontaneous release was calculated by incubating labeled targets with medium alone. Specific lysis was calculated as follows: [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100. For all experiments, the specific lysis of targets incubated without peptide was <4%.
RESULTS
Retroviral sequence alignment of the capsid region encoding the p11C, C-M CTL epitope.
The p11C, C-M-specific CTL response is immunodominant in SIV- and SHIV-infected Mamu-A*01+ rhesus monkeys. As such, this epitope-specific immune response should exert significant selective pressure on replicating viruses in these animals. However, viral mutations in this epitope appear to be rare and, when they occur, are associated with a flanking mutation. This observation suggests that the p11C, C-M region of the viral capsid may tolerate very little sequence variation. To explore this possibility, we first evaluated the known sequences of the p11C, C-M region of the capsid proteins of SIVs and other retroviruses. We aligned full-length Gag protein sequences from consensus SIVs, consensus HIVs, and 10 additional retroviruses (Fig. 1). Four of the six consensus SIV Gag sequences show complete conservation of the p11C, C-M epitope, while SIVtan shares seven of the nine residues and SIVlst shares four of the nine residues. The consensus HIV-1 Gag sequences show conservation of five of the nine p11C, C-M epitope residues, and the consensus HIV-2 Gag sequences show complete conservation of the p11C, C-M epitope. An alignment of the Gag sequences of 10 more distantly related retroviruses with the consensus SIVmac sequence showed significant sequence identity or similarity. This sequence similarity of the p11C, C-M CTL epitope among related retroviruses suggests that the epitope is located in a structurally important region of the capsid protein.
FIG. 1.
Sequence alignment of the p11C, C-M region from various retrovirus capsid proteins. Consensus SIV and HIV sequences were obtained from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov), and the additional protein sequences of other retroviruses were obtained from the National Center for Biotechnology Information Entrez database. Sequences were aligned with DNAStar MegAlign by the ClustalW method and were visualized with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). Black shading indicates sequence identity; gray shading indicates sequence similarity. The following viruses were used: FIV, feline immunodeficiency virus (accession no. P16087); EIAV, equine infectious anemia virus (accession no. AAK21105); ovine lentivirus (accession no. NP_041249); Visna virus (accession no. NP_040839); BIV, bovine immunodeficiency virus (accession no. NP_040562); Jembrana disease virus (accession no. NP_042684); HTLV-1, human T-lymphotropic virus type 1 (accession no. NP_057862); HTLV-2 (accession no. P03346); STLV-3, simian T-lymphotropic virus type 3 (accession no. AAO62100); MLV, murine leukemia virus (accession no. P29167).
p11C, C-M epitope mutations impair Gag protein expression and virion release.
We then sought to determine why mutations in this dominant epitope are infrequent. To this end, we constructed a series of SIVs with alanine point mutations at each residue of the epitope. SIVs were also constructed for these studies with substitutions in the p11C, C-M epitope consistent with those changes seen in HIV-1 viruses, with the reasoning that these changes should be quite conservative. To assess the effects of these mutations on particular steps in the viral life cycle, we measured (i) Gag protein expression and virion release, (ii) virus production, (iii) core stability, and (iv) replication kinetics (Table 2).
TABLE 2.
Effects of p11C, C-M epitope point mutations on SIV viabilitya
| Virus | Gag expression | Virion release (%) | Virus production (%) | Core stability (%) | Infectivity |
|---|---|---|---|---|---|
| Wild type | +++ | 100 | 100 | 100 | + |
| C46A | +++ | 84 | 102 | 126 | + |
| T47A | ++ | 97 | 7 | 84 | + |
| P48A | + | 93 | 26 | 48 | − |
| Y49A | +++ | 82 | 59 | 11 | − |
| D50A | + | 84 | 36 | 13 | − |
| I51A | + | 102 | 17 | 7 | − |
| N52A | + | 136 | 21 | 0 | − |
| Q53A | +++ | 101 | 98 | 139 | + |
| M54A | + | 76 | 17 | 5 | − |
| Y49Q | ++ | 97 | 26 | 42 | − |
| I51L | +++ | 93 | 6 | 135 | − |
| Q53T | +++ | 112 | 20 | 124 | + |
Comparisons to the wild-type virus were made by normalizing the values in the assays and establishing the values for the wild-type virus as 100%.
