Abstract
Analogous to cellular glycoproteins, viral envelope proteins contain N-terminal signal sequences responsible for targeting them to the secretory pathway. The prototype foamy virus (PFV) envelope (Env) shows a highly unusual biosynthesis. Its precursor protein has a type III membrane topology with both the N and C terminus located in the cytoplasm. Coexpression of FV glycoprotein and interaction of its leader peptide (LP) with the viral capsid is essential for viral particle budding and egress. Processing of PFV Env into the particle-associated LP, surface (SU), and transmembrane (TM) subunits occur posttranslationally during transport to the cell surface by yet-unidentified cellular proteases. Here we provide strong evidence that furin itself or a furin-like protease and not the signal peptidase complex is responsible for both processing events. N-terminal protein sequencing of the SU and TM subunits of purified PFV Env-immunoglobulin G immunoadhesin identified furin consensus sequences upstream of both cleavage sites. Mutagenesis analysis of two overlapping furin consensus sequences at the PFV LP/SU cleavage site in the wild-type protein confirmed the sequencing data and demonstrated utilization of only the first site. Fully processed SU was almost completely absent in viral particles of mutants having conserved arginine residues replaced by alanines in the first furin consensus sequence, but normal processing was observed upon mutation of the second motif. Although these mutants displayed a significant loss in infectivity as a result of reduced particle release, no correlation to processing inhibition was observed, since another mutant having normal LP/SU processing had a similar defect.
Secreted or membrane-anchored glycoproteins contain signal sequences targeting them to the secretory pathway (reviewed in reference 12). These so-called signal peptides (SP) can be removed co- or posttranslationally by the cellular membrane-bound signal peptidase complex (SPC). If not cleaved, they may serve as membrane anchors for proteins with distinct membrane orientations. In most cases SP cleavage is thought to occur cotranslationally. However, for some proteins, in particular retroviral glycoproteins (e.g., the human immunodeficiency virus type 1), SP cleavage occurs very late after translation (7).
Spumaretroviruses, or foamy viruses (FVs), use a replication pathway with features distinctive from orthoretroviruses (reviewed in reference 17). The particle-associated glycoprotein of FV is unique compared to other retroviral envelope proteins because its coexpression is strictly required for the FV particle release process and its function cannot be replaced by heterologous viral glycoproteins (reviewed in reference 9). The FV envelope precursor protein seems to initially have a type III protein configuration with both its N and C terminus located intracytoplasmically (10). During its transport to the cell surface, it is posttranslationally processed by cellular proteases into at least three subunits. The N-terminal signal or leader peptide (LP) has a type II conformation, whereas the C-terminal transmembrane (TM) subunit has a type I conformation. The internal surface (SU) subunit presumably associates with extracellular domains of TM on the luminal side (10, 20). For the FV budding process at least two essential interactions between Env and Gag are required (10, 14). One of these is the contact of the N-terminal cytoplasmic region of the FV Env LP, the so-called budding domain, with the N terminus of the FV Gag protein (10, 20). The LP of prototype FV (PFV) is glycosylated and cleavage products are viral particle associated (10). The type II protein configuration of the PFV LP suggested by previous data (10, 20) has recently been experimentally confirmed for the feline FV (FFV) LP (6). In the present study, we intended to determine the exact LP/SU cleavage site, analyze the requirement of its cleavage for FV particle release and infectivity, and get an idea which cellular protease might be responsible for PFV Env LP/SU cleavage.
MATERIALS AND METHODS
Cells.
The human kidney cell line 293T (4) and the human fibrosarcoma cell line HT1080 (16) were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics.
Expression constructs.
