Abstract
Cathepsin F, which is encoded by CTSF, is a cysteine proteinase ubiquitously expressed in several tissues. In a previous study, novel transcripts of the CTSF gene were identified in the crab-eating monkey deriving from the integration of an Alu element–AluYRa1. The occurrence of AluYRa1-derived alternative transcripts and the mechanism of exonization events in the CTSF gene of human, rhesus monkey, and crab-eating monkey were investigated using PCR and reverse transcription PCR on the genomic DNA and cDNA isolated from several tissues. Results demonstrated that AluYRa1 was only integrated into the genome of Macaca species and this lineage-specific integration led to exonization events by producing a conserved 3′ splice site. Six transcript variants (V1–V6) were generated by alternative splicing (AS) events, including intron retention and alternative 5′ splice sites in the 5′ and 3′ flanking regions of CTSF_AluYRa1. Among them, V3–V5 transcripts were ubiquitously expressed in all tissues of rhesus monkey and crab-eating monkey, whereas AluYRa1-exonized V1 was dominantly expressed in the testis of the crab-eating monkey, and V2 was only expressed in the testis of the two monkeys. These five transcript variants also had different amino acid sequences in the C-terminal region of CTSF, as compared to reference sequences. Thus, species-specific Alu-derived exonization by lineage-specific integration of Alu elements and AS events seems to have played an important role during primate evolution by producing transcript variants and gene diversification.
Keywords: Alternative splicing, Alu, CTSF, exonization, primate
INTRODUCTION
Alternative splicing (AS) is an important and ubiquitous molecular mechanism that increases eukaryotes genome diversity and complexity by generating different isoforms of a single gene without significantly increasing genome size (Kim et al., 2014; 2016). High-throughput sequencing data revealed that over 90% of human genes undergo AS, and that this process is more frequent in higher than in lower eukaryotes (Ast, 2004; Pan et al., 2008). There are four major types of AS events, including exon creation or loss (skipping), alternative 5′ splice sites, alternative 3′ splice sites, and intron retention (Ast, 2004; Park et al., 2015a). In the human genome, 40% of the new exons are generated by AS, and most of these are cassette exons (inclusion or skipping of a single exon) (Zhang and Chasin, 2006). In addition, over 90% of the primate-specific cassette exons (recently generated exons) overlap with transposable elements (TEs) and 62% overlap with Alu elements (Amit et al., 2007; DeBarry et al., 2006; Zhang and Chasin, 2006). Alu elements, a primate-specific class of short interspersed nuclear elements (SINE), are the most abundant type of TEs and comprise more than 10% of the human genome (Lander et al., 2001). The internal sequence of Alu elements contains potential splicing donor (GT) and acceptor sites (AG) that can be recognized by spliceosomes (Ast, 2004; Lev-Maor et al., 2003). A previous study indicated that more than 5% of the newly AS-generated exons in the human genome originate from Alu elements (Sorek et al., 2002). Therefore, Alu elements are a major source of exon generation, and Alu-derived exonization events might have an important role in increasing gene diversity by producing novel protein isoforms in human and non-human primates.
Cathepsin F, a protein that is encoded by the Cathepsin F gene (CTSF) mapped to the human chromosome 11q13, is a papain family cysteine proteinase that plays a major role in the lysosomal proteolytic system (Chapman et al., 1997). In the human genome, 11 different cysteine cathepsins have been characterized (cathepsins B, C, F, H, K, L, O, S, V, X, and W) via bioinformatics analysis (Rossi et al., 2004). Among them, CTSF has an extended N-terminal proregion, which contains a cystatin-like domain (Ahn et al., 2009; Jeric et al., 2013). Similar to cathepsins B, C, H, L, O, and Z, CTSF is ubiquitously expressed in widespread tissues, whereas cathepsins J, K, S, and W are expressed in restricted tissues or cell types (Tang et al., 2006; Turk et al., 2012). However, CTSF expression levels were higher in several human cancer cell lines than in normal cells (Vazquez-Ortiz et al., 2005) suggesting that CTSF could play an important role in carcinogenesis. Moreover, previous studies have shown that mutations in the CTSF gene are associated with Type B Kufs disease, an adult form of neuronal ceroid lipofuscinosis (Peters et al., 2015; Smith et al., 2013). The present study aimed to identify and characterize the Alu-derived exonization events in the CTSF gene of the rhesus monkey (Macaca mulatta) and the crab-eating monkey (Macaca fascicularis). Additionally, we validated the concept that lineage-specific Alu-integration lead to lineage and tissue-specific AS events in the CTSF gene during primate evolution.
MATERIALS AND METHODS
Ethics statement
Animal preparation and study design were conducted according to the Guidelines of the Institutional Animal Care and Use Committee (KRIBB-AEC-16067) of the Korea Research Institute of Bioscience and Biotechnology (KRIBB). Rhesus and crab-eating monkeys were provided by the National Primate Research Center of Republic of Korea or imported from China using a Convention on International Trade in Endangered Species of Wild Fauna and Flora permit.