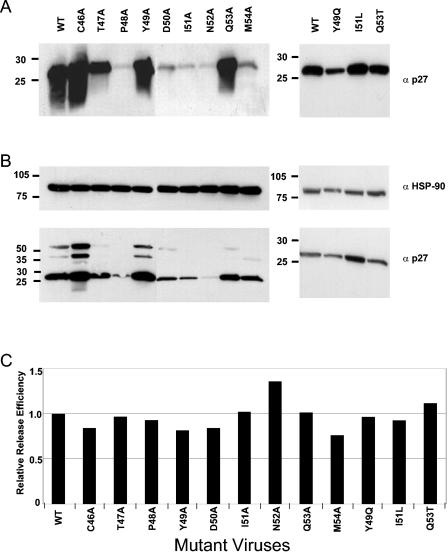
We first explored the effects of various single amino acid substitutions in the p11C, C-M epitope on Gag protein expression. We transfected 293T cells with full-length proviral plasmids and 48 h later assessed Gag p27 expression in the supernatants (Fig. 2A) and soluble cell lysates (Fig. 2B). Cells transfected with wild-type SIVmac239 proviral DNA had readily detectable levels of Gag p27 expression in both the supernatants and cell lysates (Fig. 2A and B). Alanine substitutions at positions 1, 4, and 8 (C46A, Y49A, and Q53A) as well as HIV-1-like substitutions at positions 6 and 8 (I51L and Q53T) of the p11C, C-M epitope did not significantly alter Gag p27 expression in either the supernatants or cell lysates (Fig. 2A and B). An alanine substitution at position 2 (T47A) and an HIV-1-like substitution at position 4 (Y49Q) of the p11C, C-M epitope resulted in a moderate reduction in Gag protein expression in the supernatants but did not affect Gag p27 expression in the soluble cell lysates. Importantly, alanine substitutions at positions 3, 5, 6, 7, and 9 (P48A, D50A, I51A, N52A, and M54A) of the p11C, C-M epitope reduced the levels of Gag p27 expression to near background levels in both the supernatants and soluble cell lysates. To ensure that equal quantities of samples were loaded into the gel, we normalized the supernatants to the total protein concentration (data not shown) and blotted soluble cell lysates with anti-HSP-90 (Fig. 2B). Thus, while some single amino acid substitutions in the Gag p11C, C-M epitope were tolerated, mutations of residues 2, 3, 5, 6, 7, and 9 resulted in significant decreases in Gag p27 expression.
FIG. 2.
Mutations in the p11C, C-M epitope decrease protein expression but do not affect virion release. (A and B) 293T cells were transfected with 10 μg of full-length proviral plasmid DNA containing the indicated Gag point mutations. Forty-eight hours later, supernatants (A) and cell lysates (B) were analyzed by Western blotting. (C) Virion release efficiencies were calculated as the total amounts of Gag in the supernatants divided by the total amounts of Gag in both cell lysates and supernatants, as determined for the gels shown in panels A and B. The values used for these calculations were arbitrary intensity units per square millimeter that were derived by using Quantity One image analysis software.
We next assessed the effects of these mutations on virion release. We performed a densitometric analysis of the gels shown in Fig. 2A and B and calculated the relative release efficiency as the total amount of Gag protein expression in the supernatants (Fig. 2A) divided by the total amount of Gag protein expression in both the supernatants and the soluble cell lysates (Fig. 2A and B). We observed no substantial differences in virus release efficiency between cells that were transfected with the wild-type proviral plasmid and those that were transfected with p11C, C-M epitope mutant proviral plasmids (Fig. 2C). These results are consistent with the fact that the determinants of virus budding are localized to the Gag p6 and not the Gag p27 protein (5, 22, 46). Therefore, while mutations to the p11C, C-M epitope reduced Gag protein expression, the observed reduction was comparable in both supernatants and cell lysates (Fig. 2A and B). This finding suggests that these amino acid substitutions do not result in decrements in virus particle release.