The replication-deficient PFV vector pczDWP001 is a variant of the previously described PFV Gag/Pol and the enhanced green fluorescent protein (EGFP)-neo fusion protein (EGN)-expressing vector pDL01 (15), having all translation initiation codons of the env open reading frame overlapping the pol open reading frame inactivated by ATG to ACG mutations. Thereby the pol amino acid sequence remained unchanged, but expression of residual Env coding sequences was completely prevented. The glycoprotein expression constructs used in this study are shown schematically in Fig. 1A. The expression construct pczHFVenvEM015 is a variant of the wild-type PFV Env expression construct pczHFVenvEM002 (11), having an additional BsmBI restriction site immediately upstream of the translation start for cloning purposes. The pczHFVenvEM058, -EM077, and -EM078 constructs for expression of PFV Env mutants having the first, second, or third potential N glycosylation site inactivated by a N→Q mutation were described previously (10). The furin cleavage site mutants EM119 to EM123 are based on the pczHFVenvEM015 wild-type construct and were generated by recombinant PCR techniques. The individual cloning strategies and mutagenesis primers are available on request. In constructs pczHFVenvEM119 (R122A), -EM120 (R123A), -EM121 (R126A), and -EM122 (R129A), individual arginine residues within the overlapping minimal furin consensus sequences RXXR or close to it were changed to alanine, whereas in pczHFVenvEM123 (R122A, R123A, R126A, and R129A), all four arginine residues were mutated simultaneously. The PFV Env immunoadhesin construct pAD05 is based on the murine leukemia virus-derived vector pczCFG5IEGZ (3), containing a polylinker upstream of an encephalomyocarditis virus internal ribosomal entry site driving a EGFP zeocin (EGZ) fusion protein cassette. The immunoadhesin itself comprises the extracellular domains of the PFV Env (amino acids [aa] 1 to 936) and constant domains (hinge, CH2, and CH3) of mouse immunoglobulin G2a (IgG2a) (Fig. 1B) that were cloned by PCR from BALB/c total spleen mRNA. The amino acid sequence of the chimeric protein at the fusion boundary in single letter amino acid code is N-SALQGIgseprg-C, with PFV Env sequences in upper case and mouse IgG2a (mIgG2a) sequences in lower case.
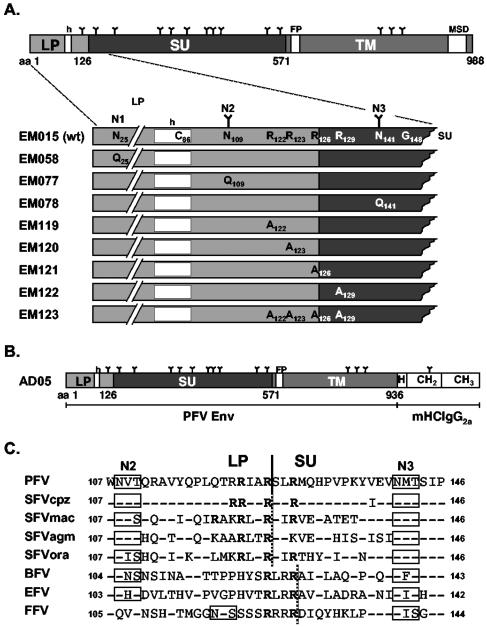
FIG. 1.
Schematic illustration of the PFV Env proteins. (A) Schematic outline of the PFV Env domain organization. The LP-SU region is enlarged below the full-length precursor protein and altered residues of the individual mutants are indicated in comparison to the wild-type sequence. (B) Schematic outline of the PFV Env immunoadhesin domain organization. (C) Sequence alignment of the LP/SU cleavage site and adjacent regions. The arginine residues of the minimal furin cleavage site consensus sequences are in bold letters. A solid black line indicates the boundaries of the PFV LP and SU subunits experimentally determined in this study. A dotted black line indicates the potential boundaries of the glycoproteins of other FV species. N glycosylation sites are boxed. Identical amino acids, except arginine residues, are indicated by hyphens. nsrsid9847724\delrsid9847724 FP, fusion peptide; MSD, membrane-spanning domain; h, hydrophobic domain of the leader peptide; mHCIgG2a, mouse IgG2a heavy-chain constant region; H, hinge region; CH2 or CH3, constant Ig heavy-chain domain 2 or 3; N1 to N3, potential N glycosylation site 1 to 3; N-linked carbohydrate chains are indicated as Y. GenBank accession numbers for the cited viral genomes are U21247 for PFV, U04327 for simian FV-chimpanzee (SFVcpz), X54482 for simian FV-macaque (SFVmac), M74895 for simian FV-African green monkey (SFVagm), AJ544579 for simian FV-orangutan (SFVora), U94514 for bovine FV (BFV), AF201902 for equine FV (EFV), and U85043 for FFV.
