Abstract
We performed a genetic suppressor element screen to identify genes whose inhibition bypasses cellular senescence. A normalized library of fragmented cDNAs was used to select for elements that promote immortalization of rat embryo fibroblasts. Fragments isolated by the screen include those with homology to genes that function in intracellular signaling, cellular adhesion and contact, protein degradation, and apoptosis. They include mouse Tid1, a homologue of the Drosophila tumor suppressor gene l(2)tid, recently implicated in modulation of apoptosis as well as gamma interferon and NF-κB signaling. We show that GSE-Tid1 enhances immortalization by human papillomavirus E7 and simian virus 40 T antigen and cooperates with activated ras for transformation. Expression of Tid1 is upregulated upon cellular senescence in rat and mouse embryo fibroblasts and premature senescence of REF52 cells triggered by activated ras. In accordance with this, spontaneous immortalization of rat embryo fibroblasts is suppressed upon ectopic expression of Tid1. Modulation of endogenous Tid1 activity by GSE-Tid1 or Tid1-specific RNA interference alleviates the suppression of tumor necrosis factor alpha-induced NF-κB activity by Tid1. We also show that NF-κB sequence-specific binding is strongly downregulated upon senescence in rat embryo fibroblasts. We therefore propose that Tid1 contributes to senescence by acting as a repressor of NF-κB signaling.
As normal cells proliferate in vitro, they progressively lose the potential to divide until they reach a state of irreversible growth arrest or senescence. The number of divisions a cell can undergo in vitro, or the so-called Hayflick limit, is particular to each cell type and varies between species (19). Senescent cells remain metabolically active, have a distinct enlarged flat morphology, and express a number of senescence-associated markers, such as senescence-associated β-galactosidase activity, plasminogen activator inhibitor 1 (PAI-1), p53, p21cip1, p19ARF, and p16INK4A (45).
Proteins of the p16INK4A/Rb and p19ARF/p53 pathways are crucial for establishing the senescence phenotype (for a review see reference 31). Disruption of these pathways through mutations or expression of viral oncogenes, such as human papillomavirus (HPV) E6 and E7, simian virus 40 (SV40) T antigen, or adenovirus E1A, leads to elimination of senescence and extension of life span in human cells and immortalization in rodent cells (31). There are intrinsic differences between human and rodent cells regarding the upstream signals that trigger senescence. Telomere shortening or maintenance of the telomere structure at the ends of chromosomes is the most likely candidate to signal replicative senescence in human cells (32), and reconstitution of telomerase activity in most human diploid fibroblasts is sufficient to bypass senescence and confer immortality (3). In contrast, rodent fibroblasts have long telomeres and can undergo senescence upon serial passaging without evidence of telomere shortening, suggesting that signals other than telomere dysfunction induce senescence (2). Furthermore, bypass of senescence in human keratinocytes, epithelial cells, and adult mammary fibroblasts and endothelial cells requires inactivation of the RB-p16 pathway in addition to telomerase activity (12, 22, 35).
It has been proposed that the telomere-independent mechanisms that limit replicative life span observed in rodent or human cells are either a stress response or a form of premature senescence triggered by inappropriate in vitro culture conditions (tissue culture stress) (6, 33, 45). Premature senescence can be induced by DNA damage (38), oxidative stress (8), or overexpression of oncogenic ras (43). Premature senescence induced by oncogenic ras is provoked by excessive mitogenic signaling through the MEK/mitogen-activated protein kinase cascade and depends on the p19ARF/p53 tumor suppressor pathway (13, 29). However, very little is known about the signaling mechanisms that trigger the so-called stress-associated senescence upon serial passaging.
A number of genetic screens were performed to identify genes that allow bypass of senescence, either triggered with oncogenic ras or induced in conditionally immortal cell lines upon temperature shift (4, 37, 46). Genes identified in these screens are mainly involved in modulation of the p19ARF or Rb-E2F1 pathway. To identify genes involved in the onset of cellular senescence upon serial passaging, we used a genetic suppressor elements (GSE) approach, a functional genetic methodology based on the expression of cDNA fragments encoding either peptides that act as inhibitors of protein function or antisense RNA segments that inhibit gene expression (17, 40). It has been successfully used for the identification of genes involved in negative control of cell growth and cell survival, including drug sensitivity and candidate tumor suppressor genes (15, 34, 41). Inactivation of genes that are crucial for the onset of cellular senescence by corresponding GSEs would lead to bypass of senescence. Rat embryo fibroblasts (REFs) were chosen as the recipient cells, because they senesce within four to five passages in vitro and do not readily undergo spontaneous immortalization but can be immortalized by a single genetic event.
Here we present the isolation of eight functional GSEs, seven of which correspond to known genes. They include genes that function in signal transduction, intracellular contacts, protein degradation, and apoptosis. Two of them, Tid1 and KIAA1389, a RapGAP-related protein, are homologous to the product of well-characterized tumor suppressor gene Drosophila lethal(2)tumorous imaginal disks [l(2)tid] (24) and tuberous sclerosis 2 (TSC2) (25).
We show that GSE-Tid1, derived from a C-terminal fragment of mouse Tid1, interferes with the activity of endogenous Tid1 proteins, facilitates cell immortalization, and promotes cell survival. We found that endogenous Tid1 was upregulated upon senescence in both REFs and mouse embryonic fibroblasts (MEFs), and ectopic expression of recombinant Tid1S in primary REFs suppressed spontaneous immortalization. Modulation of endogenous Tid1 activity by GSE-Tid1- or Tid1-specific small interfering RNAs (siRNAs) alleviated the suppression effect of Tid1 on NF-κB activity. We also found that sequence-specific DNA binding of NF-κB protein complex was gradually decreased upon serial passaging. Therefore, we propose that Tid1 contributes to senescence by acting as a repressor of NF-κB signaling.
MATERIALS AND METHODS
Cell lines and primary cultures.
REFs prepared from 13-day-old Sprague-Dawley rat embryos and MEFs isolated from 13-day-old C57BL/6 mice were maintained and serially passaged as described previously (20). All cell lines, including primary cells, were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (10% donor calf serum was used for NIH 3T3 cells). All media and supplements were from Invitrogen.
Plasmids and libraries.
