Abstract
World coffee production has faced increasing challenges associated with ongoing climatic changes. Several studies, which have been almost exclusively based on temperature increase, have predicted extensive reductions (higher than half by 2,050) of actual coffee cropped areas. However, recent studies showed that elevated [CO2] can strongly mitigate the negative impacts of heat stress at the physiological and biochemical levels in coffee leaves. In addition, it has also been shown that coffee genotypes can successfully cope with temperatures above what has been traditionally accepted. Altogether, this information suggests that the real impact of climate changes on coffee growth and production could be significantly lower than previously estimated. Gene expression studies are an important tool to unravel crop acclimation ability, demanding the use of adequate reference genes. We have examined the transcript stability of 10 candidate reference genes to normalize RT-qPCR expression studies using a set of 24 cDNAs from leaves of three coffee genotypes (CL153, Icatu, and IPR108), grown under 380 or 700 μL CO2 L−1, and submitted to increasing temperatures from 25/20°C (day/night) to 42/34°C. Samples were analyzed according to genotype, [CO2], temperature, multiple stress interaction ([CO2], temperature) and total stress interaction (genotype, [CO2], and temperature). The transcript stability of each gene was assessed through a multiple analytical approach combining the Coeficient of Variation method and three algorithms (geNorm, BestKeeper, NormFinder). The transcript stability varied according to the type of stress for most genes, but the consensus ranking obtained with RefFinder, classified MDH as the gene with the highest mRNA stability to a global use, followed by ACT and S15, whereas α-TUB and CYCL showed the least stable mRNA contents. Using the coffee expression profiles of the gene encoding the large-subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RLS), results from the in silico aggregation and experimental validation of the best number of reference genes showed that two reference genes are adequate to normalize RT-qPCR data. Altogether, this work highlights the importance of an adequate selection of reference genes for each single or combined experimental condition and constitutes the basis to accurately study molecular responses of Coffea spp. in a context of climate changes and global warming.
Keywords: climate changes, coffee, increased air [CO2], global warming, heat stress, normalization of transcriptomic studies, quantitative real-time PCR, reference genes
Introduction
Impacts from recent climate-related extremes, such as heat and cold waves, droughts and strong rainfall events, reveal remarkable vulnerability of agricultural systems. Estimated future climate changes are expected to further amplify the existing climate-related risks and create new ones (IPCC, 2013). Within this context, it is important to assess crop ability to adapt their vital processes at a speed compatible with these environmental changes, allowing the selection and breeding of elite genotypes.
Coffee is one of the most important agricultural traded commodities, growing in more than 60 tropical countries. It is estimated that ca. 25 million farmers produce coffee in over 1 million ha, with a majority of smallholders whose livelihoods depend on this crop (Waller et al., 2007). In the last years, worldwide crop yields were above 8 million tons of green coffee beans (ICO, 2014a), generating an income of ca. US$ 173,000 million (ICO, 2014b), and involving approximately 100 million people considering the entire chain of value of coffee (Bunn et al., 2015).
Coffee production and quality are highly dependent on a regular sequence of climate events; temperature and water availability are considered the most important limiting environmental variables to this crop (DaMatta and Ramalho, 2006). Also, several modeling studies on the global impact of climate changes, mostly focused on increased air temperatures, have predicted reductions of suitable areas for Arabica coffee (up to ca. 50%) by 2,050 (Bunn et al., 2015; Magrach and Ghazoul, 2015), compromising the livelihoods of millions of small householders.
Under field conditions, the superimposition of environmental events is the most common situation, as is expected to be the case of elevated [CO2] and enhanced temperatures predicted along this century. In fact, depending on anthropogenic greenhouse gas emission scenarios, [CO2] might reach between 421 and 936 μL CO2 L−1 by 2,100, accompanied by a global warming up to between 2.6 and 4.8°C relative to 1986–2005 (IPCC, 2013, 2014). However, it was recently shown that increased [CO2] in the atmosphere strengthens the coffee plant by reinforcing its photosynthetic performance (Ramalho et al., 2013; Ghini et al., 2015; DaMatta et al., 2016). Furthermore, enhanced [CO2] has a crucial role in the mitigation of heat impact, and, therefore, in the resilience of coffee plant to supra-optimal temperatures, with positive repercussions ranging from mineral nutrition (Martins et al., 2014) to the triggering of defense mechanisms and altered gene expression (Martins et al., 2014; Rodrigues et al., 2016). Therefore, the catastrophic predictions on the future of the coffee crop, that are based almost exclusively on temperature drift (Rodrigues et al., 2016), should be reconsidered. In addition, results from Rodrigues et al. (2016) strongly suggest the need to study the single and superimposed effects of elevated [CO2] and supra-optimal temperatures at all plant levels, from morphology up to the molecular assessment.
The release of Coffea sp. expressed sequence tag (EST) databases (e.g., Poncet et al., 2006; Vieira et al., 2006; Mondego et al., 2011) and Coffea canephora genome (http://coffee-genome.org/) has greatly prompted the study of genes involved in important agronomic and stress tolerance traits, making marker-assisted selection a straight forward approach. Gene expression analysis is an important tool to elucidate the complex regulatory networks of the genetic, signaling, and metabolic mechanisms that underlie plant-environmental interactions (Mallona et al., 2010). Monitoring differential gene expression and validating high-throughput RNA sequencing (RNA-seq) data is ideally achieved through quantitative real-time PCR (RT-qPCR) analysis. However, accuracy and reliability of RT-qPCR relies on the normalization of gene expression data (Artico et al., 2010; Die et al., 2010). To avoid severe pitfalls in data analysis and interpretation, this implies the selection and systematic validation of suitable reference genes to be used as internal controls. In fact, expression stability should be validated for each particular plant tissue/organ, cell, and experimental conditions, particularly when involving environmental stressful conditions (Die et al., 2010; El Kelish et al., 2014; Imai et al., 2014; da Costa et al., 2015). In addition, the stability of reference gene transcripts is also species-dependent (Andersen et al., 2004; Gutierrez et al., 2008a,b), as in the case of coffee (Cruz et al., 2009; Goulao et al., 2012). A number of housekeeping genes (e.g., b-actin, elongation factor1a, 18S ribosomal RNA, and polyubiquitin) involved in general cell metabolism pathways are widely used to calibrate RT-qPCR studies in biological systems (Willems et al., 2008; Goulao et al., 2012; Imai et al., 2014; da Costa et al., 2015; Llanos et al., 2015). However, their expression levels may vary with the experimental conditions (Petriccione et al., 2015), and their use as internal reference genes should be taken with caution (Gutierrez et al., 2008a,b). For instance, GAPDH has been indicated as one of the most stable genes during single abiotic stresses (Tian et al., 2015), whereas it appeared to be the least stable gene during the plant development (Wang et al., 2016). In addition, the species-dependent stability of reference gene transcripts was observed, for example, in TUB-A, which is the most stable reference gene during celery development (Li et al., 2016) in contrast with what has been observed during the development of cherry (Ye et al., 2015).
