Abstract
Hydrophobic fusion peptides (FPs) are the most highly conserved regions of class I viral fusion-mediating glycoproteins (vFGPs). FPs often contain conserved glycine residues thought to be critical for forming structures that destabilize target membranes. Unexpectedly, a mutation of glycine residues in the FP of the fusion (F) protein from the paramyxovirus simian parainfluenza virus 5 (SV5) resulted in mutant F proteins with hyperactive fusion phenotypes (C. M. Horvath and R. A. Lamb, J. Virol. 66:2443-2455, 1992). Here, we constructed G3A and G7A mutations into the F proteins of SV5 (W3A and WR isolates), Newcastle disease virus (NDV), and human parainfluenza virus type 3 (HPIV3). All of the mutant F proteins, except NDV G7A, caused increased cell-cell fusion despite having slight to moderate reductions in cell surface expression compared to those of wild-type F proteins. The G3A and G7A mutations cause SV5 WR F, but not NDV F or HPIV3 F, to be triggered to cause fusion in the absence of coexpression of its homotypic receptor-binding protein hemagglutinin-neuraminidase (HN), suggesting that NDV and HPIV3 F have stricter requirements for homotypic HN for fusion activation. Dye transfer assays show that the G3A and G7A mutations decrease the energy required to activate F at a step in the fusion cascade preceding prehairpin intermediate formation and hemifusion. Conserved glycine residues in the FP of paramyxovirus F appear to have a primary role in regulating the activation of the metastable native form of F. Glycine residues in the FPs of other class I vFGPs may also regulate fusion activation.
The class I viral fusion-mediating glycoproteins (vFGPs) of retroviruses, lentiviruses, filoviruses, coronaviruses, orthomyxoviruses, and paramyxoviruses have evolved similar domain architectures to cause the coalescence of viral and host cell membranes. These vFGPs are synthesized as type I integral membrane proteins which are folded into homotrimers, posttranslationally modified by the addition of carbohydrate chains, and then in most cases cleaved by a protease into a metastable complex of membrane-distal and membrane-anchored subunits. For example, the paramyxovirus precursor fusion protein (F0) is cleaved into membrane-distal F2 and membrane-anchored F1 subunits (Fig. 1) which have an integrated and not separate domain structure in the atomic structure of F (10). The membrane-distal subunits from many class I vFGPs have insertions of receptor-binding domains (12, 64); however, the paramyxoviruses have evolved separate receptor-binding proteins to regulate fusion activation (33). The membrane-proximal subunits contain two hydrophobic domains, the fusion peptide (FP) and the transmembrane (TM) domain. The FP is located at or near the new N terminus of the membrane-proximal subunit and inserts into the target cell membrane after being triggered by low pH and/or receptor binding (12, 64). The TM domain serves as an anchor for the ectodomain to the viral membrane and is required for fusion pore formation (1, 39). Adjacent to the FP and TM domains in the ectodomain are two heptad repeat (HR) regions designated HRA and HRB, respectively. Upon triggering, HRA forms a triple-stranded coiled coil that mediates insertion of the FP into the target membrane, thus forming the so-called prehairpin intermediate (12, 64). Subsequently, HRB binds into the grooves between adjacent HRA monomers in an antiparallel orientation, forming a hairpin structure that brings the FP and TM domains into juxtaposition (12, 64). Peptides derived from the HR regions inhibit viral entry by binding specifically to their complementary regions in the vFGP (8, 54, 58), thereby preventing formation of the hairpin structure which couples protein refolding directly to membrane fusion (5, 38, 42, 57). While most class I vFGPs have short linker regions between their HR regions, the paramyxovirus F protein has a two-domain insertion of approximately 250 residues (10) which may serve as a molecular scaffold to help control the release and refolding of the FP and HR regions. The cytoplasmic tails of class I vFGPs can be critical to virus assembly (59) and often help regulate activation of the ectodomain in an “inside-out” manner. Activation of fusion may occur by cleavage of associated proteins (40, 76), by cleavage of the cytoplasmic tail (7, 53), or by dissociation of a cytoplasmic tail structure (63, 71). The hydrophobic FP domains tend to be the most highly conserved domains within a given virus family, suggesting that their structures and functions are more highly constrained by selective pressures than other domains.
FIG. 1.
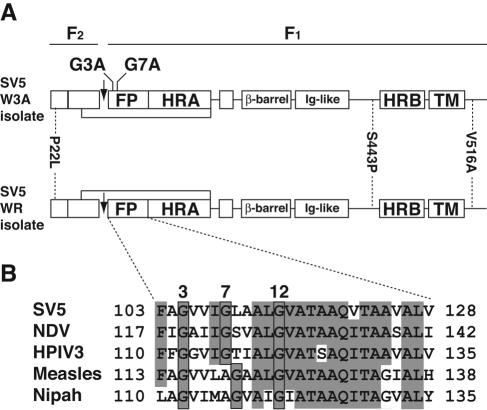
(A) Schematic diagram of the paramyxovirus F protein. The positions of the FP, HRA, β-barrel domain, immunoglobulin-like domain (Ig-like), HRB, and TM domain are shown. Identification of the folds of the F protein domains is done according to the NDV F crystal structure (10). The arrows indicate the site of cleavage of the F0 precursor protein into the F2 and F1 subunits. The lines between the F2 and F1 subunits show the location of the disulfide bond between the subunits. The locations of the three amino acid residues in the SV5 F protein that differ between the W3A and WR isolates (positions 22, 443, and 516) are identified. (B) Comparison of the amino acid sequences of the fusion peptides from the F proteins of representative members of the family Paramyxoviridae. The boxed regions indicate the conserved glycine residues. Gray shading denotes identity. In this study, FP residues of the F protein are numbered from the N terminus of the F1 subunit. The FP residues numbered by using a conventional numbering scheme are shown to the left and right of the sequences starting with the first residue of the unprocessed F0 protein. Shown are SV5 (genus Rubulavirus; strains W3A, GenBank accession number AF052755, and WR, GenBank accession number AB021962), NDV (genus Avulavirus; strain Australia-Victoria, GenBank accession number M21881), HPIV3 (genus Respirovirus; strain NIH 47885, Protein Data Bank accession number P06828), measles (genus Morbillivirus; strain Edmonston B, GenBank accession number U03655), and Nipah virus (genus Henipavirus, GenBank accession number NC_002728).
In addition to containing a high percentage of nonpolar residues, the hydrophobic FPs of the class I vFGPs often contain numerous conserved glycine residues spaced at regular intervals. For example, the FPs of influenza virus hemagglutinin (HA) and human immunodeficiency virus (HIV) gp41 have glycine contents of 29 and 26%, respectively. Several mutational studies on intact vFGPs are consistent with some conserved FP glycine residues being critical for membrane fusion activity while others are dispensable. A mutation of glycine 1 at the N terminus of HA2 to alanine is tolerated for fusion activity, while mutations to the nonpolar residues valine, leucine, isoleucine, and phenylalanine or to the polar or charged residues glutamine, histidine, glutamate, and lysine are not active for fusion, despite the mutant HA proteins having wild-type (wt)-like cell surface expression levels, trypsin cleavage, and pH values of conformational changes (23, 50, 65). The HA2 mutation G1S leads to wt-like cell surface expression, trypsin cleavage, pH of conformational change, and lipid mixing but does not cause pore formation or content mixing and has a propensity to revert to the wt sequence (15, 50). The HA2 mutants G4A, G4E, and G8A increase the pH threshold of conformational changes, but only G4E and G8A abolish fusion activity, while G4A causes fusion at levels similar to those of the wt (23, 65). For HIV gp41, G10V and G13V mutations lead to wt-like levels of gp160 synthesis, gp160 processing, and gp120 shedding but greatly reduce cell-cell fusion and virus infectivity, whereas G3V, G5V, and G20V mutations are better tolerated (18). For Moloney murine leukemia virus p15E, mutations to G13 lead to low cell surface expression, low cell-cell fusion, and greatly reduced titers, whereas mutations to G24 have wt-like expression levels but are defective for cell-cell fusion and virus rescue (79). Not all class I vFGPs have an absolute requirement for multiple critical glycine residues. For example, avian sarcoma and leukosis virus EnvA has an internal fusion peptide containing a central proline residue adjacent to its only glycine residue, G30. EnvA proteins containing G30P and G30V mutations have wt-like expression levels, have reduced Env-Gag ratios of 2/3, and yield virus titers 34 and 84% of that of the wt, respectively (4). Biophysical studies of short peptides (<25 residues) derived from the FP regions of class I vFGPs are consistent with these peptides having some membrane-perturbing activities (21, 41). A larger construct of the HA2 ectodomain (HA2 residues 1 to 127) causes pH-dependent lipid mixing which is inhibited by the G1E mutation (35). An emerging consensus view is that glycine residues in the FP are highly conserved because they maintain specific structures in target membranes necessary for perturbing lipid packing and facilitating lipid mixing (25).
The role of the three conserved glycine residues at positions 3, 7 (8 for the morbilliviruses and henipaviruses), and 12 in the FP of paramyxovirus F1 (cf. Figure 1B) is unclear. For the simian parainfluenza virus 5 (SV5) W3A isolate F protein, the FP mutations G3A, G7A, and G12A promote syncytium formation to extents 6 to 16 times greater than that of wt F, despite having cell surface expression levels of 74, 15, and 23% of wt expression levels, respectively (27). Moreover, the SV5 W3A G3A F protein causes lipid mixing at a higher initial rate than wt F (2). In contrast, for Newcastle disease virus (NDV), the comparable FP mutations G3K and G7K were reported to greatly inhibit syncytium formation (extents of 10 to 1% of wt syncytium formation, respectively), despite the F proteins having cell surface expression levels of 87 and 216% of that of the wt, respectively (62). A G12L mutation to NDV F1 causes a complete loss of cell surface expression. Syncytium formation and β-galactosidase reporter gene assays also show that the NDV F1 mutations G7A and G12A cause cell-cell fusion to extents <2% of that of the wt, despite having cell surface expression levels approximately 70 and 50% of wt levels, respectively (60).
In a separate avenue of experimentation, biophysical studies on 33-residue peptides derived from the FP of Sendai virus F are consistent with these peptides inserting into membranes with high α-helical contents, inducing bilayer perturbations, and causing lipid mixing (48, 52). G7A and G12A mutations to these peptides cause increases in α-helicity and fusogenic activity, suggesting that the increased fusogenicity of intact F proteins with GxA mutations may be due to enhanced target bilayer disruptions by the membrane-inserted FP (48). A high-resolution X-ray crystal structure of the ectodomain of NDV F does not contain interpretable density for the FP region (10); however, the core structure of the six-helix bundle (6HB) consisting of HR-derived peptides from SV5 F shows that the last seven residues of the FP continue the central triple-stranded coiled coil formed by the HRA peptides (3).