Generation of viral supernatants and analysis of transduction efficiency.
FV supernatants containing recombinant viral particles were generated essentially as described previously (8, 11), by cotransfection of 293T cells with the Gag/Pol-expressing vector pczDWP001 and an Env expression plasmid as indicated. Extra- and intracellular viral particles were harvested as described previously (10). Transductions of recombinant EGFP-expressing PFV vector particles were performed by infection of 2 × 104 target cells plated 24 h in advance in 12-well plates for 4 h using 1 ml of viral supernatant or dilutions thereof. The amount of EGFP-positive cells was determined by fluorescence-activated cell sorter analysis 72 h after infection. All transduction experiments were performed at least three times, and in each independent experiment the values obtained with wild-type PFV Env (EM015) were arbitrarily set to 100.
Antisera and Western blot expression analysis.
Western blot expression analysis of cell- and particle-associated viral proteins was performed essentially as described previously (10). The polyclonal antisera used were specific for PFV Gag (2) or the LP of PFV Env, aa 1 to 86 (10). Furthermore, a hybridoma supernatant (clone P3E10) specific for the SU subunit of PFV Env (S. W. Eastman and M. L. Linial, unpublished data) was employed in some experiments. This antibody recognizes an epitope between aa 170 and 262 in PFV SU (A. Duda and D. Lindemann, unpublished data).
Protein A precipitation and N-terminal protein sequencing.
For N-terminal sequencing of the PFV Env processing products, the pAD05 construct was transfected into 293T cells. Forty-eight hours posttransfection, cell-free supernatant containing the secreted PFV Env immunoadhesin was harvested after pelleting cellular debris by centrifugation for 5 min at 1,700 × g. The protein was subsequently purified from the supernatant by immunoprecipitation using protein A Sepharose either at a small scale (0.5 to 1.0 ml of supernatant) or medium scale (5.0 ml) for Western blot or Coomassie staining, respectively. For N-terminal sequencing, proteins were blotted onto polyvinylidene difluoride membranes, Coomassie stained for 10 min in 40% methanol, 10% acetic acid, and 0.1% Coomassie blue R250, and subsequently destained in 40% methanol and 10% acetic acid for 3 h. N-terminal protein sequencing was performed at Toplab (Martinsried, Germany).
RESULTS
Reactivity of PFV Env precursor cleavage products with domain-specific antisera.
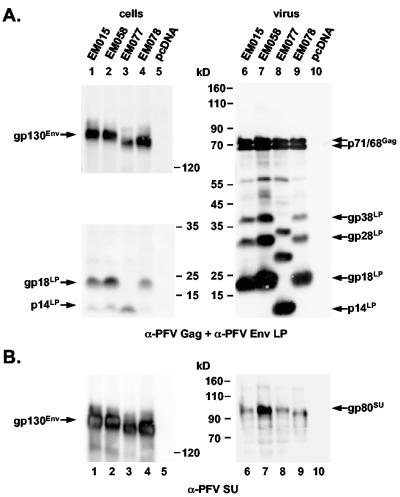
Potential PFV envelope LP cleavage by the cellular signal peptidase is predicted to occur after C86 (19). However, in previous experiments we demonstrated that this potential signal peptidase-processing site is apparently not used, because its inactivation by mutagenesis had no influence on PFV Env LP processing (10). In contrast to that result, mutagenesis of G148 resulted in almost complete inhibition of PFV Env LP processing and loss of infectivity and particle release (10). Interestingly, this residue of the PFV Env protein is part of a conserved tetrapeptide motif found in all known isolates of different FV species, which fits well the requirements of a signal peptidase cleavage site. This pointed to a role of this conserved sequence in PFV Env proteolysis and suggested that this stretch of amino acids may represent the actual processing site itself. However, the apparent molecular weight of the PFV Env LP protein backbone of about 14,000 (10) was in disagreement with the predicted mass of 17.4 kDa for a peptide comprising aa 1 to 148 of PFV Env and fitted more closely to a peptide of about 120 aa in length. Furthermore, endo H (endo-β-N-acetylglucosaminidase H)-resistant uncleaved PFV Env precursor