In a context of advancing our knowledge regarding the plant responses to estimated climate changes and global warming, this work examined, for the first time, the transcript stability of candidate reference genes to accurately perform the normalization of RT-qPCR data from expression studies of Coffea spp. plants exposed to conditions mimicking predicted future environmental conditions. For that, single and combined impacts of elevated [CO2] and heat were considered.
Materials and methods
Plant material and growth conditions
Plant materials and experimental design was previously described in detail (Rodrigues et al., 2016). Briefly, three widely cropped coffee genotypes from the two main producing species were used: Coffea arabica L. cv. Icatu (an introgressed variety from C. canephora Pierre ex A. Froehner), C. arabica L. cv. IPR108, and C. canephora cv. Conilon Clone 153 (CL153). Potted plants of 1.5 years in age were transferred from a greenhouse (ambient [CO2]) into walk-in growth chambers (EHHF 10,000, ARALAB, Portugal) differing in air [CO2] supply: 380 μL CO2 L−1 (380-plants) or 700 μL CO2 L−1 (700-plants). Both groups of plants were then grown for 10 months in 28 L pots for ca. 10 months under controlled environmental conditions of temperature (25/20°C, day/night), RH (75%), irradiance (ca. 700–800 μmol m−2 s−1), photoperiod (12 h), without limitations of nutrients (Ramalho et al., 2013), water and space for root growth, the latter evaluated by visual examination at the end of the experiment, after removing the plants from their pots.
After that 10 months period, air temperature was gradually increased at a rate of 0.5°C day−1, from 25/20°C up to 42/34°C, with a 7 day stabilization at 31/25, 37/30 and 42/34°C to perform several evaluations (Martins et al., 2014, 2016; Rodrigues et al., 2016). Leaf material was collected from newly matured leaves, of both plagiotropic and orthotropic branches of the upper (illuminated) part of the plant, from five to eight plants per genotype and immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Total RNA isolation and cDNA synthesis
Total RNA was isolated and quantified as described in Goulao et al. (2012) for each of the following 24 conditions: three genotypes (CL153, Icatu, IPR108), four temperature regimes (25/20°C, 31/25°C, 37/30°C, 42/34°C), and two [CO2] (380, 700 μL CO2 L−1), using the RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer's protocol. In order to avoid eventual genomic DNA contaminations, RNA samples were digested with Ambion® DNA-free™ DNase (Ambion, USA). RNA integrity was verified in 1.5% agarose—TAE gel electrophoresis containing GelRed Nucleic Acid Gel Stain (Biotium, USA). All RNA samples were individually analyzed for the possible presence of DNA contamination by standard PCR reactions (35 cycles) using primers designed for ubiquitin (UBQ) gene (Table 1), in the absence of cDNA synthesis. Total RNA concentration and purity were further verified through BioDrop Cuvette (BioDrop, UK) measurements to guarantee the same amount of starting material in subsequent cDNA synthesis. RNA concentrations ranged between 333 and 1,056 ng/μl, with OD 260:280 ratios always above 1.98.
Table 1.
Primer sequences and amplicon characteristics for each of the 10 candidate reference genes under evaluation.
| Gene symbol | Primer sequence (5′–3′) | Gene name | Acession number | Amplicon size (bp) |
|---|---|---|---|---|
| UBQ2 | GATGATACTTGGCCCTGCAC CCTTCCCAGCTTGTCAATGT |
Ubiquitin-conjugating enzyme E2 | GR984245 | 142 |
| α-Tub | GTGCATCTCCATCCACATTG GGTGTTGAAAGCGTCGTCT |
Alpha-tubulin | GT009437 | 146 |
| S15 | CAATAGGGGCTTGAAGAGG TACGCTGCCAATCATCTCAG |
40S ribosomal protein S15 | GR987196 | 146 |
| DNAJ | GGTGAAGCTGATGAAGCAC CCGGGCTGGGATTTAATAAG |
Plant DNA J protein | GR986679 | 157 |
| MDH | CCTGATGTCAACCACGCAACT GTGGTTATGAACTCTCCATTCAACC |
Malate dehydrogenase | GW464198.1 | 100 |
| ACT | AAGCTTGCCTATGTGGCTCTTG TCACTTGTCCATCTGGCAATTC |
Actin | GT000704 | 100 |
| ELF-4A | GGTTATGCGTGCTCTGGGTGAC ATGAACCCCACTGGAAAGAATG |
Eukaryotic initiation factor 4α | GT71729 | 103 |
| CYCL | AGCTCTACGCAGACACGACTCC GGTCGATCCTTTGAAGTGCAAG |
Cyclophilin | GT007167 | 115 |
| EF1A | CATTGTGGTCATTGGTCATGTC ACACGCTTGTCAATTCCTCCA |
Elongation factor 1α | GR996930 | 87 |
| GAPDH | AGGCTGTTGGGAAAGTTCTTC ACTGTTGGAACTCGGAATGC |
Glyceraldehyde 3-phosphate dehydrogenase | DV692958 | 100 |
One microgram of DNA-free total RNA was used to synthesize first-strand cDNAs using oligo-(dT)18 primer and the SuperScript II first-strand synthesis system (Invitrogen, USA).
Selection of reference gene sequences and primer design
A set of 10 candidate genes were selected, comprising several frequently used reference genes, based on previous reports in (a) coffee plants (Cruz et al., 2009; Goulao et al., 2012; Carvalho et al., 2013): actin (ACT), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), eukaryotic initiation factor 4α (Elf4A), elongation factor 1α (EF1A), cyclophilin (CYCL), and malate dehydrogenase (MDH); and (b) other plants (Cseke et al., 2009; El Kelish et al., 2014; Watanabe et al., 2014; Yan et al., 2014): 40S ribosomal protein subunit 15A (S15), DNAJ-like (DNAJ), ubiquitin-conjugating enzyme E2(UBQ2), and a alpha tubulin (α-TUB). The corresponding gene sequences were retrieved from published literature and the NCBI databases (Cruz et al., 2009; Vidal et al., 2010; Mondego et al., 2011; Goulao et al., 2012; Carvalho et al., 2013). Primers were designed using Primer3 software (Rozen and Skaletsky, 2000). The length of the primers was set to be between 19 and 26 bp, with a GC content ranging from 45 to 60% and a melting temperature (Tm-value) between 62° and 65°C. Amplicon length ranges were set to be between 100 and 150 bp. The probability of formation of hairpin structures and primer dimerization was subsequently checked using the Oligo Calculator (version. 3.26) algorithm (Kibbe, 2007). Primer sequences are given in Table 1.