Total RNA and genomic DNA sample preparation
Total RNA samples extracted from Homo sapiens bone marrow, whole brain, fetal brain, fetal liver, heart, kidney, liver, lung, placenta, prostate, skeletal muscle, spleen, testis, thymus, trachea, uterus, colon, small intestine, spinal cord, and stomach were purchased from Clontech Laboratories, Inc. Twelve tissue samples were collected from the cerebellum, cerebrum, kidney, heart, large intestine, liver, lung, pancreas, small intestine, spleen, stomach, and testis of specific pathogen free adult male and female rhesus monkeys (both 10-years-old) and one adult male crab-eating monkey (seven-years-old). Animals were deeply anesthetized with ketamine (5 mg/kg) by intramuscular injection, and a perfusion with diethylpyrocarbonate-treated cold phosphate-buffered saline was conducted via the common carotid artery with RNase inhibitors to prevent blood contamination and promote the recovery of intact RNA molecules from the tissue samples. Total RNA was extracted from the tissue samples using the RNeasy® Plus Mini kit (Qiagen), according to the manufacturer’s instructions. Furthermore, RNase-free DNase (Qiagen) was used to eliminate DNA contamination from the total RNA preparations. The RNA concentration and the absorbance ratio at 260 nm and 280 nm (A260/A280) were determined with a NanoDrop® ND-1000UV-Vis Spectrophotometer (NanoDrop Technologies).
Using a standard protocol, genomic DNA from heparinized blood samples was extracted from the following species: (1) HU: human (Homo sapiens); (2) hominoid: CH: chimpanzee (Pan troglodytes); (3) Old World monkeys (OWM): JA: Japanese monkey (Macaca fuscata), RH: rhesus monkey (Macaca mulatta), CR: crab-eating monkey (Macaca fascicularis), PI: pig-tail monkey (Macaca nemestrina), BO: bonnet monkey (Macaca radiata); MA: mandrill (Mandrillus sphinx), AF: African green monkey (Chlorocebus aethiops), LA: langur (Trachypithecus sp.); (4) New World monkeys (NWM): MAR: marmoset (Callithrix jacchus), TA: tamarin (Saguinus midas); and (5) prosimian: RL: ring-tailed lemur (Lemur catta).
Reverse transcription-PCR (RT-PCR) and PCR amplifications
Complementary DNA was generated using the GoScript Reverse Transcription (RT) System (Promega). Following the manufacturer’s instructions, 500 ng total RNA, 1 μl oligo (dT)15 primer, 1 μl random primer, 4 μl GoScript 5× reaction buffer, 2 μl MgCl2, 1 μl nucleotides mix, 0.5 μl recombinant RNasin® ribonuclease inhibitor (Promega), 1 μl GoScript reverse transcriptase, and nuclease-free water (up to 20 μl) were added to a microcentrifuge tube, thoroughly mixed, and incubated for 1 h at 42°C. The expression levels of CTSF on humans (NM_003793.3), rhesus monkey, and crab-eating monkey were obtained from RT-PCR amplifications using specific primer pairs (Supplementary Table S1), which were carried out for 30–33 cycles of 94°C for 30 s, 58–60°C for 30 s, and 72°C for 30 s. To validate the AluYRa1-exonized transcripts, RT-PCR experiments were performed using three validation primer pairs in rhesus and crab-eating monkeys (Supplementary Table S1). PCR amplifications of pure mRNA samples without RT were also performed to demonstrate that mRNA samples did not contain genomic DNA (data not shown). As a standard control, GAPDH was amplified from human, rhesus monkey, and crab-eating monkey.
Genomic DNA from the several primates mentioned above was amplified using primer pairs specifically designed from highly conserved sequences in human and non-human primates (Supplementary Table S1). The PCR amplification of genomic DNA was carried out for 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s.
Molecular cloning and sequencing
PCR products were separated on a 1.5% agarose gel, purified with the Gel SV Extraction kit (GeneAll), and cloned into a pGEM-T-easy vector (Promega). The cloned DNA was isolated using the Plasmid DNA Mini-prep kit (GeneAll). Primate DNA samples and alternative transcripts were sequenced by Macrogen, Inc.
AluYRa1 integration time
To estimate the integration time of AluYRa1 in CTSF (CTSF_AluYRa1), we estimated the time to most recent ancestor in NETWORK version 5.0 (Fluxus Technology Ltd.). The divergence time of AluYRa1 elements was also estimated based on 97 intact rhesus and crab-eating monkey specific AluYRa1 elements, selected from the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu). A phylogenetic tree was constructed for the 97 AluYRa1 of each monkey species using the neighbor-joining method in the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 (Tamura et al., 2013). The number of nucleotide substitutions was also calculated in MEGA 6.0, by the pairwise distance method. All polyadenylation (poly(A)) tails were excluded from the sequences before the analysis.
