Abstract
Objective. To design and implement a pharmacogenomics course that focuses on analysis and integration of pharmacogenomic data into clinical practice and to explore how participation in the course influences student self-confidence.
Design. The Basic and Clinical Pharmacogenomics course content was divided into three modules: genetic-based didactic sessions, genomic techniques and self-genotype/phenotype laboratory exercise, and clinical-based case studies. Student learning assessment included knowledge- and application-based tests and performance on a group project.
Assessment. Effectiveness of the course was evaluated using results of student performance on coded test questions, student perceptions on pre- and post-course self-assessments, performance on a group project, and course evaluation results. Student pharmacists successfully demonstrated competency in pharmacogenomics knowledge-based learning, demonstrated their abilities to apply learned skills in clinical-based scenarios, and reported improved confidence in analyzing patient-based genomic testing results.
Conclusions. This course appears to have contributed to student learning and positively influenced student self-confidence in pharmacogenomics.
Keywords: pharmacogenomics, pharmacogenetics, pharmacy curriculum, assessment, genomic testing
INTRODUCTION
Pharmacogenomics is altering the way drugs are developed and changing how medications are prescribed and administered to patients. Pharmacogenomics uses information from an individual’s genome and evaluates drug pharmacology to select drugs and drug dosages that are likely to be most effective. Pharmacogenomics is anticipated to expand the potential for personalized medication for patients and could transform pharmacy practice. The American Society of Health-System Pharmacists (ASHP) published a statement on the role of the pharmacist in clinical pharmacogenomics stating that “pharmacists have a responsibility to take a prominent role in the clinical applications of pharmacogenomics.”1 In a study of community pharmacies in North Carolina, the provision of pharmacogenetic testing was evaluated with the authors concluding that this new service line is feasible but there is a need for pharmacist training.2 With this monumental change in medication therapy options, pharmacy educators need to provide future pharmacists with in-depth, practical education in pharmacogenomics.3,4 In this regard, the Accreditation Council for Pharmacy Education (ACPE) stresses the importance of pharmacogenomics and pharmacogenetics for the doctor of pharmacy (PharmD) curriculum in the ACPE Standards 2016.5 Appendix 1 of the standards states that students have a knowledge base and ability to provide patient care in the “genetic basis for disease and individual differences in metabolizing enzymes, transporters, and other biochemical impacting drug disposition and action that underpin the practice of personalized medicine.”5 Additionally, the use of pharmacogenomics as an emerging approach to drug therapy aligns with the Center for the Advancement of Pharmacy Education (CAPE) Outcomes 2013, which emphasize the provision of patient-centered care.6
Although the importance of pharmacogenomics is recognized, curricular coverage in pharmacy education varies and gaps in practice implementation are apparent. A survey by Murphy and colleagues in 2010 demonstrated that 92% of 75 responding PharmD programs taught pharmacogenomics at their institutions at the PharmD or graduate level, but the educational approach and content varied.7 The study found the most common approach to teaching pharmacogenomics within PharmD programs (n=67) was to embed content within other required coursework (72.5%). Other less common approaches included standalone courses (21.7%) and electives (34.8%). The depth of content coverage was less than 30 didactic hours for more than 80% of the institutions. The breadth of the pharmacogenomics content covered varied dramatically between programs as indicated by responses to questions regarding two content domains: genetic basis of disease, and ethical, social, and economic implications. Sixty-one percent of faculty members responded that they believed the status of pharmacogenomics instruction at other colleges was poor or inadequate.7 With regards to practice, McCullough and colleagues in 2011 surveyed practicing pharmacists and reported that 63% of 303 respondents disagreed while only 14% agreed with the following statement: “I can accurately apply the results of a pharmacogenomics test to drug therapy selection, dosing, or monitoring.”8
To assist in the implementation of pharmacogenomics content within pharmacy and health-science curriculums, a number of recommendations and core competencies have been published including those by the American College of Clinical Pharmacy,9 the Core Competency Working Group of the National Coalition for Health Professional Education in Genetics,10 the Inter-Society Coordinating Committee for Physician Education in Genomics,11 and the American Nurses Association.12 In addition, numerous online resources and training modules have been developed to aid in the clinical implementation of pharmacogenomics. One online resource includes the Genetics/Genomics Competency Center (G2C2) funded by the National Institutes of Health National Human Genome Research Institute (http://g-2-c-2.org/). Another online, pharmacy-specific resource is the Pharmacogenomics Education Program (PharmGenEd) organized by the University of California San Diego Skaggs School of Pharmacy and Pharmaceutical Sciences.13
Recognizing the need to educate and prepare pharmacists to meet the future demand for patient care services related to pharmacogenomics, Washington State University (WSU) College of Pharmacy developed a standalone pharmacogenomics course for the PharmD curriculum in spring 2015. The WSU College of Pharmacy is a long-standing pharmacy education program in a public, research-intensive university. The pharmacogenomics course was added as a required course in the first professional year (PY1). The pharmacogenomics course was designed to include content for the development of foundational knowledge and also included required innovative application activities through which students could apply their knowledge. The primary aims and curricular goals of the course were: ensure that students demonstrate competence in the knowledge-based aspects of pharmacogenomics; prepare students to confidently apply knowledge of basic genetics in a clinical setting; provide students the opportunity to analyze and evaluate pharmacogenomic data for possible application in a clinical setting.
The purpose of this manuscript is to describe the design and implementation of a pharmacogenomics course in a PharmD curriculum. In addition, student perception data and performance data on assessments related to pharmacogenomics content and application is reported.