Researchers have recently identified two residues adjacent to HRB in SV5 F, L447 and I449, that have distinct, dual-functional roles: they regulate fusion activation and promote 6HB formation (58). Residues in influenza virus HA and human T-cell leukemia virus type 1 gp21 that have distinct roles in prefusion and fusion-activated states have also been identified (16, 75). Based on these results, we have proposed that some, if not many, conserved residues in the HR and FP regions of class I vFGPs that have functional roles in target membrane interactions and 6HB formation may also have critical roles in regulating the activation of the native states of these proteins. We wished to test our hypothesis that the conserved G3 and G7 residues in the FP of paramyxovirus F are involved in stabilizing native F, with mutations to alanine residues destabilizing the native state and thus resulting in hyperactive fusion. We constructed the FP G3A and G7A mutations into the backgrounds of the F proteins of SV5 (W3A and WR isolates), NDV, and human parainfluenza virus type 3 (HPIV3). All of the mutant F proteins except NDV G7A caused increased cell-cell fusion when the F proteins were coexpressed with the homotypic hemagglutinin-neuraminidase (HN) receptor-binding protein. Moreover, the SV5 WR F proteins containing G3A and G7A mutations became fusion active in the absence of HN-promoted fusion activation, consistent with these mutations destabilizing native F. Finally, dye transfer assays showed that the mutations lowered the energy required to activate native F, providing strong evidence that these FP residues are critical for the regulation of F protein activation in addition to any roles they might have in target membrane interactions.
MATERIALS AND METHODS
Cells, virus, and plasmids.
Monolayer cultures of Vero, BHK-21F, BSR T7/5, CV-1, and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Monolayers of BHK-21F cells were grown in DMEM supplemented with 10% FBS and 10% tryptose phosphate broth. The recombinant vaccinia virus vTF7-3, which expresses bacteriophage T7 RNA polymerase, was grown in CV-1 cells as described previously (22). pGEM2X and pCAGGS plasmids encoding SV5 W3A F, SV5 WR F, the SV5 W3A FR3 cleavage site mutation, SV5 HN, and influenza virus HA (A/Udorn/72) have been described previously (46). SV5 F proteins containing the G3A and G7A mutations were constructed by using consecutive rounds of four-primer PCR with Tgo DNA polymerase and subsequently cloned into pGEM2X SV5 W3A F, pGEM2X SV5 WR F, pCAGGS SV5 W3A F, and pCAGGS SV5 WR F. pGEM3X plasmids encoding NDV F, NDV HN, HPIV3 F, and HPIV3 HN, described previously (28), were subcloned into the pCAGGS plasmid. NDV and HPIV3 F proteins containing the G3A, G7A, and L289A mutations were constructed by using consecutive rounds of four-primer PCR with Tgo DNA polymerase and subsequently cloned into pGEM3X NDV F, pCAGGS NDV F, pGEM3X HPIV3 F, and pCAGGS HPIV3 F. The nucleotide sequences of the resulting constructs were determined by using a BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.) and an Applied Biosystems 3100-Avant automated DNA sequencer.
Expression of viral envelope glycoproteins.
Viral envelope glycoproteins were expressed by using two different expression systems: (i) Vero, BHK-21F, and HeLa cells transfected with pCAGGS DNA and (ii) CV-1 cells infected with the recombinant vaccinia virus vTF7-3 and transfected with pGEM2X or pGEM3X DNA. For expression of viral envelope glycoproteins from pCAGGS DNA, Vero, HeLa, and BHK-21F cells were transiently transfected by using the Lipofectamine Plus expression system (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Transfected Vero, BHK-21F, and HeLa cells were incubated for 4 h at 37°C before the addition of DMEM containing 10% FBS and incubation for 16 h at 37°C. For expression of viral envelope glycoproteins from pGEM2X and pGEM3X DNA, CV-1 cells in 6-well dishes with or without glass coverslips were infected with the modified vaccinia virus vTF7-3, which expresses bacteriophage T7 RNA polymerase (22), at an MOI of 10 PFU/cell. After a 30-min incubation at 37°C, appropriate amounts of pGEM2X or pGEM3X DNA were transfected into the vTF7-3-infected cells by using liposomes prepared in our laboratory (55). Infected-transfected CV-1 cells were incubated for 4 h at 37°C before the addition of DMEM containing 10% FBS and incubation for 16 h at 33°C.
Flow cytometry.
To quantify cell surface expression levels of the mutant SV5 F proteins, monolayers of Vero or CV-1 cells in 6-well dishes (70 to 80% confluent) were transfected by using 1.0 μg of F DNA as described above. At 16 h posttransfection (p.t.), the 6-well dishes were washed five times with phosphate-buffered saline containing calcium and magnesium at 0.1 g/liter (PBS+), overlaid with PBS+ solution containing primary antibody, and incubated at 4°C for 30 min. For SV5 F surface expression, the monoclonal antibody (MAb) F1a (51) was used at a 1:200 dilution. To quantify the extent to which the F protein mutations promoted conformational changes related to fusion activation, MAbs 6-7 and 21-1 (68) were used at a 1:100 dilution. The 6-well dishes were subsequently washed five times with PBS+, overlaid with PBS+ solution containing fluorescein isothiocyanate goat anti-mouse secondary antibody at a 1:100 dilution, incubated at 4°C for 30 min, and washed five times with PBS+. Cells were removed from 6-well dishes with PBS containing 50 mM EDTA and were fixed in suspension by the addition of methanol-free formaldehyde to a final concentration of 0.5%. The cell surface fluorescence of 10,000 cells was measured by using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.). Mean fluorescence intensity values were normalized to the mean fluorescence intensity value of the SV5 W3A wt F protein.
Cell surface biotinylation.
For cell surface biotinylation experiments with SV5, NDV, and HPIV3 F-expressing cells, monolayers of either HeLa cells or CV-1 cells in 6-cm-diameter dishes (70 to 80% confluent) grown in DMEM supplemented with 10% FBS were transfected as described above. At 16 h p.t., the 6-cm-diameter dishes were washed two times with PBS+ and then starved with methionine (Met)- and cysteine (Cys)-deficient DMEM for 30 min. The samples were then labeled for 15 min with 100 μCi of 35S-Promix (Amersham Pharmacia Biotech, Piscataway, N.J.) in 1 ml of DMEM (Met and Cys deficient; 20 mM HEPES buffer [pH 7.3]) before being washed once with PBS+ and chased for 2 h with 3 ml of DMEM (2 mM Met, 2 mM Cys, 20 mM HEPES buffer [pH 7.3]) to allow newly synthesized F proteins to reach the cell surface. The samples were subsequently biotinylated twice for 15 min at 4°C with 2 mg of EZ-Link Sulfo-NHS-SS-biotin (Pierce) in 1 ml of PBS+ (pH 8.0). Following washes with PBS+, PBS+ containing 50 mM Gly, and PBS+, the samples were lysed with 1 ml of ice-cold radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris [pH 7.4], 2% deoxycholate, 2% Triton X-100, 0.2% sodium dodecyl sulfate [SDS]) containing 0.15 M NaCl, 100 mM iodoacetamide, and protease inhibitors (45) and clarified by centrifugation for 10 min at 55,000 rpm with a Beckman Optima TLX ultracentrifuge. Samples were incubated overnight (18 to 22 h) at 4°C with antibodies. Antisera used were as follows: 25 μl of rabbit polyclonal anti-F2 peptide antiserum for SV5 F, 25 μl of rabbit polyclonal anti-F1 F6 antiserum for NDV F, and 4 μl of mouse MAbs C110 and C215 for HPIV3 F protein (kindly provided by Brian Murphy, National Institutes of Health). Immune complexes were adsorbed to protein A or G conjugated to Sepharose beads (protein A for SV5 F and NDV F and protein G for HPIV3 F) for 3 h at 4°C. Samples were washed three times with RIPA buffer containing 0.3 M NaCl, three times with RIPA buffer containing 0.15 M NaCl, and once with 50 mM Tris buffer (0.25 mM EDTA, 0.15 M NaCl [pH 7.4]). The samples were resuspended in 100 μl of 50 mM Tris buffer (0.5% SDS, pH 7.4), boiled for 5 min, and centrifuged for 1 min at 14,000 rpm with an Eppendorf centrifuge 5417R. The supernatants were split into two 50-μl fractions. One fraction was saved for direct loading onto the SDS-polyacrylamide gel electrophoresis (PAGE) gel so that it would include the total radiolabeled F protein. The other fraction was diluted to 1 ml with 20 mM Tris buffer (0.15 M NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% bovine serum albumin [pH 8.0]) and incubated with streptavidin-agarose for 3 h at 4°C. The samples were washed as described above for the immunoprecipitations, resuspended in SDS-PAGE sample buffer containing 2.5% (wt/vol) dithiothreitol, and fractionated on SDS-15% polyacrylamide gels (45). Quantitation of the F1 bands in the surface fractions was performed by using a Fuji (Stanford, Conn.) BioImager 1000.
Syncytium formation.
To assay the abilities of the mutant F proteins to cause syncytium formation, monolayers of BHK-21F cells in 6-well dishes (70 to 80% confluent) were transfected with 1.0 μg of SV5, NDV, or HPIV3 F DNA with or without 1.0 μg of SV5, NDV, or HPIV3 HN DNA, respectively, as described above. At 20 or 24 h p.t., cells were fixed and stained with a Hema 3 staining system (Fisher) according to the manufacturer's instructions. Representative fields were photographed with a Kodak DCS 760 (Eastman Kodak Company, Rochester, N.Y.) digital camera attached to a Nikon (Garden City, N.Y.) Diaphot inverted phase-contrast microscope.
Luciferase reporter gene assay for content mixing.
To quantify cell-cell fusion, a luciferase reporter gene assay was performed as described previously (58). Briefly, 6-well dishes containing Vero cells (70 to 80% confluent) were transfected with 1.0 μg of luciferase control DNA (Promega), 1.0 μg of pCAGGS F, and 1.0 μg of pCAGGS HN (or 1.0 μg of pCAGGS HA). At 16 h p.t., BSR T7/5 cells (expressing T7 RNA polymerase) were overlaid onto the Vero cells. Following a 6-h incubation at 37°C, the monolayers were washed, lysed, and clarified by centrifugation. From each clarified lysate, 40 μl of solution was pipetted onto a 96-well plate. The luciferase activity resulting from the fusion of the two cell populations was quantified by using luciferase assay substrate (Promega) and an Lmax luminescence microplate reader (Molecular Devices).