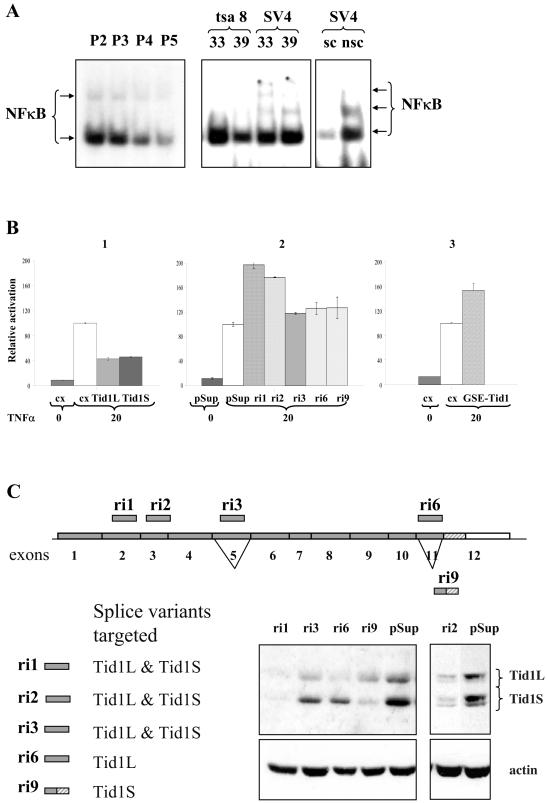
A normalized retroviral GSE library of randomly fragmented NIH 3T3 cDNAs was made in pLNCX vector (17). GSE-p53 (GSE56) was previously isolated in a GSE screen (36). Human Ha-Ras in pEJ6.6, HPV16 E7 in pMoE7, and pSE encoding the SV40 whole early region were used in immortalization assays. Tid1S cDNA was isolated from a REF λ-phage cDNA library and was inserted into pcDNA3-1 expression vector (Invitrogen). Tid1Sex3 and Tid1Sex5 cDNAs were produced by reverse transcription-PCR (RT-PCR) by using oligonucleotides designed to overlap the borders of corresponding exons (52). Human Tid1L and Tid1S cDNAs amplified from a human fetal brain cDNA library (48) were recloned into pLHCX vector. To generate knockdown vectors, hairpin oligonucleotides capable of producing siRNAs to target Tid1L and Tid1S were designed and inserted into the pRetroSuper vector (Oligoengine). The sequences targeted were GTGCTTCTTTGGCCAAAGA (ri1), GGATGATCCCAAAGCCAAG (ri2), CGAGCCTGGAACCAAAGTG (ri3), GCAAGGATAGGCGAGAGGC (ri6), and GCACTGGAAAGCGGTCAAC (ri9).
Retrovirus infection and DNA transfections.
Retroviral vectors were transfected into Bosc23 ecotropic packaging cells by using FuGene 6 reagent (Roche). Infection was accomplished by incubating cells in virus-containing medium supplemented with 8 μg of polybrene (Sigma)/ml. To induce silencing, pSuperRetro constructs were transfected into GSE1 cells by using Lipofectamine 2000 reagent (Invitrogen). All other transfections and cotransfections of primary cells and cell lines were conducted with FuGene 6.
Library screening for GSEs capable of bypassing senescence.
Bosc23 cells (7 × 106) were transfected with 12 μg of GSE library or pLNCX vector. Virus-containing medium was collected 48 h posttransfection and filtered. Virus titer was determined by infection of NIH 3T3 cells. REFs at passage 2 (total of 108 cells) were infected with the GSE library, and 48 h postinfection cells were split 1:10 for G418 selection (300 μg/ml). GSE inserts were retrieved from immortal clones by PCR using orientation-specific primers 5′-CCAAGCTTTGTTTACATCGATGGATG-3′ (sense) and 5′-ATGGCGTTAACTTAAGCTAGCTAGCTTGC-3′ (antisense), sequenced, and recloned back into pLNCX vector in both orientations.
Analysis of RNA and protein expression.
The level of Tid1 RNA expression was analyzed in MEFs and REFs by RT-PCR using several sets of primers that discriminate between the splice variants. Protein extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor cocktail (Sigma) and were analyzed by immunoblotting using standard procedures (18). Antibodies Tid-1 Ab-2 (Clone RS13) (NeoMarkers), Tid-1s (S-9) (sc-5874) (Santa Cruz), and anti-actin clone AC-40 (Sigma) were used.
EMSA.
Nuclear extracts prepared as described previously (26) were used for electrophoretic mobility shift assay (EMSA) with P32 end-labeled NF-κB consensus oligonucleotides (E3291; Promega). Poly(dI-dC) (Amersham Biosciences) was used as a nonspecific DNA competitor. In competition assays, unlabeled NF-κB oligonucleotide was used as a specific competitor (sc), and AP1 oligonucleotide (E3201; Promega) was used as a nonspecific competitor (nsc).
NF-κB luciferase reporter assay.
Reporter assays were carried out by cotransfection of 0.4 μg of NF-κB-Luc (Stratagene) with 2 μg of corresponding expression vector. Forty-eight hours after transfection, cells were treated with 20 ng of rat tumor necrosis factor alpha (TNF-α) (Insight Biotechnology)/ml for 4 h and luciferase activity was measured by using a luciferase detection kit (Promega).
RESULTS
Isolation of GSEs capable of promoting immortalization of REFs.
To identify GSEs that can bypass cellular senescence, we retrovirally transduced REFs with a normalized GSE library derived from NIH 3T3 cells and selected for geneticin-resistant colonies (Fig. 1). Second-passage REFs prepared from day 13 Sprague-Dawley rat embryos were used for the screen, because they have a high proliferation rate that ensures high transduction efficiencies upon infection with recombinant retroviruses. To achieve maximal representation of GSEs, the normalized retroviral NIH 3T3 GSE library (17) was transiently transfected into the highly transfectable ecotropic packaging cell line BOSC23 to prepare high-titer virus stocks. Empty pLNCX control vector was used as a negative control, and pLNCX GSE56, which encodes a C-terminal fragment of p53 and is known for its strong immortalization activity, was used as a positive control (36).
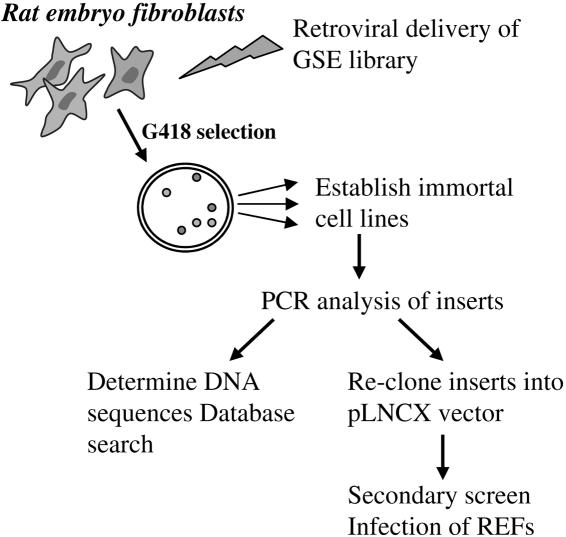
FIG. 1.