RESULTS
CTSF gene structure and comparison among human, rhesus monkey, and crab-eating monkey genomes
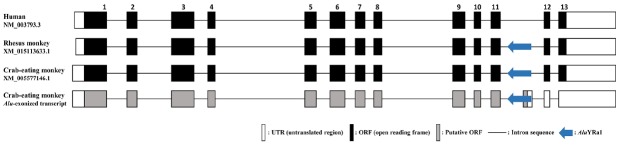
In a previous study, and based on the large-scale transcriptome sequencing and genetic analyses of 16 tissues from male and female crab-eating monkey, we identified a specific AS event (relative to the human genome) corresponding to the integration of the AluYRa1 element on the CTSF gene (Huh et al., 2012). This antisense-oriented AluYRa1 element was inserted on the 11th intron region of CTSF gene and provided a canonical splicing donor site that could produce a new AluYRa1-derived exon by exonization events. Therefore, we compared the structure of the CTSF gene among human (NM_003793), rhesus monkey (XM_015113633), and crab-eating monkey (Alu-exonized transcript XM_005577146) (Fig. 1). The CTSF transcripts of the three species comprised 13 exons and their transcription started at different positions. However, open reading frame (ORF) regions and splicing donor and acceptor sites were well conserved. Whereas the human CTSF transcript (NM_003793) encoded 484 amino acids, rhesus monkey (XM_015113633) and crab-eating monkey (XM_005577146) CTSF transcripts encoded 490 amino acids.
Fig. 1. Structural analysis of human, rhesus monkey, and crab-eating monkey CTSF gene transcripts.
The antisense-oriented AluYRa1 element (blue arrow) is located on the 11th intron of CTSF in rhesus monkey and crab-eating monkey. Open, black, and gray boxes represent exon’s untranslated region (UTR), open reading frame (ORF; protein-coding region), and putative protein-coding region, respectively. The horizontal line represents intron sequences. This figure is a structural illustration and is not drawn to scale.
Evolutionary analysis of the AluYRa1 integration in the CTSF gene
Our comparative structure analysis indicated that the integration of the AluYRa1 element led to a new exon in the CTSF gene of the crab-eating monkey, whereas the AluYRa1 element was not integrated in the human CTSF gene. Therefore, the integration time of the AluYRa1 element into primate lineages was evaluated through the PCR amplification of the CTSF gene in 13 primates, including hominoids (human and chimpanzee), OWM (Japanese monkey, rhesus monkey, crab-eating monkey, pig-tail monkey, bonnet monkey, mandrill, African-green monkey, and langur), NWM (marmosets and tamarins), and prosimians (ring-tailed lemurs) (Fig. 2). These results, together with the sequence analysis of all primate samples (Supplementary Fig. S1), demonstrated that AluYRa1 was only integrated in Macaca species (Japanese monkey, rhesus monkey, crab-eating monkey, pigtail monkey, and bonnet monkey). Both the splice acceptor site in the left arm of AluYRa1 and the splice donor site on the 3′ flanking region of the AluYRa1 element were well conserved in in Macaca species (Supplementary Fig. S1).
Fig. 2. Integration of the AluYRa1 element in the CTSF gene during primate evolution.
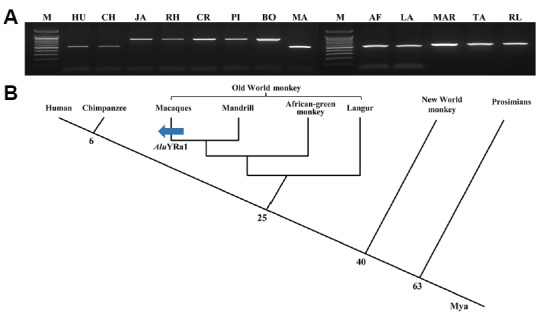
(A) PCR amplification of AluYRa1 in several primates. M indicates the molecular size marker. Primate DNA samples are abbreviated as follows: (1) HU: human; (2) hominoids: CH: chimpanzee; (3) Old World monkeys: JA: Japanese monkey, RH: rhesus monkey, CR: crab-eating monkey, PI: pigtail monkey, BO: bonnet monkey, MA: mandrill, AF: African green monkey, LA: langur; (4) New World monkeys: MAR: Marmoset, TA: tamarin; and (5) prosimian: RL: ringtailed lemur. (B) Schematic representation of the timing of AluYRa1 integration in Macaca CTSF genes. The blue arrow indicates the integration event of AluYRa1. Mya, million years ago.