proteins were detectable in cell lysates (10). Therefore, a processing by the endoplasmic reticulum resident signal peptidase seemed highly unlikely. To localize the LP processing site in more detail we analyzed viral particle-associated processing products of wild-type PFV Env and three mutants ΔN1, ΔN2, and ΔN3, inactivating the first (N25), second (N109), and third (N141) potential N glycosylation sites, respectively, with respect to their reactivity with antisera specific to subdomains of the PFV Env protein. From previous analysis (10) and unpublished data (D. Lüftenegger and D. Lindemann, unpublished data) we knew that the PFV Env protein is indeed N glycosylated at N109 and N141, whereas N25 is inaccessible to N glycosylation, suggesting that, analogous to the FFV Env LP, the PFV Env LP has a type II protein configuration and N25 is located intracytoplasmically. In agreement with this, the gp130Env precursor protein in lysates of 293T cells transfected with the ΔN1 mutant protein (EM058) comigrated with that of wild-type PFV Env (EM015) upon Western blot analysis using either an LP-specific rabbit antiserum raised against aa 1 to 86 (Fig. 2A, lanes 1 and 2) or a PFV SU-specific monoclonal antibody (Fig. 2B, lanes 1 and 2). In contrast to that, the precursor of the ΔN2 (EM077) and ΔN3 (EM078) mutants showed a mobility shift as a result of the missing sugar chains (Fig. 2A and B, lanes 3 and 4). Intriguingly, probing of the cell lysates and the corresponding viral particle lysates with anti-LP revealed a mobility shift of the LP cleavage products only for the ΔN2 mutant (EM077) (Fig. 2A, lanes 3 and 8), whereas the mobility of the cleavage products of the ΔN1 (EM058) (Fig. 2A, lanes 2 and 7) and ΔN3 (EM078) (Fig. 2A, lanes 4 and 9) mutants was unchanged compared to the wild-type protein (EM015) (Fig. 2A, lanes 1 and 6). This indicated that the LP is only glycosylated at N109 (N2) and suggested that the third N glycosylation site at N141 is already contained in SU. Consistent with this, a mobility shift of the processed SU in viral particles was observed only for the ΔN3 (EM078) mutant (Fig. 2B, lane 9) but not for ΔN2 (EM077) (Fig. 2B, lane 8) upon probing of the particle lysates with the anti-SU monoclonal antibody. Thus, our previous observation that the potential signal peptidase cleavage site at C86 is not used was confirmed by these data. Furthermore they demonstrated N glycosylation of PFV Env LP cleavage products at N109 and of the PFV Env gp80SU domain at N141. Taken together, these results indicated that LP/SU cleavage of the precursor protein must occur somewhere between aa 109 and 141. In addition, the occurrence of a mobility shift of the particle-associated higher-molecular-weight LP cleavage products gp28LP and gp38LP only upon inactivation of the N109 and not by mutation of the N141 N glycosylation site suggested that these forms were not generated by alternative proteolytic processing of the gp130Env precursor protein. Indeed we recently discovered that they are derived by posttranslational modification of gp18LP (N. Stanke and D. Lindemann, unpublished data).
FIG. 2.
Analysis of PFV Env N glycosylation mutants. Western blot analysis of 293T cell (cells) and purified PFV particle (virus) lysates using (A) anti-PFV Gag- and anti-PFV Env LP (aa 1 to 86)-specific polyclonal rabbit antisera or (B) anti-PFV-SU (P3E10) monoclonal hybridoma supernatant. The individual PFV proteins are indicated. 293T cells were cotransfected with the PFV Gag/Pol-expressing, replication-defective retroviral vector pczDWP001 and the respective PFV Env expression construct as indicated. Lanes 1 to 5, cell lysates; lanes 6 to 10, the corresponding viral particle lysates.
N-terminal sequencing of PFV Env processing products.