Gag p11C, C-M epitope mutations result in decreased virus production.
We then examined the effects of these p11C, C-M mutations on virus production. We transfected 293T cells with wild-type or p11C, C-M epitope mutant proviral plasmids and assessed virus production (Fig. 3A and B). Cells transfected with wild-type SIVmac239 proviral plasmid DNA produced readily detectable levels of virus, as measured by both RT activity and p27 expression. Cells transfected with proviral plasmid DNAs encoding alanine substitutions at positions 1 and 8 of the p11C, C-M epitope (C46A and Q53A) produced virus at levels comparable to that of the wild-type proviral plasmid DNA, as measured by both RT activity and p27 expression. However, cells transfected with proviral plasmid DNAs encoding p11C, C-M epitope alanine substitutions at positions 2, 3, 4, 5, 6, 7, and 9 (T47A, P48A, Y49A, D50A, I51A, N52A, and M54A) as well as HIV-1-like mutations at positions 4, 6, and 8 (Y49Q, I51L, and Q53T) produced significantly lower levels of virus, as measured by RT activity.
FIG. 3.
Mutations in the p11C, C-M epitope decrease virus production. 293T cells were transfected with 10 μg of full-length proviral SIVmac239 plasmid DNA (A and B) or single-round SIV.GFP plasmid DNA (C) containing the indicated Gag point mutations. Forty-eight hours later, the supernatants were harvested and filtered through a 0.45-μm-pore-size filter. Cell-free supernatants were analyzed for virus production by a colorimetric RT assay (A) and SIV p27 ELISA (B and C). The values illustrated are the means ± standard errors of duplicates.
These findings were confirmed by transferring the p11C, C-M epitope mutations into a single-round SIV.GFP plasmid and assessing virus production after the transfection of 293T cells (Fig. 3C). As observed with the SIVmac239 proviral plasmids, cells transfected with proviral plasmid DNAs encoding alanine substitutions at positions 1 and 8 of the p11C, C-M epitope (C46A and Q53A) as well as the HIV-1-like mutation at position 6 (I51L) produced virus at levels comparable to that of the wild-type SIV.GFP plasmid DNA, as measured by p27 expression. Moreover, cells transfected with proviral plasmid DNAs encoding alanine substitutions at positions 2, 3, 4, 5, 6, 7, and 9 (T47A, P48A, Y49A, D50A, I51A, N52A, and M54A) as well as HIV-1-like mutations at positions 4 and 8 (Y49Q and Q53T) produced significantly lower levels of virus, as measured by p27 expression.
The relative magnitudes of RT activity and Gag p27 expression were similar for nearly all of the mutant viruses. However, two of the mutant viruses (I51L and Q53T) showed elevated Gag p27 levels compared to their levels of RT activity, suggesting that the mutant viral particles produced by these cells may contain lower than normal quantities of RT and may therefore not be infectious. Therefore, alanine mutations at positions 2, 3, 4, 5, 6, 7, and 9, as well as an HIV-1-like mutation at position 4, of the p11C, C-M epitope decrease the level of virus production.
Capsid point mutations in the p11C, C-M epitope alter core stability.
The formation of an optimally stable core is critical for HIV-1 replication, since cores that dissociate too quickly or too slowly have been shown to be associated with reduced viral infectivity (11). To determine if point mutations in the p11C, C-M epitope alter core stability, we evaluated the mutant viruses by using a core stability assay (11). We first examined the properties of wild-type SIVmac239 cores with this assay. Virus pellets that were concentrated and purified by ultracentrifugation through a 20% sucrose cushion were layered onto a linear 30 to 70% sucrose gradient. Upon ultracentrifugation, virions entered into the linear sucrose gradient and sedimented at a density of 1.15 to 1.18 g/ml (Fig. 4A), a density that is similar to that of HIV-1 and other retroviruses (11). When concentrated wild-type SIVmac239 was ultracentrifuged through a layer of 0.5% NP-40 on top of the linear 30 to 70% sucrose gradient, the lipid bilayer was removed and mature cores sedimented at a density of 1.20 to 1.23 g/ml (Fig. 4B), a density distinct from that of intact virions.