Dye transfer assays for lipid and aqueous content mixing.
To measure the temperature dependence of cell-cell fusion mediated by the SV5, NDV, and HPIV3 F proteins, dye transfer assays were performed as described previously (58). Briefly, human erythrocytes (RBCs) were single labeled with aqueous 6-carboxyfluorescein (CF; Molecular Probes) or dual labeled with CF and the lipid probe octadecyl rhodamine B chloride (R18; Molecular Probes) as described previously (58). Monolayers of CV-1 cells grown on glass cover slides in 6-well dishes (70 to 80% confluent) were infected with vTF7-3 and transfected with pGEM DNA as described above. The CV-1 cells were incubated for 4 h at 37°C followed by 16 h at 33°C. To allow the binding of the dye-labeled RBCs but to prevent temperature-induced fusion, 1 ml of ice-cold 0.1% hematocrit dye-labeled RBCs was added to each sample, and the CV-1-RBC (effector-target) complexes were incubated on ice at 4°C for 1 h. The effector-target cell complexes were incubated at 37°C for 10 min to allow dye transfer to take place before being washed five times with ice-cold PBS+ to remove unbound RBCs. Random microscopic fields were visualized by scanning confocal microscopy with a Zeiss (Thornwood, N.Y.) LSM 410. Membrane fusion was quantified by counting CF or R18 dye transfer events from labeled RBCs to CV-1 cells. To study syncytium formation and F-dependent retention of RBCs, CV-1 effector cells were labeled with 1 μM SYTO-17 nucleic acid dye (Molecular Probes) at 37°C for 30 min, and single-labeled CF-RBCs were used as fusion targets. For the temperature dependence of dye transfer, effector-target complexes were incubated for 10 min at the reported temperatures before being washed with ice-cold PBS+. To study dye transfer mediated by the SV5 W3A FR3 mutants containing the trypsin-inducible cleavage site mutation of five Arg residues to three Arg residues (47), CV-1 effector cells were incubated in the presence or absence of TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (10 μg/ml) at 37°C for 1 h before coincubation with single-labeled CF-RBCs.
RESULTS
The G3A and G7A mutations decrease SV5 F protein cell surface expression.
Introduction of the G3A, G7A, and G12A mutations into the FP of the SV5 W3A F protein increases syncytium formation despite reducing cell surface expression levels of the mutant F proteins to 74, 15, and 23% of wt F protein expression levels (27). The G3A and G7A mutant F proteins have intracellular transport rates to the medial Golgi apparatus similar to that of wt F, and both mutant F proteins are cleaved by a host cell protease. In contrast, the G12A mutant F protein does not become resistant to endoglycosidase H digestion and has a low to undetectable level of F0 cleavage. Therefore, we decided to focus this study on the effects of the G3A and G7A mutations exclusively. The SV5 WR F protein requires coexpression of its homotypic HN receptor-binding protein to mediate cell-cell fusion under physiological conditions (30), whereas the SV5 W3A F protein causes cell-cell fusion in the absence of HN coexpression (28, 44), albeit to a lesser extent than when HN is coexpressed (46). The amino acid sequences of the F proteins from the W3A and WR isolates differ at positions 22, 443, and 516 (Fig. 1A) (30, 43). The identity of the amino acid residue at position 516 has negligible effects on cell-cell fusion, whereas proline residues at positions 22 and 443 contribute to a lower temperature required for fusion activation, faster fusion kinetics, and HN-independent fusion (30, 46, 58). The SV5 W3A F P22L protein (no proline residues at positions 22 and 443) does not cause cell-cell fusion under physiological conditions, whereas the SV5 W3A F S443P protein (two proline residues at positions 22 and 443) causes cell-cell fusion with faster kinetics, at lower temperatures, and to greater extents than SV5 W3A F wt (46, 58).
We introduced the G3A and G7A mutations into the backgrounds of both SV5 W3A and WR F proteins. If the G3A and G7A mutations to WR F destabilize the native metastable form of the F protein, one might anticipate that a large enough decrease in the fusion activation energy of WR F would abolish its requirement of HN coexpression for fusion activation. We expressed the F protein mutants using both the pGEM-VacT7 and pCAGGS expression systems and measured F protein cell surface expression levels by surface biotinylation and flow cytometric analyses (see Materials and Methods). The expression levels of individual F mutants did not vary significantly between the two expression systems and did not vary significantly between the biotinylation and flow cytometry assays (Table 1). As has been found previously (46, 58), SV5 W3A wt, SV5 WR wt, SV5 W3A S443P, and SV5 W3A P22L all had similar cell surface expression levels. The G3A mutations caused reductions in cell surface expression of approximately 70 to 95% of the wt surface expression, and the G7A mutations caused reductions of approximately 20 to 50% of that of wt F protein (Table 1). The large reduction in cell surface expression of the G7A mutants may be due to decreased F protein stability, as SV5 W3A G7A has been shown to be degraded more rapidly than wt F protein (27). None of the mutants had cell-cell surface expression levels greater than those of the SV5 W3A or WR wt F proteins.
TABLE 1.
SV5 F protein cell surface expression and MAb reactivitiesi
| Construct | Surface expression (% biotinylated F1)a | MAb F1a
|
MAb 6-7 reactivity (MFI)d | MAb 21-1 reactivity (MFI)e | |
|---|---|---|---|---|---|
| Expression (% positive cells)b | Reactivity (MFI)c | ||||
| pCAGGSf W3A wtg | 100 | 49.1 | 100 | 100 | 100 |
| pCAGGS W3A P22L | 92.6 | 47.6 | 94.5 | 1.9 | 8.4 |
| pCAGGS W3A S443P | 100.4 | 43.6 | 98.0 | 391.0 | 289.6 |
| pCAGGS W3A G3A | 85.6 | 50.9 | 85.5 | 171.1 | 153.9 |
| pCAGGS W3A G7A | 31.9 | 38.8 | 19.7 | 743.7 | 384.5 |
| pCAGGS WR wt | 96.4 | 43.0 | 106.2 | 20.3 | 19.0 |
| pCAGGS WR G3A | 100.9 | 51.6 | 73.3 | 55.5 | 52.0 |
| pCAGGS WR G7A | 54.0 | 46.1 | 39.9 | 532.3 | 104.0 |
| pGEM2Xh W3A wtg | 100 | 81.7 | 100 | 100 | 100 |
| pGEM2X W3A P22L | 96.4 | 86.1 | 98.6 | 9.7 | 8.2 |
| pGEM2X W3A S443P | 98.2 | 87.3 | 103.7 | 270.8 | 253.4 |
| pGEM2X W3A G3A | 85.0 | 76.4 | 94.5 | 195.1 | 135.6 |
| pGEM2X W3A G7A | 30.8 | 46.4 | 33.3 | 1,275.3 | 395.1 |
| pGEM2X WR wt | 94.1 | 84.7 | 99.1 | 14.1 | 6.8 |
| pGEM2X WR G3A | 66.8 | 83.7 | 69.4 | 37.9 | 37.0 |
| pGEM2X WR G7A | 41.3 | 76.7 | 46.1 | 337.2 | 92.0 |
Cell surface expression level of the SV5 F1 polypeptide as determined by biotinylation, immunoprecipitation of radiolabeled lysates, and streptavidin-agarose recovery. SV5 F DNA was transfected into HeLa cells.
The percentage of cells expressing the SV5 F protein as determined by flow cytometry using MAb F1a.
Cell surface expression level measured by flow cytometry using MAb F1a.
Reactivity of cell surface-expressed F mutants to the conformation-specific MAb 6-7 as determined by flow cytometry.
Reactivity of cell surface-expressed F mutants to the conformation-specific MAb 21-1 as determined by flow cytometry.
pCAGGS DNA transfected into Vero cells.
Values were normalized to SV5 W3A wt F protein.
pGEM2X DNA transfected into CV-1 cells, vaccinia virus vTF7-3 coinfection.
MFI, mean fluorescence intensity.
The SV5 F proteins containing fusion peptide mutations have altered reactivities to conformation-sensitive MAbs.
The conformation-specific MAbs 6-7 and 21-1 were raised to and are highly reactive with the fusion-hyperactive SV5 WR L22P F protein (equivalent to the W3A F protein S443P/V516A, which is functionally equivalent to the W3A F protein S443P) (68). Additionally, MAbs 6-7 and 21-1 show little reactivity to the HN-dependent WR F protein and the fusion-defective W3A F protein P22L unless the F proteins are briefly heated to 47°C and returned to 4°C prior to the addition of antibody. The epitopes to MAbs 6-7 and 21-1 map to the N-terminal half of F2 and the β-sheet domain in F1, regions located outside of the FP and the region adjacent to HRB where S443P is located. Thus, MAbs 6-7 and 21-1 can be used as sensitive probes for F protein conformational changes related to heightened or decreased fusion activation. The SV5 G3A and G7A mutant F proteins were expressed in Vero and CV-1 cells in the absence of HN coexpression, and surface-expressed F protein was tested for reactivity to MAbs 6-7 and 21-1. The SV5 F proteins containing G3A and G7A mutations had significantly higher MAb reactivities than their corresponding wt F proteins, and those in the W3A background had much higher MAb reactivities than those in the WR background (Table 1). The results are consistent with the G3A and G7A mutations increasing the extents of partial F protein triggering in the absence of HN coexpression or target cell binding.
The G3A and G7A mutations increase the fusogenicity of the SV5 F protein and convert SV5 WR F into an HN-independent fusion-mediating glycoprotein.