Isolation of GSEs that bypass senescence. A GSE library was delivered to REFs by retroviral infection. Transduced REFs were selected for colony formation in the presence of G418. GSEs were rescued by PCR from genomic DNA after colonies were isolated and serially propagated for 5 or 6 passages, recloned into pLNCX vector, and subjected to a secondary screen.
REFs (108) were infected with the GSE retrovirus library, equivalent to 5 × 106 independent infectious events as determined by simultaneous infection of NIH 3T3 cells, and geneticin-resistant colonies were isolated. The frequency of colony formation was about 1 in 104. Colony growth was continually monitored microscopically, and colonies that had grown larger than 5 mm in diameter after 2 weeks and were not composed of large, flat, phenotypically senescent cells were selected and isolated for expansion into cell lines. Fifteen percent of the picked colonies established cell lines, whereas none of the colonies isolated after infection with pLNCX vector alone expanded to yield cell lines. Genomic DNA was extracted from the established cell lines, and the GSE inserts were amplified using primers corresponding to the adaptor and flanking sequences within the pLNCX vector (17). Twenty-three inserts were isolated, sequenced, and recloned into pLNCX. DNA sequence analysis revealed homology to both known and unknown genes. They included genes encoding proteins that function in intracellular signaling, cellular adhesion and contacts, protein degradation, and apoptosis (Table 1).
TABLE 1.
Analysis of GSE library inserts amplified from immortalized rat fibroblasts
| Clone(s) | Insert size (bp) | Open reading frame | Sequence homology | Function |
|---|---|---|---|---|
| 3A, 4D, 12D | 239 | Unknown | Unknown, mouse chromosome 11 (100%) | Unknown |
| 16B | 284 | Antisense | Matrix protein PRELP (100%) | Attachment to extracellular matrix |
| 21A | 240 | Antisense | mTid-1 (100%), homologue of Drosophila tumor suppressor l(2)tid | Tumor suppression, apoptosis |
| 24D | 248 | Antisense | NPC p62 (100%), nuclear pore complex glycoprotein | Nuclear transport, signal transduction |
| 38D | 335 | Antisense | KIAA1389 (97%), RapGap domain protein; E6TP1 (60%), HPV E6 targeted protein; TSC2 (33%), tuberous sclerosis protein 2 | Signal transduction, tumor suppression |
| 68E | 282 | Sense | Mouse homologue of rat Sca1 (ataxin1), associated with neurodegenerative diseases | Unknown |
| 80A | 334 | Sense | Plectin (100%), hemodesmosome-associated protein, changed in basal cell carcinomas | Cytoskeleton linker protein |
| 96A | 342 | Antisense | hZimp10 (97%), PIAS family protein, inhibitor of activated STAT | Transcriptional corepressor |
GSE 21A corresponds to the 3′ end of cDNA encoding mTid1, a DnaJ domain protein known to be a modulator of apoptosis (48) and a homologue of l(2)Tid, the lethal tumorous imaginal disk protein, a tumor suppressor in Drosophila spp. GSE 38D is a fragment of cDNA for the human tuberin family protein KIAA1389 with homology to another tumor suppressor protein, tuberous sclerosis protein 2 (TSC2). GSE 16B is homologous to exon 3 of the extracellular matrix leucine-rich repeat protein PRELP, while GSE 24D is homologous to the central domain of the nuclear pore complex glycoprotein p62. GSE 80A, one of two GSEs expressed in sense orientation, encodes the C-terminal fragment of plectin, an important cytoskeletal cross-linking protein, that was recently reported to be a major early substrate for caspase-8, required for actin reorganization during apoptosis (47).
The ability of the isolated GSEs to immortalize primary cells was analyzed in a secondary screen. GSE inserts were cloned into pLNCX in both orientations and were tested individually for REF immortalization. REFs transduced with viruses carrying GSEs in the original orientation showed only a two- to threefold increase in the number of colonies compared to that of REFs infected with viruses transducing empty vector or with GSE inserts cloned in the opposite orientation (data not shown). However, when representative colonies were isolated for the original orientation, the frequency with which cell lines were established was ≥50% for most of the tested GSEs (Table 2). Colonies resulting from infection with empty vector or GSEs cloned in the wrong orientation generally failed to expand into cell lines. To further test the immortalization potential, GSEs were assessed for transformation of REFs in cooperation with oncogenic ras. We found that GSEs 4D, 16B, 21A, 24D, and 96A were able to cooperate (data not shown).
TABLE 2.
Immortalization potential of isolated GSEs
| Clonea | Orientation | Homology | Establishment of cell lines (no. positive/no. examined) |
|---|---|---|---|
| 4D | + | Unknown | 5/10 |
| 16B | + | PRELP | 8/10 |
| 16B | − | PRELP | 0/9 |
| 21A | + | Tid-1 | 5/6 |
| 24D | + | NPC p62 | 3/10 |
| 68E | + | ataxin1 | 4/10 |
| 96A | + | hZimp10 | 5/10 |
| pLNCX | NA | Empty vector | 1/10 |
| GSE 56 | NA | p53 | 10/10 |
Data on clones 38D and 80A are not included because insufficient number of colonies were obtained for further analysis.
To determine whether expression of the genes corresponding to the isolated GSEs was altered in cellular senescence, we examined their expression profiles in young and senescent MEFs and REFs by RT-PCR using primers specific to the GSE or to the corresponding cDNA. Tid1 and plectin mRNA were increased upon senescence, whereas the level of mRNA for the others was either decreased or remained unaltered (data not shown). This raised the possibility that GSEs for Tid1 and plectin cause alterations in senescence-associated activities of these genes, leading to bypass of senescence.
GSE-Tid1 promotes cell immortalization.
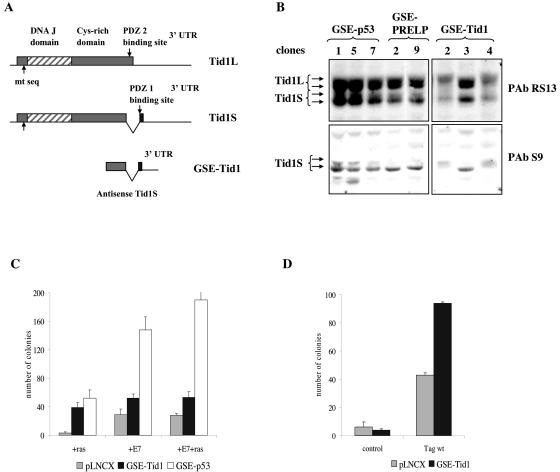
GSE-Tid1 comprises a 272-bp sequence from the 3′ end of Tid1S cDNA (Fig. 2A). Tid1S (referred to as Tid1I in reference 50) is the short splice variant of mouse Tid1 generated by alternative splicing of exon 11, resulting in deletion of the 34 carboxy-terminal amino acids and extension of the open reading frame into part of exon 12 (52). Thus, alternative splicing results in different C termini for Tid1L and Tid1S, creating two types of PDZ domain binding sites, PDZ1 and PDZ2, that could be crucial for their opposite roles in apoptosis (Fig. 2A). GSE-Tid1 corresponds to the translated and untranslated sequences at the 3′ end of Tid1S expressed in the antisense orientation. Analysis of the protein expression profile of Tid1 in representative cell lines established with either GSE-Tid1 or other GSEs by blotting with PabRS13, an antibody that detects both Tid1 L and S isoforms, revealed differences in levels and representation of Tid1 isoforms in two out of three GSE-Tid1 cell lines (Fig. 2B). Blotting with PabS9, a Tid1S-specific antibody, showed a significant decrease in Tid1S protein expression in these two cell lines (Fig. 2B), indicating that GSE-Tid1 may selectively interfere with production of Tid1S.