The NETWORK analysis performed revealed that the integration time of AluYRa1 in the CTSF gene (CTSF_AluYRa1) ranged from 5.2 to 6.59 million years (myrs) in Japanese monkey, 4.42 to 5.4 myrs in rhesus monkey, 5.53 to 7.23 myrs in crab-eating monkey, 4.91 to 5.89 myrs in pig-tail monkey, and 5.2 to 6.59 myrs in bonnet monkey. The integration time of CTSF_AluYRa1 was also calculated using the divergence time from other AluYRa1 elements in rhesus monkey and crab-eating monkey. Based on the phylogenetic tree, we selected the most similar AluYRa1 (msAluYRa1) elements among the 97 intact AluYRa1 sequences in rhesus monkey (48th AluYRa1 element) and crab-eating monkey (26th AluYRa1 element) (Supplementary Fig. S2). We then calculated the pairwise distances between CTSF_AluYRa1 and consensus AluYRa1 (dAluYRA1 = 0.037), and CTSF_AluYRa1 and msAluYRa1 (dCTSF_AluYRA1 = 0.037) in rhesus monkey, and CTSF_AluYRa1 and consensus AluYRa1 (dAluYRA1 = 0.048), and CTSF_AluYRa1 and msAluYRa1 (dCTSF_AluYRA1 = 0.044) in crab-eating monkey. Previous study indicated that AluYRa1 was integrated 9.5 million years ago (t) in the macaque genome (Han et al., 2007a). Thus, using this t value and, divergence time (t1 = (dCTSF_AluYRa1/dAluYRa1)t; Chou et al., 2002) was calculated, revealing that AluYRa1 was integrated in the CTSF gene region about 9.5 million years ago in the rhesus monkey, and about 8.7 million years ago in the crab-eating monkey (Chou et al., 2002).
Experimental validation of the AluYRa1-derived exonization event and expression patterns of the CTSF transcripts
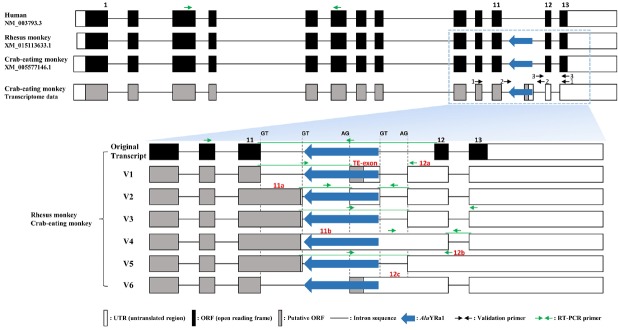
To validate the AluYRa1-derived exonization event on the CTSF gene, we performed RT-PCR experiments and sequencing analysis using three validation primer pairs in 11 different crab-eating monkey tissues (Supplementary Figs. S3 and S4). Five transcript variants (V1-V5) were generated by AS events including AluYRa1-derived exonization, intron retention, and different 5′ and 3′ splice sites (Fig. 3). The V6 transcript was obtained from RT-PCR of the V1 transcript. Only V1, V2, and V6 transcripts had an AluYRa1-derived exon (TE-exon and 12c) generated by splicing the acceptor site on the left arm of the AluYRa1 element. Comparative analysis of the transcript variants and originals identified from rhesus and crab-eating monkeys revealed that: V1 transcript had an AluYRa1-derived exon (TE-exon) and an elongated 12 exon (12a); V2 transcript had an AluYRa1-derived exon (TE-exon) and elongated 11 and 12 exons (11a and 12a); V3 transcript had elongated 11 and 12 exons (11a and 12a); V4 transcript had an intron retained exon (11b); V5 transcript had elongated 11 exon (11a) and an intron retained exon (12b); and V6 had a TE-derived and an intron retained exon (12c). To investigate the expression patterns of original and variant transcripts, RT-PCR experiments were performed transcript-specific primer pairs in 20 human tissues and 11 rhesus monkey and crab-eating monkey tissues (Figs. 4 and 5). Results showed that the original transcript of the CTSF gene was ubiquitously expressed in all human, rhesus monkey, and crab-eating monkey tissues (Fig. 4). Moreover, V3, V4, and V5 transcripts also showed a ubiquitous expression pattern in rhesus and crab-eating monkeys (Figs. 5E–5L). On the other hand, V1 and V6 transcripts showed very low expression levels in all rhesus monkey and crab-eating monkey tissues (Figs. 5A and 5B) and the V2 transcript was only expressed in the testis of rhesus and crab-eating monkeys (Figs. 5C and 5D).
Fig. 3.
Structural analysis of the CTSF gene transcripts in rhesus and crab-eating monkeys using reverse transcription PCR and sequence analysis. In both species, the antisense-oriented AluYRa1 element is located on the 11th intron of the CTSF gene and six transcript variants were identified from various tissues. Open, black, and gray boxes represent exon’s untranslated region (UTR), open reading frame (ORF; protein-coding region), and putative protein-coding region, respectively. Vertical dashed lines represent the 3′ (GT) and 5′ (AG) splice sites, and the horizontal line represents intron sequences. Black and green arrows represent validation primers and RT-PCR primers, respectively. The transposable element exon (TE-exon), 12a, 11a, 11b, 12b, and 12c, exons indicated in red font correspond to new AS-derived exons in the six variants compare with original transcript. This figure is a structural illustration and is not drawn to scale.
Fig. 4. Reverse transcription-PCR amplification of CTSF reference sequences.