FIG. 4.
p11C, C-M epitope mutations alter SIV core stability. 293T cells were transfected with 10 μg of full-length proviral plasmid DNA containing the indicated Gag point mutations. Forty-eight hours later, the viruses were harvested and pelleted through a 20% sucrose cushion. The wild-type virus was layered onto 30 to 70% linear sucrose gradients with (B) and without (A) a layer of 0.5% NP-40. After ultracentrifugation at 100,000 × g for 20 h at 16°C, 1-ml fractions were collected from the top of the gradient and analyzed by p27 ELISA (filled columns). The density of each fraction was determined by refractometry (open diamonds). (C) Wild-type and mutant viruses were concentrated and ultracentrifuged through a 0.5% NP-40 layer into a linear 30 to 70% sucrose gradient. Percent yield of cores, percentage of p27 detected in the core fraction, as determined in panel B. The values illustrated in panel C are means ± standard deviations of three experiments. WT, wild type.
To explore the effect of single amino acid mutations in the p11C, C-M epitope on core stability, we subjected wild-type and mutant SIVs to the same procedure and calculated core yields by determining the amounts of capsid protein in the dense core fractions as percentages of the total capsid protein in the gradient. Similar to that observed for HIV-1, the yield of cores for wild-type SIVmac239 was approximately 20% (Fig. 4C). Alanine substitutions at positions 1 and 8 of the p11C, C-M epitope (C46A and Q53A) as well as HIV-1-like substitutions at positions 6 and 8 (I51L and Q53T) resulted in increased core yields of 25, 27, 27, and 24%, respectively. However, alanine substitutions at positions 2, 3, 4, 5, 6, 7, and 9 (C46A, T47A, P48A, Y49A, D50A, I51A, N52A, and M54A) as well as the HIV-1 like mutation at position 4 (Y49Q) of the p11C, C-M epitope significantly reduced core yields, to 16, 9, 2, 2, 1, 0, 1, and 8%, respectively. Therefore, while mutations to the p11C, C-M epitope did not alter the gradient profiles of these mutant cores (data not shown), mutations at residues 2, 3, 4, 5, 6, 7, and 9 affected the stability of these cores.
Gag p11C, C-M epitope mutations affect viral replication.
Because mutations of all of the Gag p11C, C-M epitope residues except for those at positions 1 and 8 resulted in decreased core stability, protein expression, and virus production, we sought to determine the effects of these abnormalities on the replication of the virus. CEMx174 cells were infected with these SIVmac239 mutants, and the supernatants were analyzed for Gag p27 expression and RT activity (Fig. 5A to D). Cells infected with wild-type SIVmac239 exhibited a peak in Gag p27 production and RT activity on day 7 of culture. Cells infected with a virus containing alanine at position 1 of the p11C, C-M epitope also exhibited a peak in Gag p27 expression and RT activity on day 7. Cells infected with viruses containing either an alanine or HIV-1-like substitution at position 8 of the p11C, C-M epitope displayed a peak in Gag p27 expression and RT activity on day 14. Cells infected with viruses containing alanine at position 2 or 7 (T47A and N52A) exhibited a peak in Gag p27 expression and RT activity on days 21 and 35, respectively. The Gag p27 protein was detected in supernatants of cells infected with the position 4 (Y49A) epitope mutation only on day 21, and the level of Gag p27 was >3 log lower in the cultures than in the supernatants of cells infected with the wild-type virus. RT activity was never detected in the supernatants of these infected cells. Importantly, cells infected with viruses containing p11C, C-M alanine substitutions at positions 3, 5, 6, 7, and 9 (P48A, D50A, I51A, N52A, and M54A) as well as HIV-1-like mutations at positions 4 and 6 (Y49Q and I51L) never generated the Gag p27 protein or RT activity. Therefore, alanine substitutions and HIV-1-like mutations at positions 2, 3, 4, 5, 6, 7, 8, and 9 of the Gag p11C, C-M epitope appeared to impair viral replication.
FIG. 5.