DESIGN
The Basic and Clinical Pharmacogenomics course was implemented in 2015 as a required course for first-year PharmD students at WSU. The pharmacogenomics course was created following the recommendations of an ad hoc curriculum review committee to increase the focus of the core curriculum on future pharmacy practice needs such as pharmacogenomics and biotechnology. These recommendations resulted in several curricular revisions, which resulted in the consolidation of some courses and the creation of new courses including the pharmacogenomics course.
The course was designed to meet the following WSU College of Pharmacy Curriculum Outcomes: basic principles of drug response based on the study of the whole genome (pharmacogenomics) and its pertinence to biotechnology; and personal, ethical values and belief systems regarding emerging biotechnologies in order to understand the complexities underlying decisions of patient care. Course content and activities were developed to align with ACPE 2016 Standards, Appendix 1 on pharmacogenomics/genetics, and CAPE Outcome 2.1, which states students develop the ability to provide patient-centered care as the medication expert.5,6
The course format was one two-hour lecture per week. The course was divided into three modules that were team-taught by multiple instructors from the Department of Pharmaceutical Sciences. The course used Pharmacogenomics: Applications to Patient Care, second edition, as a required textbook.14 While no specific genetics prerequisites were required for the course, it was assumed that student pharmacists would have learned foundational genetics in biochemistry and general biology prerequisites.
The spring PY1 pharmacogenomics course aligns well with the curriculum by providing a needed introduction and overview of pharmacogenomics early on. During their first semester in the PharmD program, students enroll in pharmacokinetics, pharmacy calculations, an integrated pharmacology course series, Top 200 drugs, and a communications course. As a result, prior to taking the pharmacogenomics course, students have demonstrated understanding of dose-response relationships, dose calculations, and mechanism of a number of drugs, and they have demonstrated patient-counseling skills. In addition, the PY1 curriculum focuses on drug metabolizing enzymes and transporters responsible for drug absorption, distribution, and metabolism, which aligns well with the discussion of the pharmacogenomics of these enzymes and transporters.
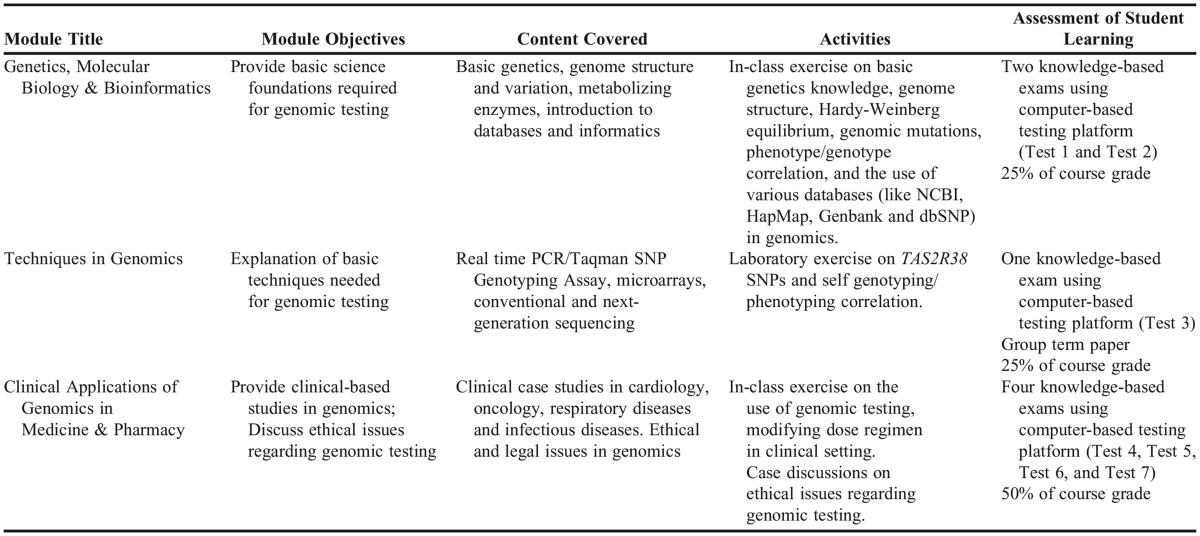
Content in the course was delivered via three modules to achieve competency outcomes as recommended by the NIH Genetics/Genomics Competency Center. The first module included content that covered basic genetics (genetics, molecular biology, and bioinformatics). The second module addressed issues related to the practical application of genomics including sound laboratory techniques. The third module was devoted to clinical implementation of pharmacogenomics in patient care settings and dose management of drug therapy for various disease states through discussion of in-class patient cases. Additionally, the third module addressed ethical issues regarding genomic testing such as privacy obligations, responsibilities concerning incidental findings, balancing collective and individual interests and well-being in research, and the potential dangers of misleading rhetoric about genetic essentialism and personalized medicine. Discussion of ethical issues focused on analyzing and responding to scenarios (based on real cases) in which clinicians are faced with ethically significant decisions. Students were invited to reflect on how they might balance competing ethical values in clinical and genomics research contexts. Table 1 includes detailed information about the objectives, content, activities, and student learning assessments within each module, whereas Appendix 1 outlines weekly activities in the course.
Table 1.
Overview of the Three Modules Within the Basic and Clinical Pharmacogenomics Course
The WSU College of Pharmacy uses a competency-based assessment grading model in which students demonstrate achievement of clearly defined learning objectives and curriculum outcomes by attainment of a score of at least an 80% on all course assessments. The college’s competency-based assessment model uses a three-tier honors/satisfactory/fail (HSF) grading model. In didactic courses, such as this one, students are given three attempts to achieve the 80% required competency on each assessment. Students not reaching the 80% competency complete a retest within one week with nonidentical question items that cover the same learning objectives as the initial test. Students not reaching the 80% competency on the retest must complete their third attempt, known as an extended learning experience (ELE), at the end of the semester. The ELE tests include questions that are worded differently but that assess the same learning objectives used in the initial tests and retests. Students needing to complete second and third attempts are encouraged to seek remediation help from the instructor. Instructors will often review material and clarify misconceptions either through one-on-one meetings or group review sessions. The purpose of the retest and ELE test is to allow students to demonstrate sufficient knowledge of the academic material to meet the established standard of competency. Accordingly, the maximum score recorded for any retest or ELE test is 80%, regardless of the actual score a student received. To receive an honors designation for this course, students need to earn a cumulative average of greater than or equal to 90%.