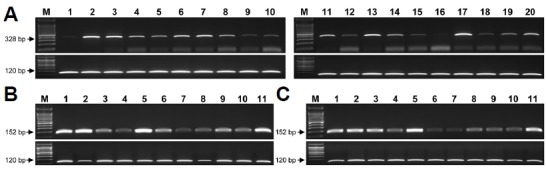
(A) Human, (B) rhesus monkey, and (C) crab-eating monkey. The GAPDH gene was used as the positive control (indicated in the 120 bp). M indicates the molecular size marker. Numbers indicate cDNA tissue samples. Human – 1: bone marrow; 2: brain (whole); 3: fetal brain; 4: colon; 5: small intestine; 6: heart; 7: kidney; 8: liver; 9: fetal liver; 10: lung; 11: placenta; 12: prostate; 13: skeletal-muscle; 14: spinal cord; 15: spleen; 16: stomach; 17: testis; 18: thymus; 19: trachea; 20: uterus. Rhesus monkey and crab-eating monkey–1: cerebellum; 2: cerebrum; 3: kidney; 4: large intestine; 5: liver; 6: lung; 7: pancreas; 8: small intestine; 9: spleen; 10: stomach; 11: testis. RT-PCR products were validated by sequence analysis.
Fig. 5. Reverse transcription-PCR amplification of reference sequence of the six transcript variants of the CTSF gene in rhesus and crab-eating monkeys.

(A and B) V1 (157 bp) and V6 (266 bp), (C and D) V2 (145 bp), (E and F) V3 (186 bp), (G and H) V4 (234 bp), (I and J) V5 (187 bp), and (K and L) GAPDH (120 bp), used as the positive control. M indicates the molecular size marker. Numbers indicate rhesus and Crab-eating monkeys cDNA tissue samples. 1: cerebellum; 2: cerebrum; 3: kidney; 4: large intestine; 5: liver; 6: lung; 7: pancreas; 8: small intestine; 9: spleen; 10: stomach; 11: testis. Transcripts were validated by sequence analysis of the reverse transcription-PCR products.
DISCUSSION
Approximately 45% of the human genome comprises TEs, which are mostly (about 90%) retroelements such as human endogenous retrovirus (about 8%), long interspersed elements (about 20%), and SINEs (about 13%) (Bannert and Kurth, 2004; Schmitz and Brosius, 2011). Alu elements, primate-specific SINEs, are the most abundant retroelements in the human and non-human primate genomes, and a high number of Alu elements might cause several genetic disorders resulting from the influence of Alu-mediated recombination events on functional genes (Deininger and Batzer, 1999). On the other hand, Alu elements can influence the transcription and the biological function of adjacent functional genes by providing polyadenylation sites, promoters, enhancers, and silencers (Han et al., 2007b; Lee et al., 2009; Park et al., 2015a). Moreover, Alu elements have many potential splice sites that can lead to new AS and exonization events in intragenic regions. Thus, although Alu elements contribute to a significant portion of human genetic diseases, they might have an important role in genome diversification, as they generate novel transcript variants and protein isoforms in human and non-human primate genomes.
In our previous transcriptome study, we identified AluY-Ra1-exonized CTSF transcripts in the crab-eating monkey (Huh et al., 2012), which were not found in the human genome, as the AluYRa1 element was not integrated in the human CTSF gene (Fig. 2). During primates’ evolution, about 110,000 Alu elements were specifically integrated in OWMs and 14 different OWM lineage-specific AluY subfamilies were grouped into four lineages: AluYRa1-4, AluYRb1-4, AluYRc1-2, and AluYRd1-4 (Han et al., 2007a). In AluYRa1, the oldest subfamily, elements in the first Alu subfamily belonging to lineage a account for about 30% of OWMs-specific AluYs. In addition, Alu elements are good genetic markers to study the phylogeny of Macaca and, within this genus, four species groups were clearly distinguished based on 358 Alu insertion polymorphisms (Li et al., 2009): sylvanus (M. sylvanus), silenus (M. nigra, M. silenus, and M. nemestrina), sinica (M. radiata, M. thibetana, and M. arctoides) and fascicularis (M. fascicularis, M. fuscata, and M. mulatta). Our integration analysis of the CTSF_AluYRa1 element revealed that it was restricted to the members of the silenus (pig-tail monkey), sinica (bonnet monkey), and fascicularis (Japanese monkey, rhesus monkey, and crab-eating monkey) groups studied here (Fig. 2 and Supplementary Fig. S1), suggesting CTSF_AluYRa1 is also a good genetic marker for Macaca phylogenetic studies. However, as we were not able to validate the integration of CTSF_AluYRa1 in all Macaca species groups, additional validation is necessary before applying tis Alu element as a genetic marker for investigating phylogenetic relationships within the genus Macaca.