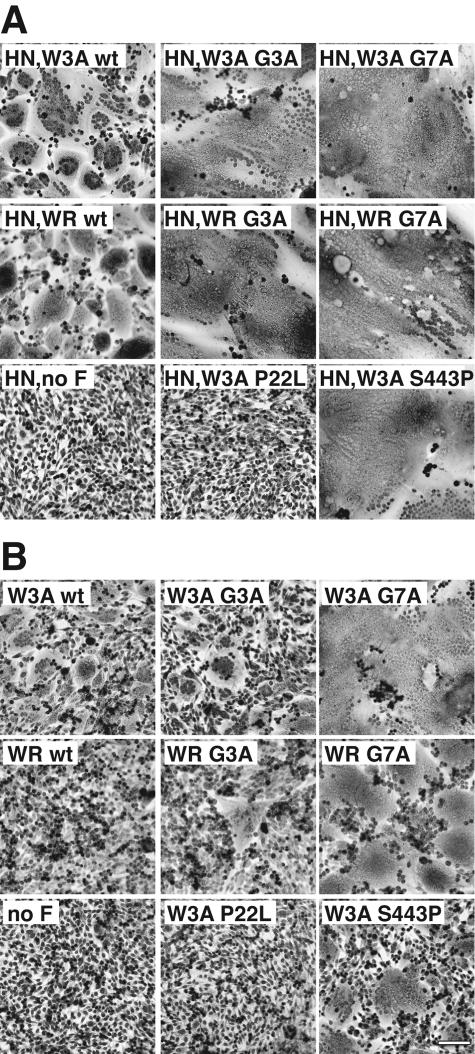
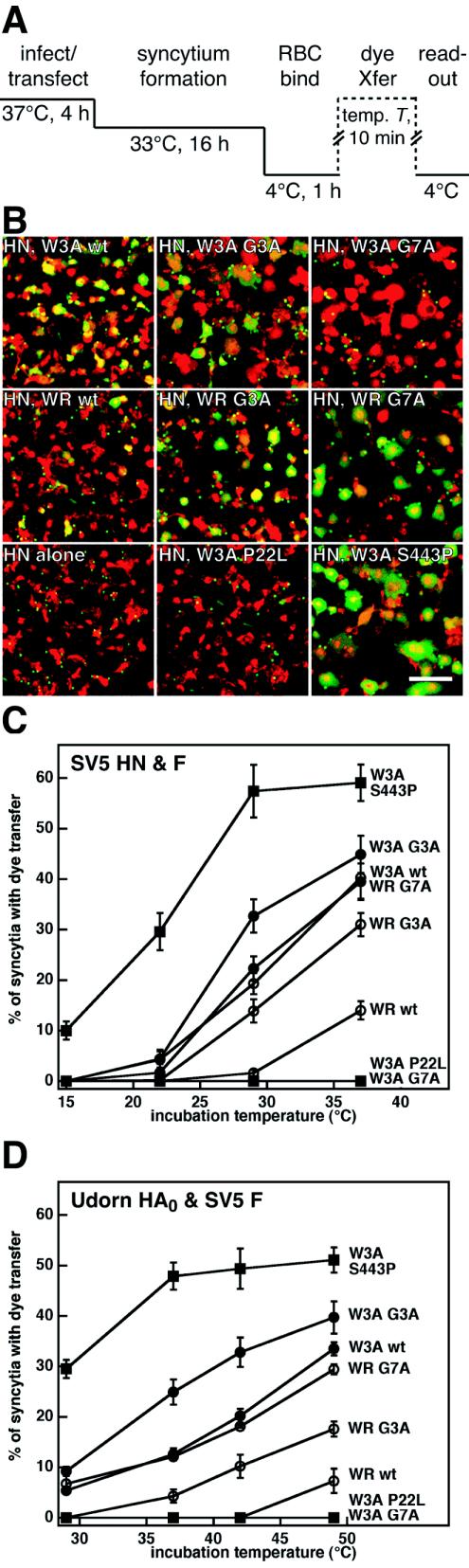
To examine the abilities of the mutant F proteins to cause cell-cell fusion, both syncytium formation and quantitative luciferase reporter gene assays were performed. Representative photomicrographs of syncytia formed between BHK-21F cells cotransfected with the SV5 F protein mutants and SV5 HN are shown in Fig. 2A. In the presence of HN coexpression, the G3A and G7A mutations increased syncytium formation in comparison to syncytium formation on expression of the SV5 W3A and WR wt F proteins. In the absence of HN coexpression, the SV5 W3A F proteins containing G3A and G7A mutations caused more extensive syncytium formation than SV5 W3A wt F protein (Fig. 2B). The SV5 WR F proteins containing G3A and G7A mutations caused extensive syncytium formation in the absence of HN coexpression, whereas the SV5 WR wt F protein did not cause detectable syncytium formation (Fig. 2B). Thus, the G3A and G7A mutations cause the WR F protein to be activated in the absence of HN coexpression, consistent with the FP mutations affecting an early intermediate in fusion activation.
FIG. 2.
Representative photomicrographs of syncytia formed between BHK-21F cells expressing the SV5 W3A and WR F proteins containing G3A and G7A mutations at 20 h p.t. in the presence of SV5 HN coexpression (A) or at 24 h p.t. in the absence of SV5 HN coexpression (B). For both panels, 1.0 μg of pCAGGS SV5 F DNA was used. For panel A, 1.0 μg of pCAGGS SV5 HN DNA was used. Bar, 200 μm.
To quantify cell-cell fusion mediated by the SV5 F protein mutants, we used a luciferase reporter gene assay (Fig. 3A). In the presence of HN coexpression, the W3A and WR F proteins containing G3A and G7A mutations exhibited greater extents of fusion than their corresponding wt F proteins (Fig. 3B) despite having lower cell surface expression levels than the wt F proteins (Table 1). To measure fusion activity in the absence of HN coexpression, the F protein mutants were also coexpressed with a surrogate receptor-binding protein, influenza virus HA0. In this experiment, HA0 serves as an attachment protein only and does not contribute directly to fusion activity as it is uncleaved and the pH is held at neutral. HA0 is coexpressed as a binding protein in this assay to maintain a constant level of effector-target cell binding during the time course of incubation. As shown in Fig. 3C, SV5 W3A G3A and G7A F proteins caused increased fusion in the absence of HN coexpression. As in the syncytium assays, the SV5 WR F proteins containing G3A and G7A mutations caused cell-cell fusion in the absence of HN coexpression, whereas SV5 WR F wt did not (Fig. 3C).
FIG. 3.
(A) Schematic diagram of the luciferase reporter gene assay to measure cell-cell fusion mediated by the SV5 W3A and WR F proteins containing G3A and G7A mutations. (B) A total of 1.0 μg of pCAGGS SV5 F DNA and 1.0 μg of pCAGGS SV5 HN DNA wasused. (C) A total of 1.0 μg of pCAGGS SV5 F DNA and 1.0 μg of pCAGGS influenza virus (A/Udorn/72) HA0 DNA was used. Uncleaved influenza virus HA0 protein at a neutral pH was used as a surrogate receptor-binding protein and did not contribute directly to cell-cell fusion (data not shown). (D) Cell-cell fusion measured as a function of cell surface expression levels of SV5 W3A and WR wt F proteins coexpressed with either 1.0 μg of pCAGGS SV5 HN DNA (closed bars) or 1.0 μg of pCAGGS influenza virus HA0 DNA (open bars). Cell surface expression of the SV5 W3A and WR wt F proteins were varied by transfecting different amounts of pCAGGS F DNA (μg DNA) and were measured by flow cytometry using MAb F1a (express). The data in all three panels are normalized to 100% fusion corresponding to the luciferase activity resulting from coexpression of SV5 HN and W3A wt F (1.0 μg of F and 1.0 μg of HN DNA), which had an average luciferase activity of 411 ± 14 relative light units (RLU). Error bars represent standard deviations of triplicate experiments.
It was previously shown that the SV5 W3A wt F protein causes increased cell-cell fusion at increased cell surface expression levels (58). As the G3A and G7A mutations decrease cell surface expression levels (Table 1), it did not seem likely that the lower cell surface expression levels of the mutant F proteins would cause the hyperactive fusion phenotypes. Nonetheless, to test the effect of lowering SV5 F wt expression levels to those of the G3A and G7A mutants, we titrated decreasing amounts of SV5 W3A and WR wt F DNA into transfection mixtures and measured the resulting cell surface expression levels and luciferase activities. The resulting reductions in SV5 W3A and WR wt F protein cell surface expression levels caused the expected decreases in cell-cell fusion (Fig. 3D), indicating that the F proteins containing G3A and G7A mutations have even greater fusion activities relative to wt F when controlling for cell surface expression levels. These results are consistent with the results from the MAb 6-7 and 21-1 binding analyses (Table 1): the F proteins containing G3A and G7A mutations have increased MAb reactivities and cell-cell fusion activities.
Fusion by the SV5 WR wt F protein is limited at a step preceding both hemifusion and prehairpin intermediate formation.
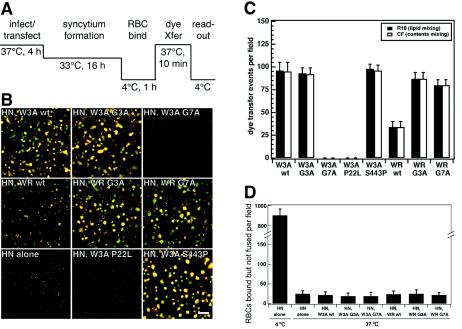
Fusion mediated by the SV5 F protein includes the following steps: F0 cleavage, HN binding to target cells, HN-promoted activation of F, insertion of the FP into target membranes (prehairpin intermediate formation), lipid mixing, and content mixing (reference 57 and references therein). As the G3A and G7A mutations convert the SV5 WR wt F protein into an F protein that causes cell-cell fusion in the absence of homotypic HN coexpression, we decided to test whether the WR wt is inefficient at causing fusion because it is blocked at a late or intermediate step in fusion, such as hemifusion or prehairpin intermediate formation. To determine if the WR wt F protein is blocked at hemifusion (positive for lipid mixing but negative for content mixing), we colabeled target RBCs with both the lipidic probe R18 and the aqueous probe CF, and we performed dye transfer assays with CV-1 effector cells coexpressing SV5 HN and F proteins (Fig. 4A). For all of the mutant F proteins, both lipid mixing and content mixing were coincidental (Fig. 4B and C). The overall extent of dye transfer promoted by the WR wt F protein was lower than that of W3A wt F, WR G3A F, and WR G7A F. However, for each individual lipid mixing event caused by WR wt F, there was a corresponding content mixing event; thus, WR wt F mediates full fusion and is not blocked at hemifusion. The addition of the lipid stalk destabilizing agent chlorpromazine did not cause increased dye transfer for any of the SV5 F proteins, indicating that they are not blocked at the restricted hemifusion stage either (data not shown). The small aqueous fluorescent dye CF (molecular weight, 376) can readily pass through small, nonenlarging pores such that it may not reveal all differences in the abilities of mutant vFGPs to induce hemifusion and pore formation. The luciferase data shown in Fig. 3 shows that fusion pores formed by WR wt F enlarge enough to allow the transfer of T7 RNA polymerase from BSR T7/5 cells to Vero cells containing the luciferase plasmid. The SV5 W3A G7A F protein did not cause any dye transfer (Fig. 4B and C), despite having the highest fusion activity as measured by syncytium formation (Fig. 2) and luciferase activity (Fig. 3). Such a phenotype is reminiscent of SV5 W3A F proteins containing aromatic mutations at residues L447 and I449 (58). These mutants have a “do-or-die” phenotype and cause rapid cell-cell fusion upon reaching the cell surface (e.g., in syncytium formation and luciferase reporter gene assays) but become inactivated in the dye transfer assay during the 16-h incubation period before the addition of the RBC target cells (Fig. 4A). The W3A G7A F protein was confirmed to have a similar do-or-die phenotype (see below).