Quantitative real-time polymerase chain reaction conditions
RT-qPCR reactions were performed in 96-well plates using iQ™ SYBR® Green Supermix (BioRad, USA). Reactions were prepared in volumes of 25 μl containing 150 ng of cDNA and 3 μM each primer, in 1 × iQ™ SYBR® Green Supermix. Reactions were run on the iQ™ 5 Real-Time Detection System (BioRad, USA) using the following parameters: hot start activation of the TaqDNA polymerase at 95°C for 10 min, followed by 40 cycles of denaturation 95°C for 15 s; annealing at 60°C for 30 s; elongation at 72°C for 30 s; and plate read. To verify the specificity of each amplification, and the absence of primer dimers, dissociation curves were obtained for each amplicon at the end of the PCR run, by continuous fluorescence measurement from 55° to 95°C, with sequential steps of 0.5°C for 15 s. Three technical replicates were used for each biological replicate and the mean Ct was used for data analyses. The full sample set was always included in each technical replicate to exclude any artifacts consequential of between-run variations. No signals were detected in non-template controls run in parallel for each primer set. Two negative controls were included for each primer pair, in which cDNA was replaced by water or total RNA.
Calculation of PCR efficiencies
Five-fold serial dilutions (1:1–1:625) of pooled cDNAs, which included equal-molar quantities of all samples independently for each genotype, were quantified in triplicates to generate standard curves for each primer pair. Based on the slopes of the standard curves, the PCR efficiency of each gene, for each genotype, was determined from the respective logarithm of the cDNA dilution, plotted against the mean threshold cycle (Ct) values. The reaction efficiency was calculated using the equation: E (%) = (10−(1/slope)−1) × 100 (da Costa et al., 2015), where E is the efficiency, in percentage, and slope is the gradient of the best-fit line in the linear regression.
Selection of reference genes
The global transcript variability of each candidate reference gene was analyzed through statistical parameters using Box Plot. The variation of gene expression was calculated by means of coefficient of variation (CV), where CV = (standard deviation mean) × 100. These analyzes were performed using the statistical program R Core Team (2016). In order to select the best reference genes for the experimental conditions, expression stability was analyzed using geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004) and BestKeeper (Pfaffl et al., 2004). The final rank of reference genes was determined with the RefFinder program (Xie et al., 2012), a web-based user-friendly comprehensive tool that integrates geNorm, Normfinder, BestKeeper, and the comparative Ct method.
Determination of the minimum number of reference genes for data normalization
The pairwise variations (V-values) were calculated using the final ranking determined by RefFinder, through pairwise variation (Vn/Vn+1) of two consecutively serial log2-transformed normalization factor (NF) ratios, based on stepwise addition of the subsequent more stable reference gene (NFn relative to NFn+1 reference genes; Vandesompele et al., 2002). The SD of the array generated by the log2-transformed NF ratios was calculated for each NF combination (Vn/Vn+1). Vn/Vn+1 were then plotted, representing changes in expression stability of NF. The optimal number of reference genes to exact standards was determined by calculating values as a pair of variation (Vn/Vn + 1) between two normalization factors consecutively classified (NC) after the gradual addition of more subsequent stable reference gene (NFN and NFN + 1), as proposed by Vandesompele et al. (2002), included in GeNorm package. A cut off value <0.15 was adopted (da Costa et al., 2015; Niu et al., 2015; Petriccione et al., 2015; Yang et al., 2015).
Validation of reference gene analysis
The evaluation of the transcript levels from the RLS gene, encoding the ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) large subunit, along the imposed temperatures (25/20°C, 31/25°C, 37/30°C, 42/34°C), and under both growth [CO2] (380, 700 μL CO2 L−1) was analyzed by RT-qPCR, using the most stable vs. the least stable genes (using one, two or three). The RT-qPCR amplification conditions were the same as described above. The relative expression data and statistical analysis were performed using the REST 2,009 software developed by Pfaffl (Technical University Munich) and Qiagen (Germany). Rest analysis takes into account PCR efficiencies and normalization gene, using the test hypothesis P (H1), i.e., the probability of difference between target and control groups be by chance. P (H1) performs at least 2,000 times of random reallocations of target samples and controls between the groups. Statistical differences were considered significant when P < 0.001 (***), P < 0.01 (**), and P < 0.05 (*).
Results and discussion
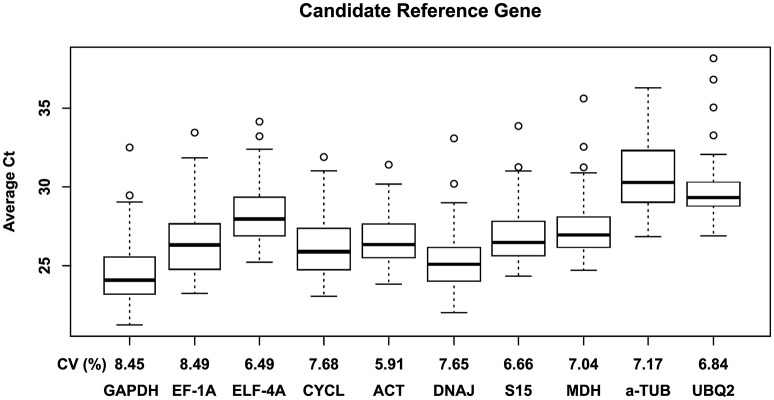
A total of 10 candidate genes (CG) to obtain an accurate validation of RT-qPCR data were assessed considering the individual comparison groups, i.e., genotype, [CO2] and temperature, multiple stress interaction ([CO2] and temperature) and total stress interaction (genotype, [CO2] and temperature). To determine the specificity of the primer pairs used in this study, melting curve analysis was performed after 40 cycles of amplification. The single peak obtained confirmed the specificity of the amplicon. No signal was detected in the negative controls of all tested primers. The standard curve method using a pool of all cDNA samples was performed to calculate RT-qPCR efficiency (E) and the correlation coefficient (R2) of each primer pair (Table 2). The raw quantification cycle (Ct) values ranged from 21.2 to 38.2 across all of the tested samples, with their minimal and maximal means falling between 24.1 ± 1.8 and 34.1 ± 2.1 (Figure 1). The results showed that GAPDH and α-TUB were the most and least abundantly expressed genes with mean Ct of 24.4 and 30.7, respectively. EF-1A showed the most variable levels of expression. Collectively, these results indicate that the transcript levels of the several candidate genes (CG) varied across different experimental samples confirming the need to screen the most appropriate reference genes for each experimental condition to accurately normalize RT-qPCR gene expression data. This result is common in plants given that most reference genes are involved in functions other than basal cell metabolism (Goulao et al., 2012; da Costa et al., 2015).
Table 2.
Efficiency values and correlation coefficient for each candidate reference genes under evaluation.
| Gene | RT-qPCR efficiency% (R2) | ||
|---|---|---|---|
| CL153 | Icatu | IPR108 | |
| GAPDH | 93.3(0.99) | 95.7(0.98) | 93.9(0.98) |
| EF-1A | 96.3(0.99) | 82.7(0.99) | 97.3(0.96) |
| ELF-4A | 90.8(0.99) | 90.9(0.98) | 93.3(0.96) |
| CYCL | 87.1(0.96) | 80.3(0.98) | 80.3(0.98) |
| ACT | 91.4(0.93) | 98.7(0.93) | 97.9(0.91) |
| DNAJ | 82.5(0.99) | 86.4(0.99) | 97.7(0.98) |
| S15 | 99.9(0.96) | 98.8(0.98) | 90.0(0.95) |
| MDH | 80.3(0.99) | 92.0(0.96) | 92.1(0.97) |
| a-TUB | 99.3(0.96) | 93.4(0.98) | 94.0(0.99) |
| UBQ2 | 98.1(0.97) | 91.2(0.93) | 92.2(0.94) |
Figure 1.