Reverse transcription-PCR results revealed six transcript variants of the CTSF gene in rhesus and crab-eating monkeys (Fig. 3), being V1, V2, and V6 generated by the AluYRa1 element. The CTSF_AluYRa1 element found here was integrated with antisense-orientation in the 11th intron region of the CTSF gene. The Alu element comprises two similar monomers (left and right arms), an A-rich linker, and a poly(A) tail (Gal-Mark et al., 2008). Many Alu-derived exonization events were identified from antisense-orientated Alu element, and were more frequently observed in the right than in the left arm (Gal-Mark et al., 2008; Lev-Maor et al., 2003; Park et al., 2015a; Sorek et al., 2002). Our results revealed that, in the rhesus and crab-eating monkeys, the TE-exon (an AluYRa1-derived exon) started on the left arm of the AluYRa1 element (polypyrimidine tract (PPT) adjacent AG sequence) and ended on its 3′ flanking region (Fig. 3 and Supplementary Fig. S1). Each left arm and 3′ flanking region of the AluYRa1 elements provided 3′ and 5′ splice sites. Interestingly, a previous study showed that the AluYRa2-derived exonization event in the BCS1L gene started in the same region as the CTSF_AluYRa1-derived event found here (Park et al., 2015a). These results suggest that although the left arm is considered a minor component when considering the occurrence exonization events, the PPT adjacent AG sequence (3′ splicing site) might be an important region for exonization events. Moreover, the CTSF_AluYRa1 boundary sequences in Japanese, pigtail, and bonnet monkeys also had well-conserved 3′ and 5′ splice sites. Therefore, AluY-Ra1-exonized CTSF transcripts may also occur in these monkeys, as spliceosomes can be recognized. However, further RT-PCR experiments need to be performed to validate AluY-Ra1-derived exonization events in other monkeys.
To investigate the expression level of the original and of the six variant transcripts of the CTSF gene, RT-PCR was performed using several human, rhesus monkey, and crab-eating monkey tissues (Figs. 4 and 5). Whereas the original, V3, V4, and V5 transcripts were ubiquitously expressed in the tissues of the three species, the only transcript variants including the AluYRa1-derived exon (V1, V2, and V6) presented low expression levels and were not detected in all tissues of rhesus monkey and crab-eating monkey. The V2 transcript, in particular, showed a testis-specific expression. Thus, the integration of the lineage-specific AluYRa1 element might lead to lineage and tissue-specific AS events. Previous studies showed that tissue-specific AS events could derive from histone modifications and tissue-specific splicing factors (Chen and Manley, 2009; Luco et al., 2010) and, in human tissues, a few Alu-exonized transcripts showed tissue-specific expression (Mersch et al., 2007). However, tissue-specific AS mechanisms caused by Alu elements are still unclear and, therefore, to understand the correlation between Alu-insertion and tissue-specific AS events, functional studies need to be performed.
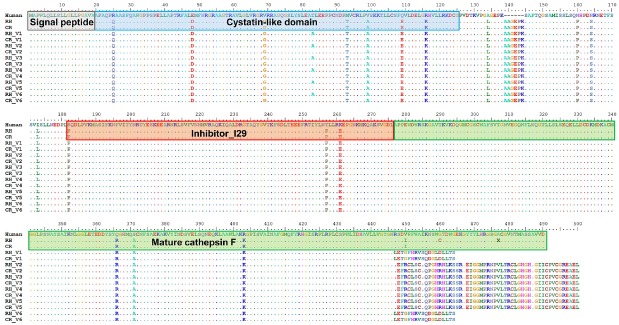
Human CTSF propeptide consists of a signal peptide, a cystatin-like domain, an I29 inhibitor domain, and a mature form of cathepsin F (Jeric et al., 2013). Previous studies have revealed the cysteine-cathepsin-related activation of programmed cell death (apoptosis) (Guicciardi et al., 2004; Repnik et al., 2012), but the physiological functions of CTSF have not been thoroughly investigated. The analysis of the translated sequences of the six transcript variants performed in the present study revealed that V1 and V6 transcripts encoded 464 amino acids, whereas V2–V5 transcripts encoded 500 amino acids. The C-terminal end region of these transcripts also differed from those of human (484 amino acids), rhesus monkey (490 amino acids), and crab-eating monkey (490 amino acids) reference genes (Fig. 6). These different C-terminal sequences were derived from several AS events, including AluYRa1-derived exonization, intron retention, and different 5′ and 3′ splicing sites (Fig. 3). Previous studies indicated that TE-derived AS events increase the functional diversification of a gene (Dagan et al., 2004; Cowley and Oakey, 2013). For example, the human ATRN gene encodes both the membrane-bound and the soluble isoforms of attractin, and the soluble form has short C-terminal region compare with membrane-bound form. This soluble form encoded by the long interspersed nuclear element 1 (LINE1)-exonized alternative transcript, which is involved in the basic inflammatory response and is released by activated T lymphocytes (Duke-Cohan et al., 1998). The C-terminal sequence of the Alu-derived casein kinase 2 (CK2) α″ subunit, which differs from that of the original isoform, is associated with determination of nuclear localization (Hilgard et al., 2002) but the functions of most alternative transcript variants remain unknown (Mola et al., 2007). Thus, the small differences in the C-terminal sequences found among the six transcript variants and reference CTSF gene might not have affected protein function. However, further studies are needed to validate whether the six CTSF transcript variants are functional or not. Although this issue was not investigated in the present study, the specific integration of the AluY-Ra1 element in Macaca might have led to lineage- and tissue-specific AS events, which might have produced the different CTSF gene transcripts found in Macaca species. Thus, Alu elements appear to be a major source of genome diversity and complexity in non-human primates.