Mutations in the p11C, C-M epitope decrease the SIV replication rate. Five micrograms of p239SpSp5′ containing the noted gag point mutations and 5 μg of p239Sp3′ were digested, extracted with phenol-chloroform, ethanol precipitated, ligated together, and transfected into CEMx174 cells by the DEAE-dextran method. Cell-free supernatants were monitored for the SIV p27 antigen (A and B) and for RT activity (C and D). The values illustrated are means ± standard deviations of triplicates. (E) Sequence analysis of five to eight clones determined at the peak of viral replication for viruses that replicated (WT [wild type], C46A, T47A, N52A, Q53A, and Q53T). The deduced amino acid sequences for 60 amino acids are shown. The Mamu-A*01-restricted p11C, C-M epitope is bracketed by vertical lines. Amino acids that differ from the wild type are shown in bold and underlined. Numbers in parentheses indicate the number of mutant clones divided by the total number of clones analyzed for each mutant.
Stability of epitope mutations in replicating virus.
Having demonstrated that mutation of each of the residues of the p11C, C-M epitope except for that at position 1 affects at least one step in the replication of SIVmac239, we sought to determine the stability of each introduced mutation in a replicating virus. We sequenced the virus produced in each of the cultures at the time of the peak of Gag p27 and RT production to determine if compensatory or reversion mutations were acquired that facilitated virus replication (Fig. 5D). Since viruses with p11C, C-M epitope mutations at positions 3, 4, 5, 6, and 9 did not replicate, no viruses from the supernatants of these infected cells were available for sequencing. The sequence of the virus obtained from the culture of cells infected with the position 7 p11C, C-M epitope substitution (N52A) on day 35 revealed only wild-type virus. This suggests that mutation at position 7 of the p11C, C-M epitope significantly decreased viral fitness and that reversion to the wild-type sequence was required for viral replication. A mutation at position 2 (T47A) of the epitope required additional compensatory mutations or reversion back to the wild-type sequence to yield replication-competent virus. In fact, the virus sequenced from the culture established with the T47A mutant on day 21 revealed four different virus species. Three of the seven sequenced clones were wild-type virus, two contained the T47A mutation in addition to an isoleucine-to-valine substitution at position 26, one contained an additional leucine-to-serine substitution at position 25, and one contained the T47A mutation alone. The fact that the majority of the sequenced clones either were wild-type virus or contained additional compensatory mutations supports the observation that mutations at position 2 of the p11C, C-M epitope impair viral fitness (12, 38, 43). Viruses that were sequenced from cells infected with position 1 or 8 epitope mutations revealed the presence of only those p11C, C-M epitope mutations. The fact that we observed neither sequence reversion nor the emergence of additional extra-epitopic mutations suggests that at least some mutations at positions 1 and 8 (C46A, Q53A, and Q53T) of the p11C, C-M epitope are structurally tolerated and can yield replication-competent viruses. Therefore, mutating the p11C, C-M epitope at any position other than position 1 or 8 appears to have a significant fitness cost for the virus, and compensatory mutations or reversion to the wild-type sequence is required for the generation of replication-competent virus.
Position 1 and position 8 mutations are recognized by Gag p11C, C-M-specific CTL.
Viral escape from the p11C, C-M-specific CTL response occurs predominantly at position 2 of the epitope and only occasionally at position 1 in vivo (1-3, 7, 9, 33). While the only viable p11C, C-M substitutions that we observed were at positions 1 and 8 of the epitope, these specific mutations at these residues of the virus have not been documented in vivo (1-3, 7, 10, 33). We reasoned that for successful viral escape from CTL to occur, mutations to the CTL epitope must be tolerated by the virus. Moreover, a selective advantage must exist for a mutant virus to become predominant in the viral quasispecies. An absence of recognition by wild-type epitope-specific CTL confers such a selective advantage. Since mutations at positions 1 and 8 of the p11C, C-M epitope were viable but are not observed in vivo, we hypothesized that these mutant epitopes may be recognized by p11C, C-M-specific CTL. To explore this possibility, we stimulated PBL from a Mamu-A*01+ SIV-infected monkey with the wild-type p11C, C-M peptide and 17 days later tested the ability of these cells to recognize target cells pulsed with the wild-type or position 1 or 8 (C46A, Q53A, and Q53T) mutant peptide in a standard 51Cr release assay. Target cells pulsed with the wild-type p11C, C-M peptide or the C46A, Q54A, or Q54T mutant peptide were comparably recognized and lysed at multiple peptide concentrations and multiple effector-to-target ratios (Fig. 6). Therefore, because they are still recognized by p11C, C-M-specific CTL, viruses with amino acid substitutions at position 1 or 8 (C46A, Q53A, and Q53T) of the p11C, C-M epitope do not have a selective advantage over the wild-type virus.