All knowledge-based assessments within this course were created and administered via a proprietary computer-based testing platform, Examsoft (ExamSoft Worldwide, Inc., Boca Raton, FL). The question formats most commonly used in the course included multiple-choice and true/false questions. Through this system, question items were coded to categories including WSU College of Pharmacy Curriculum Outcomes and course-level learning objectives. Coded question items were used to generate data and longitudinal reports to evaluate student cohort level data regarding student learning and depth and breadth of content covered.
In this course, student learning was evaluated through seven knowledge-based assessments, and students were given a maximum of three attempts to achieve competency in each. These knowledge-based assessments tested foundational knowledge of pharmacogenetics and pharmacogenomics as well as application of this knowledge. Additionally, student learning within module 2 was evaluated through a self-genotyping/phenotyping group project as described in the following section.
A unique aspect of this course was the laboratory exercise and mandatory group term paper. A self-genotyping/phenotyping laboratory exercise was designed to provide students experience with application of pharmacogenomic knowledge. The laboratory exercise was structured to emphasize the principle that the identification of polymorphic genetic variation among patients (genotype) serves as an important marker in patient care and dose management of various disease states (phenotype). Furthermore, this exercise allowed students to discuss the impact of intrinsic factors such as race, ethnicity, and gender on the observed genotype-phenotype correlation. Self-genotyping/phenotyping exercises have been used previously in other pharmacogenomics courses, but typically center on medically or pharmacologically relevant genes such as ACE or CYP450 enzymes.15-19 For the purpose of this course, a medically innocuous gene (the bitter taste receptor gene, TAS2R38) was selected, which had known single-nucleotide polymorphisms (SNPs) and testable phenotype (bitter taste perception with the compound phenylthiocarbamide, PTC).
The PY1 class of 133 students was randomly separated into 22 groups of approximately six students each. Volunteer students from each group (total = 48) donated a buccal mouthwash for DNA isolation from cheek cells using PTC extraction, amplification and an electrophoresis kit (Carolina Biological Supply Company, Whitsett, NC). For this purpose, the second module content (Table 1) introduced students to various genomic techniques including those used to isolate their DNA from cheek cells and determine their genotype using the polymerase chain reaction (PCR)-based assay. Students also had the opportunity to observe the processing of their samples by a technician and teaching assistants. The WSU Office of Research Assurances determined that the project/laboratory exercise was exempt from IRB review.
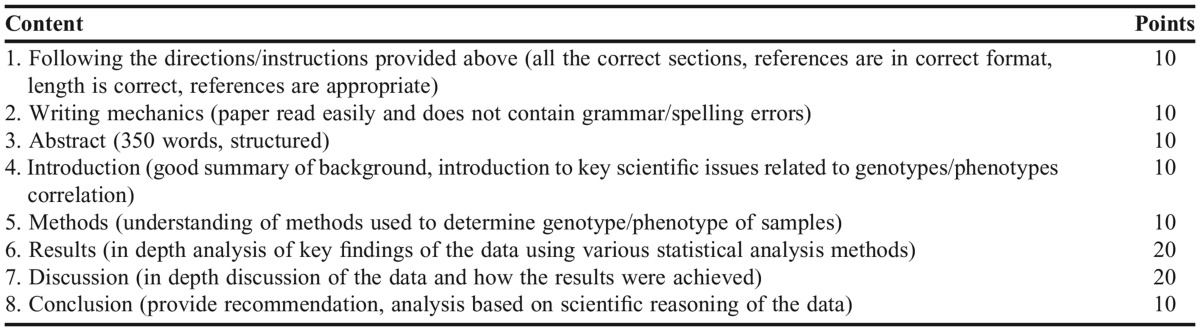
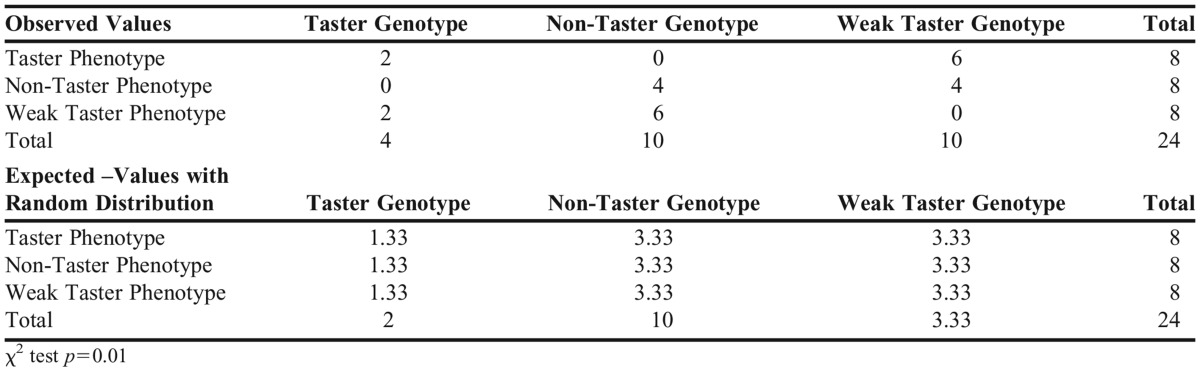
Genotypes and phenotypes were identified and recorded by the anonymous sample ID number. The aggregate data of 24 samples were then distributed to the entire class, serving as data for the group term paper. Requirements for the group term paper included elements of an actual scientific publication including a six sections layout: abstract, introduction, methods, results, discussion, and cited references. Each group was expected to submit a 10-page (excluding references), 12-point, double-spaced document with their reflections on this laboratory exercise. The group term paper was evaluated using a rubric (Table 2) that assessed the students’ ability to examine how genotypes compare with phenotypes, and on their ability to apply knowledge learned in the class in the explanation of DNA isolation techniques, genotype determination, linkage with regard to the predominant and rare haplotypes, and the correlation between SNPs in TAS2R38 gene and receptor function. Students were also expected to discuss how genotype/phenotype correlation may or may not impact “lifestyle choices” such as food preference or alcohol consumption and smoking preferences. It was expected that students discuss how genotype/phenotype may be associated with race or gender and to compare findings of the data to established knowledge in the literature. Appendix 2 shows the results of phenotypic/genotypic correlation as reported by a student group. Since the chi-square test was <.05, students concluded that there was a significant correlation between phenotype and genotype with respect to PTC taste.