Fig. 6. Multiple sequence alignment of CTSF amino acid sequences.
The six transcript variants of CTSF identified from rhesus and crab-eating monkeys were aligned with CTSF reference sequences from human, rhesus monkey, and crab-eating monkey. Dots indicate amino acids are identical to those found in the human sequence. The gray, blue, red, and green box indicate the signal peptide, cystatin-like domain, I29 inhibitor, and mature cathepsin F, respectively. RH, rhesus monkey; CR, crab-eating monkey; V1–V6, CTSF variants.
Non-human primates are the most valuable animal model species for biomedical research in microbiology, vaccine development, biochemistry, and neuroscience (Park et al., 2015a; Rhesus Macaque Genome et al., 2007), as they have more biological and behavioral similarities and closer genetic relationship to humans than other animal models such as rodents, rabbits, and dogs (Carlsson et al., 2004; Huh et al., 2012; Park et al., 2015b). Rhesus and crab-eating monkeys are the most widely and frequently used study species among non-human primates (Huh et al., 2012; Rhesus Macaque Genome et al., 2007). Previous studies demonstrated that missense mutations in the CTSF gene caused Type B Kufs disease. Patients showed progressive neurodegeneration and accumulation of abnormal lipopigments in the brain and presented dementia and motor disturbances (Peters et al., 2015; Smith et al., 2013). However, disease mechanisms linking mutations in the CTSF gene to neurodegeneration and intralysosomal storage are still unclear. Therefore, the identification of transcript variants and the assessment of their expression patterns obtained in the present study could provide basic and useful information for investigating CTSF gene-related diseases using rhesus and crab-eating monkeys as models.
Supplementary Information
ACKNOWLEDGMENTS
This research was supported by a Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program grant (KGM4241743 & KGM5161712).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Ahn S.J., Kim N.Y., Seo J.S., Je J.E., Sung J.H., Lee S.H., Kim M.S., Kim J.K., Chung J.K., Lee H.H. Molecular cloning, mRNA expression and enzymatic characterization of cathepsin F from olive flounder (Paralichthys olivaceus) Comp Biochem Physiol B Biochem Mol Biol. 2009;154:211–220. doi: 10.1016/j.cbpb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Amit M., Sela N., Keren H., Melamed Z., Muler I., Shomron N., Izraeli S., Ast G. Biased exonization of transposed elements in duplicated genes: A lesson from the TIF-IA gene. BMC Mol Biol. 2007;8:109. doi: 10.1186/1471-2199-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Bannert N., Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson H.E., Schapiro S.J., Farah I., Hau J. Use of primates in research: a global overview. Am J Primatol. 2004;63:225–237. doi: 10.1002/ajp.20054. [DOI] [PubMed] [Google Scholar]
- Chapman H.A., Riese R.J., Shi G.P. Emerging roles for cysteine proteases in human biology. Annu Rev Physiol. 1997;59:63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.H., Hayakawa T., Diaz S., Krings M., Indriati E., Leakey M., Paabo S., Satta Y., Takahata N., Varki A. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci USA. 2002;99:11736–11741. doi: 10.1073/pnas.182257399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M., Oakey R.J. Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 2013;9:e1003234. doi: 10.1371/journal.pgen.1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T., Sorek R., Sharon E., Ast G., Graur D. AluGene: a database of Alu elements incorporated within protein-coding genes. Nucleic Acids Res. 2004;32:D489–492. doi: 10.1093/nar/gkh132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBarry J.D., Ganko E.W., McCarthy E.M., McDonald J.F. The contribution of LTR retrotransposon sequences to gene evolution in Mus musculus. Mol Biol Evol. 2006;23:479–481. doi: 10.1093/molbev/msj076. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Duke-Cohan J.S., Gu J., McLaughlin D.F., Xu Y., Freeman G.J., Schlossman S.F. Attractin (DPPT-L), a member of the CUB family of cell adhesion and guidance proteins, is secreted by activated human T lymphocytes and modulates immune cell interactions. Proc Natl Acad Sci USA. 1998;95:11336–11341. doi: 10.1073/pnas.95.19.11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mark N., Schwartz S., Ast G. Alternative splicing of Alu exons--two arms are better than one. Nucleic Acids Res. 2008;36:2012–2023. doi: 10.1093/nar/gkn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi M.E., Leist M., Gores G.J. Lysosomes in cell death. Oncogene. 2004;23:2881–2890. doi: 10.1038/sj.onc.1207512. [DOI] [PubMed] [Google Scholar]
- Han K., Konkel M.K., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., et al. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007a;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Han K., Lee J., Meyer T.J., Wang J., Sen S.K., Srikanta D., Liang P., Batzer M.A. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007b;3:1939–1949. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgard P., Huang T., Wolkoff A.W., Stockert R.J. Translated Alu sequence determines nuclear localization of a novel catalytic subunit of casein kinase 2. Am J Physiol Cell Physiol. 2002;283:C472–483. doi: 10.1152/ajpcell.00070.2002. [DOI] [PubMed] [Google Scholar]