Box plot representing the expression profiling variations of each candidate reference gene. The primer pairs of each gene were used to examine all samples (n = 24; Table 1). Boxes indicate the 25th and 75th percentiles; lines across the box represent the median Ct-values; whisker caps represent the minimum and maximum values; and spots represent the outliers.
According to the Coefficient of Variation (CV) method, the most stable genes were ACT (for genotype and total stress, CV = 5.91 in both cases), UBQ2 (for temperature, CV = 5.50 and [CO2], CV = 5.71), and S15 (for multiple stress, CV = 5.58; Table 3). EF-1A was ranked as the least stable gene for genotype (CV = 8.49), temperature (CV = 8.34) and overall stress (CV = 8.49), ranking in the 6th place for [CO2] and multiple stress. DNAJ and TUB were the less stable genes for [CO2] (CV = 9.68) and multiple stress treatments (7.95). Because the CV method alone is not sufficiently accurate to estimate the stability of the mRNA levels (Goulao et al., 2012), a complementary analysis was performed based on three algorithms, geNorm, NormFinder, and BestKeeper. According to the geNorm algorithm, the average expression stability (M-value) of all CG was below the default limit of 1.5 for genotype, temperature, [CO2] and their respective interactions, therefore indicating a considerable high stability level (Table 4). GAPDH was consistently classified as the most stable gene for the five comparison groups while CYCL was the least stable gene for all groups with the exception of genotype, where MDH was the least sable gene. On the other hand, the ranking obtained with NormFinder was clearly dependent on the comparison group: MDH was the most stable for genotype, UBQ2 for temperature, GAPDH for [CO2] and ELFA for multiple and total stress groups. Such differences are due probably to different stability measures, i.e., while geNorm calculates gene expression stability based on the average pairwise expression ratio, NormFinder estimates the overall expression variation to provide individual stability values (Kozera and Rapacz, 2013). Nevertheless, similarly to geNorm, NormFinder also ranked CYCL as the least stable gene for all groups, with the exception for genotype (where α-TUB was the least stable CG).
Table 3.
Ranking of candidate reference genes according to its coefficient of variation (CV%), considering the variables genotype, temperature, [CO2] and their interaction.
| Rank | Genotypes | Temperature | [CO2] | Multiple stress | Total stress | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | CV | Gene | CV | Gene | CV | Gene | CV | Gene | CV | |
| 1 | ACT | 5.91 | UBQ2 | 5.50 | UBQ2 | 5.71 | S15 | 5.58 | ACT | 5.91 |
| 2 | ELF-4A | 6.49 | S15 | 5.82 | ACT | 5.73 | ELF-4A | 5.72 | ELF-4A | 6.49 |
| 3 | S15 | 6.66 | ACT | 5.87 | ELF-4A | 5.76 | ACT | 6.19 | S15 | 6.66 |
| 4 | UBQ2 | 6.84 | MDH | 6.15 | MDH | 6.64 | MDH | 6.25 | UBQ2 | 6.84 |
| 5 | MDH | 7.04 | ELF-4A | 6.38 | S15 | 6.65 | DNAJ | 6.69 | MDH | 7.04 |
| 6 | α-TUB | 7.17 | DNAJ | 6.54 | EF-1A | 6.86 | EF-1A | 7.15 | a-TUB | 7.17 |
| 7 | DNAJ | 7.65 | CYCL | 6.75 | α-TUB | 6.96 | CYCL | 7.33 | DNAJ | 7.65 |
| 8 | CYCL | 7.68 | GAPHD | 6.93 | CYCL | 7.52 | UBQ2 | 7.44 | CYCL | 7.68 |
| 9 | GAPHD | 8.45 | α-TUB | 7.22 | GAPHD | 8.94 | GAPHD | 7.74 | GAPHD | 8.45 |
| 10 | EF-1A | 8.49 | EF-1A | 8.34 | DNAJ | 9.68 | α-TUB | 7.95 | EF-1A | 8.49 |
Table 4.
Stability of candidate reference genes according to GeNorm, NormFinder and BestKeeper, considering the variables genotype, temperature, [CO2], and their interaction.
| Group | Ranking | geNorm | NormFinder | BestKeeper | |||
|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | R | ||
| Genotype | 1 | GAPDH | 0.24 | MDH | 0.005 | ACT | 0.97*** |
| 2 | ACT | 0.26 | DNAJ | 0.009 | MDH | 0.97*** | |
| 3 | EF-1A | 0.27 | UBQ2 | 0.010 | ELF-4A | 0.96*** | |
| 4 | ELF-4A | 0.28 | S15 | 0.011 | S15 | 0.95*** | |
| 5 | UBQ2 | 0.29 | ELF-4A | 0.013 | DNAJ | 0.93*** | |
| 6 | α-TUB | 0.32 | ACT | 0.016 | UBQ2 | 0.93*** | |
| 7 | S15 | 0.33 | EF-1A | 0.020 | GAPDH | 0.93*** | |
| 8 | CYCL | 0.42 | CYCL | 0.026 | CYCL | 0.85*** | |
| 9 | DNAJ | 0.53 | GAPDH | 0.027 | α-TUB | 0.82*** | |
| 10 | MDH | 0.62 | α-TUB | 0.030 | EF-1A | 0.81** | |
| Temperature | 1 | GAPDH | 0.14 | UBQ2 | 0.041 | MDH | 0.98*** |
| 2 | UBQ2 | 0.14 | ELF-4A | 0.057 | DNAJ | 0.97*** | |
| 3 | ELF-4A | 0.16 | GAPDH | 0.060 | UBQ2 | 0.96*** | |
| 4 | ACT | 0.16 | DNAJ | 0.071 | ACT | 0.96*** | |
| 5 | S15 | 0.17 | EF-1A | 0.076 | S15 | 0.95*** | |
| 6 | α-TUB | 0.17 | α-TUB | 0.082 | ELF-4A | 0.94*** | |
| 7 | DNAJ | 0.18 | ACT | 0.083 | EF-1A | 0.92*** | |
| 8 | EF-1A | 0.18 | MDH | 0.084 | GAPDH | 0.91*** | |
| 9 | MDH | 0.19 | S15 | 0.086 | CYCL | 0.86*** | |
| 10 | CYCL | 0.37 | CYCL | 0.246 | α-TUB | 0.84*** | |
| [CO2] | 1 | GAPDH | 0.15 | GAPDH | 0.019 | CAS15 | 1.00*** |
| 2 | UBQ2 | 0.16 | ELF-4A | 0.030 | ACT | 0.99*** | |
| 3 | ELF-4A | 0.17 | UBQ2 | 0.036 | MDH | 0.99*** | |
| 4 | S15 | 0.18 | S15 | 0.070 | ELF-4A | 0.98*** | |