Table 2.
Rubric Utilized for Grading of the Group Project Noting Requirements for the Seven Sections of the Group Term Paper
EVALUATION AND ASSESSMENT
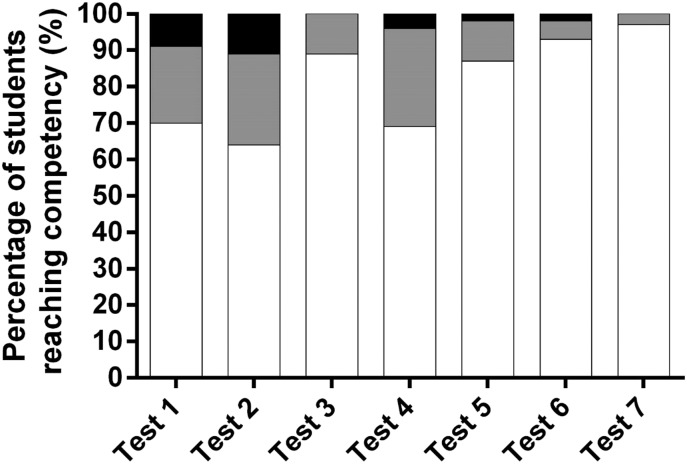
Ninety-eight percent of the enrolled students met competency at an 80% level or greater on each of the seven knowledge-based assessments and the group term paper. Figure 1 indicates the percentage of students reaching competency on the three attempts provided on the seven knowledge-based assessments. The percentage of the class (N =133 students) reaching competency on the first attempt for each of the seven tests ranged from 64% on test 2 to 97% on test 7. Following the second attempt, the percentage of students reaching competency ranged from 89% on test 2 and 100% on test 3. The average score of the class on the group term paper was 88% with only one group out of 22 having to resubmit the project in order to meet competency. Fifty-six students earned an honors grade for the course while 77 earned a grade of satisfactory.
Figure 1.
Percentage of students attaining competency of an 80% or greater for each of the three attempts allowed on the seven knowledge-based assessments given in the course. White bars indicate the percentage of students demonstrating competency on first attempt. Gray bars indicate the percentage of students demonstrating competency on second attempt. Black bars indicate the percentage of students demonstrating competency on the third attempt.
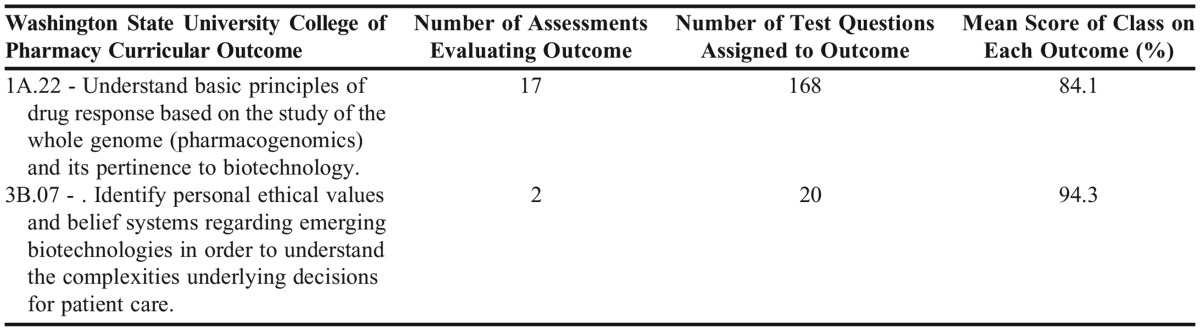
Performance on the knowledge-based assessments was further evaluated by analyzing achievement of the WSU College of Pharmacy Curriculum Outcomes assigned to test items. The data in Table 3 establish that the student cohort, in aggregate, demonstrated competence, with students earning a mean score of greater than 80% on the assigned curricular outcomes. Student performance on knowledge-based assessments and the group term paper indicate that the majority of students achieved competency in understanding and applying the basic science concepts relevant to pharmacogenomics.
Table 3.