FIG. 4.
(A) Schematic diagram of the dye transfer (dye Xfer) assay used to monitor cell-cell fusion mediated by the SV5 W3A and WR F proteins containing G3A and G7A mutations. Effector CV-1 cells were infected with vaccinia virus vTF7-3 and transfected with SV5 F DNA and SV5 HN DNA for 4 h at 37°C. The CV-1 effector cells were incubated overnight at 33°C to decrease cytopathic effects due to vaccinia virus vTF7-3 infection. Syncytium formation occurred during the overnight incubation. Effector CV-1 cells were coincubated with target cell RBCs for 1 h at 4°C, incubated for 10 min at 37°C, and incubated on ice before visualization by confocal microscopy. (B) Representative images of dye transfer caused by the mutant SV5 F proteins coexpressed with SV5 HN. RBCs were labeled with the lipidic probe octadecyl rhodamine (R18, red) and the aqueous probe CF (green). The effector-target cell complexes were incubated at 37°C for 10 min. Cell-cell fusion is observed as the transfer of red R18 and green CF from the small erythrocytes to the larger red-labeled syncytia formed by CV-1 effector cells. Bar, 200 μm. (C) Quantification of lipid and content mixing by the SV5 F protein mutants. The means and standard errors are from six microscopic fields. (D) Binding of RBCs to CV-1 cells expressing SV5 F protein mutants and SV5 HN. CV-1 cells were labeled with the fluorescent dye SYTO-17, and RBCs were labeled with the fluorescent dye CF. Samples were washed six times both before and after the 37°C incubation to remove unbound RBCs. The numbers of RBCs bound to effector CV-1 cells but not fused with CV-1 cells are reported. The means and standard errors are from three microscopic fields.
When RBCs are first bound at 4°C to cells expressing SV5 HN and the cells are then warmed to 37°C, most of the RBCs are released (57). The loss of RBC-binding activity by SV5 HN at 37°C is likely due to either the intrinsic neuraminidase activity of HN or a decrease in the affinity of HN for sialic acid as temperature is increased. The intrinsic neuraminidase activity of HPIV3 HN causes a similar release of RBCs as temperature is increased (49), and mutant NDV HN molecules with mutations at the dimer interface have been reported to have decreased affinity for the receptor sialic acid upon heating to 37°C (13). Regardless, the SV5 F protein acquires the ability to bind directly to target RBCs after the formation of the prehairpin intermediate (57). As a result, the acquisition of F-dependent (HN-independent) retention of target cell RBCs at 37°C can be used as a test for prehairpin intermediate formation (49, 57). As shown in Fig. 4D, neither WR wt F nor any of the F protein FP mutants retained significantly larger numbers of bound but unfused RBCs than the control when HN was expressed alone. For all of the F protein FP mutants, a majority of RBCs were released before F activation, a variable amount of RBCs had fused to effector cells (as shown in Fig. 4C), and there was a constant low level of bound but unfused RBCs (∼25 per field). The data strongly suggest that the F proteins were not blocked at prehairpin intermediate formation. Overall, the data are consistent with untriggered SV5 WR wt F molecules being blocked at a step before both hemifusion and prehairpin intermediate formation.
The G3A and G7A mutations decrease the energy required to activate the SV5 F protein.
Two alternative types of cell-cell fusion can occur in the dye transfer assay. First, upon reaching the cell surface, the F proteins can cause fusion between CV-1 effector cells (i.e., syncytium formation). Second, after coincubation with target RBCs, the F proteins can cause dye transfer from labeled RBCs to CV-1 effector cells. We performed another version of the dye transfer assay by using CF-labeled RBCs and SYTO-17-labeled CV-1 effector cells expressing the mutant F proteins (Fig. 5A). When the assay is performed in such a manner, both syncytium formation between CV-1 effector cells and dye transfer from RBCs can be observed. The observed trend of increased syncytium size between BHK-21F cells expressing F proteins containing G3A and G7A mutations (Fig. 2A) was recapitulated upon expression of these mutants in CV-1 cells (Fig. 5B). W3A G3A F and W3A G7A F proteins caused larger CV-1 syncytia than W3A wt F, and WR G3A F and WR G7A F caused larger syncytium formation than WR wt F. Despite causing more extensive syncytia, the SV5 W3A G7A F protein failed to cause dye transfer between the CV-1 effector cell syncytia and the RBC target cells, presumably because all of the cell surface-expressed W3A G7A F proteins had become inactivated during or after syncytium formation. The do-or-die phenotype of W3A G7A was confirmed (see below). To quantify the temperature dependence of F-mediated fusion, the dye transfer assay was repeated at various effector-target cell coincubation temperatures in both the presence and absence of HN coexpression (Fig. 5C and D, respectively). Aside from W3A G7A F being inactive for dye transfer after incubation at any of the temperatures tested, the temperature dependence of F-mediated fusion recapitulates the results from the luciferase assay. The rank order for temperature required to activate F in both the presence and absence of homotypic HN coexpression was as follows: W3A S443P F < W3A G3A F < W3A wt F ≈ WR G7A F < WR G3A F < WR wt F < W3A P22L F. HN coexpression resulted in slightly larger maximum dye transfer extents and a shift of approximately 12°C in the fusion activation temperatures. The observed decrease in temperature required to activate WR F due to the G3A and G7A mutations is consistent with these mutations destabilizing the native metastable conformation of the F protein such that lower thermal energy is required to activate enough trimers to cause dye transfer.
FIG. 5.
(A) Schematic diagram of the dye transfer (dye Xfer) assay used to monitor cell-cell fusion mediated by the SV5 W3A and WR F proteins containing G3A and G7A mutations. Effector CV-1 cells were infected with vaccinia virus vTF7-3 and transfected with SV5 F DNA and either SV5 HN or influenza virus HA0 DNA for 4 h at 37°C. The CV-1 effector cells were incubated overnight at 33°C to decrease cytopathic effects due to vaccinia virus vTF7-3 infection. Syncytium formation occurred during the overnight incubation. Effector CV-1 cells were subsequently labeled with the fluorescent dye SYTO-17 (red) for 1 h at 37°C, coincubated with CF-labeled RBCs (green) for 1 h at 4°C, incubated for 10 min at the indicated temperature, and incubated on ice before visualization by confocal microscopy. (B) Representative cropped one-quarter-field images of dye transfer caused by the mutant SV5 F proteins coexpressed with SV5 HN. The effector-target cell complexes were incubated at 37°C for 10 min. Cell-cell fusion is observed as the transfer of CF (green) from the small erythrocytes to the larger red-labeled syncytia formed by CV-1 effector cells, resulting in a yellow appearance in the merge image. Bar, 200 μm. (C) Temperature dependence of dye transfer caused by the SV5 W3A and WR mutant F proteins coexpressed with SV5 HN. After coincubation of CV-1 effector cells with erythrocyte target cells at 4°C, the effector-target cell complexes were incubated for 10 min at the reported temperatures. The extents of dye transfer are reported as the percentages of CV-1 syncytia containing CF dye. The mean values and error bars are from statistical analyses of three to five microscopic fields. (D) Temperature dependence of dye transfer caused by the SV5 W3A and WR mutant F proteins coexpressed with uncleaved influenza virus HA0. Conditions were identical to those described above for panel C.
The SV5 W3A F protein containing a G7A mutation has a do-or-die phenotype.
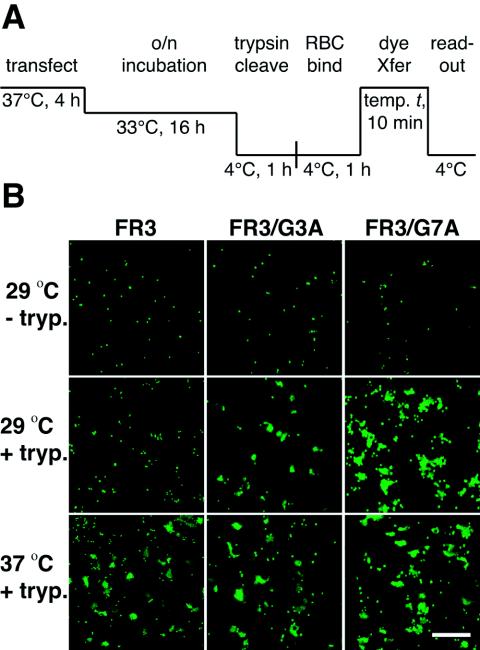
In the RBC dye transfer assays, the SV5 W3A G7A F protein caused the formation of large syncytia in CV-1 cells (Fig. 5B) but did not cause dye transfer (Fig. 4 and 5). It was previously found that SV5 W3A F proteins containing aromatic mutations at residues L447 and I449 had similar phenotypes (58). We hypothesized that the G7A mutation in the context of the SV5 W3A F protein not only destabilized the mutant F protein upon surface expression (thereby enhancing syncytium formation) but also inactivated the F protein relatively quickly (thereby preventing dye transfer from RBC target cells coincubated with CV-1 effector cells hours after cell surface expression of the W3A G7A F protein). To prevent the suspected inactivation of W3A G7A F in the absence of CF-labeled target cells, we performed the dye transfer assay using W3A G7A F constructed into the background of an F protein cleavage site mutant (i.e., FR3) that contains three arginine residues in the F protein cleavage-activation site. FR3 can be cleaved only by the addition of exogenous trypsin, and FR3/G7A is not activated (and therefore not inactivated) until after the addition of exogenous trypsin (47, 72). Here, we observed that cleavage of the SV5 W3A F protein double mutant FR3/G7A at 4°C followed by coincubation with CF-labeled RBCs at 4°C and a warm-up to 29 or 37°C resulted in efficient dye transfer (Fig. 6). In contrast, the pseudo-wt FR3 mutant (with only the two-arginine residue deletion in the cleavage site) did not cause efficient dye transfer at 29°C but instead required heating to 37°C for efficient dye transfer. The data are consistent with W3A G7A F having a do-or-die phenotype similar to those of W3A L447F, L447W, I449F, and I449W F proteins (58).