FIG. 6.
The p11C, C-M epitope of replication-competent mutant SIVs is recognized by epitope-specific CTL. PBL from a SIVmac251-infected, Mamu-A*01+ rhesus monkey were stimulated in vitro with the SIV Gag p11C, C-M peptide. On day 17 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr-release CTL assay. The target cells were 721.221 cells stably expressing the Mamu-A*01 molecule that had been incubated for 1.5 h with the indicated peptides at concentrations of 1, 10, 100, and 1,000 pM.
DISCUSSION
Almost all of the documented examples of viral escape from p11C, C-M-specific CTL are the result of position 2 epitope mutations that alter peptide binding to the Mamu-A*01 molecule (2, 3, 7, 35). From a total of 48 SIVsm-, SIVmac-, and SHIV-89.6P-infected rhesus monkeys that have been carefully evaluated, 14 animals showed the emergence of p11C, C-M epitope mutations, with 13 of these animals having mutations only at position 2 of the epitope. The results of the present study support these in vivo observations and provide an explanation for this finding. Mutations at positions 3, 4, 5, 6, 7, and 9 of the p11C, C-M epitope had a significant fitness cost for the virus as a result of decreased Gag protein expression, virus production, and core stability. In addition, these viruses were unable to replicate. Therefore, even if viruses with these mutations were to occur in vivo, they could not predominate in the quasispecies. Only position 1, 2, and 8 p11C, C-M epitope mutations yielded replication-competent viruses. However, peptides containing position 1 and 8 mutations were recognized by wild-type p11C, C-M-specific CTL. Therefore, the only mutation that is both tolerated by the virus and not recognized by p11C, C-M-specific CTL is the position 2 mutant. However, additional compensatory mutations are required to overcome the fitness costs associated with these epitope mutations (12, 38).
Our analysis in the present study was restricted to alanine substitutions and three other selected substitutions. We chose to restrict our analysis to these substitutions for two reasons. First, the use of alanine substitutions is a well-defined approach to studying the importance of individual amino acids in CTL recognition. Second, in light of the extensive sequence conservation between the capsid proteins of SIV and HIV-1, we reasoned that the three HIV-1-like mutations (Y49Q, I51L, and Q53T) were likely to be tolerated by the virus. The substitutions we made ranged from radical (D50A and Y49A) to conservative (I51A and Y49Q), with one substitution being highly conservative (I51L). While the physicochemical properties of the amino acid substitutions that we introduced into Gag had significant fitness costs for the virus, we cannot rule out the possibility that additional substitutions may exist that facilitate escape from p11C, C-M-specific CTL recognition with no associated fitness cost for the virus.
The Gag p11C, C-M epitope is located in a highly conserved region of the capsid protein, beginning amino terminal to and ending within helix 3 (amino acid residues 46 to 54). This region plays a central role in capsid assembly and early postentry events that are critical for the generation of infectious virus (26, 36, 47). Studies of the HIV-1 capsid crystal structure have shown that this region of the capsid is essential for the generation of infectious virus (43, 51) and for capsid maturation and multimerization (25, 26). More recently, this region has been shown to interact with host factors that are central to the uncoating of the capsid after entry into cells (36, 47). Thus, the p11C, C-M epitope is located in a structurally and functionally important region of the capsid protein that cannot tolerate much sequence variation.