FIG. 2.
GSE-Tid1 promotes immortalization of primary REFs. (A) GSE-Tid1 corresponds to the 3′ end of the cDNA for the short splice variant of mouse Tid1. (B) Protein expression analysis using Tid1 (L+S)-specific antibody PAbRS13 or Tid1S-specific antibody PAbS9 revealed changes in protein level and representation of isoforms in cells expressing GSE-Tid1 compared to those of cells immortalized with other GSEs. (C) GSE-Tid1 promotes immortalization of REFs with HPV E7 and cooperates with oncogenic ras for transformation. P2 REFs were transfected with either pLNCX (2.5 μg), GSE-Tid1 (2.5 μg), or GSE-p53 (2.5 μg) in conjunction with ras (2.5 μg) or HPV E7 (1 μg) or in triple transfections with both HPV E7 and ras. GSE-p53, the positive control, produced the highest level of immortalization in all cotransfection experiments. (D) GSE-Tid1 promotes immortalization of REFs with SV40 T antigen (Tag). REFs (passage 2) were transfected with pLNCX (2.5 μg) or GSE-Tid1 (2.5 μg) either alone or with pSE (SV40 whole early region, 2.5 μg; Tag wt). 3′UTR, 3′ untranslated region.
The fact that GSE-Tid1 did not affect colony formation but increased the frequency at which cell lines were produced (Table 2) suggested that GSE-Tid1 may immortalize REFs in cooperation with other cellular events. To investigate this possibility, GSE-Tid1 was cotransfected with other oncogenes into primary REFs. Cotransfection of GSE-Tid1 with oncogenic ras strongly stimulated colony formation, whereas oncogenic ras alone led to a reduction in colony number (Fig. 2C). Similarly, cotransfection of GSE-Tid1 with HPV16 E7 or triple transfections with HPV16 E7 and activated ras increased the frequency of immortalized clones (Fig. 2C). Cotransfection of GSE-Tid1 with SV40 T antigen also yielded two times more colonies than T antigen alone (Fig. 2D). Together these results suggest that GSE-Tid1 promotes survival and facilitates immortalization of primary REFs.
RNA and protein expression profile of Tid1 isoforms upon cellular senescence.
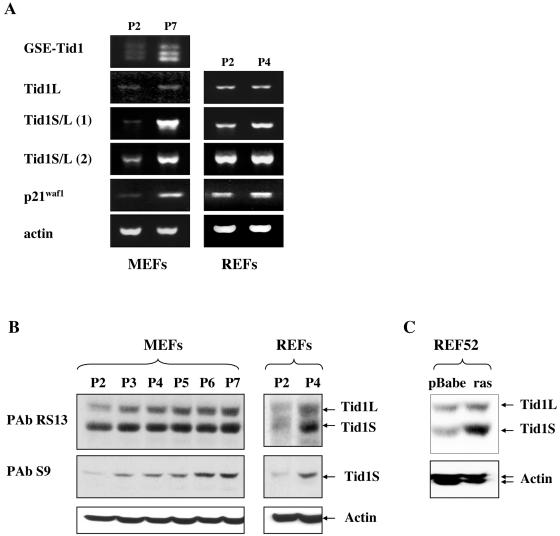
To study the role of endogenous Tid1 in senescence, we analyzed mRNA and protein expression in MEFs and REFs upon serial passaging. RT-PCR on RNA extracted from early (P2)- and late (P7)-passage MEFs, using primers for 5′ and 3′ ends of GSE-Tid1, produced three bands, with the smallest band being the size of GSE-Tid1 (Fig. 3A). The intensity of all these bands was considerably higher in P7 MEFs than in P2 MEFs.
FIG. 3.
Tid-1 is upregulated at both RNA and protein levels upon cellular senescence. (A) Tid1 RNA levels were analyzed by RT-PCR in young (P2) and senescent (P7 or P4) MEFs and REFs using sets of primers that discriminate between the long and short isoforms. (B and C) Tid-1 proteins accumulate in primary MEFs and REFs upon serial passaging and in REF52 cells upon premature senescence triggered by activated ras. Western blots were probed with the Tid1-specific monoclonal antibody PAb RS13 or the Tid1S specific polyclonal antibody PAb S9. Equal loading was confirmed with a monoclonal antiactin antibody.
To discriminate between the two major splice variants Tid1L and Tid1S, sets of primers were designed such that the 3′ primer would anneal either within or outside of exon 11. RT-PCR with the 3′ primer within exon 11, which only recognizes Tid1L transcripts, produced faint bands that were not significantly different in intensity between P2 and P7 MEFs (Fig. 3A). In contrast, when sequences present in both Tid1L and Tid1S were amplified with a reverse primer that mapped upstream of exon 11, there was a significant increase in the level of expression upon senescence in both MEFs and REFs [Fig. 3A, Tid1S/L (1)]. When a 3′ primer derived from sequences downstream of exon 11, and therefore expected to give rise to fragments of different lengths for Tid1L and Tid1S, was used, an increase in expression was observed (most evident in MEFs), but the fragments were indistinguishable [Fig. 3A, Tid1S/L (2)]. However, when the PCR band was cloned and sequenced, clones corresponding to both the short and long isoforms were identified. Together these results indicate that Tid1 RNA and, in particular, the RNA for the Tid1S splice variant is upregulated upon senescence.
To determine whether there were changes in protein expression of the Tid1 isoforms upon senescence, total extracts were prepared from cultures of MEFs at each passage and were analyzed by Western blotting. To ensure that only changes in expression due to senescence were being examined, extracts were prepared from subconfluent cultures that had been passaged the previous day and, thus, should not contain quiescent cells. Western blots with antibody PAb RS13 revealed a steady increase in the level of both Tid1L and Tid1S isoforms in MEFs upon serial passaging (Fig. 3B). A similar accumulation of both isoforms was observed in REFs when senescent P4 cells were compared with actively proliferating P2 cells. Accumulation of the Tid1S isoform upon passaging was confirmed by blotting the extracts with PAb S9, which specifically recognizes only this isoform (Fig. 3B).