FIG. 6.
(A) Schematic diagram of the dye transfer (dye Xfer) assay using effector CV-1 cells coexpressing the SV5 proteins HN and FR3 (trypsin-inducible cleavage mutation of five Arg residues to three Arg residues at the cleavage site). Conditions were similar to those described in the legend of Fig. 4 except that following overnight (o/n) incubation at 33°C, the CV-1 effector cells were incubated at 4°C for 1 h in either the presence (+tryp.) or absence (−tryp.) of 10 μg of TPCK-trypsin/ml before coincubation with target RBCs at 4°C for 1 h. (B) Representative cropped one-quarter-field images of dye transfer caused by the mutant SV5 FR3 proteins coexpressed with SV5 HN. The effector-target cell complexes were incubated at either 29 or 37°C for 10 min. Cell-cell fusion is observed as the transfer of CF (green) from the small erythrocytes to the larger CV-1 effector cells. Bar, 200 μm.
G3A and G7A mutations in the NDV F and HPIV3 F proteins cause little or no reductions in cell surface expression.
There are conflicting reports on the roles of the conserved glycine residues in the FP regions of other paramyxovirus F proteins. For example, NDV F proteins containing G3K, G7K, and G7A mutations were reported to cause syncytium formation to only 1 to 10% of the extent of wt F syncytium formation (60, 62). Alternatively, a G7A mutation in the context of a 33-residue peptide derived from the FP of Sendai virus F is reported to increase peptide fusogenic activity (48). To test the effects of G3A and G7A mutations on the F proteins from other paramyxoviruses, we built these mutations into the backgrounds of the NDV F and HPIV3 F proteins. We chose the F proteins from NDV and HPIV3 because these paramyxoviruses are significantly different enough from SV5, and each other, to be classified into three different genera within the paramyxovirus family but are still similar enough to have HN receptor-binding proteins that are expected to be similar in structure to that of SV5 HN (14, 34). Additionally, a high-resolution structure of most of the NDV F ectodomain has been reported (10). We also built an L289A mutation into the background of NDV F for use as a positive control for hyperactive fusion by NDV F, as NDV L289A has been reported previously to cause syncytium formation more efficiently than NDV wt F and also to cause a limited extent of syncytium formation in the absence of NDV HN coexpression (61).
We expressed the NDV and HPIV3 F proteins containing G3A and G7A mutations by using both the pCAGGS and pGEM-VacT7 expression systems and measured their cell surface expression levels by surface biotinylation (see Materials and Methods). The FP mutations caused little or no reductions in the cell surface expression levels of both NDV F (Fig. 7A) and HPIV3 F (Fig. 7B). The relative amounts of the F1 subunit in the surface biotinylated fractions were quantified as markers for overall F protein cell surface expression. The NDV G3A and G7A F mutations caused small reductions in cell surface expression (approximately 95 and 92% of the wt, respectively). The HPIV3 G7A mutation reduced cell surface expression to approximately 80% of that of the wt, whereas the HPIV3 G3A mutation did not significantly alter cell surface expression.
FIG. 7.
Cell surface expression of the NDV F proteins (A) and HPIV3 F proteins (B) containing G3A and G7A mutations. HeLa cell cultures were labeled with 35S-Promix for 15 min and chased for 2 h to allow newly synthesized F proteins to reach the cell surface. Cultures were subsequently biotinylated at 4°C with NHS-SS-biotin. Following immunoprecipitation, the samples were split into two fractions. One fraction was directly loaded onto the SDS-PAGE gel to show the total radiolabeled F protein (T). The other fraction was bound to streptavidin-agarose to recover the fraction of the F protein that was biotinylated on the cell surface (S). All of the samples were boiled under reducing conditions and analyzed by SDS-PAGE. The reported cell surface expression efficiencies (Surface F1) were determined by measuring the intensities of the F1 bands in the surface (S) fractions and normalizing them to a wt level of 100. Similar results were obtained by expression of the NDV and HPIV3 F protein mutants in CV-1 cells by using pGEM3-X DNA and infection with the recombinant vaccinia virus vTF7-3 (data not shown). F0, uncleaved F protein; F1, cleaved F1 subunit; F2, cleaved F2 subunit.
G3A and G7A mutations increase NDV F and HPIV3 F protein fusogenic activity only when the F proteins are coexpressed with their homotypic HN receptor-binding proteins.
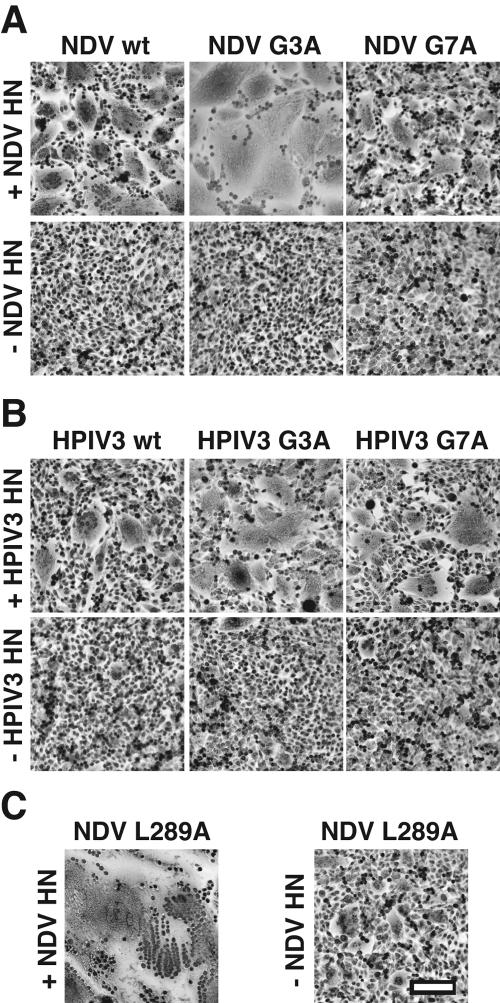
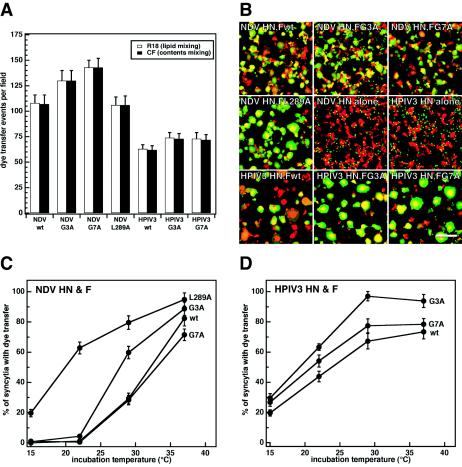
Syncytium assays were performed to qualitatively inspect the abilities of the mutant NDV and HPIV3 F proteins to cause cell-cell fusion. These assays were performed in the presence and absence of homotypic HN coexpression for both the NDV F protein mutants (Fig. 8A) and the HPIV3 F protein mutants (Fig. 8B). In the presence of NDV HN coexpression, the NDV F protein containing the G3A mutation caused more extensive syncytium formation than wt F, whereas the NDV F protein containing the G7A mutation caused syncytium formation to a slightly reduced extent compared to that of wt F. Both of the HPIV3 F proteins containing G3A and G7A mutations caused more extensive syncytium formation than wt F in the presence of HPIV3 HN coexpression. However, neither NDV F wt, HPIV3 F wt, nor any of the G3A or G7A mutant F proteins caused significant syncytium formation when expressed in the absence of coexpression of the homotypic HN receptor-binding protein. A limited extent of syncytium formation occurred between BHK-21F cells expressing NDV F L289A in the absence of NDV HN (Fig. 8C), confirming previous results (61). Moreover, the NDV F L289A mutation caused much more extensive syncytium formation in the presence of homotypic HN coexpression than NDV wt, G3A, and G7A, showing that the L289A mutation, which is located in the core of the NDV F protein (10), has a greater destabilizing effect than the G3A and G7A mutations in the FP region.
FIG. 8.
Representative photomicrographs of syncytia formed between BHK-21F cells expressing the NDV F proteins containing G3A and G7A mutations (A), HPIV3 F proteins containing G3A and G7A mutations (B), and NDV F protein containing an L289A mutation (C). Photomicrographs were taken at 20 h p.t. for cells coexpressing the F and HN proteins (+NDV HN and +HPIV HN) and at 24 h p.t. for cells expressing only the F proteins (−NDV HN and −HPIV3 HN). For all three panels, 1.0 μg of pCAGGS F protein DNA was used, and 1.0 μg of pCAGGS HN protein DNA was used where noted (+NDV HN and +HPIV3 HN). Bar, 200 μm.
We quantified cell-cell fusion mediated by the NDV and HPIV3 F protein mutants by using the luciferase reporter gene assay. The results recapitulate those obtained in the syncytium assays. In the presence of homotypic HN coexpression, NDV G3A F, HPIV3 G3A F, and HPIV3 G7A F caused small yet significant increases in cell-cell fusion (Fig. 9). NDV G7A F reduced cell-cell fusion to approximately 33% of that of NDV wt F, whereas NDV L289A F increased cell-cell fusion by approximately 200% of that of NDV wt F. In the absence of HN coexpression, none of the wt or mutant F proteins (including NDV L289A) promoted detectable cell-cell fusion as measured by the luciferase assay (data not shown). Therefore, even though the NDV G3A F, HPIV3 G3A F, and HPIV3 G7A F mutations cause an increase in cell-cell fusion, their effect on F protein triggering is not sufficient to trigger NDV F or HPIV3 F in the absence of HN coexpression.
FIG. 9.
Cell-cell fusion mediated by the NDV and HPIV3 F proteins containing G3A and G7A mutations as measured by the luciferase reporter gene assay. Conditions are identical to those described in the legend of Fig. 3B except that 1.0 μg of pCAGGS NDV or HPIV3 F DNA and 1.0 μg of pCAGGS NDV or HPIV3 HN DNA were used. Cell-cell fusion is expressed as luminescence arising from luciferase activity, with background luminescence due to NDV HN and HPIV3 HN expression serving as a negative control. None of the NDV or HPIV3 F protein mutants (even NDV L289A) caused luciferase activity in the absence of coexpression with its homotypic HN receptor-binding protein (data not shown). Error bars represent standard deviations from triplicate experiments.
The NDV F G3A and HPIV3 F G3A and G7A mutations lower the energy required to activate the F proteins.