Assessment of Curriculum Outcome Coverage in a Pharmacogenomics Course and Mean Score Performance on Curricular Outcomes
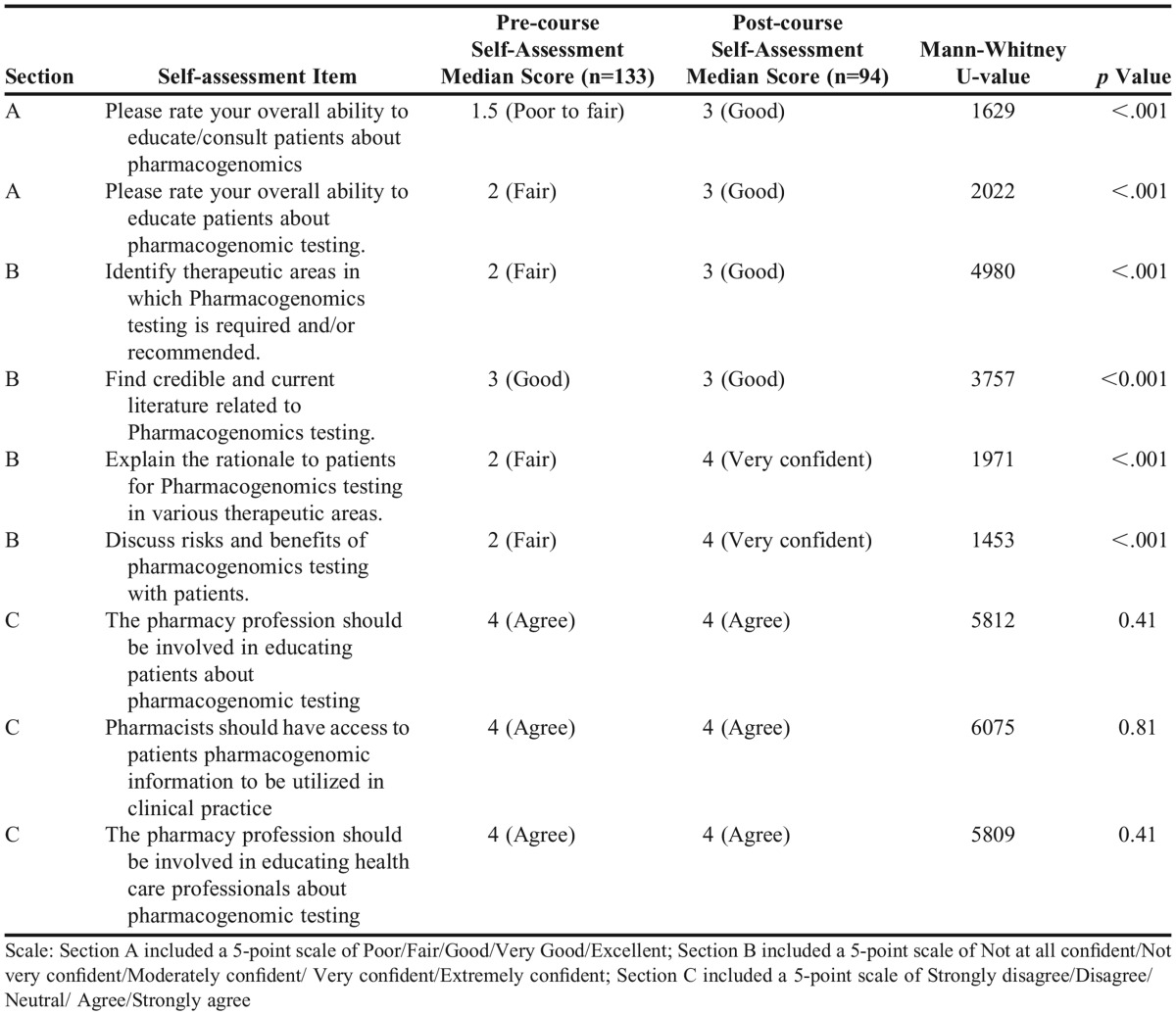
In addition to performance data from the student learning assessments, changes in student perceptions regarding different aspects of pharmacogenomics were evaluated from the results of two anonymous student self-assessment questionnaires (Appendix 3) administered via an electronic survey platform, Qualtrics (Qualtrics Provo, UT). To assess the changes in student perception, one self-assessment was completed pre-course and the other was completed post-course. Response rates for the pre-course and post-course student self-assessment were 100% and 70%, respectively, for the class of 133 students. The decrease in response rate likely reflects a small decrease in class attendance from the first week of the semester to the last as well as likely survey fatigue with numerous course and instructor evaluations occurring at the same time.
The items within the self-assessment were divided into three different sections (Appendix 3). In section A, each student rated their overall ability to educate/consult patients about pharmacogenomics (item 1) and to educate patients about pharmacogenomics testing (item 2) using a five-tier “ability” self-measure that ranged from poor to excellent. In section B, students rated their confidence in four different aspects of pharmacogenomics testing (items 3-7). In section C, students were asked about their perceptions regarding the role of pharmacists in relation to pharmacogenomics testing (items 8-10). As these self-measures of abilities, confidence, and perceptions are ordinal with no natural neutral value, similar to a Likert-type scale, a Mann-Whitney test was used to compare the distributions of answers between the two unpaired samples using Graphpad Prism 6 (GraphPad Software, Inc., La Jolla, CA). The results of the Mann-Whitney U tests are summarized in Table 4.
Table 4.