Premature senescence triggered by ectopic expression of activated ras induces a pattern of gene expression similar to that of cells undergoing replicative senescence upon serial passaging, and these patterns are distinct from those in quiescent cells (44). To determine whether Tid1 isoforms are also upregulated upon premature senescence, oncogenic ras was introduced into REF-52 cells by retroviral infection. Seven days postinfection with activated ras, REF52 cells underwent growth arrest and phenotypic changes consistent with senescence. A parallel culture of REF52 cells infected with the backbone retrovirus did not undergo growth arrest but continued to proliferate until the cells became contact inhibited. Western blotting of lysates prepared from the REF52 cultures with the anti-Tid1 monoclonal antibody PAb RS13 showed that both Tid1 isoforms were upregulated; however, the Tid1S isoform accumulated to a much higher level, thereby altering the ratio between the two proteins (Fig. 3C).
Thus, both replicative senescence and premature senescence triggered by oncogenic ras were associated with upregulation of Tid1 proteins. While the protein levels of both isoforms showed a steady increase upon serial passaging of MEFs and REFs, ras-induced premature senescence of REF52 was accompanied by a predominant accumulation of Tid1S.
Ectopic expression of recombinant Tid1S in primary cells.
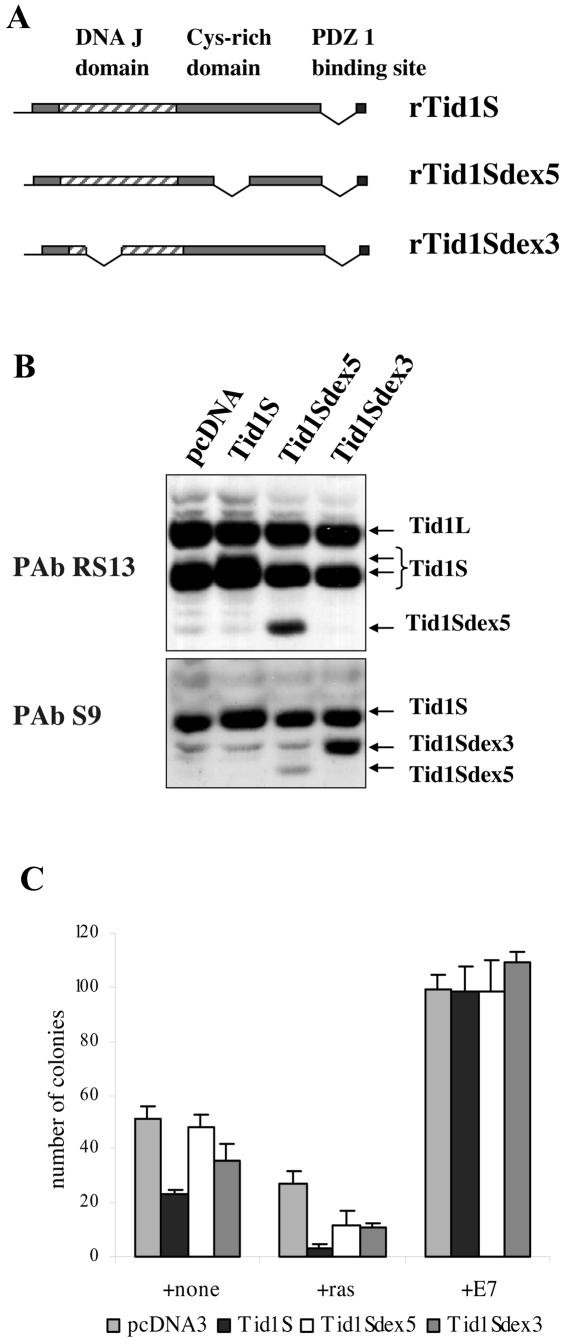
To determine the role of Tid1S proteins in immortalization, we isolated a rat Tid1S cDNA from a REF cDNA library and generated two splice variants: Tid1Sex5, lacking exon 5, which corresponds to the low-abundance splicing variant described previously (50), and Tid1Sex3, missing exon 3, which corresponds to a novel splice variant identified by a BLAST search of the rat expressed sequence tag database (Fig. 4A). Protein expression from these constructs was monitored by transient transfection in SAOS2 cells (Fig. 4B). Even though the Western blot analysis was complicated by the presence of the endogenous proteins, the Tid1S expression vector clearly led to an increase in Tid1S protein, altering the ratio between two isoforms (Fig. 4B). Both Tid1Sex5 and Tid1Sex3 gave rise to proteins that migrated faster than Tid1S, and they were detected by a combination of PAb RS13 and PAb S9. PAb RS13 does not recognize the Tid1Sex3 splicing variant due the absence of the epitope (Fig. 4B).
FIG. 4.
Ectopic expression of Tid1S suppresses spontaneous immortalization. (A) A cDNA corresponding to Tid1S was isolated from a REF cDNA λ-phage library and cloned into the pcDNA3.1 expression vector. Expression constructs with deletions of exon 5 or exon 3 were also generated. (B) Tid1S constructs were transiently transfected into SAOS2 cells, and protein expression was analyzed by Western blotting with PAb RS13 and PAb S9. (C) Tid1S constructs were transfected into REFs (passage 2) alone or with ras or with HPV E7, and immortal cells were selected in presence of G418.
To determine the effect of ectopic expression of Tid1S upon immortalization, we introduced the Tid1S constructs into REFs alone or in conjunction with oncogenic ras. We found that Tid1S suppresses colony formation compared to that of the control vector (Fig. 4C). Cotransfection of Tid1S with activated ras decreased the number of colonies obtained compared to that with ras alone; thus, Tid1S enhances the prosenescent activity of activated ras in primary cells. Cotransfection of the deletion constructs, where Tid1S lacked either exon 3 or exon 5, with oncogenic ras did not have the same inhibitory affect as intact Tid1S, suggesting that both the DnaJ and cystine-rich domains are essential for the suppression of growth. This suppression of colony formation by Tid1S was overcome by the HPV16 E7 gene, indicating that the immortalizing functions of HPV16 E7 are dominant to the inhibitory effects of Tid1S.
NF-κB is a potential target of Tid1 in senescence.
Previously published data has indicated that Tid1 may have multiple cellular targets and may modulate activity of proteins involved in the ras, gamma interferon, and NF-κB signaling pathways (10, 42, 50). Because ectopic expression of GSE-Tid1 promoted cell immortalization and cell survival, we explored the possibility that GSE-Tid1 may act through modulation of NF-κB signaling, a pathway known to be linked to inhibition of apoptosis and promotion of cell survival.