To test the temperature dependence of fusion mediated by the NDV and HPIV3 F protein mutants, we performed dye transfer assays similar to those performed with the SV5 F protein mutants. First, we measured dye transfer by using RBC target cells dual labeled with lipidic R18 and aqueous CF. For all of the F protein mutants coexpressed with their homotypic HN receptor-binding proteins, both lipid mixing and content mixing were coincident: either both R18 and CF dye transfer occurred together or no transfer occurred at all (Fig. 10A). None of the NDV or HPIV3 F proteins (not even NDV L289A) caused dye transfer in the absence of homotypic HN coexpression (data not shown). NDV G7A F caused a greater number of dye transfer events per microscopic field than NDV wt F despite causing reduced syncytium formation and luciferase activity. Observation of the phase-contrast images revealed that NDV wt F caused more extensive syncytium formation between CV-1 cells than NDV G7A F in the dye transfer assay, thus reducing the total number of possible dye transfer events per field. Therefore, we decided to repeat the dye transfer assay with CF-labeled RBCs and SYTO-17-labeled CV-1 effector cells so that syncytium formation and dye transfer could be visualized simultaneously (Fig. 10B). Additionally, the extents of dye transfer could be normalized to the maximum possible number of dye transfer events per microscopic field, thus controlling for differential syncytium formation. The percentages of syncytia with dye transfer are reported as the function of incubation temperature for the NDV and HPIV3 F protein mutants in Fig. 10C and D, respectively. The data show that the NDV G3A and L289A mutations decrease the thermal energy required to activate NDV F, while the NDV G7A mutation causes a slight increase in the fusion activation energy. For HPIV3 F, both G3A and G7A mutations decrease the fusion activation energy. None of the wt or mutant NDV or HPIV3 F proteins (including NDV L289A) caused dye transfer in the absence of homotypic HN coexpression (data not shown). The fact that NDV L289A F causes a limited amount of syncytium formation but no luciferase activity or dye transfer in the absence of HN coexpression is consistent with the syncytium assay being a more sensitive assay for poorly fusogenic F proteins, perhaps since its incubation time is longer (24 h p.t.) and its readout of heterokaryons may be less dependent on synchronous events. Despite the inabilities of the NDV and HPIV3 F proteins containing G3A and G7A mutations to cause cell-cell fusion in the absence of homotypic HN coexpression, these mutations (except NDV G7A F) cause an increase in F protein fusogenicity in the presence of HN coexpression due to a decrease in energy required for the activation of the F protein for fusion.
FIG. 10.
Dye transfer assay used to monitor cell-cell fusion mediated by the NDV and HPIV3 F proteins containing G3A and G7A mutations. (A) Quantification of lipid and content mixing by the NDV and HPIV3 F protein mutants. The assay was performed as described in the legend of Fig. 4A by using RBC target cells colabeled with the lipidic dye R18 (red) and the aqueous dye CF (green). The means and standard errors are from three microscopic fields. (B) Representative cropped one-quarter-field images of dye transfer caused by the mutant NDV F proteins coexpressed with NDV HN and the mutant HPIV3 F proteins coexpressed with HPIV3 HN. The assay was performed as described in the legend of Fig. 5A with CF-labeled target RBCs and SYTO-17-labeled CV-1 effector cells. The effector-target cell complexes were incubated at 37°C for 10 min. Cell-cell fusion is observed as the transfer of green CF from the small erythrocytes to the larger red-labeled syncytia formed by CV-1 effector cells, resulting in a yellow appearance in the merge image. Bar, 200 μm. (C) Temperature dependence of dye transfer caused by the NDV mutant F proteins coexpressed with NDV HN. After coincubation of CV-1 effector cells with erythrocyte target cells at 4°C, the effector-target cell complexes were incubated for 10 min at the reported temperatures. The extents of dye transfer are reported as the percentages of CV-1 syncytia containing CF dye. The mean values and error bars are from statistical analyses of three to five microscopic fields. (D) Temperature dependence of dye transfer caused by the HPIV3 mutant F proteins coexpressed with HPIV3 HN. Conditions are identical to those described above for panel C. None of the NDV or HPIV3 F protein mutants (even NDV L289A) caused dye transfer in the absence of coexpression with its homotypic HN receptor-binding protein (data not shown).
DISCUSSION
The data presented here are consistent with the conserved glycine residues at positions 3 and 7 in the FP region of the paramyxovirus F protein serving critical roles in regulating the activation of the native metastable form of the F protein. In the contexts of both the W3A and WR SV5 F proteins, G3A and G7A mutations cause increases in both F-mediated cell-cell fusion and F protein reactivity to conformation-sensitive MAbs. The G3A and G7A mutations also result in SV5 WR F proteins that become triggered for fusion in the absence of the receptor-binding protein HN, unlike SV5 WR wt F that requires HN coexpression for triggering under biological conditions (30, 46). The paramyxovirus F protein adopts a number of unique structures that are important in the membrane fusion process: (i) a fusion-inactive uncleaved F0 structure (20, 29, 32, 69), (ii) a native metastable F1 plus F2 structure (46, 73), (iii) an activated intermediate triggered by HN binding its receptor (24, 57, 67), (iv) a prehairpin intermediate in which the fusion peptide interacts with target membranes (57, 58), and (v) a fusogenic-postfusogenic form that has a 6HB formed by HRA and HRB as its core (3, 78). The G3A and G7A mutations in the FP could affect any or all of these F protein conformations. Previous studies have shown that SV5 G3A and G7A F proteins are synthesized, processed, and cleaved in a manner similar to that of SV5 wt F (27, 72). Another study showed that the fusogenicity and α-helicity of 33-residue peptides derived from the FP of Sendai virus F are increased by G7A and G12A substitutions (48). These results suggest that fusion hyperactivity by intact F proteins with GxA mutations may be due to increased membrane penetration and disruption by the FP in late fusion intermediates. If the effects of the G3A and G7A mutations were due solely to more favorable interactions between the FP and target membranes in late fusion intermediates, one would predict that such mutations would have no effect on the fusion activity of F proteins blocked at early steps in the fusion process before the FP inserts into target membranes. Results of previous studies are consistent with the decreased fusion activity of SV5 WR F compared to SV5 W3A F being due to a kinetic block in the activation of an early fusion intermediate (31, 46, 58). Moreover, the data reported here show that preactivated SV5 WR F is blocked at an early step in the fusion process, preceding both prehairpin intermediate formation and lipid mixing. The G3A and G7A mutations help overcome the kinetic block of WR F, presumably by destabilizing native F, and thus contribute to increased membrane fusion. Moreover, the destabilizing effects of the G3A and G7A mutations are large enough to lead to WR F triggering and membrane fusion even in the absence of HN coexpression. Thus, in the context of SV5 WR F, the primary effects of the G3A and G7A mutations are to more readily trigger the preactivated native F protein. The present work does not address secondary effects of the mutations due to target membrane interactions.
The paramyxoviruses have evolved a number of strategies to precisely regulate the activation of their protein machines to cause membrane fusion for virus-to-cell entry and, in some cases, cell-to-cell spread. First, the paramyxoviruses have evolved separate receptor-binding proteins (HN, H, and G) that trigger the F proteins after target cell binding. The F proteins in naturally occurring paramyxoviruses are triggered for fusion in the presence of their homotypic receptor-binding protein, and most paramyxovirus F proteins require coexpression of their homotypic HN protein for fusion. Protein-protein interactions between HN and F have been observed biochemically (66, 77), and mutations that disrupt those interactions also decrease membrane fusion (19, 24). Second, the paramyxoviruses have evolved metastable F proteins that couple the energy released from protein refolding to the work of membrane fusion (57). To activate the F protein at the right time and place, the native metastable form of the F protein must maintain a kinetic barrier which is large enough to prevent indiscriminant inactivation yet small enough to allow triggering after target cell binding. Several F protein residues and regions have been identified as being critical for the regulation of the kinetic barrier of the F protein. Mutations to these residues cause increased activation, and in some cases indiscriminant inactivation, of the F protein. These residues include SV5 WR L22 (three residues from the amino terminus of F2 after signal sequence cleavage) (30), SV5 WR K132 in HRA (31), NDV L289 in the core of the F protein (61), a stretch of residues adjacent to HRB in SV5 W3A F (including S443, L447, and I449) (46, 58), and cytoplasmic tail extensions in SV5 SER F (including L539 and L548) (63, 71). The present data show that residues G3 and G7 in the FP regions of SV5, NDV, and HPIV3 also help regulate the kinetic barrier of the paramyxovirus F protein.
In some cases, paramyxovirus F proteins can be triggered artificially in the absence of the homotypic receptor-binding protein by the presence of a closely related heterotypic receptor-binding protein (6, 70), by receptor-binding activity of the F protein (11, 26), by destabilizing agents such as heat and urea (46, 58, 73), or by destabilizing mutations (30, 31, 46). In such cases, triggering in the absence of the receptor-binding protein is less efficient than in its presence. The present data show that G3A and G7A mutations in the FP circumvent the SV5 WR F protein's requirement for its homotypic HN protein for fusion activation, presumably by destabilizing the native form of F. Naturally occurring destabilizing residues in SV5 F that contribute to HN-independent fusion activity in cell-cell fusion assays are P22, E132, and P443 (30, 31, 46). Here, we show that NDV G3A F, HPIV3 G3A F, and HPIV3 G7A F do not cause cell-cell fusion in the absence of HN coexpression, despite causing small yet significant increases in membrane fusion compared to wt F in the presence of homotypic HN coexpression. One possible reason for the G3A and G7A mutations circumventing the requirement of HN coexpression for activation of SV5 WR F but not for activation of NDV F or HPIV3 F is that the kinetic barriers of NDV F and HPIV3 F may be much greater than those of SV5 WR F. If such is the case, combinations of destabilizing mutations may result in mutant NDV F and HPIV3 F proteins that can cause cell-cell fusion in the absence of HN coexpression. Alternatively, the mechanisms by which the HN protein promotes activation of the F protein may differ between the SV5 F protein and other paramyxovirus F proteins that may have absolute requirements for HN coexpression (i.e., NDV F and HPIV3 F). For NDV F, it has been proposed that NDV HN clamps down the F protein and activates the F protein by releasing it after target cell binding (37, 67). If such is the case, then the NDV F and HPIV3 F proteins would be expected to be inactivated in the absence of HN coexpression, and destabilizing mutations to F would not lead to HN-independent fusion activation. In the absence of NDV HN coexpression, the NDV L289A F protein has been shown previously (61) and here to cause a limited extent of cell-cell fusion in syncytium assays but not in reporter gene and dye transfer assays. Further experiments will be required to better characterize the NDV L289A F protein and to differentiate between the two proposed models for HN-promoted fusion activation.