Statistical Analysis of the Change in Student Abilities, Confidence, and Perceptions as Assessed From a Pre-course and Post-course Self-assessment
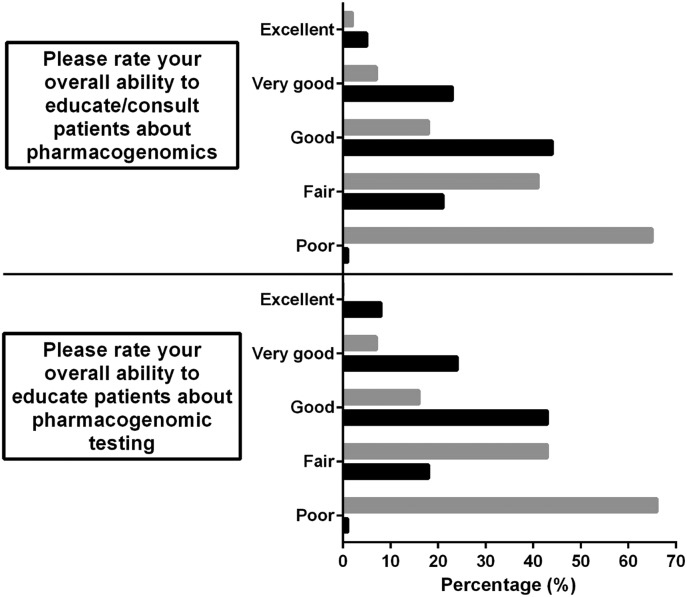
For section A, the distribution of the responses was similar to both questions for each administration of the survey (Figure 2). Pre-coursework student self-assessment measures of ability to educate/consult patients about pharmacogenomics (item 1) were 82% poor to fair and to educate patients about pharmacogenomics testing (item 2) were 80% poor to fair. On the post-course self-assessment, ability measures improved specifically 68% good to excellent on item 1 and 63% good to excellent on item 2.
Figure 2.
Histograms of student responses of their perceptions for their abilities to educate patients about pharmacogenomics overall and to provide education about pharmacogenomic testing as collected through a pre- and post-course self-assessment. Gray bars indicate results from the pre-course self-assessment. Black bars indicate results from the post-course self-assessment.
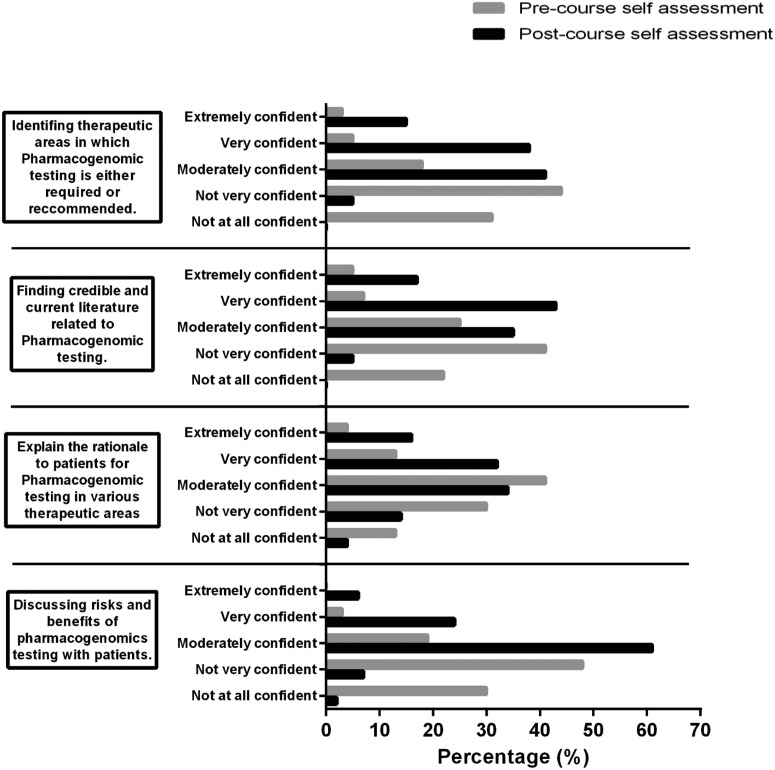
In section B of the self-assessment, students rated their confidence in pharmacogenomics testing using a scale ranging from not at all confident to extremely confident (Figure 3). Students’ confidence levels on the pre- and post-course self-assessment were statistically different, indicating that student confidence in performing aspects of pharmacogenomic testing increased following completion of the course.
Figure 3.
Histograms of student responses of their confidences to perform various elements of pharmacogenomics testing as assessed through a self-assessment given pre-course and post-course. Gray bars indicate results from the pre-course self-assessment. Black bars indicate results from the post-course self-assessment.
In section C of the self-assessment, students were asked their opinions on the role of pharmacists in pharmacogenomics testing. On the pre-course self-assessment, 71% of respondents agreed or strongly agreed that pharmacists should be involved with patient pharmacogenomics testing education (item 8); 72% of the respondents agreed or strongly agreed that pharmacists should have access to patient clinical practice pharmacogenomics information (item 9); and 67% of the respondents agreed or strongly agreed that pharmacists should be involved in educating health care professionals about pharmacogenomics testing (item 10). The post-coursework self-assessments showed increased agreement with the items above, with 80% agree or strongly agree, 82% agree or strongly agree, and 82% agree or strongly agree, respectively.
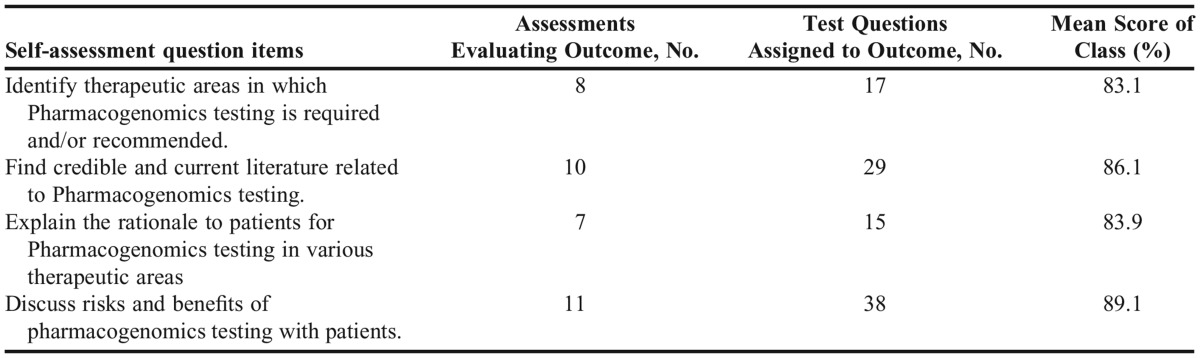
To determine if student perceptions of their own confidence in pharmacogenomics translated to demonstration of competency on the knowledge-based assessments, the self-assessment items from section B were coded to related test items within Examsoft. The longitudinal reports of performance on test questions coded to self-assessment items are reported in Table 5. Overall, these data support that student perceptions of confidence on the self-assessment items did translate to attainment of competency on the material, with competency demonstrated between 83.1% and 89.1% across the four self-assessment items.