We first examined NF-κB DNA binding activity in REFs upon serial passaging, and we found that the sequence-specific DNA binding activity of the NF-κB transcription factor complex gradually decreased (Fig. 5A). It was recently reported that the Tid1L protein suppresses NF-κB signaling by repressing IκBα phosphorylation and nuclear translocation of NF-κB (10). The reduction in NF-κB activity in cells undergoing senescence is in accordance with the increase in Tid1 protein expression (Fig. 3B) and may reflect a functional link between Tid1 and the NF-κB complex. NF-κB DNA binding activity was also reduced in REFs conditionally immortalized with a temperature-sensitive SV40 tsA58 T antigen when they enter a senescence-like state upon inactivation of T antigen (Fig. 5A). SV4, a REF line immortalized with wild-type T antigen and used as a control, did not show a reduction in NF-κB DNA binding activity.
FIG. 5.
NF-κB is a potential target of Tid1 in senescence. (A) NF-κB sequence-specific DNA binding activity was downregulated upon senescence. Nuclear extracts from serially passaged REFs (P2 to P5) or tsa8 and SV4 cells upon temperature shift were analyzed. As controls, unlabeled NF-κB oligonucleotide was used as a specific competitor (sc) and AP-1 oligonucleotide was used as a nonspecific competitor (nsc). (B) Modulation of Tid1 activity with ectopic expression of human Tid1L and Tid1S (1), Tid1 specific siRNAs (2), or GSE-Tid1 (3) altered NF-κB transcriptional activation activity as measured by NF-κB luciferase reporter assay. (C) Hairpin oligonucleotides capable of producing siRNAs to target Tid1L and Tid1S were designed and inserted into the pRetroSuper vector (pSup). Four out of five siRNAs transiently expressed in GSEp53-1 cells efficiently silenced Tid1 isoforms as revealed by Western blot with PAb RS13 antibody.
We determined whether ectopic overexpression of Tid1S can also affect NF-κB activity, as has been reported for Tid1L (10). U2OS cells were cotransfected with an NF-κB luciferase reporter, and human Tid1 expression vectors and luciferase activity were examined 4 h after treatment of transfected cells with TNF-α. Ectopic expression of human Tid1S resulted in a more than twofold reduction of TNF-α-induced NF-κB activity (Fig. 5B, panel 1). The reduction was similar to that observed with human Tid1L. This suppression of NF-κB activity by ectopic expression of human Tid1S was also observed in rat GSEp53-1 cells, with Tid1S being a stronger repressor than Tid1L (data not shown).
To modulate expression of endogenous Tid1 proteins, we designed siRNA oligonucleotides that would either silence both major isoforms or selectively suppress expression of one of them (Fig. 5C). The corresponding oligonucleotides were cloned into pRetroSuper vector and introduced into GSEp53-1 cells. Western blot analysis confirmed siRNA-mediated knockdown of endogenous Tid1 proteins (Fig. 5C). Coexpression of NF-κB luciferase reporter vector with the siRNA constructs ri1 and ri2, which target both Tid1 isoforms, resulted in a significant increase in TNF-α-induced NF-κB activity, while selective silencing of one isoform by ri6 or ri9 had only a slight effect (Fig. 5B, panel 2). The siRNA construct ri3, which induced only a partial reduction of protein expression of both isoforms, showed a reduced effect on NF-κB activity. These results indicate that alterations in the level of endogenous Tid1 proteins can directly affect TNF-α-induced NF-κB activity.
Finally, we cotransfected the NF-κB reporter with a GSE-Tid1 expression vector and observed an approximately 50% increase in TNF-α-induced NF-κB activity (Fig. 5B, panel 3), confirming that GSE-Tid1 modulates endogenous Tid1 proteins and reduces their inhibitory effect on NF-κB. Thus, the ability of GSE-Tid1 to promote immortalization and cell survival could be due to the activation of the NF-κB complex. However, interaction between Tid1 proteins and other cellular targets may also be important for the role of Tid1 in senescence and for the immortalization properties of GSE-Tid1, but they remain to be clarified.
DISCUSSION
Here we employed a functional genetic screen to identify new genes involved in cellular senescence. A normalized GSE library generated from mRNA of NIH 3T3 cells (17) was delivered into REFs by retroviral infection, and cells that bypassed senescence were selected. A secondary screen was employed to ensure that the isolated GSEs were truly capable of bypassing senescence. This involved recloning the GSEs into a retroviral vector in both sense and antisense orientations and repeating the REF immortalization assay. The stringency of the secondary screen was further increased by isolating colonies and assessing their ability to establish cell lines.
We identified eight GSEs corresponding to eight independent genes. Several of these genes, like nuclear pore protein p62 or cytoskeleton protein plectin, are well characterized, while the functions of other proteins, such as KIAA1389 and hZimp10, are predicted on the basis of their sequence and remain to be elucidated. Most of the isolated GSEs correspond to genes involved in signal transduction, consistent with the view of rodent cell senescence as a stress-related phenomenon (31, 33). Interestingly, one of the identified genes, PRELP, was recently proposed to be involved in the development of Hutchinson-Gilford progeria, accelerated aging syndrome (27).
Two of the eight isolated GSEs show a high level of homology to known tumor suppressor genes. The protein product of KIAA1389 harbors a putative RapGAP domain and is closely related to RapGTPase-activating protein E6TP1 (1), which is targeted for degradation by the E6 protein of high-risk human papilloma viruses (14). Both KIAA1389 and E6TP1 are also related to tuberin, the product of the tumor suppressor TSC2, whose inactivation is associated with familial benign tumors (25). Tuberin controls mTOR signaling by acting as part of the GTPase-activating complex for Rheb (Ras homologue enriched in brain) (49).
We also isolated a GSE fragment from a candidate tumor suppressor gene of Tid1. The tumor suppressor function of the Drosophila homologue l(2)tid is well established, as homozygous mutations in this gene result in hyperproliferation and tumor formation in imaginal disks of Drosophila larvae. Several facts point towards a similar role for mammalian Tid1 in tumor development: expression of endogenous Tid1 was found altered in basal cell carcinomas (7) while ectopic expression of human Tid1 in tumor cell lines led to suppression of their transformed phenotype (9). Our results indicate that Tid1 has the capacity to promote growth arrest and suppress immortalization, because GSE-Tid1 promoted immortalization and ectopic expression of Tid1S cDNA in REFs reduced spontaneous immortalization.