| 5 | α-TUB | 0.18 | EF-1A | 0.076 | UBQ2 | 0.98*** | |
| 6 | ACT | 0.19 | α-TUB | 0.077 | GAPDH | 0.97*** | |
| 7 | EF-1A | 0.19 | ACT | 0.082 | DNAJ | 0.94** | |
| 8 | MDH | 0.23 | MDH | 0.105 | α-TUB | 0.91* | |
| 9 | DNAJ | 0.31 | DNAJ | 0.180 | EF-1A | 0.91* | |
| 10 | CYCL | 0.53 | CYCL | 0.357 | CYCL | 0.47 | |
| Multiple stress | 1 | GAPDH | 0.16 | ELF-4A | 0.028 | MDH | 0.97*** |
| 2 | ELF-4A | 0.16 | UBQ2 | 0.053 | DNAJ | 0.96*** | |
| 3 | α-TUB | 0.17 | α-TUB | 0.060 | S15 | 0.96*** | |
| 4 | ACT | 0.18 | GAPDH | 0.062 | ELF -4A | 0.96*** | |
| 5 | UBQ2 | 0.18 | DNAJ | 0.074 | ACT | 0.95*** | |
| 6 | S15 | 0.18 | ACT | 0.077 | EF-1A | 0.90*** | |
| 7 | DNAJ | 0.20 | S15 | 0.082 | UBQ2 | 0.90*** | |
| 8 | MDH | 0.21 | MDH | 0.094 | GAPDH | 0.88*** | |
| 9 | EF-1A | 0.27 | EF-1A | 0.164 | CYCL | 0.83*** | |
| 10 | CYCL | 0.43 | CYCL | 0.288 | α-TUB | 0.50* | |
| Total stress | 1 | GAPDH | 0.16 | ELF-4A | 0.040 | MDH | 0.97*** |
| 2 | UBQ2 | 0.18 | UBQ2 | 0.044 | S15 | 0.96*** | |
| 3 | ELF-4A | 0.18 | GAPDH | 0.055 | ELF-4A | 0.95*** | |
| 4 | S15 | 0.19 | S15 | 0.085 | ACT | 0.95*** | |
| 5 | ACT | 0.19 | ACT | 0.095 | DNAJ | 0.94*** | |
| 6 | α-TUB | 0.22 | DNAJ | 0.105 | UBQ2 | 0.93*** | |
| 7 | DNAJ | 0.23 | MDH | 0.111 | GAPDH | 0.92*** | |
| 8 | MDH | 0.23 | α-TUB | 0.112 | CYCL | 0.82*** | |
| 9 | EF-1A | 0.24 | EF-1A | 0.123 | EF-1A | 0.79*** | |
| 10 | CYCL | 0.44 | CYCL | 0.295 | α-TUB | 0.73*** | |
For sake of simplicity, only significant differences were indicated (
P < 0.001;
P < 0.01;
P < 0.05) related to the BestKeeper test.
In order to consolidate the data obtained with geNorm and NormFinder, we have included in the analysis the BestKeeper algorithm, which calculates the expression stabilities of the CG in accordance with SD and CV values. The calculated results classified ACT as the most stable gene for the genotype (together with MDH) and S15 for the [CO2] group. For temperature, multiple stress and overall stress groups the most stable gene was MDH. α-TUB, EF-1A, and CYCL were ranked as the least stable genes, always below the 6th place and usually in the three last positions. Despite the differences between the ranking obtained with the CV method and the three algorithms, which are related to the individual classification criteria, CYCL and α-TUB were consistently ranked in the bottom five positions.
The consensus ranking obtained with RefFinder (Table 5), classified MDH as the most stable gene for all comparison groups, therefore constituting a good choice for the studies of Coffea spp. under future estimated conditions of increased [CO2] and temperature. Furthermore, ACT and S15 were always ranked in the top five positions, while α-TUB and CYCL were amongst the least stable genes, as previously found in previous tests (Tables 3, 4). MDH and ACT have been previously described as being amongst the reference genes with higher transcript stability for RT-qPCR studies in Coffea spp. subjected to N starvation, salt and heat stresses (Carvalho et al., 2013) and to cold and drought stresses (Goulao et al., 2012), respectively. However, there was no consensus for the majority of candidate reference genes, i.e., the expression stability strongly varied according to the environmental stressful condition, again highlighting the importance of a proper and accurate selection of reference genes for each single experimental condition.
Table 5.
Overall ranking of the most stable genes within each treatment made through RefFinder program, considering the variables genotype, temperature, [CO2] and their interaction.
| Genotypes | Temperature | [CO2] | Multiple stress | Total stress | |||||
|---|---|---|---|---|---|---|---|---|---|
| MDH | 1.68 | MDH | 1.86 | MDH | 1.68 | MDH | 1.41 | MDH | 1.41 |
| S15 | 1.86 | UBQ2 | 1.86 | S15 | 1.78 | ELF-4A | 2.21 | ACT | 1.57 |
| ACT | 2.99 | DNAJ | 2.11 | UBQ2 | 2.71 | DNAJ | 2.63 | S15 | 2.78 |
| DNAJ | 3.34 | ACT | 3.46 | ACT | 2.83 | S15 | 2.78 | DNAJ | 3.94 |
| ELF-4A | 3.83 | S15 | 3.98 | ELF-4A | 4.40 | ACT | 4.40 | ELF-4A | 4.95 |
| UBQE2 | 5.05 | ELF-4A | 6.24 | GAPDH | 6.82 | GAPDH | 6.64 | UBQ2 | 5.05 |
| GAPDH | 7.00 | GAPDH | 6.74 | EF-1A | 7.00 | UBQ2 | 6.74 | GAPDH | 7.00 |
| CYCL | 8.00 | EF-1A | 8.24 | a-TUB | 8.00 | CYCL | 8.00 | CYCL | 8.00 |
| EF-1A | 9.00 | CYCL | 8.74 | CYCL | 8.80 | EF-1A | 9.15 | a-TUB | 9.24 |
| a-TUB | 10.00 | a-TUB | 10.00 | DNAJ | 9.00 | a-TUB | 9.24 | EF-1A | 9.74 |
For all comparison groups at V2/V3 the Pairwise variation (V) value was already below 0.15 (Figure 2), indicating that the use of two genes guarantee a strong accuracy for the normalization of RT-qPCR data. It is noteworthy to mention that for all treatments the pairwise variation values were <0.10 (except for the use of just one reference gene), indicating low expression instability of the selected genes tested for these environmental conditions.
Figure 2.
Pairwise variation analysis to determine the optimal number of reference genes for RT-qPCR data normalization.