Table 5.
Results of Longitudinal Report of Self-assessment Items Linked to Test Questions
At the end of the semester, instructor and course evaluations were released to the students using WSU’s course evaluation program, Explorance Blue (Explorance, Montréal, Canada). The response rate for the course evaluation was 33% (44 of 133 students responded). Although low, this response rate is typical for most courses during this semester. Selected student comments are reported in Appendix 4.
DISCUSSION
In response to the anticipated health care demand for pharmacists who are competent in designing patient-specific drug therapy using pharmacogenomics, the Basic and Clinical Pharmacogenomics course was added to the WSU College of Pharmacy curriculum in spring 2015 as a required course for PY1 students. The primary aims and curricular goals of the course were: to ensure that students demonstrate competence in the knowledge-based aspects of pharmacogenomics; to prepare students to confidently apply knowledge of basic genetics/genomics in a clinical setting; and to provide students the opportunity to synthesize an informed therapeutic action plan for dose management and personalized patient care through analysis and evaluation of pharmacogenetic data.
As a cohort, students successfully achieved competency on test question items related to the assigned WSU COP curricular outcomes related to pharmacogenomics. This indicates that students successfully learned pharmacogenomics content as assessed through seven knowledge-based examinations. It is important to note that students appeared to struggle more with the content on initial tests that focused on foundations of genomic testing as fewer students reached competency on the first attempt (Figure 1).
Student perceptions of their overall abilities to educate patients about pharmacogenomics, in general, and pharmacogenomics testing, specifically, improved based on the results of the pre- and post-course self-assessment. For both question items related to patient education, over 60% of respondents rated their abilities between good and excellent in the post-course survey compared to approximately 50% reporting poor abilities in the pre-course survey. These results suggest that the content and activities within this course helped students improve their perceived abilities to educate patients on their clinical test results.
Student confidence in several clinical aspects of pharmacogenomics patient care skills improved between the pre- and post-course self-assessment. The majority of student respondents reported in the pre-course self-assessment that they were not confident in three of the four skills areas. In the post-course survey, student respondents reported they were moderately to extremely confident in their clinical pharmacogenomics skills. The average performance of the student cohort on test question items coded to the four clinical skills areas ranged from 83% correct to 89% correct indicating that student perception of confidence aligned with their actual performance on assessments. Participation in this course appears to have positively influenced cohort confidence about their clinical skills in pharmacogenomics while at the same time students were able to demonstrate competency in pharmacogenomics content and clinical skill areas. Additionally, the self-assessments indicated that students believed the role of pharmacists in pharmacogenomics testing is highly important.
A number of lessons were learned with implementation of the pharmacogenomics course. First, some students found the content areas of basic genetics and molecular biology challenging. Basic genetics and molecular biology are not prerequisites for admission to WSU College of Pharmacy. To address this gap, the instructors in the pharmacogenomics course were able to adjust the basic genetics content so that it was at an appropriate level for the students. The majority of students view the pharmacogenomics course as essential for their professional practice, and they were excited about the knowledge they gained. Table 6 highlights examples of students’ feedback demonstrating their excitement about the course and their abilities to synthesize an informed therapeutic action plan for dose management and personalized patient care through analysis and evaluation of a patient’s genotype/phenotype data analysis. Thus, the program is committed to optimizing this course in order to improve students’ genomic knowledge and confidence in applying basic genetics in a clinical setting.
The evaluation of effectiveness of the Basic and Clinical Pharmacogenomics course has several limitations. First, the responses to the pre- and post-course self-assessments were not linked, which prevents any analysis of individual student responses, and therefore, only aggregate data can be reported. Second, the group project was conducted in groups of approximately six students, so group performance does not necessarily verify individual student competencies. Finally, few IPPE and APPE sites currently exist for student pharmacists to practice and apply pharmacogenomics in “real” patient care settings.
As pharmacogenomics becomes part of health care practice, pharmacogenomics in pharmacy education will need to expand by establishing coursework that lays the basic foundation necessary for developing competency in knowledge and clinical application. This course can serve as a model for other institutions that are in need to develop a pharmacogenomics course to prepare future pharmacists to provide patient-centered care through pharmacogenomics.
SUMMARY
The Basic and Clinical Pharmacogenomics course was added to the WSU College of Pharmacy curriculum in spring 2015 for PY1 students. The primary aim of the course was for students to demonstrate competency in their knowledge of pharmacogenomics, which would prepare them to confidently apply the concepts in a clinical setting. Furthermore the course was designed with the intention that it would provide students with the opportunity to synthesize therapeutic action plans through analysis and evaluation of a patient’s genomics test results. These aims/goals were evaluated using coded question items, student perceptions gathered through pre- and post-course self-assessments, and a group project using a self-genotyping/phenotyping laboratory exercise. As a cohort, PY1 students successfully demonstrated competency on seven independent knowledge-based tests. Student perceptions of their overall abilities to educate patients about pharmacogenomics in general and genomic testing in particular improved based on the results of the pre- and post-course self-assessment. The group project appeared to be a useful approach for helping students synthesize knowledge-based content and clinical application of genomic testing. Overall, students demonstrated competency, gained confidence, and verified their abilities to apply learned skills in clinical-based scenarios. Thus, participation in this course had a positive impact on student pharmacist education.