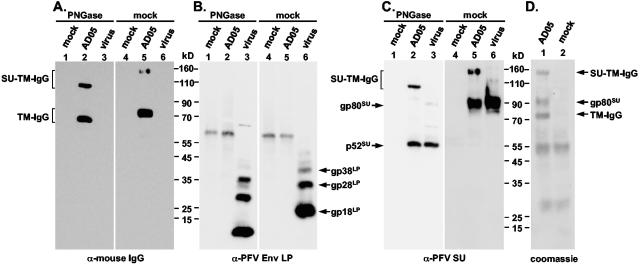
In order to isolate sufficient amounts of PFV Env processing products for analysis by Edman degradation, we generated a PFV Env immunoadhesin (AD05) comprising the extracellular domains of PFV Env (aa 1 to 936) and the constant domain of the mouse IgG2a heavy chain (hinge, CH2, and CH3 domain) (Fig. 1B). By Western blot analysis of protein A Sepharose pelleted supernatant of transfected 293T cells and incubation with various PFV Env-specific or mouse IgG-specific antisera, reactive bands with molecular weights of about 150,000, 90,000, and 75,000 were detected (Fig. 3A to C, lane 5). The 150-kDa protein and its deglycosylated 110-kDa form reacted with anti-mouse IgG- (Fig. 3A, lanes 2 and 5) and anti-PFV-SU-specific (Fig. 3C, lanes 2 and 5) antibodies but not anti-PFV-LP-specific antibodies (Fig. 3B, lanes 2 and 5). The 90-kDa protein and its deglycosylated 52-kDa form were detected by anti-PFV-SU antibodies (Fig. 3C, lanes 2 and 5) but not by anti-mouse IgG- (Fig. 3A, lanes 2 and 5) or anti-PFV-LP-specific (Fig. 3B, lanes 1 and 5) antibodies. Finally, the 75-kDa protein and its deglycosylated 68-kDa form were stained only by anti-mouse IgG-specific antibodies (Fig. 3A, lanes 2 and 5). Taken together, the differential reactivity to the antisera used suggested that the 150-kDa protein is a SU-TM-IgG processing intermediate having only the LP cleaved off, the 90-kDa protein is the mature SU subunit, and the 75-kDa protein is the TM-IgG subunit of the immunoadhesin. Comigration of native (Fig. 3C, lanes 5 and 6) or peptide:N-glycosidase F (PNGase F)-treated (Fig. 3C, lanes 2 and 3) SU subunits of the immunoadhesin and of PFV particles indicated that N- and C-terminal processing of the SU subunit of the immunoadhesin is identical to that of PFV Env incorporated into PFV particles. For N-terminal sequence determination the immunoadhesin, purified by protein A Sepharose precipitation of transfected 293T supernatant (Fig. 3D, lane 1), was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis along with mock-purified control supernatant of cells transfected with the empty expression vector (Fig. 3D, lane 2). Coomassie staining revealed proteins of about 150, 90, and 75 kDa specific for the immunoadhesin (Fig. 3D, lane 1). Proteins of approximately 55 and 28 kDa were stained in protein A precipitates of both samples (Fig. 3D, lanes 1 and 2) and most probably are bovine Ig heavy- and light-chain proteins copurified from the fetal calf serum in the growth medium. N-terminal sequencing of the immunoadhesin-specific 150-, 90-, and 75-kDa proteins revealed the amino acid sequences SLRM, SLRMQH, and SVDN, respectively. Taken together, these data suggested that PFV Env LP/SU cleavage occurs after R126 of the gp130Env precursor protein. Furthermore, they indicated that the PFV Env SU/TM furin cleavage site predicted by sequence (19) and mutagenesis (1, 15) analysis is indeed used and the PFV Env SU subunit is processed after R571. These two processing events of the precursor protein generate PFV Env SU and TM subunits with N-terminal amino acid sequences of SLRMQH and SVDNNY, respectively.
FIG. 3.