In order to validate the consensus ranking, we have examined the relative expression of RLS, encoding the large-subunit of RuBisCO. The choice of this enzyme was related to its central role in the C-assimilation pathway, considering that improved crop photosynthetic efficiency under high temperature might be achieved through the replacement of a number of candidate amino acid substitutions to improve RuBisCO performance (Orr et al., 2016). Moreover, the RLS gene was selected based on the fact that, according to structural-functional analyses, most of the important amino acid residues necessary for catalysis are present in the large subunits (Morita et al., 2014). The consensus ranking established by RefFinder identified MDH, UBQ2, and DNAJ as the best stable genes to temperature, whereas a-TUB, CYCL and EF-1A where the least stable ones. Given that the increase in atmospheric temperature has proven to be the most disturbing variable to metabolism performance, mineral nutrition and acclimation limitation under predicted future environmental conditions (Martins et al., 2014, 2016; Rodrigues et al., 2016), with particular strong negative implications to RuBisCO catalytic ability (Rodrigues et al., 2016), we decided to use the most/least stable genes to temperature for RLS validation.
The expression profiles were normalized with the two and three most and least stable genes (Tables 6, 7), being observed that the expression patterns of RLS varied with the comparison group. Noticeably, due to the high SD of Ct values in many cases variations in RLS expression along the stress imposition were not reported as statistically significant. The patterns of RLS expression were nearly the same in RT-qPCR reactions calibrated with two or three genes. Independently of the stability of RLS mRNA, the increase in temperature generally resulted in up-regulation of RLS expression at standard [CO2] of 380 μL CO2 L−1 in all genotypes, except in IPR108 at 31/25° and 37/30°C where RLS expression was nearly the same as the control. For the 700 μL CO2 L−1 comparison group, the use of the most stable reference genes produced different results from those obtained with the least stable genes at 25/20° and 31/25°C. Thus, when reactions were normalized with the most stable genes, a consistent decrease in the steady-state amounts of RLS mRNA (which is the net result of transcription and degradation) was observed at these temperatures in all genotypes, while the use of the least stable genes indicated a positive regulation of RLS expression. For 37/30°C and 42/34°C, normalization of RT-qPCR with the most or the least stable genes produced similar up-regulated expression patterns of RLS. Notably, such an up-regulation occurred when RuBisCO activity begin to decrease (37/30°C) or was severely depressed (42/34°C) (Rodrigues et al., 2016). Such apparent discrepancy was also noted in chloroplastic ascorbate peroxidase (APX). APX activity was the most reduced (to about 10% of the control) at 42/34°C among the studied enzymes, contrasting to the largest increase in APX transcripts amongst all studied genes (Martins et al., 2016). Such large increases in gene transcripts may be related to an attempt of the plants to compensate the severe decline of enzyme activity, and further highlights the need of integrated experiments that include gene expression and enzyme activity studies.
Table 6.
Analysis of RLS expression by RT-qPCR, calibrated with the two most (MDH and UBQ2) and the two least (a-TUB and CYCL) genes for the temperature group/treatment.
| Reference genes | MDH and UBQ2 | a-TUB and CYCL | ||
|---|---|---|---|---|
| Genotype | Temperature | [CO2] | RLS expression | RLS expression |
| CL 153 | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.361 | 1.768 | ||
| 31/25°C | 380 | 1.353 | 5.665 | |
| 700 | 0.504 | 3.559 | ||
| 37/30°C | 380 | 1.581 | 12.556 | |
| 700 | 1.467 | 12.86 | ||
| 42/34°C | 380 | 4.565 | 25.922 | |
| 700 | 8.889 | 25.114 | ||
| Icatu | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.781 | 1.553 | ||
| 31/25°C | 380 | 1.900** | 3.604* | |
| 700 | 1.505 | 2.546* | ||
| 37/30°C | 380 | 3.423* | 6.957* | |
| 700 | 2.776* | 5.742* | ||
| 42/34°C | 380 | 5.220* | 19.923* | |
| 700 | 7.097* | 32.153* | ||
| IPR 108 | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.249* | 0.603 | ||
| 31/25°C | 380 | 0.778 | 1.345 | |
| 700 | 0.801* | 1.720* | ||
| 37/30°C | 380 | 0.844 | 1.851*** | |
| 700 | 1.070 | 2.172 | ||
| 42/34°C | 380 | 4.940* | 21.732** | |
| 700 | 1.399 | 4.01 | ||
For sake of simplicity, only significant differences were indicated (
P < 0.001;
P < 0.01;
P < 0.05).
Table 7.
Analysis of RLS expression by RT-qPCR, calibrated with the three most (MDH, UBQ2 and DNAJ) and the three least (a-TUB, CYCL and EF-1A) genes for the temperature group/treatment.
| Reference genes | MDH, UBQ2, and DNAJ | α-TUB, CYCL and EF-1A | ||
|---|---|---|---|---|
| Genotype | Temperature | [CO2] | RLS expression | RLS expression |
| CL 153 | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.410 | 1,347 | ||
| 31/25°C | 380 | 1,424 | 2,780 | |
| 700 | 0.522 | 1,747 | ||
| 37/30°C | 380 | 1,730 | 5,701 | |
| 700 | 1,435 | 6,357 | ||
| 42/34°C | 380 | 4,214 | 10,853 | |
| 700 | 6,527 | 10,782 | ||
| Icatu | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.744 | 1, 449 | ||
| 31/25°C | 380 | 1.608* | 2.807** | |
| 700 | 1,215 | 2.170* | ||
| 37/30°C | 380 | 2.726* | 5.250* | |
| 700 | 2.219* | 3.955* | ||
| 42/34°C | 380 | 4.087* | 9.637* | |
| 700 | 5.389* | 12.979* | ||
| IPR 108 | 25/20°C | 380 | 1.00 | 1.00 |
| 700 | 0.249* | 1, 059 | ||
| 31/25°C | 380 | 0.697* | 1.307* | |
| 700 | 1,123 | 2.865* | ||
| 37/30°C | 380 | 0.759 | 1,495 | |
| 700 | 1, 093 | 3,453 | ||
| 42/34°C | 380 | 4.950* | 11.012* | |
| 700 | 1,759 | 4,645 | ||
For sake of simplicity, only significant differences were indicated (
***P < 0.001;
P < 0.01;
P < 0.05).
Given the facts described above, the reference genes used in this study were quite stable under the applied experimental conditions, as pairwise variation values were <0.10. The results obtained in the groups 700-25/20°C and 700-31/25°C call up for the importance to select the best reference genes. Altogether, despite the low instability of a-TUB, CYCL, and EF-1A, the use of reference genes showing a high stability of their mRNA amount is the best option for the correct interpretation of data from transcriptional studies (Roy et al., 2011; Kozera and Rapacz, 2013). Also importantly, this corroborates with previous studies in Coffea sp., which indicate that the selection of reference genes for an accurate RT-qPCR normalization should be performed accordingly to each for specific condition (Cruz et al., 2009; Goulao et al., 2012; Carvalho et al., 2013; Yuyama et al., 2016).
Conclusions
In summary, our study reinforces the importance of the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009) to normalize RT-qPCR data. The consensus ranking obtained with RefFinder showed that the transcript stability of the evaluated reference candidate genes changed according to the conditions/interactions, although MDH was the best one in all treatments, therefore, constituting a good common basis for RT-qPCR data normalization. However, the Pairwise variation analysis showed the need to use two reference genes, with the second most stable gene changing according to the genotypes and imposed environmental conditions: S15 for genotype and [CO2], UBQ2 for temperature, EJF-4A for multiple stress, and ACT for multiple stress. Using the expression profiles of coffee RLS, results from the in silico aggregation and experimental validation confirmed that the use of two reference genes is an adequate procedure to normalize RT-qPCR data.