ACKNOWLEDGMENTS
The authors acknowledge Danielle Teague for her assistance in the preparation of this manuscript; Ryan Maynard for his assistance in using Qualtrics; and Sihan Wang and Annette Myers for their technical assistance in preparing and analyzing buccal samples. Finally, the authors thank the WSU College of Pharmacy Class of 2018 for participating in the self-assessment and laboratory exercises used within the course.
Appendix 1. Lectures Presented in Basic and Clinical Pharmacogenomics Course

Appendix 2. Results of Genotypic/Phenotypic Correlation of DNA Samples Extracted (N=24) for Group Term Paper
Appendix 3. Student Self-assessment Questionnaire Administered Pre- and Post-course Delivery
Appendix 4. Selected Student Comments From Course Evaluations Highlighting the Importance of the Pharmacogenomics Course
REFERENCES
- 1.American Society of Health-System Pharmacists. ASHP statement on the pharmacist’s role in clinical pharmacogenomics. Am J Health Syst Pharm. 2015;75:579–581. doi: 10.2146/sp150003. [DOI] [PubMed] [Google Scholar]
- 2.Moaddeb J, Mills R, Haga SB. Community pharmacists’ experience with pharmacogenetic testing. JAPhA. 2015;55(6):587–594. doi: 10.1331/JAPhA.2015.15017. [DOI] [PubMed] [Google Scholar]
- 3.Frueh FW, Lesko LJ, Goodsaid F, Rudman A, Huang SM, Lesko LJ. The need for education in Pharmacogenomics: a regulatory perspective. Pharmacogenomics J. 2005;5(4):218–220. doi: 10.1038/sj.tpj.6500316. [DOI] [PubMed] [Google Scholar]
- 4.McKinnon R, Anderson C. Transforming pharmaceutical education to accelerate the acceptance and implementation of personalized medicine. Am J Pharm Educ. 2011;75(6):Article 107. doi: 10.5688/ajpe756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Accreditation Council for Pharmacy Education (ACPE) Accreditation Standards and Key Elements for the Professional Program in Pharmacy Leading to the Doctor of Pharmacy Degree. 2015. https://www.acpe-accredit.org/pdf/Standards2016FINAL.pdf. Accessed July 22, 2015.
- 6.Medina MS, Plaza CM, Stowe CD, et al. Center for the Advancement of Pharmacy Education 2013 educational outcomes. Am J Pharm Educ. Oct 14 2013;77(8):Article 162. doi: 10.5688/ajpe778162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy JE, Green JS, Adams LA, Squire RB, Kuo GM, McKay A. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am J Pharm Educ. 2010;74(1):1–10. doi: 10.5688/aj740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCullough KB, Formea CM, Berg KD, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75(3):Article 51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavallari LH, Overholser BR, Anderson D, et al. Recommended basic science foundation necessary to prepare pharmacists to manage personalized pharmacotherapy Pharmacother. 2010306228e–235e [Google Scholar]
- 10.Jenkins J, Blitzer M, Boehm K, et al. Recommendations of core competencies in genetics essential for all health professionals. Genetics Med. 2001;3(2):155–159. doi: 10.1097/00125817-200103000-00011. [DOI] [PubMed] [Google Scholar]
- 11.Korf BR, Berry AB, Limson M, et al. Framework for development of physician competencies in genomic medicine: report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genetics Med. 2014;16(11):804–809. doi: 10.1038/gim.2014.35. [DOI] [PubMed] [Google Scholar]
- 12. Essential of Genetic and Genomic Nursing: Competencies, Curricula Guidelines and Outcome Indicators. 2nd ed. Silver Spring, Md.: American Nurses Association; 2008.
- 13.Kuo GM, Ma JD, Lee KC, et al. Institutional profile: University of California San Diego pharmacogenomics education program (PharmGenEd): bridging the gap between science and practice. Pharmacogen. 2011;12(2):149–153. doi: 10.2217/pgs.10.213. [DOI] [PubMed] [Google Scholar]
- 14. McLeod HL. Pharmacogenomics: Applications to Patient Care. 2nd ed. Lenexa, KS: American College of Clinical Pharmacy; 2009.
- 15.Brazeau DA, Brazeau GA. A required course in human genomics, pharmacogenomics, and bioinformatics. Am J Pharm Educ. 2006;70(6):Article 125. doi: 10.5688/aj7006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoell DL, Johnston JS, Bao S, Kelley KA. A genotyping exercise for pharmacogenetics in pharmacy practice. Am J Pharm Educ. 2009;73(3):Article 43. doi: 10.5688/aj730343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krynetskiy E, Calligaro IL. Introducing pharmacy students to pharmacogenomic analysis. Am J Pharm Educ. 2009;73(4):Article 71. doi: 10.5688/aj730471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickola TJ, Munson AM. Pharmacogenomics primer course for first professional year pharmacy students. Pharmacogenomics. 2014;15(1):39–48. doi: 10.2217/pgs.13.197. [DOI] [PubMed] [Google Scholar]
- 19.O’Brien TJ, Goodsaid F, Plack M, et al. Development of an undergraduate pharmacogenomics curriculum. Pharmacogenomics. 2009;10(12):1979–1986. doi: 10.2217/pgs.09.145. [DOI] [PubMed] [Google Scholar]