Characterization of a PFV Env-mIgG2a immunoadhesin. The AD05 PFV Env-IgG (AD05) immunoadhesin was purified in small (A-C) or medium scale (D) by protein A precipitation from transfected 293T cell supernatant. As control, protein A precipitate from equal amounts of supernatant from 239T transfected with the empty expression vector pczCFG5IEGZ (mock) was used. Purified PFV particles (virus) were harvested by ultracentrifugation through 20% sucrose from the supernatant of 293T cells cotransfected with pczDWP001 and pczHFVenvEM015 expression constructs. Prior to separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, samples were deglycosylated using PNGase F (PNGase) or mock (mock) incubated. After transfer of the proteins to polyvinylidene difluoride membranes, they were analyzed by Western blot using (A) anti-mouse IgG-, (B) anti-PFV Env LP-, and (C) anti-PFV Env SU-specific antibodies or directly Coomassie stained (D). The individual PFV proteins are indicated on the sides of the autoradiograms.
Mutagenesis of LP/SU furin consensus cleavage sites.
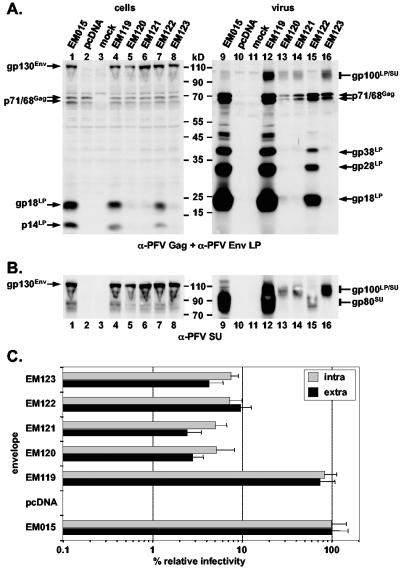
Sequence alignment of the regions of other primate, bovine, equine, and feline FV glycoproteins corresponding to the experimentally determined PFV Env LP/SU cleavage site revealed the presence of one to three minimal furin consensus cleavage sites (18) in all proteins (Fig. 1C). In PFV Env two furin cleavage sites overlap each other (Fig. 1C). The N-terminal protein sequence data of the AD05 immunoadhesin clearly showed that only the first cleavage site (RIAR) is used. To support the protein sequencing data and analyze the function of LP/SU cleavage for PFV replication, R123, R126, and R129 of the two overlapping furin consensus cleavage sites and R122 immediately upstream of the first were mutated individually or all together to alanine (Fig. 1A). The mutant expression constructs (EM119 to EM123) were cotransfected with the PFV Gag/Pol and EGFP-Neo marker protein expressing PFV vector pczDWP001 into 293T cells. Western blot analysis of the corresponding 293T cell lysates revealed a similar expression level of all PFV Env proteins, except for R123A (EM120) showing a slightly reduced expression (Fig. 4A, lanes 1 to 8). For R123A (EM120), R126A (EM121), and the quadruple mutant (EM123), an almost complete inhibition of cellular LP/SU cleavage was observed (Fig. 4A, lanes 5, 6, and 8). The cellular processing of the R122A (EM119) and the R129A (EM122) mutant was slightly reduced compared to the wild type (EM015) (Fig. 4A, lanes 1, 4, and 7). Infectivity analysis of the respective cell supernatants or freeze-thaw cell lysates revealed a 10- to 15-fold reduction for the R129A mutant (EM122), a 13- to 25-fold reduction for the quadruple mutant (EM123), and a 20- to 40-fold reduction for the R123A (EM120) and R126A (EM121) mutants (Fig. 4C). The R122A mutant (EM119) displayed an infectivity similar to the wild type (EM015) (Fig. 4C). Western blot analysis of particle preparations purified from cell culture supernatant revealed a strongly reduced particle release for all mutants except R122A (EM119), evident by the anti-PFV Gag immunostaining (Fig. 4A, lanes 9 to 16). Analysis using the LP-specific polyclonal rabbit antiserum or the SU-specific monoclonal antibody revealed upon prolonged exposure the presence of small amounts of LP-SU protein fragments in wild-type (EM015) particle preparations in addition to SU subunit processing products (Fig. 4A and B, lane 9). This form was more prominent in the R122A mutant (EM119) but was the only SU reactive form in particle preparations of R123A (EM120), R126A (EM121), and the quadruple mutant (EM123) (Fig. 4A and B, lanes 12, 13, 14, and 16). In particle preparations of the R129A mutant (EM122), no LP-SU and only small amounts of SU protein fragments were detectable (Fig. 4A and B, lane 15). These data supported the finding of the protein sequencing data, indicating that only the first potential furin cleavage site is used. Furthermore, they suggested that mutations around the cleavage site interfere with particle release and particles containing only LP-SU subunits seemed to retain at least some of their infectivity.