Recent studies have implicated mammalian Tid1 proteins in several signaling pathways, including those of ras, gamma interferon, and NF-κB (10, 42, 50). Our data suggest that the role of Tid1 proteins as positive regulators of cellular senescence may be linked to their ability to modulate NF-κB signaling. Cheng et al. (10) showed that hTid1 antagonizes activities of various NF-κB activators, including human T-cell leukemia virus (HTLV) Tax, TNF-α, and Bcl10, by repressing IκB kinase (IKKβ) activity and enhancing the stability of IκB molecules. Consistent with these results, we found that silencing of Tid1 with Tid1-specific siRNAs considerably enhanced NF-κB activity induced by treatment of cells with TNF-α. Ectopic expression of GSE-Tid1 had a similar enhancing effect on NF-κB activity that could be responsible for its role in promoting cell survival and immortalization. The Tid1 siRNAs that enhance NF-κB activity could not be directly tested for immortalization of REFs as they were lethal, possibly due to the involvement of Tid1 in other signaling pathways. This is supported by a recent publication showing that homozygous loss of Tid1 causes embryonic lethality (30). Unlike siRNAs which silence expression of the protein, GSE fragments may only perturb the functions relevant for the genetic screen utilized.
NF-κB signaling was only recently recognized to play an important role in tumorigenesis (for recent reviews see references 21 and 28). Constitutive activation of NF-κB is found in a variety of human cancers and can contribute to the malignant phenotype by driving cell proliferation, increasing resistance to apoptosis, and promoting cell survival and migration. Multiple mechanisms could disrupt the normal regulation of NF-κB activity in cancer cells. The Tax oncoprotein of HTLV-1 that causes adult T-cell leukemia (ATL) has been shown to directly interact with the IKK complex, resulting in enhancement of IKKβ kinase activity and subsequent activation of the NF-κB complex (11). It was then reported that Tax associates with hTid1/Hsp70 and antagonizes the suppressive activity of hTid1 on IKKβ-mediated phosphorylation of IκBα (9, 10). Another tumor suppressor protein, CYLD, that is mutated in familial cylindromatosis, skin appendage cancer predisposition syndrome, was recently described to be a negative regulator of NF-κB signaling by targeting the activity of the IKK complex through binding to IKKγ (or NEMO) and deubiquitination of signaling proteins that function upstream of IKKs (5, 23, 51).
There is presently no consensus about the role of NF-κB signaling in cellular senescence. Some groups have reported rather dramatic tissue-specific increases or decreases in NF-κB activity in humans and animals upon ageing (reviewed in reference 16). Here we have found that senescence in REFs was accompanied by a steady decline in NF-κB sequence-specific DNA binding activity, and we have proposed that this was due to increased expression of Tid1. Interestingly, it was recently published that p14ARF/p19ARF proteins that are crucial for cellular senescence suppress NF-κB function and its antiapoptotic activity independently of MDM2 and p53 by direct inhibition of RelA transactivation without affecting NF-κB sequence-specific DNA binding (39). Taken together, these data suggest that downregulation of NF-κB activity upon senescence could be controlled by a number of mechanisms, including Tid1 activity in the cytoplasm and ARF activity in the nucleus, and the perturbation of these regulatory mechanisms can lead to aberrantly active NF-κB and contribute to tumorigenesis. Isolation of the Tid1 fragment capable of promoting cell survival and immortalization validates the use of the GSE approach in the search for genes involved in control of irreversible growth arrest and senescence. The role of other genes identified in this screen in regulation of cellular senescence and immortalization remains to be elucidated.
Acknowledgments
We thank Silvia Benvenuti for REF52 protein extracts.
This work was supported by ReNeuron, Ltd. (Guildford, United Kingdom), to P.S.J. and NIH grant CA60730 to A.V.G.
REFERENCES
- 1.Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta 1603:47-82. [DOI] [PubMed] [Google Scholar]
- 2.Blasco, M. A. 2003. Telomeres in cancer and aging: lessons from the mouse. Cancer Lett. 194:183-188. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. M. Kortlever, M. Lingbeek, F. Trettel, M. E. MacDonald, M. van Lohuizen, and R. Bernards. 2002. TBX-3, the gene mutated in ulnar-mammary syndrome, is a negative regulator of p19ARF and inhibits senescence. J. Biol. Chem. 277:6567-6572. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., S. M. Nijman, A. M. Dirac, and R. Bernards. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424:797-801. [DOI] [PubMed] [Google Scholar]
- 6.Campisi, J. 2001. From cells to organisms: can we learn about aging from cells in culture? Exp. Gerontol. 36:607-618. [DOI] [PubMed] [Google Scholar]
- 7.Canamasas, I., A. Debes, P. G. Natali, and U. Kurzik-Dumke. 2003. Understanding human cancer using Drosophila: Tid47, a cytosolic product of the DnaJ-like tumor suppressor gene l2Tid, is a novel molecular partner of patched related to skin cancer. J. Biol. Chem. 278:30952-30960. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Q. M., J. C. Bartholomew, J. Campisi, M. Acosta, J. D. Reagan, and B. N. Ames. 1998. Molecular analysis of H2O2-induced senescent-like growth arrest in normal human fibroblasts: p53 and Rb control G1 arrest but not cell replication. Biochem J. 332:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H., C. Cenciarelli, Z. Shao, M. Vidal, W. P. Parks, M. Pagano, and C. Cheng-Mayer. 2001. Human T cell leukemia virus type 1 Tax associates with a molecular chaperone complex containing hTid-1 and Hsp70. Curr. Biol. 11:1771-1775. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, H., C. Cenciarelli, M. Tao, W. P. Parks, and C. Cheng-Mayer. 2002. HTLV-1 Tax-associated hTid-1, a human DnaJ protein, is a repressor of IκB kinase β subunit. J. Biol. Chem. 277:20605-20610. [DOI] [PubMed] [Google Scholar]
- 11.Chu, Z. L., Y. A. Shin, J. M. Yang, J. A. DiDonato, and D. W. Ballard. 1999. IKKγ mediates the interaction of cellular IκB kinases with the tax transforming protein of human T cell leukemia virus type 1. J. Biol. Chem. 274:15297-15300. [DOI] [PubMed] [Google Scholar]
- 12.Dickson, M. A., W. C. Hahn, Y. Ino, V. Ronfard, J. Y. Wu, R. A. Weinberg, D. N. Louis, F. P. Li, and J. G. Rheinwald. 2000. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20:1436-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferbeyre, G., E. de Stanchina, A. W. Lin, E. Querido, M. E. McCurrach, G. J. Hannon, and S. W. Lowe. 2002. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 22:3497-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garkavtsev, I., A. Kazarov, A. Gudkov, and K. Riabowol. 1996. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 14:415-420. [DOI] [PubMed] [Google Scholar]