Author contributions
According to their competences, all authors contributed transversally to the several stages of the work, including its design, data acquisition, analysis and interpretation, critically review of the manuscript, and approval of the submitted version. Furthermore, they agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Funding
This work was supported by by national funds from Fundação para a Ciência e a Tecnologia through the projects PTDC/AGR-PRO/3386/2012, the research units UID/AGR/04129/2013 (LEAF) and UID/GEO/04035/2013 (GeoBioTec), as well through the grant SFRH/BPD/47563/2008 (AF) co-financed through the POPH program subsidized by the European Social Fund. Brazilian funding from CAPES (grants PDSE: 000427/2014-04, WR; 0343/2014-05, MM), CNPq and Fapemig (fellowships to FD, FP, and EC) are also greatly acknowledged.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank DELTA Cafés (Portugal) for technical assistance, and Profs. T. Sera (IAPAR) for supplying IPR108 material. Thanks are also due to M. Costa, and N. Duro, for technical help.
Glossary
Abbreviations
- RT-qPCR
quantitative real-time polymerase chain reaction.
References
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Artico S., Nardeli S. M., Brilhante O., Grossi-de-Sa M. F., Alves-Ferreira M. (2010). Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real time quantitative RT-PCR data. BMC Plant Biol. 10:49. 10.1186/1471-2229-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn C., Läderach P., Ovalle-Rivera O., Kirschke D. (2015). A bitter cup: climate change profile of global production of Arabica and Robusta coffee. Clim. Change 129, 89–101. 10.1007/s10584-014-1306-x [DOI] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Carvalho K., Bespalhok Filho J. C., dos Santos T. B., de Souza S. G., Vieira L. G., Pereira L. F., et al. (2013). Nitrogen starvation, salt and heat stress in coffee (Coffea arabica L.): identification and validation of new genes for qPCR normalization. Mol. Biotechnol. 53, 315–325. 10.1007/s12033-012-9529-4 [DOI] [PubMed] [Google Scholar]
- Cruz F., Kalaoun S., Nobile P., Colombo C., Almeida J., Barros L., et al. (2009). Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT–PCR. Mol. Breed. 23, 607–616. 10.1007/s11032-009-9259-x [DOI] [Google Scholar]
- Cseke L. J., Tsai C.-J., Rogers A., Nelsen M. P., White H. L., Karnosky D. F., et al. (2009). Transcriptomic comparison in the leaves of two aspen genotypes having similar carbon assimilation rates but different partitioning patterns under elevated [CO2]. New Phytol. 182, 891–911. 10.1111/j.1469-8137.2009.02812.x [DOI] [PubMed] [Google Scholar]
- da Costa M., Duro N., Batista-Santos P., Ramalho J. C., Ribeiro-Barros A. I. (2015). Validation of candidate reference genes for qRT-PCR studies in symbiotic and non-symbiotic Casuarina glauca Sieb. ex Spreng. under salinity conditions. Symbiosis 66, 21–35. 10.1007/s13199-015-0330-6 [DOI] [Google Scholar]
- DaMatta F. M., Godoy A. G., Menezes-Silva P. E., Martins S. C. V., Sanglard L. M. V. P., Morais L. E., et al. (2016). Sustained enhancement of photosynthesis in coffee trees grown under free-air CO2 enrichment conditions: disentangling the contributions of stomatal, mesophyll, and biochemical limitations. J. Exp. Bot. 67, 341–352. 10.1093/jxb/erv463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaMatta F. M., Ramalho J. C. (2006). Impacts of drought and temperature stress on coffee physiology and production: a review. Theor. Exp. Plant Physiol. 18, 55–81. 10.1590/S1677-04202006000100006 [DOI] [Google Scholar]
- Die J. V., Román B., Nadal S., González-Verdejo C. I. (2010). Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232, 145–153. 10.1007/s00425-010-1158-1 [DOI] [PubMed] [Google Scholar]
- El Kelish A., Zhao F., Heller W., Durner J., Winkler J. B., Behrend H., et al. (2014). Ragweed (Ambrosia artemisiifolia) pollen allergenicity: SuperSAGE transcriptomic analysis upon elevated CO2 and drought stress. BMC Plant Biol. 14:176. 10.1186/1471-2229-14-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghini R., Torre-Neto A., Dentzien A. F. M., Guerreiro-Filho O., Lost R., Patrício F. R. A., et al. (2015). Coffee growth, pest and yield responses to free-air CO2 enrichment. Clim. Chang. 132, 307–320. 10.1007/s10584-015-1422-2 [DOI] [Google Scholar]
- Goulao L. F., Fortunato A. S., Ramalho J. C. (2012). Selection of reference genes for normalizing quantitative real-time PCR gene expression data with multiple variables in Coffea spp. Plant Mol. Biol. Rep. 30, 741–759. 10.1007/s11105-011-0382-6 [DOI] [Google Scholar]
- Gutierrez L., Mauriat M., Guenin S., Pelloux J., Lefebvre J.-F., Louvet R., et al. (2008b). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. 10.1111/j.1467-7652.2008.00346.x [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Mauriat M., Pelloux J., Bellini C., van Wuytswinkel O. (2008a). Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20, 1734–1735. 10.1105/tpc.108.059774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICO (2014a). Trade Statistics. Available online at: http://www.ico.org/trade_statistics.asp (Accessed July 2016)
- ICO (2014b). World coffee trade (1963–2013): A Review of the Markets, Challenges and Opportunities Facing the Sector. ICC (International Coffee Council), 111–115 Rev. 1, 29p. Available online at: http://www.ico.org/show_news.asp?id=361 (Accessed July 2016) [Google Scholar]
- Imai T., Ubi B. E., Saito T., Moriguchi T. (2014). Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS ONE 9:e86492. 10.1371/journal.pone.0086492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2013). Climate change 2013, in The Physical Science Basis. Summary for Policymakers, Technical Summary and Frequent Asked Questions. Part of the Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds Stocker T.F., Qin D., Plattner G. K., Tignor M. M. B., Allen S. K., Boschung J., et al. (Intergovernmental Panel on Climate Change; ), 203. [Google Scholar]
- IPCC (2014). Climate change 2014, in Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds Edenhofer O., Pichs-Madruga R., Sokona Y., Farahani E., Kadner S., Seyboth K., et al. (Cambridge, New York, NY: Cambridge University Press; ), 1435. [Google Scholar]