Whether a virus can escape from antigen-specific CTL responses through particular mutations reflects a balance of fitness costs and the selective advantage for the mutant viruses. While viral escape from a CTL response can sometimes occur by mutations that lead to altered epitope processing or CTL recognition, viral escape from p11C, C-M-specific CTL occurs exclusively by sequence changes that result in reduced binding of the epitope peptides to the Mamu-A*01 molecule (2, 3, 6). An analysis of the Mamu-A*01 peptide binding motif suggested that numerous p11C, C-M epitope mutations alter peptide binding to the Mamu-A*01 molecule and therefore should facilitate viral escape (45). However, because of the extensive structural and functional constraints on the sequence of the p11C, C-M epitope, only a limited number of these mutations are tolerated in vivo (2, 3, 7, 33). This is in stark contrast to the equally dominant Mamu-A*01-restricted Tat SL8 epitope, in which single point mutations are observed at every residue of the epitope within the first 3 weeks after SIVmac239 infection (1).
CTL responses that recognize structurally constrained epitopes are associated with long-term survival (12, 19, 27, 30, 35, 38), while CTL responses specific for less-conserved regions of the virus are associated with rapid progression (9, 18, 31, 48-50). SIVmac239-infected Mamu-A*01+ rhesus monkeys develop very high-frequency CTL responses against the immunodominant Gag p11C, C-M epitope (24) and demonstrate better control of virus replication and delayed disease progression compared to Mamu-A*01− animals (9, 30). A major contributing factor to the delayed disease progression observed in SIVmac239-infected Mamu-A*01+ rhesus monkeys may in fact be the inability of the virus to accumulate mutations in the region of the capsid encoding the p11C, C-M epitope and thereby to escape recognition by CTL. Thus, CTL responses directed against structurally conserved epitopes may provide the largest degree of protection against clinical disease progression.
Acknowledgments
We thank Karen Hershberger for a critical review of the manuscript.
This work was supported by NIH grants T32-AI07387 and AI-20729.
REFERENCES
- 1.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z. W., A. Craiu, L. Shen, M. J. Kuroda, U. C. Iroku, D. I. Watkins, G. Voss, and N. L. Letvin. 2000. Simian immunodeficiency virus evades a dominant epitope-specific cytotoxic T lymphocyte response through a mutation resulting in the accelerated dissociation of viral peptide and MHC class I. J. Immunol. 164:6474-6479. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z. W., L. Shen, M. D. Miller, S. H. Ghim, A. L. Hughes, and N. L. Letvin. 1992. Cytotoxic T lymphocytes do not appear to select for mutations in an immunodominant epitope of simian immunodeficiency virus gag. J. Immunol. 149:4060-4066. [PubMed] [Google Scholar]
- 8.Couillin, I., B. Culmann-Penciolelli, E. Gomard, J. Choppin, J. P. Levy, J. G. Guillet, and S. Saragosti. 1994. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 180:1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. T., L. A. Knapp, P. Jing, J. L. Mitchen, M. Dykhuizen, D. C. Montefiori, C. D. Pauza, and D. I. Watkins. 1999. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunol. Lett. 66:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 11.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P., D. Price, M. Nowak, S. Rowland-Jones, R. Phillips, and A. McMichael. 1997. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol. Rev. 159:17-29. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, J. R., S. Sutjipto, P. A. Marx, and N. C. Pedersen. 1992. Shared antigenic epitopes of the major core proteins of human and simian immunodeficiency virus isolates. J. Med. Primatol. 21:265-269. [PubMed] [Google Scholar]
- 17.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, X., X. Gao, M. Ramanathan, Jr., G. R. Deschenes, G. W. Nelson, S. J. O'Brien, J. J. Goedert, D. D. Ho, T. R. O'Brien, and M. Carrington. 2002. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+-T-cell responses for groups of HIV-1-infected individuals with different HLA-B*35 genotypes. J. Virol. 76:12603-12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kestler, H., T. Kodama, D. Ringler, M. Marthas, N. Pedersen, A. Lackner, D. Regier, P. Sehgal, M. Daniel, N. King, and R. C. Desrosiers. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109-1112. [DOI] [PubMed] [Google Scholar]