- Huh J.W., Kim Y.H., Park S.J., Kim D.S., Lee S.R., Kim K.M., Jeong K.J., Kim J.S., Song B.S., Sim B.W., et al. Large-scale transcriptome sequencing and gene analyses in the crab-eating macaque (Macaca fascicularis) for biomedical research. BMC Genomics. 2012;13:163. doi: 10.1186/1471-2164-13-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeric B., Dolenc I., Mihelic M., Klaric M., Zavasnik-Bergant T., Guncar G., Turk B., Turk V., Stoka V. N-terminally truncated forms of human cathepsin F accumulate in aggresome-like inclusions. Biochim Biophys Acta. 2013;1833:2254–2266. doi: 10.1016/j.bbamcr.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Kim T., Kim J.O., Oh J.G., Hong S.E., Kim do H. Pressure-overload cardiac hypertrophy is associated with distinct alternative splicing due to altered expression of splicing factors. Mol Cells. 2014;37:81–87. doi: 10.14348/molcells.2014.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H., Choe S.H., Song B.S., Park S.J., Kim M.J., Park Y.H., Yoon S.B., Lee Y., Jin Y.B., Sim B.W., et al. Macaca specific exon creation event generates a novel ZKSCAN5 transcript. Gene. 2016;577:236–243. doi: 10.1016/j.gene.2015.11.051. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee J.R., Huh J.W., Kim D.S., Ha H.S., Ahn K., Kim Y.J., Chang K.T., Kim H.S. Lineage specific evolutionary events on SFTPB gene: Alu recombination-mediated deletion (ARMD), exonization, and alternative splicing events. Gene. 2009;435:29–35. doi: 10.1016/j.gene.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Li J., Han K., Xing J., Kim H.S., Rogers J., Ryder O.A., Disotell T., Yue B., Batzer M.A. Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements. Gene. 2009;448:242–249. doi: 10.1016/j.gene.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R.F., Pan Q., Tominaga K., Blencowe B.J., Pereira-Smith O.M., Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersch B., Sela N., Ast G., Suhai S., Hotz-Wagenblatt A. SERpredict: detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mola G., Vela E., Fernandez-Figueras M.T., Isamat M., Munoz-Marmol A.M. Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol. 2007;366:1055–1063. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Park S.J., Kim Y.H., Lee S.R., Choe S.H., Kim M.J., Kim S.U., Kim J.S., Sim B.W., Song B.S., Jeong K.J., et al. Gain of a new exon by a lineage-specific Alu element-integration event in the BCS1L gene during primate evolution. Mol Cells. 2015a;38:950–958. doi: 10.14348/molcells.2015.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim Y.H., Nam G.H., Choe S.H., Lee S.R., Kim S.U., Kim J.S., Sim B.W., Song B.S., Jeong K.J., et al. Quantitative expression analysis of APP pathway and tau phosphorylation-related genes in the ICV STZ-induced non-human primate model of sporadic Alzheimer’s disease. Int J Mol Sci. 2015b;16:2386–2402. doi: 10.3390/ijms16022386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Rittger A., Weisner R., Knabbe J., Zunke F., Rothaug M., Damme M., Berkovic S.F., Blanz J., Saftig P., et al. Lysosomal integral membrane protein type-2 (LIMP-2/SCARB2). is a substrate of cathepsin-F, a cysteine protease mutated in type-B-Kufs-disease. Biochem Biophys Res Commun. 2015;457:334–340. doi: 10.1016/j.bbrc.2014.12.111. [DOI] [PubMed] [Google Scholar]
- Repnik U., Stoka V., Turk V., Turk B. Lysosomes and lysosomal cathepsins in cell death. Biochim Biophys Acta. 2012;1824:22–33. doi: 10.1016/j.bbapap.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Rhesus Macaque Genome, S. Analysis C., Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rossi A., Deveraux Q., Turk B., Sali A. Comprehensive search for cysteine cathepsins in the human genome. Biol Chem. 2004;385:363–372. doi: 10.1515/BC.2004.040. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Brosius J. Exonization of transposed elements: A challenge and opportunity for evolution. Biochimie. 2011;93:1928–1934. doi: 10.1016/j.biochi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Smith K.R., Dahl H.H., Canafoglia L., Andermann E., Damiano J., Morbin M., Bruni A.C., Giaccone G., Cossette P., Saftig P., et al. Cathepsin F mutations cause Type B Kufs disease, an adult-onset neuronal ceroid lipofuscinosis. Hum Mol Genet. 2013;22:1417–1423. doi: 10.1093/hmg/dds558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.H., Lee J.W., Galvez M.G., Robillard L., Mole S.E., Chapman H.A. Murine cathepsin F deficiency causes neuronal lipofuscinosis and late-onset neurological disease. Mol Cell Biol. 2006;26:2309–2316. doi: 10.1128/MCB.26.6.2309-2316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Ortiz G., Pina-Sanchez P., Vazquez K., Duenas A., Taja L., Mendoza P., Garcia J.A., Salcedo M. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer. 2005;5:68. doi: 10.1186/1471-2407-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Chasin L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc Natl Acad Sci USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.