- Kibbe W. A. (2007). OligoCalc: an online oligonucleotide properties calculator. Nucl. Acids Res. 35, W43–W46. 10.1093/nar/gkm234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozera B., Rapacz M. (2013). Reference genes in real-time PCR. J. Appl. Gen. 54, 391–406. 10.1007/s13353-013-0173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Y., Wang F., Jiang Q., Wang G. L., Tian C., Xiong A. S. (2016). Validation and comparison of reference genes for qPCR normalization of celery (Apium graveolens) at different development stages. Front. Plant Sci. 7:313. 10.3389/fpls.2016.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos A., François J. M., Parrou J. L. (2015). Tracking the best reference genes for RT-qPCR data normalization in filamentous fungi. BMC Genomics. 16:71. 10.1186/s12864-015-1224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrach A., Ghazoul J. (2015). Climate and pest-driven geographic shifts in global coffee production: implications for forest cover, biodiversity and carbon storage. PLoS ONE 10:e0133071. 10.1371/journal.pone.0133071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. (2010). Validation of reference genes for quantitative realtime PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10:4 10.1186/1471-2229-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins L. D., Tomaz M. A., Lidon F. C., DaMatta F. M., Ramalho J. C. (2014). Combined effects of elevated [CO2] and high temperature on leaf mineral balance in Coffea spp. plants. Clim. Chang. 126, 365–379. 10.1007/s10584-014-1236-7 [DOI] [Google Scholar]
- Martins M. Q., Rodrigues W. P., Fortunato A. S., Leitão A. E., Rodrigues A. P., Pais I. P., et al. (2016). Protective response mechanisms to heat stress in interaction with high [CO2] conditions in Coffea spp. Front. Plant Sci. 7:947. 10.3389/fpls.2016.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondego J. M., Vidal R. O., Carazzolle M. F., Tokuda E. K., Parizzi L. P., Costa G. G., et al. (2011). An EST-based analysis identifies new genes and reveals distinctive gene expression features of Coffea arabica and Coffea canephora. BMC Plant Biol. 11:30. 10.1186/1471-2229-11-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Hatanaka T., Misoo S., Fukayama H. (2014). Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of rubisco in rice. Plant Physiol. 164, 69–79. 10.1104/pp.113.228015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Qi J., Zhang G., Xu J., Tao A., Fang P., et al. (2015). Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 6:848. 10.3389/fpls.2015.00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr D. J., Alcβntara A., Kapralov M. V., Andralojc P. J., Carmo-Silva E., Parry M. A. J. (2016). Surveying rubisco diversity and temperature response to improve crop photosynthetic efficiency. Plant Physiol. 172, 707–717. 10.1104/pp.16.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petriccione M., Mastrobuoni F., Zampella L., Scortichini M. (2015). Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 5:16961. 10.1038/srep16961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper - excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Poncet V., Rondeau M., Tranchant C., Cayrel A., Hamon S., De Kochko A., et al. (2006). SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol. Genet. Genomics 276, 436–449. 10.1007/s00438-006-0153-5 [DOI] [PubMed] [Google Scholar]
- Ramalho J. C., Rodrigues A. P., Semedo J. N., Pais I. P., Martins L. D., Simões-Costa M. C., et al. (2013). Sustained photosynthetic performance of Coffea spp. under long-term enhanced [CO2]. PLoS ONE 8:e82712 10.1371/journal.pone.0082712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: Available online at: https://www.R-project.org (Accessed November 2016). [Google Scholar]
- Rodrigues W. P., Martins M. Q., Fortunato A. S., Rodrigues A. P., Semedo J. N., Simões-Costa M. C., et al. (2016). Long-term elevated air [CO2] strengthens photosynthetic functioning and mitigates the impact of supra-optimal temperatures in tropical Coffea arabica and C. canephora species. Glob. Change Biol. 22, 415–431. 10.1111/gcb.13088 [DOI] [PubMed] [Google Scholar]
- Roy S. J., Tucker E. J., Tester M. (2011). Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 14, 232–239. 10.1016/j.pbi.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Bioinformatics methods and protocols: methods, in Molecular Biology, Chapter 20, Primer3 on the WWW for general users and for biologist programmers, eds Krawetz S., Misener S. (Totowa, NJ: Humana; ), 365–386. [DOI] [PubMed] [Google Scholar]
- Tian C., Jiang Q., Wang F., Wang G. L., Xu Z. S., Xiong A. S. (2015). Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 10:e0117569. 10.1371/journal.pone.0117569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–research0034.11. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R. O., Mondego J. M. C., Pot D., Ambrósio A. B., Andrade A. C., Pereira L. F. P., et al. (2010). A high-throughput data mining of single nucleotide polymorphisms in coffea species expressed sequence tags suggests differential homeologous gene expression in the allotetraploid Coffea arabica. Plant Physiol. 154, 1053–1066. 10.1104/pp.110.162438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira L. G. E., Andrade A. C., Colombo C. A., Moraes A. A. H., Metha A., Oliveira A. C., et al. (2006). Brazilian coffee genome project: an EST-based genomic resource. Theor. Exp. Plant Physiol. 18, 95–108. 10.1590/S1677-04202006000100008 [DOI] [Google Scholar]
- Waller J. M., Bigger M., Hillocks R. J. (2007). Coffee pests, diseases and their management, in World Coffee Production, Chapter 2. eds Waller J. M., Bigger M., Hillocks R.J. (Egham; Surrey, BC: CAB International,) 17–40. [Google Scholar]
- Wang G. L., Tian C., Jiang Q., Xu Z. S., Wang F., Xiong A. S. (2016). Comparison of nine reference genes for real-time quantitative PCR in roots and leaves during five developmental stages in carrot (Daucus carota L.). J. Hortic. Sci. Biotechnol. 91, 264–270. 10.1080/14620316.2016.1148372 [DOI] [Google Scholar]
- Watanabe C. K., Sato S., Yanagisawa S., Uesono Y., Terashima I., Noguchi K. (2014). Effects of elevated CO2 on levels of primary metabolites and transcripts of genes encoding respiratory enzymes and their diurnal patterns in Arabidopsis thaliana: possible relationships with respiratory rates. Plant Cell Physiol. 55, 341–357. 10.1093/pcp/pct185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E., Leyns L., Vandesompele J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379, 127–129. 10.1016/j.ab.2008.04.036 [DOI] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. 10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- Yan X., Dong X., Zhang W., Yin H., Xiao H., Chen P., et al. (2014). Reference gene selection for quantitative Real-Time PCR normalization in Reaumuria soongorica. PLoS ONE 9:e104124. 10.1371/journal.pone.0104124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Pan H., Noland J. E., Zhang D., Zhang Z., Liu Y., et al. (2015). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5:18201. 10.1038/srep18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Zhang F., Tao Y., Song S., Fang J. (2015). Reference gene selection for quantitative real-time PCR normalization in different cherry genotypes, developmental stages and organs. Sci. Hortic. 181, 182–188. 10.1016/j.scienta.2014.10.027 [DOI] [Google Scholar]
- Yuyama P. M., Reis Júnior O., Ivamoto S. T., Domingues D. S., Carazzolle M. F., Pereira G. A., et al. (2016). Transcriptome analysis in Coffea eugenioides, an Arabica coffee ancestor, reveals differentially expressed genes in leaves and fruits. Mol. Genet. Genomics 291, 323–336. 10.1007/s00438-015-1111-x [DOI] [PubMed] [Google Scholar]