- 16.Giardina, C., and A. K. Hubbard. 2002. Growing old with nuclear factor-κB. Cell Stress Chaperones 7:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudkov, A. V., A. R. Kazarov, R. Thimmapaya, S. A. Axenovich, I. A. Mazo, and I. B. Roninson. 1994. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc. Natl. Acad. Sci. USA 91:3744-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Hayflick, L., and P. S. Moorehead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 20.Ikram, Z., T. Norton, and P. S. Jat. 1994. The biological clock that measures the mitotic life-span of mouse embryo fibroblasts continues to function in the presence of simian virus 40 large tumor antigen. Proc. Natl. Acad. Sci. USA 91:6448-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 22.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 23.Kovalenko, A., C. Chable-Bessia, G. Cantarella, A. Israel, D. Wallach, and G. Courtois. 2003. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424:801-805. [DOI] [PubMed] [Google Scholar]
- 24.Kurzik-Dumke, U., A. Debes, M. Kaymer, and P. Dienes. 1998. Mitochondrial localization and temporal expression of the Drosophila melanogaster DnaJ homologous tumor suppressor Tid50. Cell Stress Chaperones 3:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwiatkowski, D. J. 2003. Tuberous sclerosis: from tubers to mTOR. Ann. Hum. Genet. 67:87-96. [DOI] [PubMed] [Google Scholar]
- 26.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, M. 2003. PRELP, collagen, and a theory of Hutchinson-Gilford progeria. Ageing Res. Rev. 2:95-105. [DOI] [PubMed] [Google Scholar]
- 28.Lin, A., and M. Karin. 2003. NF-κB in cancer: a marked target. Semin. Cancer Biol. 13:107-114. [DOI] [PubMed] [Google Scholar]
- 29.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, J. F., M. Hayashi, S. Woo-Kim, B. Tian, J. F. Huang, C. Fearns, S. Takayama, J. M. Zapata, Y. Yang, and J. D. Lee. 2004. Tid1, a cochaperone of the heat shock 70 protein and the mammalian counterpart of the Drosophila tumor suppressor l(2)tid, is critical for early embryonic development and cell survival. Mol. Cell. Biol. 24:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcotte, R., and E. Wang. 2002. Replicative senescence revisited. J. Gerontol. A Biol. Sci. Med. Sci. 57:B257—B269. [DOI] [PubMed] [Google Scholar]
- 32.Masutomi, K., E. Y. Yu, S. Khurts, I. Ben-Porath, J. L. Currier, G. B. Metz, M. W. Brooks, S. Kaneko, S. Murakami, J. A. DeCaprio, R. A. Weinberg, S. A. Stewart, and W. C. Hahn. 2003. Telomerase maintains telomere structure in normal human cells. Cell 114:241-253. [DOI] [PubMed] [Google Scholar]
- 33.Mathon, N. F., and A. C. Lloyd. 2001. Cell senescence and cancer. Nat. Rev. Cancer 1:203-213. [DOI] [PubMed] [Google Scholar]
- 34.Neznanov, N., L. Neznanova, R. V. Kondratov, L. Burdelya, E. S. Kandel, D. M. O'Rourke, A. Ullrich, and A. V. Gudkov. 2003. Dominant negative form of signal-regulatory protein-alpha (SIRPα/SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-κB. J. Biol. Chem. 278:3809-3815. [DOI] [PubMed] [Google Scholar]
- 35.O'Hare, M. J., J. Bond, C. Clarke, Y. Takeuchi, A. J. Atherton, C. Berry, J. Moody, A. R. Silver, D. C. Davies, A. E. Alsop, A. M. Neville, and P. S. Jat. 2001. Conditional immortalization of freshly isolated human mammary fibroblasts and endothelial cells. Proc. Natl. Acad. Sci. USA 98:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ossovskaya, V. S., I. A. Mazo, M. V. Chernov, O. B. Chernova, Z. Strezoska, R. Kondratov, G. R. Stark, P. M. Chumakov, and A. V. Gudkov. 1996. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl. Acad. Sci. USA 93:10309-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peeper, D. S., A. Shvarts, T. Brummelkamp, S. Douma, E. Y. Koh, G. Q. Daley, and R. Bernards. 2002. A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nat. Cell Biol. 4:148-153. [DOI] [PubMed] [Google Scholar]
- 38.Robles, S. J., and G. R. Adami. 1998. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene 16:1113-1123. [DOI] [PubMed] [Google Scholar]
- 39.Rocha, S., K. J. Campbell, and N. D. Perkins. 2003. p53- and Mdm2-independent repression of NF-κB transactivation by the ARF tumor suppressor. Mol. Cell 12:15-25. [DOI] [PubMed] [Google Scholar]
- 40.Roninson, I. B., and A. V. Gudkov. 2003. Genetic suppressor elements in the characterization and identification of tumor suppressor genes. Methods Mol. Biol. 222:413-436. [DOI] [PubMed] [Google Scholar]
- 41.Sanz, G., L. Mir, and A. Jacquemin-Sablon. 2002. Bleomycin resistance in mammalian cells expressing a genetic suppressor element derived from the SRPK1 gene. Cancer Res. 62:4453-4458. [PubMed] [Google Scholar]
- 42.Sarkar, S., B. P. Pollack, K. T. Lin, S. V. Kotenko, J. R. Cook, A. Lewis, and S. Pestka. 2001. hTid-1, a human DnaJ protein, modulates the interferon signaling pathway. J. Biol. Chem. 276:49034-49042. [DOI] [PubMed] [Google Scholar]
- 43.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 44.Shelton, D. N., E. Chang, P. S. Whittier, D. Choi, and W. D. Funk. 1999. Microarray analysis of replicative senescence. Curr. Biol. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 45.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 46.Shvarts, A., T. R. Brummelkamp, F. Scheeren, E. Koh, G. Q. Daley, H. Spits, and R. Bernards. 2002. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 16:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stegh, A. H., H. Herrmann, S. Lampel, D. Weisenberger, K. Andra, M. Seper, G. Wiche, P. H. Krammer, and M. E. Peter. 2000. Identification of the cytolinker plectin as a major early in vivo substrate for caspase 8 during CD95- and tumor necrosis factor receptor-mediated apoptosis. Mol. Cell. Biol. 20:5665-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Syken, J., T. De-Medina, and K. Munger. 1999. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc. Natl. Acad. Sci. USA 96:8499-8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 50.Trentin, G. A., X. Yin, S. Tahir, S. Lhotak, J. Farhang-Fallah, Y. Li, and M. Rozakis-Adcock. 2001. A mouse homologue of the Drosophila tumor suppressor l(2)tid gene defines a novel Ras GTPase-activating protein (RasGAP)-binding protein. J. Biol. Chem. 276:13087-13095. [DOI] [PubMed] [Google Scholar]
- 51.Trompouki, E., E. Hatzivassiliou, T. Tsichritzis, H. Farmer, A. Ashworth, and G. Mosialos. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424:793-796. [DOI] [PubMed] [Google Scholar]
- 52.Yin, X., and M. Rozakis-Adcock. 2001. Genomic organization and expression of the human tumorous imaginal disc (TID1) gene. Gene 278:201-210. [DOI] [PubMed] [Google Scholar]