- 21.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, E., and H. G. Gottlinger. 1996. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J. Virol. 70:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 25.Lanman, J., T. T. Lam, S. Barnes, M. Sakalian, M. R. Emmett, A. G. Marshall, and P. E. Prevelige, Jr. 2003. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 325:759-772. [DOI] [PubMed] [Google Scholar]
- 26.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 27.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 29.Mortara, L., F. Letourneur, P. Villefroy, C. Beyer, H. Gras-Masse, J. G. Guillet, and I. Bourgault-Villada. 2000. Temporal loss of Nef-epitope CTL recognition following macaque lipopeptide immunization and SIV challenge. Virology 278:551-561. [DOI] [PubMed] [Google Scholar]
- 30.Mothe, B. R., J. Weinfurter, C. Wang, W. Rehrauer, N. Wilson, T. M. Allen, D. B. Allison, and D. I. Watkins. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 77:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhl, T., M. Krawczak, P. Ten Haaft, G. Hunsmann, and U. Sauermann. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 169:3438-3446. [DOI] [PubMed] [Google Scholar]
- 32.Nietfield, W., M. Bauer, M. Fevrier, R. Maier, B. Holzwarth, R. Frank, B. Maier, Y. Riviere, and A. Meyerhans. 1995. Sequence constraints and recognition by CTL of an HLA-B27-restricted HIV-1 gag epitope. J. Immunol. 154:2189-2197. [PubMed] [Google Scholar]
- 33.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 34.O'Connor, D. H., T. M. Allen, and D. I. Watkins. 2002. Cytotoxic T-lymphocyte escape monitoring in simian immunodeficiency virus vaccine challenge studies. DNA Cell Biol. 21:659-664. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 77:9029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peyerl, F. W., D. H. Barouch, W. W. Yeh, H. S. Bazick, J. Kunstman, K. J. Kunstman, S. M. Wolinsky, and N. L. Letvin. 2003. Simian-human immunodeficiency virus escape from cytotoxic T lymphocyte recognition at a structurally constrained epitope. J. Virol. 77:12572-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 40.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 42.Robinson, S., W. A. Charini, M. H. Newberg, M. J. Kuroda, C. I. Lord, and N. L. Letvin. 2001. A commonly recognized simian immunodeficiency virus Nef epitope presented to cytotoxic T lymphocytes of Indian-origin rhesus monkeys by the prevalent major histocompatibility complex class I allele Mamu-A*02. J. Virol. 75:10179-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rue, S. M., J. W. Roos, L. M. Amzel, J. E. Clements, and S. A. Barber. 2003. Hydrogen bonding at a conserved threonine in lentivirus capsid is required for virus replication. J. Virol. 77:8009-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 45.Sidney, J., J. L. Dzuris, M. J. Newman, R. P. Johnson, A. Kaur, K. Amitinder, C. M. Walker, E. Appella, B. Mothe, D. I. Watkins, and A. Sette. 2000. Definition of the Mamu A*01 peptide binding specificity: application to the identification of wild-type and optimized ligands from simian immunodeficiency virus regulatory proteins. J. Immunol. 165:6387-6399. [DOI] [PubMed] [Google Scholar]
- 46.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 47.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 48.Tang, J., C. Costello, I. P. Keet, C. Rivers, S. Leblanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 15:317-324. [DOI] [PubMed] [Google Scholar]
- 49.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomiyama, H., K. Miwa, H. Shiga, Y. I. Moore, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1997. Evidence of presentation of multiple HIV-1 cytotoxic T lymphocyte epitopes by HLA-B*3501 molecules that are associated with the accelerated progression of AIDS. J. Immunol. 158:5026-5034. [PubMed] [Google Scholar]
- 51.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, R., B. Leschonsky, E. Harrer, C. Paulus, C. Weber, B. D. Walker, S. Buchbinder, H. Wolf, J. R. Kalden, and T. Harrer. 1999. Molecular and functional analysis of a conserved CTL epitope in HIV-1 p24 recognized from a long-term nonprogressor: constraints on immune escape associated with targeting a sequence essential for viral replication. J. Immunol. 162:3727-3734. [PubMed] [Google Scholar]
- 53.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]