FIG. 4.
Characterization of potential PFV Env furin cleavage site mutants. Western blot analysis of 293T cell (cells) and purified PFV particle (virus) lysates using (A) anti-PFV Gag- and anti-PFV Env LP (aa1 to 86)-specific polyclonal rabbit antisera or (B) anti-PFV-SU (P3E10) monoclonal hybridoma supernatant. The individual PFV proteins are indicated. 293T cells were cotransfected with the PFV Gag/Pol-expressing, replication-defective retroviral vector pczDWP001 and the respective PFV Env expression construct as indicated. Lanes 1 to 8, cell lysates; lanes 9 to 16, the corresponding viral particle lysates; mock, lysate from cells transfected only with pcDNA vector. (C) Relative infectivity of extracellular (black bars) and intracellular (grey bars) PFV particles pseudotyped with the individual PFV Env proteins as indicated on the left in comparison to the wild type. Mean values and standard deviations of two independent experiments with a total of 12 values for each individual Env protein are shown.
DISCUSSION
The FV glycoprotein, in particular the N-terminal cytoplasmic domain of the LP, has a crucial function in the viral replication cycle since its interaction with the viral capsid is absolutely required for the budding and particle release process (10, 20). The biosynthesis of the PFV envelope protein is highly unusual for a glycoprotein. Proteolytic processing of the N-terminal LP domain, containing the signal sequence targeting the precursor to the secretory pathway, occurs only after synthesis of the full-length precursor protein. Previous mutagenesis analysis implicated the involvement of a conserved tetrapeptide motif around aa 148 in LP processing, if it is not the cleavage site itself. We provide here strong experimental evidence that the cellular signal peptidase complex does not cleave PFV LP/SU at this position, but instead processing is mediated by furin or a furin-like protease after aa R126. N-terminal sequencing of precursor processing products of a PFV Env immunoadhesin molecule and the processing analysis of N glycosylation mutants revealed an LP/SU cleavage site that matches the minimal requirements of a furin consensus sequence shown in Fig. 1C. In contrast, the SU/TM cleavage site fits the requirements of an optimal furin recognition site (18), as previously predicted and implicated by mutagenesis analysis (1, 15, 19). This processing site was confirmed in this study by N-terminal sequencing of the immunoadhesin TM-IgG subunit. These results suggest that cleavage at both processing sites is either mediated by different furin-like proteases or by the same protease with different efficiencies at the individual processing sites. In both cases this can serve a regulatory role. However, an answer to this question requires the identification of the specific individual protease(s) involved in PFV Env processing. Interestingly, all known FV isolates from different species contain at least one, sometimes up to three, overlapping minimal furin consensus sequences in the analogous regions of the precursor protein, suggesting that all of them display a similar LP/SU processing behavior. Indeed observations on the FFV LP processing provide similar results and demonstrate an in vitro cleavage of synthetic FFV Env peptides and recombinant proteins spanning the putative LP/SU cleavage site by furin (M. Löchelt, personal communication). Highly sensitive Western blot analysis using a monoclonal antibody against the PFV Env SU subunit revealed the presence of small amounts of unprocessed LP-SU subunit intermediates already in wild-type PFV particles, indicating that complete LP/SU processing may not be absolutely required for acquisition of infectivity. Different arginine point mutants in the vicinity of the cleavage site further support this assumption. Compared to the wild-type protein, the mutation of R122 (EM119) for example, lying outside of the minimal furin cleavage site, already resulted in a substantial increase in the amount of LP-SU processing intermediates relative to fully processed SU subunits. Nevertheless this mutant displayed a particle release behavior and infectivity analogous to that of wild-type PFV Env. Furthermore, in viral particles of the single mutants R123A (EM120) and R126A (EM121) or the quadruple mutant (R122A, R123A, R126A, and R129A) (EM123), only LP-SU processing intermediates but no fully processed SU subunits and only low amounts of gp18LP were detectable, indicating an almost complete block in LP/SU cleavage. Although these mutants displayed a significant FV particle release defect and infectivities measured in supernatants of transfected 293T cells were reduced 10- to 40-fold, their relative infectivities compared to wild-type PFV Env were in a similar range as for the R129A (EM122) mutant, showing a similar reduction in particle release but a normal LP/SU cleavage pattern in viral particles. Therefore, the reduction in infectivity in the supernatant of cells transfected with certain mutants seems to correlate with the particle release defect of those mutants rather than with the LP/SU processing defect. Altogether the data on the cleavage site mutagenesis indicate that LP/SU cleavage in contrast to the essential SU/TM processing (1, 15) is not a prerequisite for obtaining infectious FV particles. Apparently uncleaved LP/SU on the viral particle still allows receptor interaction and subsequent fusion of viral and cellular lipid membranes by TM.