Structural analyses of the influenza virus HA are consistent with its FP forming three critical structures: a surface loop in HA0 that exposes the cleavage site to proteases (9), a chain packed into a surface cavity in the native metastable form of HA1 plus HA2 (74) that regulates the energy required to activate HA (17, 56), and a kinked helical structure that binds to and most likely disrupts target membranes (25). In the native metastable structure, HA2 FP residues 1 to 9 form a two-turn structure that packs into a pocket of charged residues (74). In this amino-terminal portion of the FP, conserved glycine residues are located at positions 1, 4, and 8, an approximately four-residue spacing of glycine residues that is similarly found in the amino-terminal portion of the FP of the paramyxovirus F protein (Fig. 1B). The mutation of these HA FP glycine residues to alanine residues results in HA proteins that are more easily triggered but are less efficient at or incapable of causing membrane fusion (65). To accommodate the addition of a methyl group to the alpha carbons of these residues by virtue of G1A, G4A, and G8A mutations, the main chain of the FP in this region would have to alter its structure in an apparently energetically unfavorable manner. Such structural perturbations to the packing of the FP in the pocket thus lower the energy required to activate HA. G3A and G7A mutations in the FP of paramyxovirus F may have similar structural consequences in its native structure. A crystal structure of the ectodomain of NDV F does not contain interpretable density for the FP residues (10). However, the structure reveals that the carboxy-terminal residues of the F2 subunit terminate near a prominent radial channel in which the FP could be sequestered from the aqueous environment (12). A cryoelectron microscopy structure of the Sendai virus F protein ectodomain reveals a similar central cavity in which the FP might insert after proteolytic cleavage (36). The results of the present work are consistent with GxA mutations in the FP of paramyxovirus F disrupting the packing of the FP into such a cavity on the surface of the F protein, perhaps in a manner similar to that of GxA mutations in the context of influenza virus HA. Further high-resolution studies of the paramyxovirus F protein ectodomain will be required to confirm or refute this hypothesis.
Acknowledgments
We thank Karen Kantor for help with molecular cloning and George Leser for technical advice regarding the biotinylation assay. We thank Masato Tsurudome for providing the 6-7 and 21-1 MAbs, Rick Randall for providing F1a MAb, Brian Murphy for providing C110 and C215 MAbs, and Bernard Moss for providing the recombinant vaccinia virus vTF7-3.
This work was supported in part by research grant AI-23173 from the National Institute of Allergy and Infectious Disease. C.J.R. is an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1997. A glycine to alanine substitution in the paramyxovirus SV5 fusion peptide increases the initial rate of fusion. Virology 238:283-290. [DOI] [PubMed] [Google Scholar]
- 3.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 4.Balliet, J. W., K. Gendron, and P. Bates. 2000. Mutational analysis of the subgroup A avian sarcoma and leukosis virus putative fusion peptide domain. J. Virol. 74:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrego-Diaz, E., M. E. Peeples, R. M. Markosyan, G. B. Melikyan, and F. S. Cohen. 2003. Completion of trimeric hairpin formation of influenza virus hemagglutinin promotes fusion pore opening and enlargement. Virology 316:234-244. [DOI] [PubMed] [Google Scholar]
- 6.Bossart, K. N., L.-F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 11.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 13.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 15.Cross, K. J., S. A. Wharton, J. J. Skehel, D. C. Wiley, and D. A. Steinhauer. 2001. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 20:4432-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniels, P. S., S. Jeffries, P. Yates, G. C. Schild, G. N. Rogers, J. C. Paulson, S. A. Wharton, A. R. Douglas, J. J. Skehel, and D. C. Wiley. 1987. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 6:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels, R. S., J. C. Downie, A. J. Hay, M. Knossow, J. J. Skehel, M. L. Wang, and D. C. Wiley. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40:431-439. [DOI] [PubMed] [Google Scholar]
- 18.Delahunty, M. D., I. Rhee, E. O. Freed, and J. S. Bonifacino. 1996. Mutational analysis of the fusion peptide of the human immunodeficiency virus type 1: identification of critical glycine residues. Virology 218:94-102. [DOI] [PubMed] [Google Scholar]
- 19.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 20.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138-150. [DOI] [PubMed] [Google Scholar]
- 21.Epand, R. M. 2003. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 1614:116-121. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gething, M.-J., R. W. Doms, D. York, and J. M. White. 1986. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J. Cell Biol. 102:11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gravel, K. A., and T. G. Morrison. 2003. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 77:11040-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, X., J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2001. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8:715-720. [DOI] [PubMed] [Google Scholar]
- 26.Harris, J., and D. Werling. 2003. Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response. Cell. Microbiol. 5:671-680. [DOI] [PubMed] [Google Scholar]
- 27.Horvath, C. M., and R. A. Lamb. 1992. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 66:2443-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath, C. M., R. G. Paterson, M. A. Shaughnessy, R. Wood, and R. A. Lamb. 1992. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J. Virol. 66:4564-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu, M., A. Scheid, and P. W. Choppin. 1981. Activation of the Sendai virus fusion protein (F) involves a conformational change with exposure of a new hydrophobic region. J. Biol. Chem. 256:3557-3563. [PubMed] [Google Scholar]
- 30.Ito, M., M. Nishio, M. Kawano, S. Kusagawa, H. Komada, Y. Ito, and M. Tsurudome. 1997. Role of a single amino acid at the amino terminus of the simian virus 5 F2 subunit in syncytium formation. J. Virol. 71:9855-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, M., M. Nishio, H. Komada, Y. Ito, and M. Tsurudome. 2000. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81:719-727. [DOI] [PubMed] [Google Scholar]
- 32.Kohama, T., W. Garten, and H.-D. Klenk. 1981. Changes in conformation and charge paralleling proteolytic activation of Newcastle disease virus glycoproteins. Virology 111:364-376. [DOI] [PubMed] [Google Scholar]
- 33.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 34.Lawrence, M. C., N. A. Borg, V. A. Streltsov, P. A. Pilling, V. C. Epa, J. N. Varghese, J. L. McKimm-Breschkin, and P. M. Colman. 2004. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335:1343-1357. [DOI] [PubMed] [Google Scholar]
- 35.Leikina, E., D. L. LeDuc, J. C. Macosko, R. Epand, Y. K. Shin, and L. V. Chernomordik. 2001. The 1-127 HA2 construct of influenza virus hemagglutinin induces cell-cell hemifusion. Biochemistry 40:8378-8386. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig, K., B. Baljinnyam, A. Herrmann, and C. Bottcher. 2003. The 3D structure of the fusion primed Sendai F-protein determined by electron cryomicroscopy. EMBO J. 22:3761-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGinnes, L. W., K. Gravel, and T. G. Morrison. 2002. Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J. Virol. 76:12622-12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami, T., S. Ablan, E. O. Freed, and Y. Tanaka. 2004. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J. Virol. 78:1026-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nieva, J. L., and A. Agirre. 2003. Are fusion peptides a good model to study viral cell fusion? Biochim. Biophys. Acta 1614:104-115. [DOI] [PubMed] [Google Scholar]
- 42.Park, H. E., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048-1053. [DOI] [PubMed] [Google Scholar]
- 43.Paterson, R. G., T. J. R. Harris, and R. A. Lamb. 1984. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc. Natl. Acad. Sci. USA 81:6706-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson, R. G., S. W. Hiebert, and R. A. Lamb. 1985. Expression at the cell surface of biologically active fusion and hemagglutinin-neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc. Natl. Acad. Sci. USA 82:7520-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, United Kingdom.
- 46.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 47.Paterson, R. G., M. A. Shaughnessy, and R. A. Lamb. 1989. Analysis of the relationship between cleavability of a paramyxovirus fusion protein and length of the connecting peptide. J. Virol. 63:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peisajovich, S. G., R. F. Epand, R. M. Epand, and Y. Shai. 2002. Sendai virus N-terminal fusion peptide consists of two similar repeats, both of which contribute to membrane fusion. Eur. J. Biochem. 269:4342-4350. [DOI] [PubMed] [Google Scholar]
- 49.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall, R. E., D. F. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 52.Rapaport, D., and Y. Shai. 1994. Interaction of fluorescently labeled analogues of the amino-terminal fusion peptide of Sendai virus with phospholipid membranes. J. Biol. Chem. 269:15124-15131. [PubMed] [Google Scholar]
- 53.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 55.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 56.Ruigrok, R. W. H., S. R. Martin, S. A. Wharton, J. J. Skehel, P. M. Bayley, and D. C. Wiley. 1986. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 155:484-497. [DOI] [PubMed] [Google Scholar]
- 57.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 60.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seth, S., A. Vincent, and R. W. Compans. 2003. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin-neuraminidase protein requirement for membrane fusion. J. Virol. 77:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 65.Steinhauer, D. A., S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1995. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J. Virol. 69:6643-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, H. Komada, and Y. Ito. 2001. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) protein: difference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J. Virol. 75:8999-9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umino, Y., T. Kohama, T. A. Sato, A. Sugiura, H.-D. Klenk, and R. Rott. 1990. Monoclonal antibodies to three structural proteins of Newcastle disease virus: biological characterization with particular reference to the conformational change of envelope glycoproteins associated with proteolytic cleavage. Virology 176:656-657. [DOI] [PubMed] [Google Scholar]
- 70.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waning, D. L., C. J. Russell, T. S. Jardetzky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. USA 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, C. D., R. G. Paterson, and R. A. Lamb. 1995. Mutants of the paramyxovirus SV5 fusion protein: regulated and extensive syncytium formation. Virology 209:242-249. [DOI] [PubMed] [Google Scholar]
- 73.Wharton, S. A., J. J. Skehel, and D. C. Wiley. 2000. Temperature dependence of fusion by Sendai virus. Virology 271:71-78. [DOI] [PubMed] [Google Scholar]
- 74.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-375. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, K. A., A. L. Maerz, and P. Poumbourios. 2001. Evidence that the transmembrane domain proximal region of the human T-cell leukemia virus type 1 fusion glycoprotein gp21 has distinct roles in the prefusion and fusion-activated states. J. Biol. Chem. 276:49466-49475. [DOI] [PubMed] [Google Scholar]
- 76.Wyma, D. J., J. Jiang, J. Shi, J. Zhou, J. E. Lineberger, M. D. Miller, and C. Aiken. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, N. L., P. M. Cannon, D. Chen, and W. F. Anderson. 1998. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J. Virol. 72:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]