In addition, our data suggest that mutation of residues, in particular evolutionarily conserved ones, in the C-terminal domain of LP or the N-terminal domain of SU interfere with PFV particle release (10). This can explain the LP/SU processing defect of the previously characterized G148R (EM084) mutant (10). The processing defect of EM084 can result from an intracellular transport defect, as indicated by a severely impaired particle release. This mutant may, therefore, never reach the intracellular compartment in which the furin-like protease that mediates LP/SU cleavage is localized. Alternatively, it is possible that the conformation of EM084 is altered in such a way that the cleavage site becomes inaccessible for processing by the cellular protease. In summary, the results from this study show that unlike other viral glycoproteins such as human immunodeficiency virus type 1 gp160 (13) or Ebola GP (5) that are processed by cellular furin-like proteases at a single cleavage site in the precursor protein, PFV glycoprotein precursor proteolysis by this type of proteases occurs at two sites, resulting in its unusual final topology and adding another unique feature for a retroviral glycoprotein to PFV Env.
Acknowledgments
We thank Martin Löchelt for communicating results prior to publication.
This work was supported by grants from the DFG (Li621/2-1, Li621/2-3) and BMBF (01ZZ0102) to D.L. Work of M.L.L. and S.W.E. was supported by grant CA 18282 from the National Cancer Institute.
REFERENCES
- 1.Bansal, A., K. L. Shaw, B. H. Edwards, P. A. Goepfert, and M. J. Mulligan. 2000. Characterization of the R572T point mutant of a putative cleavage site in human foamy virus Env. J. Virol. 74:2949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baunach, G., B. Maurer, H. Hahn, M. Kranz, and A. Rethwilm. 1993. Functional analysis of human foamy virus accessory reading frames. J. Virol. 67:5411-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinev, D., B. W. Jordan, B. Neufeld, J. D. Lee, D. Lindemann, U. R. Rapp, and S. Ludwig. 2001. Extracellular signal regulated kinase 5 (ERK5) is required for the differentiation of muscle cells. EMBO Rep. 2:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Bridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldmann, H., V. E. Volchkov, V. A. Volchkova, U. Stroher, and H. D. Klenk. 2001. Biosynthesis and role of filoviral glycoproteins. J. Gen. Virol. 82:2839-2848. [DOI] [PubMed] [Google Scholar]
- 6.Geiselhart, V., A. Schwantes, P. Bastone, M. Frech, and M. Löchelt. 2003. Features of the Env leader protein and the N-terminal Gag domain of feline foamy virus important for virus morphogenesis. Virology 310:235-244. [DOI] [PubMed] [Google Scholar]
- 7.Li, Y., L. Luo, D. Y. Thomas, and C. Y. Kang. 1994. Control of expression, glycosylation, and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology 204:266-278. [DOI] [PubMed] [Google Scholar]
- 8.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindemann, D., and P. A. Goepfert. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111-129. [DOI] [PubMed] [Google Scholar]
- 10.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell. Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 13.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 14.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 17.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 18.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, G., and M. J. Mulligan. 1999. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J. Gen. Virol. 80:245-254. [DOI] [PubMed] [Google Scholar]
- 20.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]